Abstract
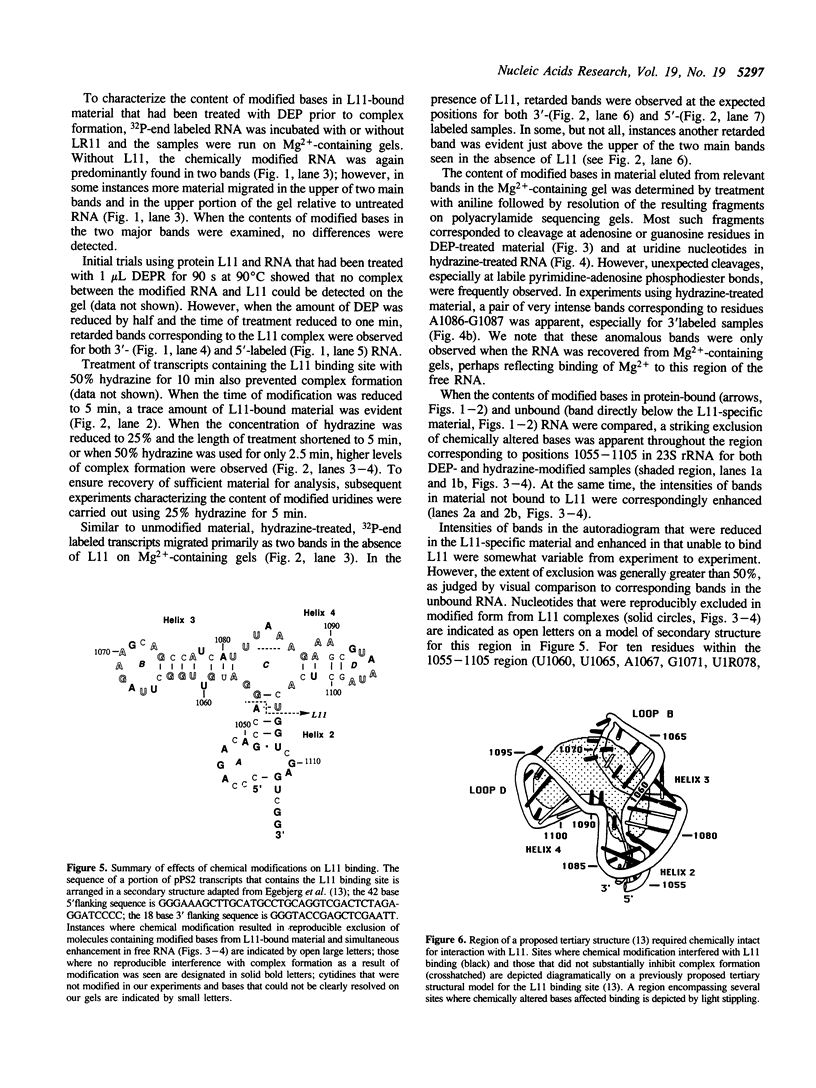
The interaction between ribosomal protein L11 from Escherichia coli and in vitro synthesized RNA containing its binding site from 23S rRNA was characterized by identifying nucleotides that interfered with complex formation when chemically modified by diethylpyrocarbonate or hydrazine. Chemically modified RNA was incubated with L11 under conditions appropriate for specific binding of L11 and the resulting protein-RNA complex was separated from unbound RNA on Mg(2+)-containing polyacrylamide gels. The ability to isolate L11 complexes on such gels was affected by the extent of modification by either reagent. Protein-bound and free RNAs were recovered and treated with aniline to identify their content of modified bases. Exclusion of RNA containing chemically altered bases from L11-associated material occurred for 29 modified nucleotides, located throughout the region corresponding to residues 1055-1105 in 23S rRNA. Ten bases within this region did not reproducibly inhibit binding when modified. Multiple bands of RNA were consistently observed on the nondenaturing gels, suggesting that significant intermolecular RNA-RNA interactions had occurred.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beauclerk A. A., Hummel H., Holmes D. J., Böck A., Cundliffe E. Studies of the GTPase domain of archaebacterial ribosomes. Eur J Biochem. 1985 Sep 2;151(2):245–255. doi: 10.1111/j.1432-1033.1985.tb09095.x. [DOI] [PubMed] [Google Scholar]
- Christensen A., Mathiesen M., Peattie D., Garrett R. A. Alternative conformers of 5S ribosomal RNA and their biological relevance. Biochemistry. 1985 Apr 23;24(9):2284–2291. doi: 10.1021/bi00330a024. [DOI] [PubMed] [Google Scholar]
- Egebjerg J., Douthwaite S. R., Liljas A., Garrett R. A. Characterization of the binding sites of protein L11 and the L10.(L12)4 pentameric complex in the GTPase domain of 23 S ribosomal RNA from Escherichia coli. J Mol Biol. 1990 May 20;213(2):275–288. doi: 10.1016/S0022-2836(05)80190-1. [DOI] [PubMed] [Google Scholar]
- Egebjerg J., Douthwaite S., Garrett R. A. Antibiotic interactions at the GTPase-associated centre within Escherichia coli 23S rRNA. EMBO J. 1989 Feb;8(2):607–611. doi: 10.1002/j.1460-2075.1989.tb03415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor M. J., Uhlenbeck O. C. Substrate sequence effects on "hammerhead" RNA catalytic efficiency. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1668–1672. doi: 10.1073/pnas.87.5.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausner T. P., Geigenmüller U., Nierhaus K. H. The allosteric three-site model for the ribosomal elongation cycle. New insights into the inhibition mechanisms of aminoglycosides, thiostrepton, and viomycin. J Biol Chem. 1988 Sep 15;263(26):13103–13111. [PubMed] [Google Scholar]
- Keith G. Optimization of conditions for labeling the 3' OH end of tRNA using T4 RNA ligase. Biochimie. 1983 Jun;65(6):367–370. doi: 10.1016/s0300-9084(83)80159-x. [DOI] [PubMed] [Google Scholar]
- Kim J., Cheong C., Moore P. B. Tetramerization of an RNA oligonucleotide containing a GGGG sequence. Nature. 1991 May 23;351(6324):331–332. doi: 10.1038/351331a0. [DOI] [PubMed] [Google Scholar]
- Kime M. J., Ratcliffe R. G., Moore P. B., Williams R. J. On the renaturation of ribosomal protein L11. Eur J Biochem. 1980 Sep;110(2):493–498. doi: 10.1111/j.1432-1033.1980.tb04891.x. [DOI] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan P. C., Draper D. E. Thermodynamics of protein-RNA recognition in a highly conserved region of the large-subunit ribosomal RNA. Biochemistry. 1989 Dec 26;28(26):9949–9956. doi: 10.1021/bi00452a012. [DOI] [PubMed] [Google Scholar]
- Schmidt F. J., Thompson J., Lee K., Dijk J., Cundliffe E. The binding site for ribosomal protein L11 within 23 S ribosomal RNA of Escherichia coli. J Biol Chem. 1981 Dec 10;256(23):12301–12305. [PubMed] [Google Scholar]
- Spacciapoli P., Doviken L., Mulero J. J., Thurlow D. L. Recognition of tRNA by the enzyme ATP/CTP:tRNA nucleotidyltransferase. Interference by nucleotides modified with diethyl pyrocarbonate or hydrazine. J Biol Chem. 1989 Mar 5;264(7):3799–3805. [PubMed] [Google Scholar]
- Stark M. J., Cundliffe E., Dijk J., Stöffler G. Functional homology between E. coli ribosomal protein L11 and B. megaterium protein BM-L11. Mol Gen Genet. 1980;180(1):11–15. doi: 10.1007/BF00267346. [DOI] [PubMed] [Google Scholar]
- Stark M., Cundliffe E. On the biological role of ribosomal protein BM-L11 of Bacillus megaterium, homologous with Escherichia coli ribosomal protein L11. J Mol Biol. 1979 Nov 15;134(4):767–769. doi: 10.1016/0022-2836(79)90485-6. [DOI] [PubMed] [Google Scholar]
- Tate W. P., Dognin M. J., Noah M., Stöffler-Meilicke M., Stöffler G. The NH2-terminal domain of Escherichia coli ribosomal protein L11. Its three-dimensional location and its role in the binding of release factors 1 and 2. J Biol Chem. 1984 Jun 10;259(11):7317–7324. [PubMed] [Google Scholar]
- Thompson J., Cundliffe E., Dahlberg A. E. Site-directed mutagenesis of Escherichia coli 23 S ribosomal RNA at position 1067 within the GTP hydrolysis centre. J Mol Biol. 1988 Sep 20;203(2):457–465. doi: 10.1016/0022-2836(88)90012-5. [DOI] [PubMed] [Google Scholar]
- Thompson J., Cundliffe E., Stark M. Binding of thiostrepton to a complex of 23-S rRNA with ribosomal protein L11. Eur J Biochem. 1979 Jul;98(1):261–265. doi: 10.1111/j.1432-1033.1979.tb13184.x. [DOI] [PubMed] [Google Scholar]
- Thompson J., Schmidt F., Cundliffe E. Site of action of a ribosomal RNA methylase conferring resistance to thiostrepton. J Biol Chem. 1982 Jul 25;257(14):7915–7917. [PubMed] [Google Scholar]
- Thurlow D. L., Ehresmann C., Ehresmann B. Nucleotides in 16S rRNA that are required in unmodified form for features recognized by ribosomal protein S8. Nucleic Acids Res. 1983 Oct 11;11(19):6787–6802. doi: 10.1093/nar/11.19.6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurlow D. L., Shilowski D., Marsh T. L. Nucleotides in precursor tRNAs that are required intact for catalysis by RNase P RNAs. Nucleic Acids Res. 1991 Feb 25;19(4):885–891. doi: 10.1093/nar/19.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann R. A. Protein-RNA interactions in the bacterial ribosome. Methods Enzymol. 1979;59:551–583. doi: 10.1016/0076-6879(79)59113-7. [DOI] [PubMed] [Google Scholar]
- el-Baradi T. T., de Regt V. C., Einerhand S. W., Teixido J., Planta R. J., Ballesta J. P., Raué H. A. Ribosomal proteins EL11 from Escherichia coli and L15 from Saccharomyces cerevisiae bind to the same site in both yeast 26 S and mouse 28 S rRNA. J Mol Biol. 1987 Jun 20;195(4):909–917. doi: 10.1016/0022-2836(87)90494-3. [DOI] [PubMed] [Google Scholar]






