Abstract
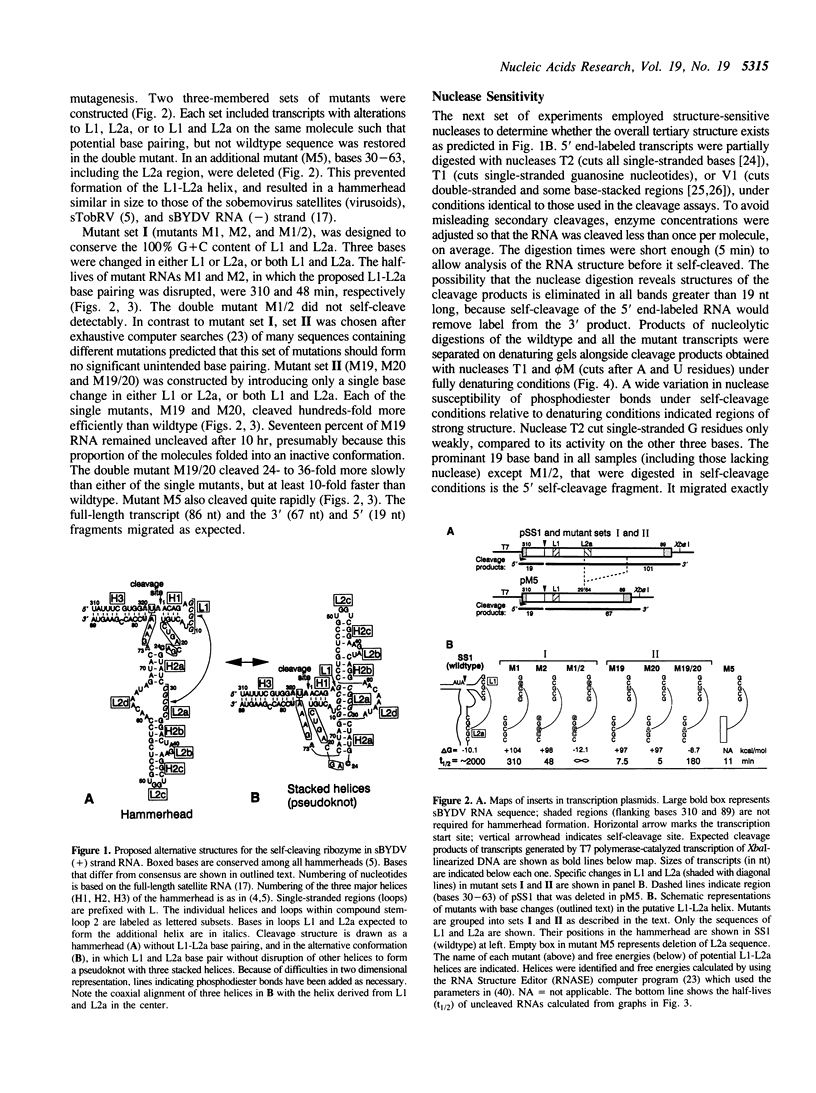
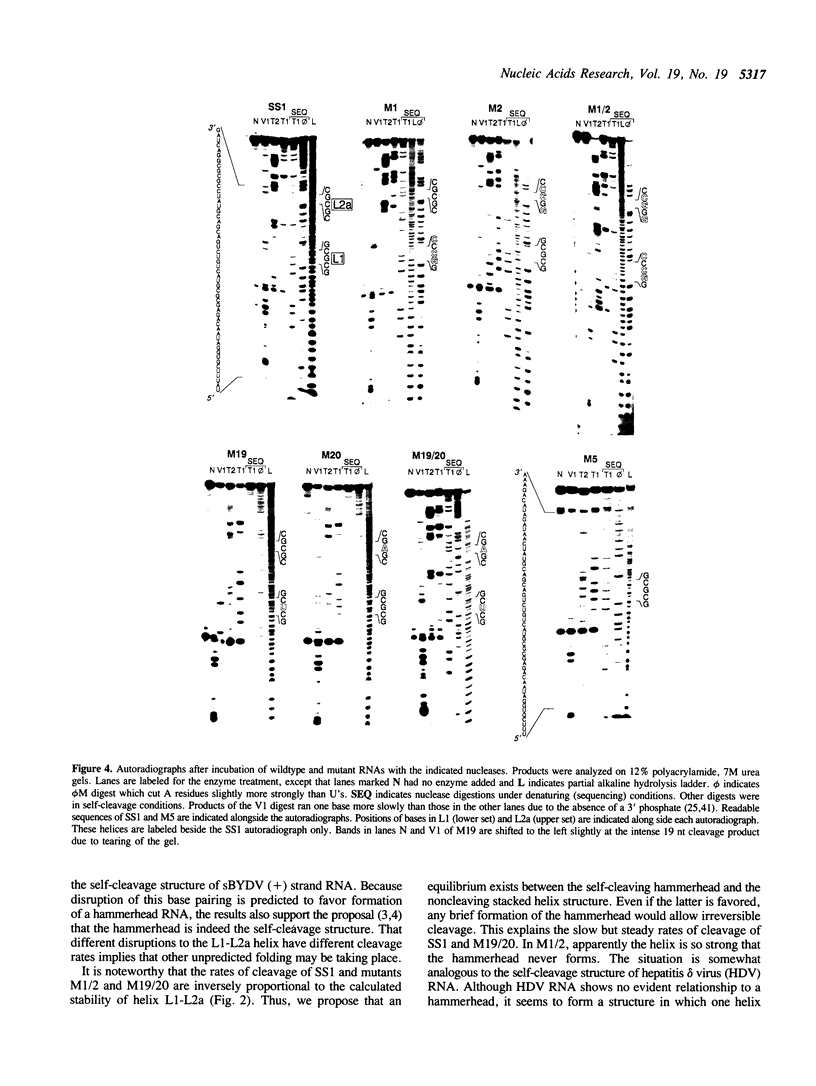
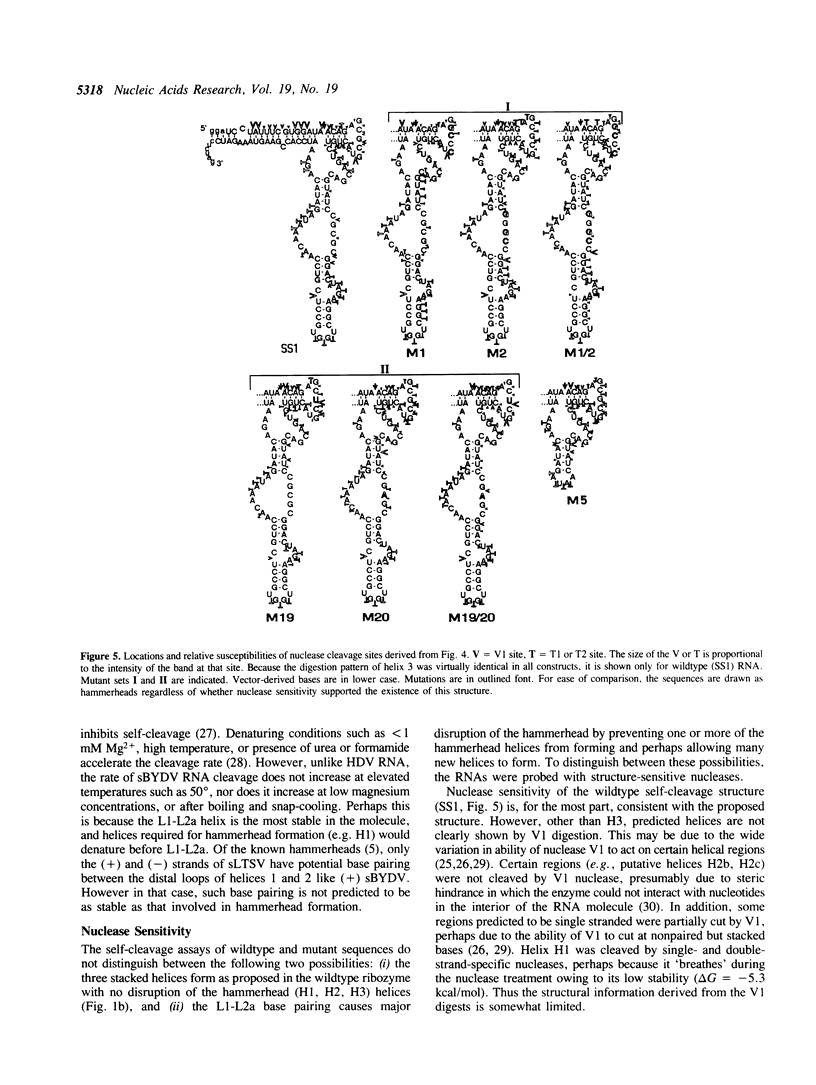
A self-cleaving satellite RNA associated with barley yellow dwarf virus (sBYDV) contains a sequence predicted to form a secondary structure similar to catalytic RNA molecules (ribozymes) of the 'hammerhead' class (Miller et al., 1991, Virology 183, 711-720). However, this RNA differs from other naturally occurring hammerheads both in its very slow cleavage rate, and in some aspects of its structure. One striking structural difference is that an additional helix is predicted that may be part of an unusual pseudoknot containing three stacked helices. Nucleotide substitutions that prevent formation of the additional helix and favor the hammerhead increased the self-cleavage rate up to 400-fold. Compensatory substitutions, predicted to restore the additional helix, reduced the self-cleavage rate by an extent proportional to the calculated stability of the helix. Partial digestion of the RNA with structure-sensitive nucleases supported the existence of the proposed alternative structure in the wildtype sequence, and formation of the hammerhead in the rapidly-cleaving mutants. This tertiary interaction may serve as a molecular switch that controls the rate of self-cleavage and possibly other functions of the satellite RNA.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bruening G. Compilation of self-cleaving sequences from plant virus satellite RNAs and other sources. Methods Enzymol. 1989;180:546–558. doi: 10.1016/0076-6879(89)80123-5. [DOI] [PubMed] [Google Scholar]
- Bruening G., Passmore B. K., van Tol H., Buzayan J. M., Feldstein P. A. Replication of a plant virus satellite RNA: evidence favors transcription of circular templates of both polarities. Mol Plant Microbe Interact. 1991 May-Jun;4(3):219–225. doi: 10.1094/mpmi-4-219. [DOI] [PubMed] [Google Scholar]
- Cedergren R., Gautheret D., Lapalme G., Major F. A secondary and tertiary structure editor for nucleic acids. Comput Appl Biosci. 1988 Mar;4(1):143–146. doi: 10.1093/bioinformatics/4.1.143. [DOI] [PubMed] [Google Scholar]
- Conway L., Wickens M. Modification interference analysis of reactions using RNA substrates. Methods Enzymol. 1989;180:369–379. doi: 10.1016/0076-6879(89)80112-0. [DOI] [PubMed] [Google Scholar]
- Davies C., Sheldon C. C., Symons R. H. Alternative hammerhead structures in the self-cleavage of avocado sunblotch viroid RNAs. Nucleic Acids Res. 1991 Apr 25;19(8):1893–1898. doi: 10.1093/nar/19.8.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H. Phy M: an RNase activity specific for U and A residues useful in RNA sequence analysis. Nucleic Acids Res. 1980 Jul 25;8(14):3133–3142. doi: 10.1093/nar/8.14.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein L. M., Gall J. G. Self-cleaving transcripts of satellite DNA from the newt. Cell. 1987 Feb 13;48(3):535–543. doi: 10.1016/0092-8674(87)90204-2. [DOI] [PubMed] [Google Scholar]
- Epstein L. M., Pabón-Peña L. M. Alternative modes of self-cleavage by newt satellite 2 transcripts. Nucleic Acids Res. 1991 Apr 11;19(7):1699–1705. doi: 10.1093/nar/19.7.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor M. J., Uhlenbeck O. C. Substrate sequence effects on "hammerhead" RNA catalytic efficiency. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1668–1672. doi: 10.1073/pnas.87.5.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster A. C., Davies C., Sheldon C. C., Jeffries A. C., Symons R. H. Self-cleaving viroid and newt RNAs may only be active as dimers. Nature. 1988 Jul 21;334(6179):265–267. doi: 10.1038/334265a0. [DOI] [PubMed] [Google Scholar]
- Forster A. C., Symons R. H. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987 Apr 24;49(2):211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- Forster A. C., Symons R. H. Self-cleavage of virusoid RNA is performed by the proposed 55-nucleotide active site. Cell. 1987 Jul 3;50(1):9–16. doi: 10.1016/0092-8674(87)90657-x. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J., Gerlach W. L. Sequences required for self-catalysed cleavage of the satellite RNA of tobacco ringspot virus. Gene. 1989 Oct 15;82(1):43–52. doi: 10.1016/0378-1119(89)90028-0. [DOI] [PubMed] [Google Scholar]
- Haseloff J., Gerlach W. L. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature. 1988 Aug 18;334(6183):585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- Heus H. A., Uhlenbeck O. C., Pardi A. Sequence-dependent structural variations of hammerhead RNA enzymes. Nucleic Acids Res. 1990 Mar 11;18(5):1103–1108. doi: 10.1093/nar/18.5.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins C. J., Rathjen P. D., Forster A. C., Symons R. H. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 1986 May 12;14(9):3627–3640. doi: 10.1093/nar/14.9.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp G. Enzymatic approaches to probing of RNA secondary and tertiary structure. Methods Enzymol. 1989;180:192–212. doi: 10.1016/0076-6879(89)80102-8. [DOI] [PubMed] [Google Scholar]
- Koizumi M., Iwai S., Ohtsuka E. Construction of a series of several self-cleaving RNA duplexes using synthetic 21-mers. FEBS Lett. 1988 Feb 15;228(2):228–230. doi: 10.1016/0014-5793(88)80004-8. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lockard R. E., Kumar A. Mapping tRNA structure in solution using double-strand-specific ribonuclease V1 from cobra venom. Nucleic Acids Res. 1981 Oct 10;9(19):5125–5140. doi: 10.1093/nar/9.19.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowman H. B., Draper D. E. On the recognition of helical RNA by cobra venom V1 nuclease. J Biol Chem. 1986 Apr 25;261(12):5396–5403. [PubMed] [Google Scholar]
- Miller W. A., Hercus T., Waterhouse P. M., Gerlach W. L. A satellite RNA of barley yellow dwarf virus contains a novel hammerhead structure in the self-cleavage domain. Virology. 1991 Aug;183(2):711–720. doi: 10.1016/0042-6822(91)91000-7. [DOI] [PubMed] [Google Scholar]
- Perrotta A. T., Been M. D. A pseudoknot-like structure required for efficient self-cleavage of hepatitis delta virus RNA. Nature. 1991 Apr 4;350(6317):434–436. doi: 10.1038/350434a0. [DOI] [PubMed] [Google Scholar]
- Pleij C. W., Rietveld K., Bosch L. A new principle of RNA folding based on pseudoknotting. Nucleic Acids Res. 1985 Mar 11;13(5):1717–1731. doi: 10.1093/nar/13.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi J. D., Wyatt J. R., Tinoco I., Jr A pseudoknotted RNA oligonucleotide. Nature. 1988 Jan 21;331(6153):283–286. doi: 10.1038/331283a0. [DOI] [PubMed] [Google Scholar]
- Rosenstein S. P., Been M. D. Self-cleavage of hepatitis delta virus genomic strand RNA is enhanced under partially denaturing conditions. Biochemistry. 1990 Sep 4;29(35):8011–8016. doi: 10.1021/bi00487a002. [DOI] [PubMed] [Google Scholar]
- Ruffner D. E., Dahm S. C., Uhlenbeck O. C. Studies on the hammerhead RNA self-cleaving domain. Gene. 1989 Oct 15;82(1):31–41. doi: 10.1016/0378-1119(89)90027-9. [DOI] [PubMed] [Google Scholar]
- Ruffner D. E., Stormo G. D., Uhlenbeck O. C. Sequence requirements of the hammerhead RNA self-cleavage reaction. Biochemistry. 1990 Nov 27;29(47):10695–10702. doi: 10.1021/bi00499a018. [DOI] [PubMed] [Google Scholar]
- Schulz V. P., Reznikoff W. S. In vitro secondary structure analysis of mRNA from lacZ translation initiation mutants. J Mol Biol. 1990 Jan 20;211(2):427–445. doi: 10.1016/0022-2836(90)90363-Q. [DOI] [PubMed] [Google Scholar]
- Sheldon C. C., Symons R. H. Mutagenesis analysis of a self-cleaving RNA. Nucleic Acids Res. 1989 Jul 25;17(14):5679–5685. doi: 10.1093/nar/17.14.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon C. C., Symons R. H. RNA stem stability in the formation of a self-cleaving hammerhead structure. Nucleic Acids Res. 1989 Jul 25;17(14):5665–5677. doi: 10.1093/nar/17.14.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoncsits A., Brownlee G. G., Brown R. S., Rubin J. R., Guilley H. New rapid gel sequencing method for RNA. Nature. 1977 Oct 27;269(5631):833–836. doi: 10.1038/269833a0. [DOI] [PubMed] [Google Scholar]
- Singleton C. K., Klysik J., Stirdivant S. M., Wells R. D. Left-handed Z-DNA is induced by supercoiling in physiological ionic conditions. Nature. 1982 Sep 23;299(5881):312–316. doi: 10.1038/299312a0. [DOI] [PubMed] [Google Scholar]
- Symons R. H. The intriguing viroids and virusoids: what is their information content and how did they evolve? Mol Plant Microbe Interact. 1991 Mar-Apr;4(2):111–121. doi: 10.1094/mpmi-4-111. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Davis P. W., Hardin C. C., Puglisi J. D., Walker G. T., Wyatt J. RNA structure from A to Z. Cold Spring Harb Symp Quant Biol. 1987;52:135–146. doi: 10.1101/sqb.1987.052.01.018. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck O. C. A small catalytic oligoribonucleotide. Nature. 1987 Aug 13;328(6131):596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- Vary C. P., Vournakis J. N. RNA structure analysis using T2 ribonuclease: detection of pH and metal ion induced conformational changes in yeast tRNAPhe. Nucleic Acids Res. 1984 Sep 11;12(17):6763–6778. doi: 10.1093/nar/12.17.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarling D. A., Calhoun C. J., Feuerstein B. G., Sena E. P. Cytoplasmic microinjection of immunoglobulin Gs recognizing RNA helices inhibits human cell growth. J Mol Biol. 1990 Jan 5;211(1):147–160. doi: 10.1016/0022-2836(90)90017-G. [DOI] [PubMed] [Google Scholar]
- Zarling D. A., Calhoun C. J., Hardin C. C., Zarling A. H. Cytoplasmic Z-RNA. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6117–6121. doi: 10.1073/pnas.84.17.6117. [DOI] [PMC free article] [PubMed] [Google Scholar]





