Abstract
The centromere is the locus that directs chromosomal inheritance at cell division. While centromeres in diverse eukaryotes are commonly found at sites of repetitive DNA, their location is epigenetically specified. The histone H3 variant CENP-A is the prime candidate for epigenetically marking the centromere, and recent work has uncovered several additional proteins that play key roles in centromere assembly and maintenance. We describe advances in the identification and characterization of proteins that form the centromere, and focus on recent findings that have advanced our understanding of the assembly of functional centromeric chromatin.
1. Introduction
The centromere is the locus that ensures proper segregation of chromosomes from one generation to the next. Centromeres are typically housed within large (e.g. megabases in mammals) repetitive DNA elements [1,2], and define the site of formation of mitotic kinetochores that mediate chromosomal attachment to the mitotic microtubule-based spindle [3]. In all eukaryotes, except for budding yeast, the centromere is thought to be maintained through an epigenetic mechanism, and the most attractive candidate to provide the centromere-specifying epigenetic mark is a histone variant that replaces H3 in centromeric nucleosomes. This variant is called centromeric protein A (CENP-A) in humans [4], Cse4 in budding yeast [5], Cnp1 in fission yeast [6], CID in fruit flies [7], HCP-3 in roundworms [8], and CenH3 in plants [9].
The CENP-A protein was originally isolated from the nuclei of calf thymus and bull sperm [10], and the tissues/cells of origin provided strong hints of its role in both somatic and germline chromosomal inheritance. Indeed, deletion of CENP-A in diverse organisms leads to severe defects in chromosome segregation and mitotic checkpoint functions [5,8,11-17]. The loss of a functioning centromere, without the formation of a de novo centromere (neocentromere), is catastrophic for the integrity of the genome in dividing cells and leads to chromosomal mis-segregation, generating aneuploidy (i.e. the gain or loss of one or more chromosomes) [18]. Aneuploidy is a hallmark of cancer cells [19], and aneuploidy generated during meiotic divisions leads to the most common form of prenatal death as well as the most common cause of mental retardation in humans [20]. The aberrant gain of an additional centromere on a chromosome leads to genomic instability via chromosomal breakage during cell division [21]. In sum, maintaining one (and only one) centromere per chromosome is key for the fidelity of the genome.
While the presence of an array of CENP-A-containing nucleosomes is broadly conserved, the surrounding chromatin environment varies from one species to the next. Centromere repeat sequences are common, but the sequences highly diverge between species and the number of repeats is extremely variable even in a single species [22]. There is also enormous diversity between species in the expanse of the chromosome covered by the centromere, deviating strongly from prototypical ‘regional’ centromeres (e.g. regional centromeres found in fission yeast, fruit flies, maize, mammals). Two particularly strong deviations are in the popular model organisms budding yeast and roundworms. The smallest centromeres are found in the budding yeast Saccharomyces cerevisiae and some related fungal species, where 125 bp defines the centromere [23-26]. Indeed, there is only one stable Cse4-containing nucleosome at budding yeast centromeres [5,27,28]. The roundworm Caenorhabditis elegans is one of many eukaryotic species that have holocentric chromosomes [29,30], where centromeres form along the entire length of the chromosome in a manner that is presumably discontinuous on the linear DNA sequence, but where spindle microtubule attachments occur at only a small subset of centromeric sites [31].
2. CENP-A and centromeric chromatin
CENP-A and H3 share sequence homology within their histone fold domains (~60% in humans), but there is no sequence identity at the N-terminus [32]. Despite the N-terminal sequence divergence found between CENP-A and H3, the information required to deposit CENP-A at centromeres is in the histone fold domain, where loop 1 and the α2 helix comprise the CENP-A Targeting Domain (CATD) [33]. Indeed, a chimeric H3 protein that contains the 22 amino acid substitutions that generate the CATD (H3CATD) targets to centromeres [33,34]. H3CATD expression rescues the lethal knockdown of endogenous CENP-A [34]. The CATD also induces conformational rigidity, as measured by hydrogen/deuterium exchange experiments that measure the polypeptide backbone dynamics of the histones, in (CENP-A/H4)2 tetramers and CENP-A-containing nucleosomes relative to the conventional counterparts containing histone H3 [33,35,36]. In budding yeast, the CATD of CENP-ACse4 was found to confer CENP-ACse4 ubiquitylation in order to ensure that excess CENP-ACse4 protein, if present, is rapidly degraded [37]. In frog egg extracts, however, the C-terminal four-to-six a.a. of CENP-A is sufficient, when substituted onto conventional histone H3 in reconstituted nucleosome arrays, to recruit functional kinetochores [38]. This recent finding, thus, describes a system where pre-assembly of CENP-A nucleosomes bypasses the requirement for the CATD and indicates a critical role for the unstructured C-terminal “tail” of CENP-A in kinetochore assembly [38]. Nonetheless, unique structural and dynamic features conferred by CENP-A are vital for propagating centromere identity and have likely importance for centromere/kinetochore assembly events in other cell types and/or eukaryotic species.
2.1 The structure of CENP-A-containing histone complexes and CENP-A-containing nucleosomes
The first high-resolution structural information on CENP-A came from the human sub-nucleosomal (CENP-A/H4)2 heterotetramer [36]. The unique features of (CENP-A/H4)2 relative to conventional complexes containing H3, revealed by crystal structures and solution studies (small angle x-ray scattering), include an overall 10 Å compaction due to rotation at the CENP-A/CENP-A interface, a bulge in the loop 1 of CENP-A of the opposite charge as on H3, and hydrophobic interactions at the CENP-A/H4 interface that provide the structural basis for the conformational rigidity measured by hydrogen/deuterium exchange [33,36]. All three of these changes in CENP-A map to the CATD, and led to the proposal that CENP-A distinguishes centromeres from the rest of the chromosome via structural deviation from within the folded octameric (i.e. [CENP-A/H4/H2A/H2B]2) core of the nucleosome. Such an octamer is not the only possibility for CENP-A-containing nucleosomes and there have been several proposals for alternative arrangements involving different histone stoichiometry, non-histone incorporation, and/or reverse handedness of DNA wrapping [36,39-47] that are discussed in detail elsewhere along with the proposal that CENP-A-containing nucleosomes may mature during the cell cycle through intermediate steps [48].
The first crystal structure of a CENP-A-containing nucleosome structure has been reported more recently [49]. While 147 bp of DNA were used to wrap the nucleosome, only the central 121 bp are visible in the structure [49]. This is consistent with CENP-A-containing nucleosomes preferring an ‘open’ conformation with transient unwrapping of the final turn(s) of DNA at the superhelical termini [50,51]. The crystal structure [49] also confirmed the surface exposure of the bulged L1 [36], as well as nearly identical side-chain orientations at the CENP-A/H4 interface that are proposed to be responsible for the conformational rigidity of CENP-A/H4 nucleosomes measured by hydrogen-deuterium exchange [35,36]. The rotation of the CENP-A/CENP-A interface could cause the H2B/H4 four-helix bundle to rotate to avoid steric clashing and result in a nucleosome with an overall altered shape relative to canonical nucleosomes [36]. Alternatively, the CENP-A/CENP-A interface could rotate upon nucleosome formation leading to an overall structure that is highly similar to canonical nucleosomes. The latter scenario with a highly similar overall shape to canonical nucleosomes is clearly what occurs in the reported crystal structure of CENP-A-containing nucleosomes [49]. On the other hand, the possibility of alternative conformations involving CENP-A/CENP-A rotation in polynucleosome arrays, such as those found at centromeres, should be explored since the C-terminal ~45 a.a. of H2A exhibit higher rates of hydrogen/deuterium exchange than in arrays containing conventional nucleosomes, perhaps indicative of an altered overall nucleosome shape [51].
2.2 Centromeric chromatin
Beyond individual nucleosomes, short stretches of CENP-A-containing nucleosomes at each centromere are interspersed linearly along the DNA with stretches of conventional nucleosomes containing canonical H3, but all CENP-A nucleosomes in an individual centromere nonetheless coalesce in three dimensions; a feature that is conserved in vertebrates, invertebrates, and plants [52,53]. Indeed, several proposals for a regular geometry of centromeric chromatin have been put forward [3,52,54,55]. Elsewhere in the genome, long-distance chromosome associations are mediated by bridging proteins bound at so-called locus control regions [56-58]. At the centromere, some form of bridging seems likely to coalesce the CENP-A-containing nucleosomes at each centromere. Potential candidates for this role come from the CENP-A Nucleosome Associated Complex (CENP-ANAC), a subset of what is collectively termed the Constitutive Centromere Associated Network (CCAN) of 16 proteins [40,59, reviewed in 60].
While a broad understanding of what particular centromere functions each of the CCAN components perform remains incomplete, there has been some recent progress in this area. For instance, CCAN component CENP-C has been shown to bind to the C-terminal ‘tail’ of CENP-A [61] and is proposed to bridge interactions between CENP-A nucleosomes and other centromere proteins [38,61-64]. CCAN components CENP-T and CENP-W form a close complex and are proposed to interact with H3 nucleosomes interspersed with CENP-A nucleosomes [40,63,64], perhaps leading to structural alterations that facilitate microtubule assembly at the kinetochore prior to mitosis [65]. In addition, CENP-T undergoes tension-dependent changes throughout the cell cycle, which could alter the shape of the inner kinetochore before or after microtubule binding [66]. Artificial tethering of CENP-C and CENP-T to an ectopic chromosomal locus is sufficient to completely bypass the requirement for CENP-A nucleosomes in mitosis [67]. In addition to these components, CCAN component CENP-N has been shown to bind directly to CENP-A-containing nucleosomes [38,68], and depletion of CENP-N decreases newly synthesized CENP-A deposition at the centromere [68].
The extensive protein/protein interactions within the CCAN [40,59-68] make components therein likely candidates to bridge CENP-A nucleosomes to each other and to other centromere/kinetochore components, ultimately linking centromeric chromatin to the outer kinetochore and spindle microtubules. Loss of these proteins could prevent important temporal and spatial contacts necessary for a centromere to function normally. Alternatively, it is also possible that self-self interactions between CENP-A-containing nucleosomes on the same chromosome (or physical features of the nucleosome that delineate it somehow from bulk chromatin) could drive coalescence in three dimensions. In addition, inter-chromosomal (or inter-chromatid) coalescence at the kinetochore has been observed in some specific biological contexts [69-72]. Centromeric chromatin organization does not appear to be random and culminates during mitosis in generating a unique chromatin surface that is competent for kinetochore assembly and passes the physical requirements for chromosome segregation.
The chromatin environment surrounding CENP-A-containing nucleosomes is also proposed to be involved in stabilizing centromeres. Several studies have shown distinct posttranslational modifications on the other core histones near CENP-A-containing chromatin. One experimental approach to assess the function of surrounding centromeric chromatin utilizes cell lines harboring human artificial chromosomes (HACs). Targeted disruption of H3K4me2 and H3K9me3, for instance, led to loss of CENP-A and HAC stability [73,74]. Further experiments identified a vital role for nucleosomes enriched with H3K4me2 and H3K36me2, as suggested by loss of CENP-A and kinetochore structure at HAC centromeric regions when these modifications were disrupted [74]. Another recent study using DT40 chicken cells found high levels of H3K9me3 and, in contrast to the HAC constructs [73,74], low levels of H3K4me2 interspersed with CENP-A nucleosomes [55]. In naturally occurring neocentromeres, some marks, such as H3K4me2 and H3K9me3, have been shown to be nearly absent [75], challenging the view that these particular chromatin modifications play a role where a relatively high stoichiometry is required. Posttranslational modifications on CENP-A itself are largely unknown beyond the mitotic phosphorylation by Aurora B [76,77] and the aforementioned ubiquitylation [37,78,79], and they represent potential additional modes of differentiating centromeric chromatin from the rest of the chromosome.
3. Centromere assembly and the cell cycle
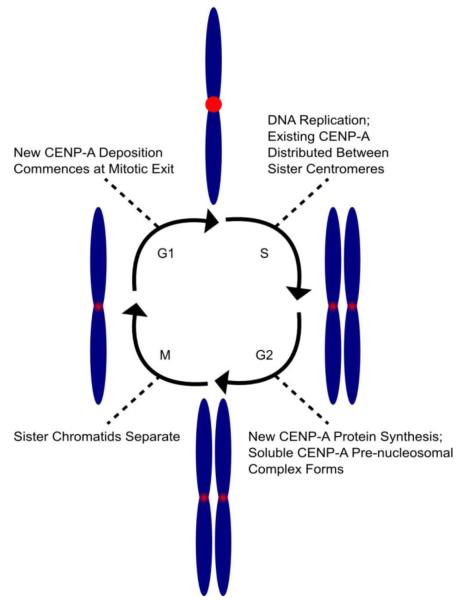
Centromeric DNA is replicated during mid-to-late S phase in Drosophila and human cells (Fig. 1) [39,80,81]. Using fluorescence pulse-chase labeling, it is clear that existing (i.e. ‘old’) CENP-A protein is equally distributed on both daughter strands in the complete absence of any new CENP-A deposition [82], diluting the amount of CENP-A to half its initial density on centromeric DNA following S phase. New CENP-A protein is synthesized after S phase in G2 and deposited later during mitotic exit and following G1 in human cells and fruit fly embryos [39,80,82-84]. This cell cycle timing is distinct from the H3 variants found in bulk chromatin, H3.1 (canonical H3), H3.2, and H3.3 [85-88]. H3.1/H3.2 are both synthesized and deposited into chromatin during S phase [85,89,90], and associate with CAF-1 [91-94], a chromatin assembly factor that associates with PCNA [95], the sliding DNA clamp used during DNA synthesis [96]. H3.3, on the other hand, is synthesized [86] and deposited [97] throughout the cell cycle with the aid of the histone chaperones HIRA [98] and DAXX [99-101]. The uncoupling of CENP-A protein synthesis/deposition from DNA synthesis [39,80,82-84] gave an early indication that dedicated mechanisms in the cell exist to duplicate the pool of CENP-A nucleosomes to refill the available sites at every centromere and avoid dilution of the centromere specifying chromatin mark through subsequent cell cycles.
Fig. 1.
Model of CENP-A containing chromatin throughout the cell cycle in animals. The levels of CENP-A at centromeres change with the cell cycle. Prior to S phase, CENP-A is fully loaded at the centromere, but upon replication, the number of CENP-A molecules present on each daughter strand are reduced to half per centromere copy as no new CENP-A protein is added. During G2, new CENP-A is synthesized and assembles into a soluble complex with its binding partner H4 and its chaperone HJURPScm3, but is not deposited at centromeres until G1. Cells progress through mitosis with half-loaded centromeres. During late anaphase/telophase, HJURPScm3 begins to deposit CENP-A to duplicate CENP-A protein levels.
3.1 Priming/licensing centromeric chromatin for CENP-A deposition
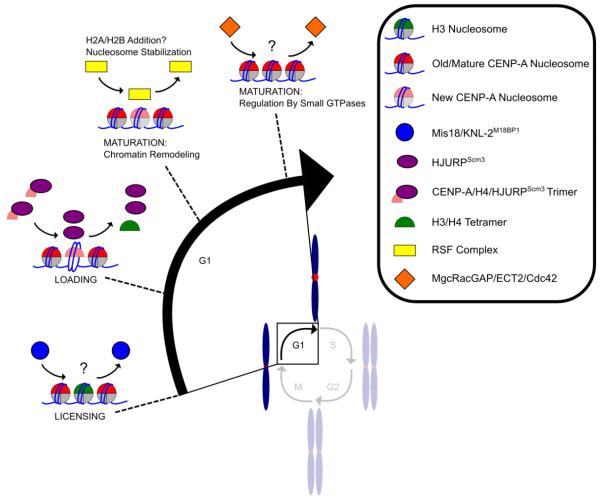
The first known step to achieve duplication of CENP-A-containing nucleosomes during mitotic exit and the subsequent G1 phase of the cell cycle is a priming or licensing step involving Mis18 (first identified in S. pombe and later in mammals, worms, and other eukaryotes) and KNL-2 (Fig. 2) [102-104]. The S. pombe proteins Mis16 and Mis18 were initially found to function in localizing CENP-ACnp1 to centromeres [102]. Mis16 is the orthologue of human RbAp46/48, chromatin assembly proteins that bind to prenucleosomal histone H4 [105-107] when it is in complex with H3 [91,108] or CENP-A [109,110]. Both S. pombe lacking Mis16 and HeLa cells lacking RbAp46/48 drastically reduce levels of CENP-A at centromeres [102], in addition to presumed major bulk chromatin disruptions. In human cell lines, knockdown of Mis18 (a two subunit complex containing hMis18α and hMis18β) prevented loading of newly synthesized CENP-A at the centromere [103]. In C. elegans, an RNAi screen for proteins required to localize CENP-A to the centromere identified KNL-2, a Myb-domain protein that localizes to centromeres throughout mitosis [104] that is the orthologue of human M18BP1 (herein referred to as KNL-2M18BP1) [103]. While depletion of KNL-2M18BP1 prevents CENP-A loading at centromeres in both worms and human cells [103,104], the cell-cycle timing of the two orthologues differ. In worms, KNL-2M18BP1 and CENP-A colocalize at centromeres throughout the cell cycle while human KNL-2M18BP1 localizes to centromeres only during late anaphase/telophase and early G1 [103,104]. Further evidence suggests that CENP-C recruits the KNL-2M18BP1 complex to centromeres as early as metaphase [111]. What exactly is the priming/licensing reaction? No members of the Mis18/KNL-2M18BP1 complex have been reported to physically contact CENP-A itself, but their loss indirectly affects CENP-A levels at the centromere. While there is some evidence that the complex may alter the state of histone acetylation at centromeres [102,103], the succulent details of centromere licensing and priming await further investigation.
Fig. 2.
Model of new CENP-A deposition and maturation during G1 in animals. During mitotic exit, centromeric chromatin is not competent for CENP-A loading until Mis18 and M18BP1KNL-2 license centromeric chromatin. Two HJURPScm3 molecules deposit newly synthesized CENP-A into the DNA. In mid/late G1, the RSF complex accumulates at centromeres and is implicated in stabilizing new CENP-A, as are MgcRacGAP, ECT2, and Cdc42, which are proposed to further stabilize the newly deposited pool of CENP-A nucleosomes.
Findings from studies in Drosophila indicate the involvement of at least two components, RCA1 (orthologous to human Emi1) and cyclin A, that regulate the anaphase promoting complex early in mitosis [112]. This regulation may provide an even earlier licensing step, since a discrete pool of cyclin A protein localizes to centromeres in flies [112]. Additionally, CENP-ACID deposition begins prior to the metaphase-to-anaphase transition in cultured fly cells [113], as opposed to the later licensing and deposition steps in cultured mammalian cells [82].
3.2 HJURPScm3: The CENP-A specific chaperone
CENP-A deposition occurs with its partner H4 and requires a histone chaperone (Fig. 2), similar to the H3 versions found elsewhere in the genome. Histone chaperones are proteins that assist in histone deposition into (and removal from) nucleosomes, while also preventing unwanted and incorrect interactions with other proteins and regions of DNA [114]. The first identified histone chaperone, nucleoplasmin, was purified from Xenopus laevis eggs and prevented precipitation of DNA and histones in solution [115]. After this initial discovery, chaperones for several histone complexes were discovered, including: CAF-1 for H3.1/H4 [116], FACT for H2A/H2B [117], HIRA for H3.3/H4 heterotetramers/heterodimers [98], and several others (reviewed in [118]). For CENP-A, multiple lines of evidence suggest that Holliday Junction Recognition Protein (HJURP) is the CENP-A-specific chaperone in many eukaryotes [110,119]. HJURP is enriched at centromeres at the end of telophase and early G1 [110,119,120], and its centromere targeting requires Mis18 [121]. Depletion of HJURP leads to the loss of new CENP-A deposition and defects in chromosome segregation [110,119,122]. HJURP is the orthologue of the yeast Scm3 protein that is essential for centromere function (herein referred to as HJURPScm3) [46,123,124]. The N-terminal 80 amino acids of HJURPScm3 are the only region that shows any detectable homology to yeast HJURPScm3 [125] and is also the domain that interacts with CENP-A [126]. Prior to the proposal that yeast HJURPScm3 is a chaperone for CENP-ACse4 [42,127], it was already known to physically associate with CENP-ACse4 [46] and provide a necessary step in CENP-ACse4 targeting to centromeres [46,123,124]. Further, HJURPScm3 can chaperone assembly of (CENP-A/H4)2 tetrasomes [126] and octameric (CENP-ACse4/H4/H2A/H2B)2 nucleosomes [121,128,129] using purified components and conventional nucleosome assembly approaches.
In budding yeast, reconstituting CENP-ACse4 nucleosomes with HJURPScm3 using the salt dialysis method resulted in HJURPScm3 incorporation into a nucleosome-like particle containing HJURPScm3, CENP-ACse4, and H4 on the extremely AT-rich DNA of budding yeast centromeres [47]. As opposed to in fission yeast where HJURPScm3 is absent from centromeres during mitosis [127], budding yeast HJURPScm3 remains stably incorporated at centromeric sites throughout the course of the cell cycle [47], but varies in absolute levels [130] suggesting that HJURPScm3 is usually a component of the S. cerevisiae centromere as opposed to a transient component during new CENP-A nucleosome assembly as in mammals.
Three high-resolution structures of HJURPScm3 in complex with CENP-A/H4 have recently been reported, using S. cerevisiae, Kluyveromyces lactis and Homo sapiens proteins, respectively [131-133]. They all show contacts between an α-helix of HJURPScm3 with the α2-helix within the CATD of CENP-A, and all show that the binding of HJURPScm3 blocks the formation of the four-helix bundle of the sub-nucleosomal (CENP-A/H4)2 heterotetramer [36,131-133]. The three studies also describe some form of occlusion of portions of the DNA binding ridge of CENP-A/H4 [36,49] by HJURPScm3 binding [131-133], but there was disagreement about the location of these regions and the mode of occlusion by HJURPScm3 [131-133]. The two yeast structures disagreed sharply in terms of overall structure and the recognition residues of CENP-ACse4 important for recognition by HJURPScm3, despite starting with nearly identical orthologues from the related budding yeasts S. cerevisiae [131] and Kluyveromyces lactis [132], with the major differences likely emerging from the inclusion and/or exclusion of sequences in the constructs used to express the constituents. In addition, the S. cerevisiae structure [131] was solved using a single-chain molecule (Cse4-Scm3-H4) wherein one of the key helices (α1) of the histone fold of H4 was omitted, which others have stated may have led to an unnatural structure [132].
In the case of the human structure, one major surprise was that despite the fact that the H3CATD chimera is capable of binding to HJURPScm3 [119,126,133], and despite the fact that the major binding surface of HJURPScm3 and CENP-A comprises the L1 and α2-helix [133] that comprise the entire CATD [33], the authors concluded that there are no good candidate residues in the CATD for specific recognition by HJURPScm3 [133]. Instead, a single residue, Ser68 in the α1-helix of CENP-A, was proposed to be the primary specificity determinant for HJURPScm3 binding [133]. This conclusion was supported by GST-pulldown experiments using single amino acid swap mutations in CENP-A and H3 (the corresponding residue in H3 is a glutamine) where loss or gain, respectively, of HJURPScm3 binding was reported [133].
In sum, despite these three recent structures [131-133], there is not yet a clear picture of how CENP-A is actually sorted by HJURPScm3 away from the vast excess of other H3 variants that are expressed at high levels to fill bulk chromatin on the rest of the chromosome. One contributing factor could be the temporal differences between H3.1 (the major form of H3) and CENP-A protein synthesis [39,80,82-84], which would allow HJURPScm3 to associate with newly made CENP-A during G2 after H3.1 synthesis occurs [85-90]. However, the H3.3 ‘replacement’ variant is synthesized throughout the cell cycle [85-88] and can be interspersed with CENP-A nucleosomes in G2 [134], so that CENP-A is not temporally sorted away from H3.3 in any simple fashion.
HJURPScm3 binding blocks CENP-A/CENP-A tetramerization, indicating that like the Asf1-mediated bulk chromatin assembly pathway [94,135,136], it deposits a heterodimer of CENP-A/H4 instead of a (CENP-A/H4)2 heterotetramer, but the final product is thought to be an octameric nucleosome harboring a sub-nucleosomal (CENP-A/H4)2 heterotetramer. Temporary tethering of HJURPScm3 at an ectopic site on a chromsome recruits CENP-A that is stably retained after subsequent removal of HJURPScm3 and assembles a de novo kinetochore at mitosis [121].
3.3 Maturing newly deposited CENP-A complexes into a stable form of centromeric chromatin
One ‘maturation’ step for conventional nucleosomes involves one or more ATP-dependent remodeling events that help complete nucleosome assembly and/or space newly deposited histone complexes [137]. At the centromere, the generic ATP-dependent remodeler, the RSF complex [138], has also been proposed to perform such a function after initial CENP-A deposition in G1 [139], but its exact role at the centromere remains unclear (Fig. 2). One possibility is that this chromatin remodeling event is when H2A/H2B heterodimers are added. Further, immunoprecipitation of KNL-2M18BP1 in HeLa cells followed by mass spectrometry identified MgcRacGAP, a GTPase activating protein (GAP) from the Rho GTPase family, as a protein interactor with KNL-2M18BP1 that has been implicated even later in G1 as an essential protein for stabilizing newly deposited CENP-A protein (Fig. 2) [140]. Based on the interaction with KNL-2M18BP1, MgcRacGAP was thought to function during the priming step, but depletion of MgcRacGAP did not alter KNL-2M18BP1 localization or CENP-A stability [140] – implying a role in nascent CENP-A nucleosome assembly/maturation downstream of priming. MgcRacGAP, along with ECT2 (a guanine nucleotide exchange factor) and the GTPase Cdc42, were proposed to create a GTPase cycle to maintain CENP-A at centromeres after it is deposited into DNA [140], but how this signaling culminates in CENP-A stabilization remains mysterious.
3.4 CENP-A dilution during S phase
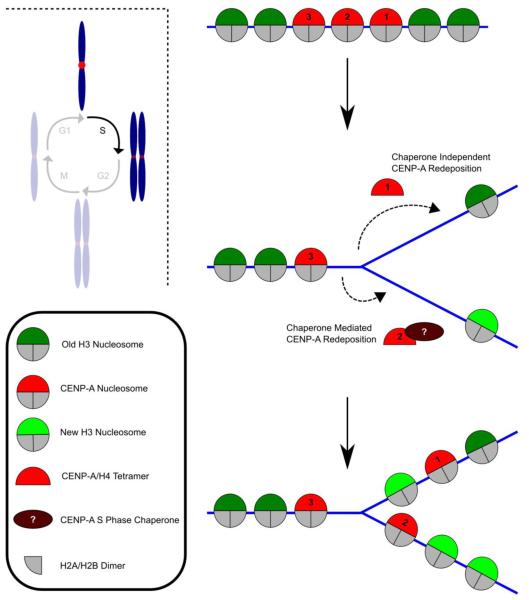
When the replication fork passes through chromatin, CENP-A is very likely to be ejected, at least temporarily, in order for DNA synthesis to occur. The mechanism for CENP-A deposition back into chromatin is unknown, as is whether or not it requires an unidentified S phase specific chaperone (Fig. 3). In either case, it is also unclear if mammalian CENP-A-complexes are retained locally and rapidly deposited behind the replication fork, a proposed model for retaining epigenetic information on H3/H4 at chromosome arm loci [141], or if it must be re-targeted back to centromeres by a dedicated pathway. Photobleaching experiments on groups of centromeres in the vicinity of PCNA-containing foci that were tracked for a few hours in S phase found only very low levels of fluorescence recovery [83], but dynamics of CENP-A at individual centromeres of metazoans have not been measured over the course of a cell cycle or complete phases therein. Since HJURPScm3 specifically acts on newly expressed CENP-A/H4 complexes [119], it is not a good candidate for an S phase chaperone, if one indeed exists. Condensin II is also reported to play a role in maintaining pre-existing CENP-A at the centromere during interphase, but it remains unclear whether or not this occurs specifically during S phase [122]. If an S phase chaperone does exist, what would restrict it to redepositing CENP-A at the same centromeric site it originated from? CENP-A could be re-targeted to a centromere on a neighboring chromosome instead, but whether or not this occurs during S phase has yet to be determined.
Fig. 3.
Model of dilution of CENP-A-containing nucleosomes during S phase in animals. During replication, nucleosomes are presumably ejected from DNA as the replication fork progresses. In the diagram, CENP-A nucleosome positions are indicated as 1-3 to follow each site after replication and redeposition steps. CENP-A could be redeposited through a passive process without the aid of a chaperone (CENP-A nucleosome position 1) or it could interact with an unidentified chaperone that deposits it back into DNA behind the moving fork (CENP-A nucleosome position 2). Because new CENP-A is not synthesized until G2, only “old” reassembled CENP-A-containing nucleosomes are deposited back into the DNA, leaving behind nucleosome assembly sites filled by deposition of a canonical H3-containing nucleosome.
On the other hand, CENP-A deposition back into chromatin could be purely mediated by a passive process independent of a specific chaperone, where a large concentration of CENP-A-complexes near the replication fork drives their selective re-assembly at adjacent chromatin via local diffusion (Fig. 3). Regardless of the mechanism and the nature of the sub-nucleosomal histone complex (likely to contain CENP-A/H4 heterodimers or [CENP-A/H4]2 heterotetramers) liberated from centromeric DNA by the replication machinery, the final product is strongly proposed to be two new daughter strands each with half the amount of CENP-A per unit length of centromeric DNA as on the mother strand (Fig. 3) [82,134].
With only half the amount of CENP-A distributed to newly synthesized DNA, there would in theory be gaps left in the DNA where no new nucleosomes are deposited. Recent evidence using stretched chromatin fibers and labeled H3.3 indicate that H3.3 is deposited in the gaps left by CENP-A dilution during S phase, but once cells enter G1 the amount of H3.3 at the centromere is reduced [134]. Therefore, H3.3 may act as a placeholder until deposition of new CENP-A occurs later in the cell cycle, but it is unclear how H3.3 is evicted from centromeric chromatin once newly expressed CENP-A arrives.
4. Concluding remarks
Many questions at the centromere remain to be answered. For instance, the primary centromere paradoxes continue to confound the field regarding (1) the rapid evolution [22] and (2) the functional dispensability [142] of the repetitive DNA sequences found at the centromeres of a preponderance of diverse eukaryotic species. In addition, it is not clear why some centromere components and chromatin features are required for efficient centromere formation but then appear to be dispensable once formed [143,144]. Why would there need to be special information solely required for new centromere establishment if the formation of a de novo centromere is not something typically encountered during the lifetime of an organism? Despite such unanswered questions, the recent studies discussed here have clearly advanced our view of how the centromere is organized and assembled over somatic cell divisions. The histone variant CENP-A is considered the key epigenetic mark of the centromere, but a variety of proteins contribute to its assembly, maintenance, and stability at the centromere. Key events throughout the cell cycle ensure that CENP-A and other centromeric proteins are present during the proper time and at the proper place. Future work is now needed to understand the precise nature of the epigenetic mechanisms that specify centromere location, as well as to understand the paradoxical relationship between epigenetic and genetic contributions to centromere function.
Highlights.
Structure of CENP-A complexes provide insight into centromere function and propagation.
The levels of CENP-A present at the centromere change with the cell cycle.
The CENP-A deposition pathway during G1 requires distinct steps.
S phase CENP-A redeposition may occur with or without the assistance of chaperones.
Acknowledgements
Work in the Black Lab is supported by a grant from the National Institutes of Health (GM082989), a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund, and a Rita Allen Foundation Scholar Award. S.J.F. is supported in part by the University of Pennsylvania Genetics Training Grant from the National Institutes of Health (5T32GM008216).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Manuelidis L, Wu JC. Homology between human and simian repeated DNA. Nature. 1978;276:92–94. doi: 10.1038/276092a0. [DOI] [PubMed] [Google Scholar]
- [2].Willard HF. Chromosome-specific organization of human alpha satellite DNA. Am. J. Hum. Genet. 1985;37:524–532. [PMC free article] [PubMed] [Google Scholar]
- [3].Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Earnshaw WC, Migeon BR. Three related centromere proteins are absent from the inactive centromere of a stable isodicentric chromosome. Chromosoma. 1985;92:290–296. doi: 10.1007/BF00329812. [DOI] [PubMed] [Google Scholar]
- [5].Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- [6].Takahashi K, Chen ES, Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- [7].Henikoff S, Ahmad K, Platero JS, van Steensel B. Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci. U.S.A. 2000;97:716–721. doi: 10.1073/pnas.97.2.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Buchwitz BJ, Ahmad K, Moore LL, Roth MB, Henikoff S. A histone-H3-like protein in C. elegans. Nature. 1999;401:547–548. doi: 10.1038/44062. [DOI] [PubMed] [Google Scholar]
- [9].Talbert PB, Masuelli R, Tyagi AP, Comai L, Henikoff S. Centromeric Localization and Adaptive Evolution of an Arabidopsis Histone H3 Variant. Plant Cell. 2002;14:1053–1066. doi: 10.1105/tpc.010425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Palmer DK, O’Day K, Trong HL, Charbonneau H, Margolis RL. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc. Natl. Acad. Sci. U.S.A. 1991;88:3734–3738. doi: 10.1073/pnas.88.9.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Howman EV, Fowler KJ, Newson AJ, Redward S, MacDonald AC, Kalitsis P, Choo KH. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl. Acad. Sci. U.S.A. 2000;97:1148–1153. doi: 10.1073/pnas.97.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chen Y, Baker RE, Keith KC, Harris K, Stoler S, Fitzgerald-Hayes M. The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol. Cell. Biol. 2000;20:7037–7048. doi: 10.1128/mcb.20.18.7037-7048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA. Functional Analysis of Kinetochore Assembly in Caenorhabditis elegans. J. Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sanyal K, Carbon J. The CENP-A homolog CaCse4p in the pathogenic yeast Candida albicans is a centromere protein essential for chromosome transmission. Proc. Natl. Acad. Sci. U.S.A. 2002;99:12969–12974. doi: 10.1073/pnas.162488299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Régnier V, Vagnarelli P, Fukagawa T, Zerjal T, Burns E, Trouche D, Earnshaw W, Brown W. CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Mol. Cell Biol. 2005;25:3967–3981. doi: 10.1128/MCB.25.10.3967-3981.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Maehara K, Takahashi K, Saitoh S. CENP-A reduction induces a p53-dependent cellular senescence response to protect cells from executing defective mitoses. Mol. Cell. Biol. 2010;30:2090–2104. doi: 10.1128/MCB.01318-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lermontova I, Koroleva O, Rutten T, Fuchs J, Schubert V, Moraes I, Koszegi D, Schubert I. Knock down of CENH3 in Arabidopsis reduces mitotic divisions and causes sterility by disturbed meiotic chromosome segregation. Plant J. 2011;68:40–50. doi: 10.1111/j.1365-313X.2011.04664.x. [DOI] [PubMed] [Google Scholar]
- [18].Cleveland DW, Mao Y, Sullivan KF. Centromeres and Kinetochores: From Epigenetics to Mitotic Checkpoint Signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- [19].Weaver BA, Cleveland DW. Does aneuploidy cause cancer? Curr. Opin. Cell Biol. 2006;18:658–667. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- [20].Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- [21].Runge KW, Wellinger RJ, Zakian VA. Effects of excess centromeres and excess telomeres on chromosome loss rates. Mol. Cell. Biol. 1991;11:2919–2928. doi: 10.1128/mcb.11.6.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Henikoff S, Ahmad K, Malik HS. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- [23].Bloom KS, Carbon J. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell. 1982;29:305–317. doi: 10.1016/0092-8674(82)90147-7. [DOI] [PubMed] [Google Scholar]
- [24].Fitzgerald-Hayes M, Clarke L, Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982;29:235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- [25].Panzeri L, Philippsen P. Centromeric DNA from chromosome VI in Saccharomyces cerevisiae strains. EMBO J. 1982;1:1605–1611. doi: 10.1002/j.1460-2075.1982.tb01362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Saunders M, Fitzgerald-Hayes M, Bloom K. Chromatin structure of altered yeast centromeres. Proc. Natl. Acad. Sci. U.S.A. 1988;85:175–179. doi: 10.1073/pnas.85.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- [28].Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc. Natl. Acad. Sci. U.S.A. 2007;104:14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Herman RK, Madl JE, Kari CK. Duplications in Caenorhabditis elegans. Genetics. 1979;92:419–435. doi: 10.1093/genetics/92.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Albertson DG, Thomson JN. The kinetochores of Caenorhabditis elegans. Chromosoma. 1982;86:409–428. doi: 10.1007/BF00292267. [DOI] [PubMed] [Google Scholar]
- [31].Maddox PS, Oegema K, Desai A, Cheeseman IM. “Holo”er than thou: chromosome segregation and kinetochore function in C. elegans. Chromosome Res. 2004;12:641–653. doi: 10.1023/B:CHRO.0000036588.42225.2f. [DOI] [PubMed] [Google Scholar]
- [32].Sullivan KF, Hechenberger M, Masri K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J. Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- [34].Black BE, Jansen LET, Maddox PS, Foltz DR, Desai AB, Shah JV, Cleveland DW. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol. Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- [35].Black BE, Brock MA, Bédard S, Woods VL, Jr, Cleveland DW. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc. Natl. Acad. Sci. U.S.A. 2007;104:5008–5013. doi: 10.1073/pnas.0700390104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Sekulic N, Bassett EA, Rogers DJ, E B. Black, The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ranjitkar P, Press MO, Yi X, Baker R, MacCoss MJ, Biggins S. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol. Cell. 2010;40:455–464. doi: 10.1016/j.molcel.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Guse A, Carroll CW, Moree B, Fuller CJ, Straight AF. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 2011;477:354–358. doi: 10.1038/nature10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Shelby RD, Vafa O, Sullivan KF. Assembly of CENP-A into Centromeric Chromatin Requires a Cooperative Array of Nucleosomal DNA Contact Sites. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Foltz DR, Jansen LET, Black BE, Bailey AO, Yates JR, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- [41].Camahort R, Shivaraju M, Mattingly M, Li B, Nakanishi S, Zhu D, Shilatifard A, Workman JL, Gerton JL. Cse4 is part of an octameric nucleosome in budding yeast. Mol. Cell. 2009;35:794–805. doi: 10.1016/j.molcel.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Williams JS, Hayashi T, Yanagida M, Russell P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol. Cell. 2009;33:287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Furuyama T, Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009;138:104–113. doi: 10.1016/j.cell.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lavelle C, Recouvreux P, Wong H, Bancaud A, Viovy J-L, Prunell A, Victor J-M. Right-handed nucleosome: myth or reality? Cell. 2009;139:1216–1217. doi: 10.1016/j.cell.2009.12.014. [DOI] [PubMed] [Google Scholar]
- [46].Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- [47].Xiao H, Mizuguchi G, Wisniewski J, Huang Y, Wei D, Wu C. Nonhistone Scm3 binds to AT-rich DNA to organize atypical centromeric nucleosome of budding yeast. Mol. Cell. 2011;43:369–380. doi: 10.1016/j.molcel.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-A nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takana Y, Oda T, Sato M, Park SY, Kimura H, Kurumizaka H. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- [50].Conde e Silva N, Black BE, Sivolob A, Filipski J, Cleveland DW, Prunell A. CENPA-containing nucleosomes: easier disassembly versus exclusive centromeric localization. J. Mol. Biol. 2007;370:555–573. doi: 10.1016/j.jmb.2007.04.064. [DOI] [PubMed] [Google Scholar]
- [51].Panchenko T, Sorensen TC, Woodcock CL, Kan Z-Y, Wood S, Resch MG, Luger K, Englander SW, Hansen JC, Black BE. Replacement of histone H3 with CENP-A directs global nucleosome array condensation and loosening of nucleosome superhelical termini. Proc. Natl. Acad. Sci. U.S.A. 2011;108:16588–16593. doi: 10.1073/pnas.1113621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Yan H, Jiang J. Rice as a model for centromere and heterochromatin research. Chromosome Res. 2007;15:77–84. doi: 10.1007/s10577-006-1104-z. [DOI] [PubMed] [Google Scholar]
- [54].Zinkowski RP, Meyne J, Brinkley BR. The centromere-kinetochore complex: a repeat subunit model. J. Cell Biol. 1991;113:1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ribeiro SA, Vagnarelli P, Dong Y, Hori T, McEwen BF, Fukagawa T, Flors C, Earnshaw WC. A super-resolution map of the vertebrate kinetochore. Proc. Natl. Acad. Sci. U.S.A. 2010;107:10484–10489. doi: 10.1073/pnas.1002325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Dekker J, Rippe K, Dekker M, Kleckner N. Capturing Chromosome Conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- [57].Tolhuis B, Palstra R-J, Splinter E, Grosveld F, de Laat W. Looping and Interaction between Hypersensitive Sites in the Active β-globin Locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- [58].Drissen R, Palstra R-J, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W. The active spatial organization of the β-globin locus requires the transcription factor EKLF. Genes Dev. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR, Desai A, Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- [60].Perpelescu M, Fukagawa T. The ABCs of CENPs. Chromosoma. 2011;120:425–446. doi: 10.1007/s00412-011-0330-0. [DOI] [PubMed] [Google Scholar]
- [61].Carroll CW, Milks KJ, Straight AF. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 2010;189:1143–1155. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Ando S, Yang H, Nozaki N, Okazaki T, Yoda K. CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol. Cell. Biol. 2002;22:2229–2241. doi: 10.1128/MCB.22.7.2229-2241.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, McEwen BF, Shang W-H, Suzuki E, Okawa K, Cheeseman IM, Fukagawa T. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135:1039–1052. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- [64].Obuse C, Yang H, Nozaki N, Goto S, Okazaki T, Yoda K. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells. 2004;9:105–120. doi: 10.1111/j.1365-2443.2004.00705.x. [DOI] [PubMed] [Google Scholar]
- [65].Prendergast L, van Vuuren C, Kaczmarczyk A, Doering V, Hellwig D, Quinn N, Hoischen C, Diekmann S, Sullivan KF. Premitotic assembly of human CENPs -T and -W switches centromeric chromatin to a mitotic state. PLoS Biol. 2011;9:e1001082. doi: 10.1371/journal.pbio.1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Suzuki A, Hori T, Nishino T, Usukura J, Miyagi A, Morikawa K, Fukugawa T. Spindle microtubules generate tension-dependent changes in the distribution of inner kinetochore proteins. J. Cell Biol. 2011;193:125–140. doi: 10.1083/jcb.201012050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Gascoigne KE, Takeuchi K, Suzuki A, Hori T, Fukagawa T, Cheeseman IM. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 2011;145:410–422. doi: 10.1016/j.cell.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Carroll CW, Silva MCC, Godek KM, Jansen LET, Straight AF. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat. Cell Biol. 2009;11:896–902. doi: 10.1038/ncb1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Appelgren H, Kniola B, Ekwall K. Distinct centromere domain structures with separate functions demonstrated in live fission yeast cells. J. Cell Sci. 2003;116:4035–4042. doi: 10.1242/jcs.00707. [DOI] [PubMed] [Google Scholar]
- [70].Li X, Dawe RK. Fused sister kinetochores initiate the reductional division in meiosis I. Nat. Cell Biol. 2009;11:1103–1108. doi: 10.1038/ncb1923. [DOI] [PubMed] [Google Scholar]
- [71].Gan L, Ladinsky MS, Jensen GJ. Organization of the Smallest Eukaryotic Spindle. Curr. Biol. 2011;21:1578–1583. doi: 10.1016/j.cub.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Takeo S, Lake CM, Morais-de-Sá E, Sunkel CE, Hawley RS. Synaptonemal Complex-Dependent Centromeric Clustering and the Initiation of Synapsis in Drosophila Oocytes. Curr. Biol. 2011;21:1845–1851. doi: 10.1016/j.cub.2011.09.044. [DOI] [PubMed] [Google Scholar]
- [73].Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, Kandels-Lewis S, Larionov V, Earnshaw WC, Masumoto H. Inactivation of a Human Kinetochore by Specific Targeting of Chromatin Modifiers. Dev. Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Bergmann JH, Rodríguez MG, Martins NMC, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LET, Earnshaw WC. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2010;30:328–340. doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Alonso A, Hasson D, Cheung F, Warburton PE. A paucity of heterchromatin at functional human neocentromeres. Epigenetics Chromatin. 2010;3 doi: 10.1186/1756-8935-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Zeitlin SG, Barber CM, Allis CD, Sullivan KF, Sullivan K. Differential regulation of CENP-A and histone H3 phosphorylation in G2/M. J. Cell. Sci. 2001;114:653–661. doi: 10.1242/jcs.114.4.653. [DOI] [PubMed] [Google Scholar]
- [77].Zeitlin SG, Shelby RD, Sullivan KF. CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J. Cell Biol. 2001;155:1147–1157. doi: 10.1083/jcb.200108125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Hewawasam G, Shivaraju M, Mattingly M, Venkatesh S, Martin-Brown S, Florens L, Workman JL, Gerton JL. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol. Cell. 2010;40:444–454. doi: 10.1016/j.molcel.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Moreno-Moreno O, Medina-Giró S, Torras-Llort M, Azorín F. The F Box Protein Partner of Paired Regulates Stability of Drosophila Centromeric Histone H3, CenH3(CID) Curr. Biol. 2011;21:1488–1493. doi: 10.1016/j.cub.2011.07.041. [DOI] [PubMed] [Google Scholar]
- [80].Shelby RD, Monier K, Sullivan KF. Chromatin assembly at kinetochores is uncoupled from DNA replication. J. Cell Biol. 2000;151:1113–1118. doi: 10.1083/jcb.151.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Sullivan B, Karpen G. Centromere identity in Drosophila is not determined in vivo by replication timing. J. Cell Biol. 2001;154:683–690. doi: 10.1083/jcb.200103001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Jansen LET, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Hemmerich P, Weidtkamp-Peters S, Hoischen C, Schmiedeberg L, Erliandri I, Diekmann S. Dynamics of inner kinetochore assembly and maintenance in living cells. J. Cell Biol. 2008;180:1101–1114. doi: 10.1083/jcb.200710052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Schuh M, Lehner CF, Heidmann S. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr. Biol. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- [85].Worcel A, Han S, Wong ML. Assembly of newly replicated chromatin. Cell. 1978;15:969–977. doi: 10.1016/0092-8674(78)90280-5. [DOI] [PubMed] [Google Scholar]
- [86].Wu RS, Tsai S, Bonner WM. Patterns of histone variant synthesis can distinguish G0 from G1 cells. Cell. 1982;31:367–374. doi: 10.1016/0092-8674(82)90130-1. [DOI] [PubMed] [Google Scholar]
- [87].Brown DT, Wellman SE, Sittman DB. Changes in the levels of three different classes of histone mRNA during murine erythroleukemia cell differentiation. Mol. Cell Biol. 1985;5:2879–2886. doi: 10.1128/mcb.5.11.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Wunsch AM, Lough J. Modulation of histone H3 variant synthesis during the myoblastmyotube transition of chicken myogenesis. Dev. Biol. 1987;119:94–99. doi: 10.1016/0012-1606(87)90210-7. [DOI] [PubMed] [Google Scholar]
- [89].Osley MA. The Regulation of Histone Synthesis in the Cell Cycle. Annu. Rev. Biochem. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- [90].Hake SB, Garcia BA, Duncan EM, Kauer M, Dellaire G, Shabanowitz J, Bazett-Jones DP, Allis DC, Hunt DF. Expression patterns and posttranslational modifications associated with mammalian histone H3 variants. J. Biol. Chem. 2006;281:559–568. doi: 10.1074/jbc.M509266200. [DOI] [PubMed] [Google Scholar]
- [91].Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome Assembly by a Complex of CAF-1 and Acetylated Histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- [92].Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- [93].Mello JA. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep. 2002;3:329–334. doi: 10.1093/embo-reports/kvf068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Natsume R, Eitoku M, Akai Y, Sano N, Horikoshi M, Senda T. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–341. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- [95].Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- [96].O’Donnell M, Kuriyan J, Kong XP, Stukenberg PT, Onrust R. The sliding clamp of DNA polymerase III holoenzyme encircles DNA. Mol. Biol. Cell. 1992;3:953–957. doi: 10.1091/mbc.3.9.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Ahmad K, Henikoff S. The Histone Variant H3.3 Marks Active Chromatin by Replication-Independent Nucleosome Assembly. Mol. Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- [98].Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- [99].Goldberg AD, Banaszynski LA, Noh K-M, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, Wen D, Chapgier A, DeKelver RC, Miller JC, Lee YL, Boydston EA, Holmes MC, Gregory PD, Greally JM, Rafii S, Yang C, Scambler PJ, Garrick D, Gibbons RJ, Higgs DR, Cristea IM, Urnov FD, Zheng D, Allis CD. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Drané P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24:1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Lewis PW, Elsaesser SJ, Noh K-M, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. U.S.A. 2010;107:14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M. Mis16 and Mis18 Are Required for CENP-A Loading and Histone Deacetylation at Centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- [103].Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev. Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- [104].Maddox PS, Hyndman F, Monen J, Oegema K, Desai A. Functional genomics identifies a Myb domain–containing protein family required for assembly of CENP-A chromatin. J. Cell Biol. 2007;176:757–763. doi: 10.1083/jcb.200701065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr. Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- [106].Song J-J, Garlick JD, Kingston RE. Structural basis of histone H4 recognition by p55. Genes Dev. 2008;22:1313–1318. doi: 10.1101/gad.1653308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Murzina NV, Pei X-Y, Zhang W, Sparkes M, Vicente-Garcia J, Pratap JV, McLaughlin SH, Ben-Shahar TR, Verreault A, Luisi BF, Laue ED. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure. 2008;16:1077–1085. doi: 10.1016/j.str.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Smith S, Stillman B. Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 1991;10:971–980. doi: 10.1002/j.1460-2075.1991.tb08031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Furuyama T, Dalal Y, Henikoff S. Chaperone-mediated assembly of centromeric chromatin in vitro. Proc. Natl. Acad. Sci. U.S.A. 2006;103:6172–6177. doi: 10.1073/pnas.0601686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- [111].Moree B, Meyer CB, Fuller CJ, Straight AF. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J. Cell Biol. 2011;194:855–871. doi: 10.1083/jcb.201106079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Erhardt S, Mellone BG, Betts CM, Zhang W, Karpen GH, Straight AF. Genomewide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J. Cell Biol. 2008;183:805–818. doi: 10.1083/jcb.200806038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Mellone BG, Grive KJ, Shteyn V, Bowers SR, Oderberg I, Karpen GH. Assembly of Drosophila centromeric chromatin proteins during mitosis. PLoS Genet. 2011;7:e1002068. doi: 10.1371/journal.pgen.1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Ellis RJ. Molecular chaperones: assisting assembly in addition to folding. Trends Biochem. Sci. 2006;31:395–401. doi: 10.1016/j.tibs.2006.05.001. [DOI] [PubMed] [Google Scholar]
- [115].Laskey RA, Honda BM, Mills AD, Finch JT. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature. 1978;275:416–420. doi: 10.1038/275416a0. [DOI] [PubMed] [Google Scholar]
- [116].Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- [117].Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- [118].Eitoku M, Sato L, Senda T, Horikoshi M. Histone chaperones: 30 years from isolation to elucidation of the mechanisms of nucleosome assembly and disassembly. Cell. Mol. Life Sci. 2007;65:414–444. doi: 10.1007/s00018-007-7305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Foltz DR, Jansen LET, Bailey AO, Yates JR, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Silva MCC, Jansen LET. At the right place at the right time: novel CENP-A binding proteins shed light on centromere assembly. Chromosoma. 2009;118:567–574. doi: 10.1007/s00412-009-0227-3. [DOI] [PubMed] [Google Scholar]
- [121].Barnhart MC, Kuich PHJL, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J. Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Bernad R, Sánchez P, Rivera T, Rodríguez-Corsino M, Boyarchuk E, Vassias I, Ray-Gallet D, Arnaoutov A, Dasso M, Almouzni G, Losada A. Xenopus HJURP and condensin II are required for CENP-A assembly. J. Cell Biol. 2011;192:569–582. doi: 10.1083/jcb.201005136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, Baker RE. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc. Natl. Acad. Sci. U.S.A. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL. Scm3 is Essential to Recruit the Histone H3 Variant Cse4 to Centromeres and to Maintain a Functional Kinetochore. Mol. Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- [125].Sanchez-Pulido L, Pidoux AL, Ponting CP, Allshire RC. Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell. 2009;137:1173–1174. doi: 10.1016/j.cell.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Shuaib M, Ouararhni K, Dimitrov S, Hamiche A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc. Natl. Acad. Sci. U.S.A. 2010;107:1349–1354. doi: 10.1073/pnas.0913709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Pidoux AL, Choi ES, Abbott JKR, Liu X, Kagansky A, Castillo AG, Hamilton GL, Richardson W, Rappsilber J, He X, Allshire RC. Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol. Cell. 2009;33:299–311. doi: 10.1016/j.molcel.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Shivaraju M, Camahort R, Mattingly M, Gerton JL. Scm3 is a centromeric nucleosome assembly factor. J. Biol. Chem. 2011;286:12016–12023. doi: 10.1074/jbc.M110.183640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Dechassa ML, Wyns K, Li M, Hall MA, Wang MD, Luger K. Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes. Nat. Commun. 2011;2:313. doi: 10.1038/ncomms1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Luconi L, Araki Y, Erlemann S, Schiebel E. The CENP-A chaperone Scm3 becomes enriched at kinetochores in anaphase independently of CENP-A incorporation. Cell Cycle. 2011;10:3369–3378. doi: 10.4161/cc.10.19.17663. [DOI] [PubMed] [Google Scholar]
- [131].Zhou Z, Feng H, Zhou B-R, Ghirlando R, Hu K, Zwolak A, Jenkins L.M. Miller, Xiao H, Tjandra N, Wu C, Bai Y. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature. 2011 doi: 10.1038/nature09854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Cho U-S, Harrison SC. Recognition of the centromere-specific histone Cse4 by the chaperone Scm3. Proc. Natl. Acad. Sci. U.S.A. 2011;108:9367–9371. doi: 10.1073/pnas.1106389108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Hu H, Liu Y, Wang M, Fang J, Huang H, Yang N, Li Y, Wang J, Yao X, Shi Y, Li G, Xu R-M. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 2011;25:901–906. doi: 10.1101/gad.2045111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Dunleavy EM, Almouzni G, Karpen GH. H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G(1) phase. Nucleus. 2011;2:146–157. doi: 10.4161/nucl.2.2.15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].English CM, Maluf NK, Tripet B, Churchill MEA, Tyler JK. ASF1 binds to a heterodimer of histones H3 and H4: a two-step mechanism for the assembly of the H3-H4 heterotetramer on DNA. Biochemistry. 2005;44:13673–13682. doi: 10.1021/bi051333h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].English CM, Adkins MW, Carson JJ, Churchill MEA, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].LeRoy G, Orphanides G, Lane WS, Reinberg D. Requirement of RSF and FACT for Transcription of Chromatin Templates in Vitro. Science. 1998;282:1900–1904. doi: 10.1126/science.282.5395.1900. [DOI] [PubMed] [Google Scholar]
- [139].Perpelescu M, Nozaki N, Obuse C, Yang H, Yoda K. Active establishment of centromeric CENP-A chromatin by RSF complex. J. Cell Biol. 2009;185:397–407. doi: 10.1083/jcb.200903088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Lagana A, Dorn JF, De Rop V, Ladouceur A-M, Maddox AS, Maddox PS. A small GTPase molecular switch regulates epigenetic centromere maintenance by stabilizing newly incorporated CENP-A. Nat. Cell Biol. 2010;12:1186–1193. doi: 10.1038/ncb2129. [DOI] [PubMed] [Google Scholar]
- [141].Radman-Livaja M, Verzijlbergen KF, Weiner A, van Welsem T, Friedman N, Rando OJ, van Leeuwen F. Patterns and Mechanisms of Ancestral Histone Protein Inheritance in Budding Yeast. PLoS Biol. 2011;9:e1001075. doi: 10.1371/journal.pbio.1001075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Eichler EE. Repetitive conundrums of centromere structure and function. Hum. Mol. Genet. 1999;8:151–155. doi: 10.1093/hmg/8.2.151. [DOI] [PubMed] [Google Scholar]
- [143].Folco HD, Pidoux AL, Urano T, Allshire RC. Heterochromatin and RNAi Are Required to Establish CENP-A Chromatin at Centromeres. Science. 2008;319:94–97. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Okada T, Ohzeki J, Nakano M, Yoda K, Brinkley WR, Larionov V, Masumoto H. CENP-B Controls Centromere Formation Depending on the Chromatin Context. Cell. 2007;131:1287–1300. doi: 10.1016/j.cell.2007.10.045. [DOI] [PubMed] [Google Scholar]