Abstract
The kinetochore is the protein machine built at the centromere that integrates mechanical force and chemical energy from dynamic microtubules into directed chromosome motion. The kinetochore also provides a powerful signaling function that is able to alter the properties of the spindle checkpoint and initiate a signal transduction cascade that leads to inhibition of the anaphase promoting complex and cell cycle arrest. Together, the kinetochore accomplishes the feat of chromosome segregation with unparalleled accuracy. Errors in segregation lead to Down’s syndrome, the most frequent inherited birth defect, pregnancy loss, and cancer. Over a century after the discovery of the kinetochore, an architectural map comprising greater than 100 proteins is emerging. Understanding the architecture and physical biology of the key components provides new insights into how this fascinating machine moves genomes.
Introduction
The kinetochore is the protein–DNA machine that links microtubules to chromosomes. The machine is central to the processes that enable the accuracy of chromosome segregation required for cell division and multicellularity. The kinetochore is able to capture and nucleate microtubules, harness force from microtubule dynamics, and provide a scaffold for a signaling network. The physical states of components within the kinetochore, including post-translational modification or configurational states, control the interaction of signaling proteins (spindle assembly checkpoint) with cell division kinases. Once sister chromatid biorientation is achieved and the checkpoint is satisfied, entry into anaphase and chromosome segregation can proceed.
The kinetochore was observed in the light microscope close to 100 years ago (reviewed in [1]) and its trilaminar plate-like ultrastructure in the electron microscope over 50 years ago [2,3]. Several decades have been devoted to the identification and characterization of kinetochore components. Building on the power of genetics and biochemistry, over 100 kinetochore proteins have been identified [4]. Through the analysis of biophysical properties and genetic interaction maps it has been established that there are approximately 6–7 subcomplexes, each of which contain between 4 and 12 protein subunits. These complexes are assembled on small (budding yeast) or large (mammals) centromeric DNA templates [5].
Our next challenge will be to understand the mechanisms that underlie function. This requires knowledge of the inner workings of the kinetochore. One of the unique features of the kinetochore with respect to other protein machines is that it physically links two very different materials. Unlike protein machines that track along a nucleic acid roadway (polymerases), or cut and splice the roadway to change topology or repair DNA damage (topoisomerases or DNA repair enzymes), centromere DNA is weaved into the fabric of the kinetochore. This fabric is able to withstand forces exerted by the spindle apparatus as well as remain stable for the duration of mitosis, which can last several hours in mammalian cells.
To appreciate the complexity of mitosis we start with a simple comparison of scale. In humans, approximately 2 meters of DNA is copied and segregated to daughter cells ~50 microns in diameter. In yeast, 3 millimeters of DNA is distributed within cells ~5 microns in diameter. DNA compaction throughout mitosis is critical to deal with the 1,000–100,000-fold difference in scale. The kinetochore is designed to hold on to an ever-shortening and growing track (the dynamic microtubule plus-end) and yet not apply so much force that DNA strand breakage and mitotic catastrophe results. A second challenge in mitosis is one of fidelity. Chromosome number varies over at least an order of magnitude (Drosophila, 8; Goldfish, 94) and doesn’t correlate with organism size, complexity, or generation time. A powerful surveillance mechanism, the spindle assembly checkpoint (SAC) is responsible for synchronizing the mechanics of segregating tens to hundreds of individual chromosomes with cell cycle progression. Maloriented or unattached chromosomes are individually able to halt the cell cycle, thereby ensuring that every last chromosome will be segregated. Finally, we need to identify the key parameters in this process. Parameters such as kinetochore or chromosome size, or microtubule number may not be critical to mitosis. A case in point is the comparison between point centromeres in budding yeast versus regional centromeres in fission yeast and larger eukaryotes. Is size or mass indicative of a feature or property that makes any mechanistic predictions? Mitosis occurs in a milieu where viscosity dominates and mass and inertia are negligible (i.e., low Reynolds number). As experimental scientists, our intuition stems from an environment where inertia and mass dominate (high Reynolds number) and thus fails inside the cell.
The View from Inside
To gain intuition and evaluate experimental data within the context of the cell we need to consider the material properties of the individual components, such as microtubules and chromosomes, and appreciate a world where motors stop dead in their tracks when starved for energy (no movement in the absence of energy) and microtubule-based motors do not ‘float away’ upon release from their microtubule track (viscosity rules and gravity can be ignored). Toward this end, we require physical definitions for macromolecular machines that rely on objective standards that will allow us to compare a given machine working under different conditions (moving fast or idling) or states (e.g., cell cycle). The paradigm for thinking about materials and quantifying their physical properties can be attributed to Robert Hooke, who wrote “Ut tensio sic uis”, which translates to “As the extension, so the force”. This is known as Hooke’s law and states that for a simple spring, extension is linearly related to force. The problem with Hooke’s law is that it doesn’t distinguish a piece of rubber from a steel spring, i.e. whether elasticity is a property of the material or whether it is a property of the geometry and dimensions of the structure. Young’s modulus is the parameter that accounts for the material properties and distinguishes elasticity from geometry (area). If we consider the pressure (force per area, known as stress) and strain (change in length or extension, ΔL/L) of a material, we can rewrite Hooke’s law as Stress/Strain. This relationship is known as Young’s modulus and is an objective measure of the springiness of a given material. Young’s modulus has units of force/area (Pascals, N/m2) as strain is a unitless dimension.
A second definition will allow us to distinguish a steel rod from a steel chain (or a microtubule from DNA). Is the structure stiff over a long (steel rod) or short (steel chain) length scale? The persistence length (Lp) defines how well a polymer resists thermal force. Intuitively, it is the length over which thermal bending is significant. In mathematical terms, persistence length is the decay length of the cosines between the tangent vectors (t(s)) along the monomers of the chain. The correlation between the tangent vectors decays exponentially as [t(0) – t(s)] ~ e−s/Lp.
The goal in understanding kinetochore function and chromosome segregation is integrating kinetochore protein structure and organization with the material properties of the constituent parts, i.e. DNA and microtubules (Figure 1). This review will evaluate the physical properties of the key molecules, the thermodynamics of individual components (i.e., equilibrium polymers such as microtubules, worm-like chains such as DNA) and the emergent principles that stem from physical biology and life in a crowded, confined environment. This will guide our efforts in deducing the mechanisms that contribute to the staggering fidelity of chromosome segregation.
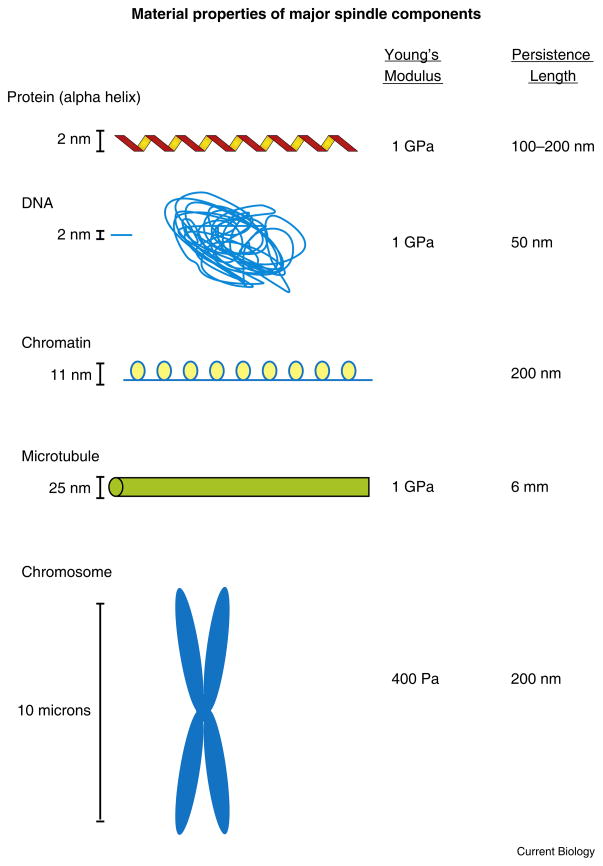
Figure 1. Material properties of the major spindle components.
The mitotic spindle is composed of proteins and DNA. The dominant protein in the spindle is tubulin, organized into an equilibrium polymer known as microtubules. DNA is wrapped around histone protein into nucleosomes, which are further compacted into the chromosome. DNA behaves as a worm-like chain and adopts a random coil due to thermal fluctuations of the chain.
Bookends of the Kinetochore: Microtubules and Chromatin
The kinetochore is a force transducer that integrates mechanical force and chemical energy into directed chromosome motion and is a powerful signaling system that coordinates the state of chromosome attachment and/or tension with cell cycle progression. As with any machine that involves mechanical force or stress, the structure and function of that machine is dictated in large part by the composition of the building materials.
Microtubules are hollow cylinders about 25 nm in diameter (Figure 1). They are stiff (Young’s modulus of 1.2 GPa) [6], meaning they are structurally rigid like plastics such as Plexiglass. The persistence length of a microtubule is roughly 6 mm. This is orders of magnitude longer than a typical eukaryotic cell; thus, any microtubule bending observed in live cells reflects active chemical processes. Spindle microtubules are endothermically self-assembled from a pool of tubulin subunits in vivo [7,8]. The free energy of assembly is thermodynamically favorable. This is counter-intuitive when considering the disorder of tubulin subunits versus the order of the microtubule polymer. However, the loss of ordered water surrounding the tubulin dimer leads to an entropically favorable polymeric state [7,8]. The change in enthalpy is about 34 mkcal/mole, entropy 0.125 e.u. (0.001 kcal/Kelvin/mole) (ΔG=ΔH–TΔS = 34 – (300×0.125), 34 − 37.5). Since the free energy difference between dimer and polymeric states is small, it makes intuitive sense that the dimer versus polymer concentration will be comparable (since neither state is energetically favored over the other). In fact, the experimental measurements bear this out [9,10]. Polymers in which approximately equal amounts of polymer and dimer exist at ambient temperature are referred to as equilibrium polymers. Another way to think about the thermodynamics is to convert the free energy to a potential force. It has recently been discussed that ~7 kcal/mole is an approximately universal number that applies to any protein subunit immobilized by protein–protein interaction [11]. Thus, the loss of ordered water is on the order of 44 kcal/mole (7 kcal/mole (300K x 0.125 e.u.) = 44 kcal/mole). We can estimate the theoretical limit of force from a growing microtubule according to the number of subunits added (N = 13), the force in pN (1 kcal/mole = 1.6 kBT, 1 kBT = 4.1 pN nm, ΔG = −3.5 kcal/mole; 1.6 x 4.1 pN nm x 3.5 kcal/mole = 20pN nm), and the change in distance upon subunit addition, 8 nm (13 subunits x 20 pN/8 nm). The force produced by a single microtubule growing a distance of 13 dimers is about 30 pN. This theoretical estimate is about 10 times the force exerted by a single microtubule against a rigid barrier using optical tweezers in vitro (2.7 pN) [12]. To evaluate whether these forces are large or small relative to chromosome movement, we need to consider the properties of the DNA cargo that microtubules are responsible for moving.
DNA is long (millimeters to meters), thin (2 nanometers) and highly compacted. When DNA is extended, it has a Young’s modulus of 0.30 GPa [13], not unlike a microtubule. However, the persistence length of DNA is on the order of 50 nm, 5 orders of magnitude shorter than a microtubule. This short persistence length means that a long-chain DNA polymer adopts a random coil, whose end-to-end distance is related to the persistence length (lp) and the contour length L of the chain (Re-e = lp√L/lp) (Figure 1). The random coil of DNA is considerably shorter than the contour length and as a consequence is much softer than extended DNA due to the compaction. The chromosome is also very soft in comparison to extended naked DNA (chromosomes, 400 Pa) [13,14]. The material property of the chromosome is dictated by both protein composition (approximately equal molecular weight of protein and DNA) and the tendency for this polymer to adopt a random coil. Unlike microtubules, DNA is not an equilibrium polymer, rather it is freely jointed and behaves as a worm-like chain. A unique property of these polymers is the elasticity arising from deformation of chains. This elasticity is known as entropic elasticity and reflects the tendency for the chains to adopt a state of highest entropy, in which they have the greatest freedom of movement. This feature of DNA accounts in part for the compliance (stretch) of the centromere relative to stiff proteins and microtubules.
DNA is complexed with proteins in the cell and is condensed several orders of magnitude to fit into the confines of a typical nucleus. The centromeric chromatin has several additional features that distinguishes it from the bulk of non-centromeric chromatin. One is the replacement of the canonical Histone H3 with a centromere-specific Histone H3 variant that is conserved throughout phylogeny (human CENPA, budding yeast Cse4, fission yeast cnp1, Drosophila cid) [15]. Second is the organization of pericentric chromatin, on the order of 20–50 kb of DNA surrounding the centromere shaped into a specific geometric arrangement [16,17]. The energetics of DNA in comparison to microtubules is a much more challenging enterprise. Naked DNA is a very weak entropic spring. The spring constant of a freely jointed chain like DNA is 3 kBT/n(2 lp)2 with n = number of segments. The spring constant is 0.036 fN/nm for a 10 kb strand of DNA. Since pericentric chromatin is complexed with histone octamers every ~200 bp and with proteins such as topoisomerase II, condensin and cohesin, the magnitude of the spring constant is likely to reflect the contribution of protein as well as that of the naked DNA. In a metaphase spindle at steady state, the magnitude of forces exerted by spindle microtubules and microtubule-based motor proteins in the spindle are likely in balance with the spring constant of DNA and the protein complexes that bind sister chromatids.
The Building Blocks: Kinetochore Protein Number and Geometry
Force has both magnitude and direction, making it a vector quantity. Before we can seriously understand how forces are transmitted and how conformational changes are exerted, we must begin with the geometry and spatial arrangement of the kinetochore components.
Attempts to understand the mechanical function of the kinetochore have been advanced by developments in digital microscopy and quantitative data analysis. Ascertaining the number of proteins in the kinetochore is a critical first step in deducing the geometry and spatial arrangement of this complex structure. Globular proteins such as those found in the kinetochore are quite rigid, on the scale of 1 GPa [18]; however, for a three-dimensional view we need to know their number and position with respect to each other, the linkages between different sets of kinetochore proteins, and their position relative to the microtubule and centromere DNA. Toward this end, protein counts have been determined in vertebrate and fungal kinetochores [19–21]. These studies reveal that the stoichiometry of kinetochore proteins corresponds to the number of microtubule binding sites. The number of proteins in a single kinetochore–microtubule attachment site ranges from 16–20 for the microtubule-associated Dam1 complex and as low as 1–2 for the centromere DNA proximal complexes.
While the protein counts do not reveal subunit organization within the kinetochore, a simplifying assumption is that they are symmetric with respect to the microtubule lattice. In this view, the hundred or so different kinetochore proteins may adopt a cylindrical geometry that would be approximately 75 nm in length and have an inner diameter of ~30 nm [22] (Figure 2). The wall of the kinetochore cylinder is about 5 nm thick. The surface area of the kinetochore at a single microtubule plus-end can be estimated from the end of the cylinder exposed on the surface of the chromosome (a circle, (πr2 = 3.14 x 15 nm2) to be 7 x 10−4 microns2. Estimates from electron microscopy studies indicate that the mammalian kinetochore has a surface area of 0.16 micron2 [23,24]. To evaluate the mammalian kinetochore with its multiple microtubule attachment sites of 20–25 microtubules relative to one yeast kinetochore with a single microtubule attachment, we consider the entire yeast spindle as a structural correlate of one mammalian kinetochore. In yeast, the 16 kinetochores are clustered into in an annular structure surrounding the spindle microtubules [17]. From this perspective, the cluster of 16 yeast kinetochores is geometrically comparable to the mammalian kinetochore. Estimates of the surface area accounting for this geometry is 0.12 microns2 (a cluster of 16 kinetochores surrounding the central 8 interpolar microtubules, πr2 (r ~200 nm) = 0.12 microns2). Therefore, the surface area of the cluster of 16 kinetochores in budding yeast is comparable to the surface area of kinetochores in Ptk rat kangaroo cells that comprise a multi-microtubule attachment site of at least 23 microtubules. The hypothesis that stems from these physical measurements is that the kinetochore is a multi-microtubule attachment site [25] that consists of repeated units of the core functional unit observed in budding yeast. The alternative hypothesis is that the mammalian kinetochore is disorganized with no repetitive subunits and no discrete microtubule binding site [26]. As discussed below, the order and conservation in kinetochore protein number and position in mammals and yeast lends additional support for a conserved, repeating-subunit model of organization.
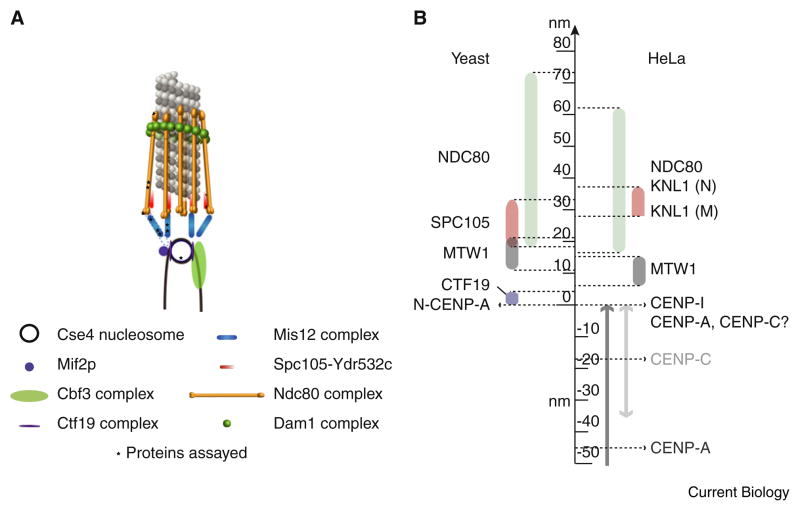
Figure 2. The protein architecture of microtubule attachment sites at the budding yeast and HeLa kinetochores is highly conserved.
(A) 3D visualization of the metaphase budding yeast kinetochore–microtubule attachment, as predicted by the protein localization data, assuming a symmetric arrangement of kinetochore protein complexes around the cylindrical microtubule lattice. Black stars indicate the positions of fluorescent labels used in distance measurements. Dashed lines indicate established biochemical interactions between two protein complexes (adapted from Figure 3 in [22]). (B) Yeast measurements are derived from kinetochore proteins labeled at the carboxyl or amino terminus with fluorescent proteins. In the case of many HeLa kinetochore measurements, antibodies recognized other regions within the proteins. The differences arising from this experimental factor have not been quantified. Letters in the bracket next to the KNL-1 protein indicate amino terminus (N) and middle protein (M).
The copy number and position of kinetochore proteins have important implications in understanding the mechanisms of kinetochore function. Microtubule attachment to the kinetochore requires the NDC80 complex [27,28]. The strength of the kinetochore–microtubule attachment is directly modulated by the Aurora B kinase, which phosphorylates many residues in the NDC80 complex [29]. Therefore, the number of NDC80 complex molecules per kinetochore is a key parameter for force generation at the kinetochore. Similarly, the Dam1/DASH complex is also an essential protein complex for kinetochore–microtubule attachments in budding yeast. It can form oligomeric rings around the microtubule lattice in vitro, although the in vivo conformation of this complex is not known [30]. The average number of Dam1/DASH complex molecules per kinetochore, however, is sufficient to form one oligomeric ring per microtubule. Interestingly, the Dam1/DASH complex, which is non-essential in fission yeast, exists in much smaller numbers at the fission yeast centromere and cannot form rings [31]. The kinetochore protein stoichiometry also reveals an insight into the assembly of the kinetochore: the numbers of centromere-bound/associated components per attachment site are much smaller than the numbers for the microtubule-binding kinetochore complexes. To build such a structure, one or more of the centromere complexes may interface with multiple copies of other kinetochore complexes. Alternatively, some of the kinetochore complexes may oligomerize, such as the KNL-1/Blinkin protein that has been shown to form tetramers or pentamers in vitro [32].
The spatial arrangement of structural and regulatory proteins within the kinetochore is integral to the mechanics of microtubule attachment, force generation, and regulation. Efforts to understand the spatial arrangement of proteins around the microtubule lattice have mostly relied on serial section electron microscopy of fixed cells [26,33]. However, in the absence of protein-specific markers, these studies provide limited information about the ultrastructure of the kinetochore. Two new studies based on fluorescence microscopy now provide a sharper image of the protein architecture of the kinetochore. Analytical microscopy techniques allow the determination of the centroid of Gaussian spots with nanometer accuracy [34,35], and can be used to measure the distance separating a pair of fluorophores of different colors [36]. This distance measurement technique can be extended to clusters of fluorophores (emitting at two wavelengths, say red and green) provided that each red fluorophore is separated from a corresponding green fluorophore by the same distance and in the same direction. This concept was used in a preliminary assay that focused on fluorescently labeled chromosomes isolated from Drosophila S2 cells [37]. It was applied in live budding yeast cells, as well as fixed and live HeLa cells, to resolve the kinetochore protein architecture along the axis of kinetochore–microtubule attachment at nanometer-scale resolution [22,38,39]. The in vivo map of the budding yeast kinetochore proteins complements the protein copy numbers and known structural information to provide the first visualization of the in vivo protein architecture of a single kinetochore–microtubule attachment (Figure 2).
These visualizations of the protein linkage extending from the centromere to the microtubule contribute to the understanding of the mechanisms that generate forces at the kinetochore. However, there are two limitations of these studies that one must be aware of while interpreting the reported distance measurements. Firstly, both studies provide protein separations only along the kinetochore–microtubule axis. Geometric models assume symmetric arrangement of kinetochores within a cluster in budding yeast and an analogous arrangement of kinetochore–microtubule attachment sites within the kinetochore in HeLa cells. Thus, protein separations either across the spindle axis or along the optical axis are unknown. Secondly, due to the inability to resolve single kinetochore–microtubule attachments, these studies also average over many microtubule attachment sites. This means that if kinetochore–microtubule attachments assume different architectures when attached to a polymerizing microtubule, as opposed to a depolymerizing microtubule, the structure observed in these studies is an average of these two states.
The protein machinery connecting centromeric chromatin to the plus-end of a microtubule in budding yeast occupies ~80 nm, which is similar to the distance measured in HeLa cells, as discussed below (also see Figure 2). From the point of view of force generation, the localization of the microtubule- binding NDC80 and Dam1/DASH complex molecules is of the most interest. In budding yeast, the NDC80 complex spans almost 55 nm. This measured length suggests that NDC80 binds to the microtubule with its axis more or less parallel to the microtubule axis. This interpretation is made possible by the persistence of kinetochore–microtubule attachments in budding yeast, which ensures invariable geometry. In the HeLa kinetochore, the head-to-tail length of the NDC80 complex was found to be 45 nm, which Wan et al. [39] used to predict a constitutively bent configuration of the NDC80 complex at the ‘hinge’ found in the coiled-coil domain of the Ndc80 subunit [40]. However, vertebrate kinetochores can typically acquire additional end-on microtubule attachments after taxol treatment, implying that not all microtubule attachments may be occupied in metaphase [41,42]. Thus, a small fraction of microtubule attachment sites are likely unoccupied at a given kinetochore, and it is not clear if unengaged NDC80 complex molecules at these sites acquire an orientation that is markedly different from the engaged molecules. Interpretation is further complicated by the flexible hinge region within the NDC80 complex, which can reduce its length by up to 15 nm. The small number of NDC80 molecules suggests that the attachment site presents an open architecture for free access to the microtubule plus-end by regulatory proteins. The Dam1/DASH complex shows a surprising location that is 10 nm inside the microtubule-binding head domain of the NDC80 complex (Figure 2). Although this distance does not help in ascertaining whether the Dam1/DASH complex at the kinetochore exists in the form of rings or partial helices, or oligomeric patches, it does propose a novel NDC80-dependent role for the Dam1/DASH complex.
The microtubule-binding NDC80 complex is linked back to the centromere DNA by the Mtw1 and Spc105 complexes. In budding yeast, the Mtw1 complex spans a ~10–20 nm gap separating the CENP-A nucleosome from the centromeric end of the NDC80 complex. The Spc105 protein likely extends from the Spc24/25 end outward (towards the microtubule). In budding yeast, the Mtw1 and Spc105 complexes undergo conformational and/or orientation changes after cell cycle transition into anaphase. Comparison of HeLa kinetochore architectures in control and taxol-treated conditions revealed that the kinetochore may contain two substructures, one containing the NDC80 and two subunits of the Mtw1 complex, and the other containing the rest of the kinetochore proteins that can be mechanically decoupled. The final linkage to centromeric chromatin is mediated by the Ctf19 complex in budding yeast and this protein complex spans~10 nm. In the absence of structural information for the Ctf19, Mtw1, and Spc105 complexes, however, the localization of single protein domains/ends along the kinetochore–microtubule axis does not provide functional insight.
Geometry as a Function of Microtubule Attachment and Force
The single CENP-A nucleosome in budding yeast allows definite demarcation of centromere DNA within the kinetochore in vivo, thus providing the total kinetochore length of ~75 nm. In vertebrates, kinetochore length deduced from CENP-A centroid position is larger, ~110 nm [39] (Figure 2). These measurements also reveal a surprising aspect of kinetochore architecture: despite a larger number of regional centromere-specific proteins, the length of the kinetochore proteins along the microtubule axis is comparable to their length in budding yeast. Sister kinetochores with large separations also have a larger distance between CENP-A (and the DNA-binding CENP-C) and NDC80 centroids. In the absence of any tension (taxol-treated cells), the centroids of CENP-A and CENP-C proteins move ~16 nm closer to the NDC80 centroids. Furthermore, at low centromere stretch, separation between centroids of CENP-I and CENP-C is ~11 nm, while CENP-I and CENP-A is ~30 nm. CENP-I and CENP-A nucleosomes are reported to have close biochemical associations [43–45], suggesting that that only a fraction of chromatin-bound CENP-A and CENP-C is exposed on the surface of the chromosome. These discrepancies indicate that not all molecules in the kinetochore may be ‘mechanically productive’. In support of this idea, it has been shown that kinetochore function is not perturbed even if 90% of centromeric CENP-A is depleted. Thus, only ~10% of the CENP-A protein at the centromere is essential for the recruitment of kinetochore proteins [46]. Under mitotic forces, a fraction of molecules may get pulled away from the remaining complexes because such complexes do not experience the same forces due to indirect connection to the force-generating machinery. The actual displacement of the kinetochore-connected CENP-A molecules will depend on several factors such as the proportion of excess CENP-A molecules and the structural organization of the kinetochore.
The compliance or flexibility within the kinetochore (indicated by movements of CENP-A and CENP-C molecules) brings two related aspects of kinetochore organization to the fore. First, what is the nature of connection between neighboring microtubule attachment sites at regional centromeres? A recent study of condensin-depleted DT40 chicken cells makes the striking observation that, in the absence of condensin, a chromosome compaction protein, the entire kinetochore structure can move away from the chromosome due to microtubule-pulling forces while retaining the appearance of a plate-like structure in electron micrographs [47]. Thus, lateral linkages among neighboring microtubule attachment sites must exist. Such linkages can get highly stretched if one kinetochore forms attachments to both spindle poles (a merotelic attachment) [48], yet they maintain a stable structure while bearing forces generated by proper kinetochore–microtubule attachments. The nature, composition, and mechanical properties of such connections remain unknown. The second aspect concerns how the DNA/chromatin-binding/associated domain of the kinetochore interacts with the microtubule-binding domain. The number of microtubule-binding proteins per microtubule within the vertebrate kinetochore will be determined by the two main functional requirements — magnitude of force and persistence of attachment. For instance, an increased requirement for force may dictate an increase in the number of binding proteins, or alternatively, an increase in the duration of binding (persistence). Structures containing the required number of microtubule-binding proteins can be assembled by following either the budding yeast plan or by following a different biochemical assembly plan. Thus, the question of whether the identity of each microtubule attachment site extends all the way down to the associated CENP-A nucleosome becomes pertinent. Protein counting data from fission yeast and vertebrates support the hypothesis that the repeat-unit in budding yeast is conserved throughout phylogeny [19,21].
Geometry Sensing
Conformational changes in kinetochore and centromere architecture can provide natural readouts of the state and strength of microtubule attachment at the kinetochore. The roles of force-induced deformations in spindle assembly checkpoint signaling and in the regulation of kinetochore–microtubule attachments have recently been addressed. One set of studies shows that the spindle assembly checkpoint monitors structural changes within the kinetochore rather than the separation between sister kinetochores as previously hypothesized [38,49,50]. In HeLa and S2 cells, concurrent measurements of inter-kinetochore distance and intra-kinetochore separation between CENP-A and NDC80 demonstrate that tension-dependent phosphorylation within the kinetochore is correlated with changes in the intra-kinetochore distance rather than the traditionally measured inter-kinetochore separation. Thus, centromere tension is not directly monitored by the spindle assembly checkpoint, but rather changes within the kinetochore to turn the checkpoint off. Yet, the separation between sister kinetochores is likely used as an indicator of correct attachments by the error-correction mechanism, as documented by Liu et al. [51]. This study measured Aurora B phosphorylation activity at different locations within the kinetochore using FRET-based phosphorylation sensors, and found a strong correlation between the strength of Aurora B-mediated phosphorylation and the position of the FRET sensor in the kinetochore [51]. As expected, localization of Aurora B closer to its targets within the kinetochore also leads to unstable kinetochore–microtubule attachments. These findings suggest that those kinetochore–microtubule attachments that do not generate sufficient force to pull the kinetochore phospho-epitopes away from the centromere-localized Aurora B kinase get selectively destabilized. Taken together, these studies show that the spindle assembly checkpoint senses microtubule attachment to the kinetochore through a structural change that occurs within the kinetochore rather than the inter-kinetochore linkage. At the same time, centromere stretching provides an indirect read-out of the magnitude of force generated at the kinetochore for the Aurora B-mediated error correction machinery.
The kinetochore distance measurement studies reveal candidate structural (or orientation) changes within the kinetochore that may result from changes in the state of microtubule attachment and that then influence the behavior of the kinetochore. The positions of checkpoint proteins within the kinetochore are also important. In HeLa kinetochores, Wan et al. [39] located the checkpoint protein Bub1 near the Spc24/Spc25 end of the NDC80 complex. The locations of other checkpoint proteins, such as Mad1 and Mad2, could not be determined in metaphase kinetochores since these proteins are stripped off from the kinetochores after the establishment of bipolar attachment. Based on a comparison of kinetochore architecture in metaphase control cells (tension high or low, checkpoint off) and taxol-treated cells (low tension, checkpoint on), Wan et al. hypothesize that the activity of the spindle assembly checkpoint may be triggered by an as yet unidentified flexible linker that connects the microtubule-binding NDC80 complex to the inner kinetochore [39]. Kinetochore architecture in metaphase and anaphase budding yeast cells reveals that the most significant distance/orientation changes occur within the Mtw1 and Spc105 complexes [22]. The NDC80 complex also shows a reduction in its length, which likely results from intra-molecular bending at the hinge region within the Ndc80 subunit [39]. The functional significance of these architectural changes remains to be determined.
Physics of Chromosome Movement
Our understanding of the physics of chromosome movement is influenced by in vitro measurements on microtubule dynamics, single molecule motor movement, and DNA stretching and recoil [52]. The attempts to make comparable measurements in vivo are less straightforward, particularly in determining the molecular driving force for chromosome segregation. The classic in vivo experiments measuring spindle forces were performed on grasshopper cells with exquisitely calibrated microneedles. Nicklas was the first to point out that relative to other protein machines, the spindle is exceedingly weak [53], as the goal is to achieve fidelity not speed. As stated above, DNA itself is an entropic spring that has a measurable, albeit small, spring constant. In addition, cellular DNA is rarely if ever without protein, which together with its compaction give rise to the extreme softness of chromosomes relative to extended DNA. The helicity of DNA has major biological consequences, namely that it can be bent and twisted, known as supercoiling. Like a telephone coil, DNA can rotate around itself. In a covalently closed circle, the topology is fixed. From a statistical mechanics perspective, it is no surprise that any set of covalently closed circular molecules are not homogeneous. Rather, they differ by the number of topological turns. Supercoiling is characterized by two types of twisting. Twist is the number of turns of the helix and writhe is the number of times the helix crosses itself on a planar projection. In comparing the material properties of microtubules and DNA, we can consider the differences in free energy for various structural states of each polymer. The free energy of supercoiling is on the order of 250 kcal/mole DNA [54,55]. This is the free energy associated with supercoiling a covalently closed circular plasmid isolated from bacteria to a superhelical density (σ) of −.05. Superhelical density is defined as the change in linking number/linking number of the relaxed molecule (ΔLk)/(Lk0). In eukaryotes, the average superhelical density of DNA in cells is −0.05. The change in enthalpy is about 560 kcal/mole and entropy is 1 kcal/K/mole. There is much more energy devoted to regulating the structure of DNA relative to that required for pushing the microtubule, an equilibrium polymer, into polymer versus dimeric states (see above).
In terms of the kinetochore, at one end the connection is to a stiff polymer that is sitting on a thermodynamic knife edge between polymerized versus depolymerized states. A mere 75 nm away, on the other side of the kinetochore, resides the DNA polymer that is far from its thermodynamically favored state. From this basic chemical and physical perspective, the structure and organization of pericentric chromatin must be considered in the generation and/or transmission of tension. Is this energy put to use during segregation? In the most extreme view, the thermodynamics of entropic springs (represented by replicated DNA strands) in a confined space has been invoked as a plausible mechanism for chromosome segregation in bacteria [56,57]. In addition, upon loss of cohesion between sister chromatids, chromatid separation ensues in the absence of microtubules [58]. The separation distances are larger than expected from thermodynamic fluctuation, suggestive of a polymer repulsion force.
Tension Distribution and Sensing across Sister Chromatids
As sister chromatids become oriented between the two microtubule organizing centers of the spindle apparatus (known as biorientation), tension is developed between sister chromatids. The monitoring of biorientation is critical to the fidelity of chromosome segregation. Whether the spindle assembly checkpoint monitors tension or attachment is subject to considerable debate [59,60]. Tension has been assumed to be linearly correlated with distance between sister chromatids. However, as discussed above, there are stiff (microtubules, extended proteins) and floppy (chromatin) elements in the kinetochore. Thus, it is no longer useful to relate change in length as a proxy for tension. Determining the absolute value of tension between sister chromatids will require measuring the Young’s modulus of the pericentric chromatin (how its length change (strain) varies as a function of pressure, i.e. stress/strain), which will depend upon the compaction of the DNA together with chromatin proteins and their higher order conformation. There are several models for the path of the DNA in the pericentric region [16,17,61,62]. However, we can examine physical principles that will guide future research. There are two major considerations from polymer physics for how tension is distributed in networks. One is the organization of the network as a series of loops upon loops. One might think about pericentric chromatin as a molecular bottle-brush. These are highly branched structures with side chains connected to a long polymer backbone. The brush-like architecture provides a mechanism to focus tension to the backbone and theoretical estimates reveal that this tension can be in the pN to nN range [62]. This is critical to our understanding of what is driving chromosome segregation. The prevailing hypothesis is that chromosomes are dragged through the cell interior by the kinetochore. An alternative view is that the stored energy in the chromosome coil is harnessed in a productive way to facilitate segregation (Figure 3). Second are excluded volume effects arising from steric repulsion that limits the conformational freedom of an individual loop (Figure 3). Contrary to cellular conditions, most biophysical studies are conducted in dilute solutions of salt water. Therefore, data from such studies must be evaluated with the caveat that they may bear little resemblance to the crowded interior of a cell [63].
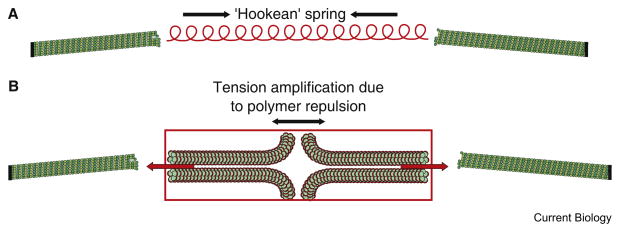
Figure 3. A schematic diagram of force generation in a mitotic spindle.
(A) The contribution of chromatin to the force balance in the spindle has been postulated as an inward force linearly related to extension (‘Hookean’ spring). (B) Based upon the distribution of cohesin [17] and pericentric chromatin [65], there may be force generation from the repulsion of polymers in a confined space (see text and [64]).
The current mechanical model for the mitotic spindle depicts microtubule dynamics connected via the kinetochore through a linear Hookean chromatin spring, and this configuration accounts for tension in the spindle (Figure 3). An alternative view comes from considering the network properties of the chromatin polymer. Depending on the solvent conditions, monomers may attract, resulting in coil shrinkage (polymer globule), or repel, resulting in coil swelling, known as the excluded volume effect (the polymer dissolves) (see [57]). The transition between these states is known as the coil–globule transition. When a large number of polymer chains are forced to share the same space, or confined to the same volume of space, the chains get extended due to repulsion between chains. The bond tension is non-uniformly distributed and, depending on the geometry, can be focused to a particular strand leading to tension amplification [64]. Thus, the configuration of pericentric chromatin may generate tension between sister chromatids (Figure 3). To evaluate this hypothesis, and to estimate the magnitude of the resulting tension, requires knowledge of the polymeric structure, solvent conditions, and crowding conditions between the two spindle poles in mitosis.
Conclusions
This decade has seen tremendous progress in resolving the molecular architecture of the kinetochore. Genetic screens designed to identify new genes involved in chromosome segregation, and proteomic approaches crafted to purify factors associated with known kinetochore and centromere proteins have assembled an extensive list of kinetochore and centromere proteins. Identification of component proteins stimulated experimental dissection of the molecular function of kinetochore and centromere components using in vivo, and more recently, in vitro approaches. The unique mode of kinetochore–microtubule attachment suggests that its molecular architecture will play an integral role in kinetochore function and regulation. Quantitative microscopy- based assays developed for studying kinetochore organization with high resolution allow direct examination of the organization of kinetochore–microtubule attachments rather than individual components of the kinetochore. These studies reveal a conserved kinetochore architecture that is necessary for incorporating structural studies of individual kinetochore complexes. Despite this new view of the kinetochore, many issues such as the structure of individual protein complexes, their interfaces with other complexes, the organization of centromeric chromatin, and the functional significance of kinetochore architecture remain unresolved. The protein architecture of the kinetochore established through quantitative studies now provides a foundation for understanding the biophysical mechanisms underlying kinetochore function and chromosome segregation.
Acknowledgments
We thank Dr. Ajit Joglekar (UNC-CH) and Ms. Marybeth Anderson (UNC-CH) for critical reading of the manuscript and Julian Haase (UNCCH) for figure illustrations.
References
- 1.Maiato H, Lince-Faria M. The perpetual movements of anaphase. Cell Mol Life Sci. 2010;67:2251–2269. doi: 10.1007/s00018-010-0327-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brinkley BR, Stubblefield E. The fine structure of the kinetochore of a mammalian cell in vitro. Chromosoma. 1966;19:28–43. doi: 10.1007/BF00332792. [DOI] [PubMed] [Google Scholar]
- 3.Jokelainen PT. The ultrastructure and spatial organization of the metaphase kinetochore in mitotic rat cells. J Ultrastruct Res. 1967;19:19–44. doi: 10.1016/s0022-5320(67)80058-3. [DOI] [PubMed] [Google Scholar]
- 4.Welburn JP, Cheeseman IM. Toward a molecular structure of the eukaryotic kinetochore. Dev Cell. 2008;15:645–655. doi: 10.1016/j.devcel.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 5.Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gittes F, Mickey B, Nettleton J, Howard J. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J Cell Biol. 1993;120:923–934. doi: 10.1083/jcb.120.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inoue S, Fuseler J, Salmon ED, Ellis GW. Functional organization of mitotic microtubules. Physical chemistry of the in vivo equilibrium system. Biophys J. 1975;15:725–744. doi: 10.1016/S0006-3495(75)85850-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salmon ED. Pressure-induced depolymerization of spindle microtubules. II Thermodynamics of in vivo spindle assembly. J Cell Biol. 1975;66:114–127. doi: 10.1083/jcb.66.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morris JR, Lasek RJ. Monomer-polymer equilibria in the axon: direct measurement of tubulin and actin as polymer and monomer in axoplasm. J Cell Biol. 1984;98:2064–2076. doi: 10.1083/jcb.98.6.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schliwa M, Euteneuer U, Herzog W, Weber K. Evidence for rapid structural and functional changes of the melanophore microtubuleorganizing center upon pigment movements. J Cell Biol. 1979;83:623–632. doi: 10.1083/jcb.83.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erickson HP. Co-operativity in protein-protein association. The structure and stability of the actin filament. J Mol Biol. 1989;206:465–474. doi: 10.1016/0022-2836(89)90494-4. [DOI] [PubMed] [Google Scholar]
- 12.Laan L, Husson J, Munteanu EL, Kerssemakers JW, Dogterom M. Force-generation and dynamic instability of microtubule bundles. Proc Natl Acad Sci USA. 2008;105:8920–8925. doi: 10.1073/pnas.0710311105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith SB, Cui Y, Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 14.Marshall WF, Marko JF, Agard DA, Sedat JW. Chromosome elasticity and mitotic polar ejection force measured in living Drosophila embryos by four-dimensional microscopy-based motion analysis. Curr Biol. 2001;11:569–578. doi: 10.1016/s0960-9822(01)00180-4. [DOI] [PubMed] [Google Scholar]
- 15.Talbert PB, Henikoff S. Histone variants–ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010;11:264–275. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 16.Pidoux AL, Allshire RC. The role of heterochromatin in centromere function. Philos Trans R Soc Lond B Biol Sci. 2005;360:569–579. doi: 10.1098/rstb.2004.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeh E, Haase J, Paliulis LV, Joglekar A, Bond L, Bouck D, Salmon ED, Bloom KS. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howard J. Mechanisms of Motor Proteins and the Cytoskeleton. Sunderland, Massachusetts: Sinauer Associates, Inc; 2001. [Google Scholar]
- 19.Joglekar AP, Bouck D, Finley K, Liu X, Wan Y, Berman J, He X, Salmon ED, Bloom KS. Molecular architecture of the kinetochore- microtubule attachment site is conserved between point and regional centromeres. J Cell Biol. 2008;181:587–594. doi: 10.1083/jcb.200803027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnston K, Joglekar A, Hori T, Suzuki A, Fukagawa T, Salmon ED. Vertebrate kinetochore protein architecture: protein copy number. J Cell Biol. 2010;189:937–943. doi: 10.1083/jcb.200912022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joglekar AP, Bloom K, Salmon ED. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr Biol. 2009;19:694–699. doi: 10.1016/j.cub.2009.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cherry LM, Faulkner AJ, Grossberg LA, Balczon R. Kinetochore size variation in mammalian chromosomes: an image analysis study with evolutionary implications. J Cell Sci. 1989;92(Pt 2):281–289. doi: 10.1242/jcs.92.2.281. [DOI] [PubMed] [Google Scholar]
- 24.McEwen BF, Ding Y, Heagle AB. Relevance of kinetochore size and microtubule-binding capacity for stable chromosome attachment during mitosis in PtK1 cells. Chromosome Res. 1998;6:123–132. doi: 10.1023/a:1009239013215. [DOI] [PubMed] [Google Scholar]
- 25.Zinkowski RP, Meyne J, Brinkley BR. The centromere-kinetochore complex: a repeat subunit model. J Cell Biol. 1991;113:1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong Y, Vanden Beldt KJ, Meng X, Khodjakov A, McEwen BF. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat Cell Biol. 2007;9:516–522. doi: 10.1038/ncb1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller SA, Johnson ML, Stukenberg PT. Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80(Hec1) Curr Biol. 2008;18:1785–1791. doi: 10.1016/j.cub.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guimaraes GJ, Dong Y, McEwen BF, Deluca JG. Kinetochore- microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr Biol. 2008;18:1778–1784. doi: 10.1016/j.cub.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 30.Westermann S, Avila-Sakar A, Wang HW, Niederstrasser H, Wong J, Drubin DG, Nogales E, Barnes G. Formation of a dynamic kinetochore- microtubule interface through assembly of the Dam1 ring complex. Mol Cell. 2005;17:277–290. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 31.Franco A, Meadows JC, Millar JB. The Dam1/DASH complex is required for the retrieval of unclustered kinetochores in fission yeast. J Cell Sci. 2007;120:3345–3351. doi: 10.1242/jcs.013698. [DOI] [PubMed] [Google Scholar]
- 32.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 33.McIntosh JR, Grishchuk EL, Morphew MK, Efremov AK, Zhudenkov K, Volkov VA, Cheeseman IM, Desai A, Mastronarde DN, Ataullakhanov FI. Fibrils connect microtubule tips with kinetochores: a mechanism to couple tubulin dynamics to chromosome motion. Cell. 2008;135:322–333. doi: 10.1016/j.cell.2008.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yildiz A, Park H, Safer D, Yang Z, Chen LQ, Selvin PR, Sweeney HL. Myosin VI steps via a hand-over-hand mechanism with its lever arm undergoing fluctuations when attached to actin. J Biol Chem. 2004;279:37223–37226. doi: 10.1074/jbc.C400252200. [DOI] [PubMed] [Google Scholar]
- 36.Churchman LS, Okten Z, Rock RS, Dawson JF, Spudich JA. Single molecule high-resolution colocalization of Cy3 and Cy5 attached to macromolecules measures intramolecular distances through time. Proc Natl Acad Sci USA. 2005;102:1419–1423. doi: 10.1073/pnas.0409487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schittenhelm R, Heeger S, Althoff F, Walter A, Heidmann S, Mechtler K, Lehner C. Spatial organization of a ubiquitous eukaryotic kinetochore protein network in Drosophila chromosomes. Chromosoma. 2007;116:385–402. doi: 10.1007/s00412-007-0103-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uchida KS, Takagaki K, Kumada K, Hirayama Y, Noda T, Hirota T. Kinetochore stretching inactivates the spindle assembly checkpoint. J Cell Biol. 2009;184:383–390. doi: 10.1083/jcb.200811028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wan X, O’Quinn RP, Pierce HL, Joglekar AP, Gall WE, DeLuca JG, Carroll CW, Liu ST, Yen TJ, McEwen BF, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang HW, Long S, Ciferri C, Westermann S, Drubin D, Barnes G, Nogales E. Architecture and flexibility of the yeast Ndc80 kinetochore complex. J Mol Biol. 2008;383:894–903. doi: 10.1016/j.jmb.2008.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cassimeris L, Rieder CL, Rupp G, Salmon ED. Stability of microtubule attachment to metaphase kinetochores in PtK1 cells. J Cell Sci. 1990;96(Pt 1):9–15. doi: 10.1242/jcs.96.1.9. [DOI] [PubMed] [Google Scholar]
- 42.McEwen BF, Heagle AB, Cassels GO, Buttle KF, Rieder CL. Kinetochore fiber maturation in PtK1 cells and its implications for the mechanisms of chromosome congression and anaphase onset. J Cell Biol. 1997;137:1567–1580. doi: 10.1083/jcb.137.7.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, 3rd, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 44.Obuse C, Yang H, Nozaki N, Goto S, Okazaki T, Yoda K. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CENcomplex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells. 2004;9:105–120. doi: 10.1111/j.1365-2443.2004.00705.x. [DOI] [PubMed] [Google Scholar]
- 45.Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR, 3rd, Desai A, Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- 46.Liu ST, Rattner JB, Jablonski SA, Yen TJ. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J Cell Biol. 2006;175:41–53. doi: 10.1083/jcb.200606020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ribeiro SA, Gatlin JC, Dong Y, Joglekar A, Cameron L, Hudson DF, Farr CJ, McEwen BF, Salmon ED, Earnshaw WC, et al. Condensin regulates the stiffness of vertebrate centromeres. Mol Biol Cell. 2009;20:2371–2380. doi: 10.1091/mbc.E08-11-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cimini D, Cameron LA, Salmon ED. Anaphase spindle mechanics prevent mis-segregation of merotelically oriented chromosomes. Curr Biol. 2004;14:2149–2155. doi: 10.1016/j.cub.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 49.Maresca TJ, Salmon ED. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J Cell Biol. 2009;184:373–381. doi: 10.1083/jcb.200808130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O’Connell CB, Loncarek J, Hergert P, Kourtidis A, Conklin DS, Khodjakov A. The spindle assembly checkpoint is satisfied in the absence of interkinetochore tension during mitosis with unreplicated genomes. J Cell Biol. 2008;183:29–36. doi: 10.1083/jcb.200801038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu D, Vader G, Vromans MJ, Lampson MA, Lens SM. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 2009;323:1350–1353. doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vale RD. Microscopes for fluorimeters: the era of single molecule measurements. Cell. 2008;135:779–785. doi: 10.1016/j.cell.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nicklas RB. The forces that move chromosomes in mitosis. Annu Rev Biophys Biophys Chem. 1988;17:431–449. doi: 10.1146/annurev.bb.17.060188.002243. [DOI] [PubMed] [Google Scholar]
- 54.Depew DE, Wang JC. Conformational fluctuations of DNA helix. Proc Natl Acad Sci USA. 1975;72:4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seidl A, Hinz HJ. The free energy of DNA supercoiling is enthalpy-determined. Proc Natl Acad Sci USA. 1984;81:1312–1316. doi: 10.1073/pnas.81.5.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jun S, Mulder B. Entropy-driven spatial organization of highly confined polymers: lessons for the bacterial chromosome. Proc Natl Acad Sci USA. 2006;103:12388–12393. doi: 10.1073/pnas.0605305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jun S, Wright A. Entropy as the driver of chromosome segregation. Nat Rev Microbiol. 2010;8:600–607. doi: 10.1038/nrmicro2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- 59.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 60.Pinsky BA, Biggins S. The spindle checkpoint: tension versus attachment. Trends Cell Biol. 2005;15:486–493. doi: 10.1016/j.tcb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 61.Marshall OJ, Marshall AT, Choo KH. Three-dimensional localization of CENP-A suggests a complex higher order structure of centromeric chromatin. J Cell Biol. 2008;183:1193–1202. doi: 10.1083/jcb.200804078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ribeiro SA, Vagnarelli P, Dong Y, Hori T, McEwen BF, Fukagawa T, Flors C, Earnshaw WC. A super-resolution map of the vertebrate kinetochore. Proc Natl Acad Sci USA. 2010;107:10484–10489. doi: 10.1073/pnas.1002325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zimmerman SB, Minton AP. Macromolecular crowding: biochemical, biophysical, and physiological consequences. Annu Rev Biophys Biomol Struct. 1993;22:27–65. doi: 10.1146/annurev.bb.22.060193.000331. [DOI] [PubMed] [Google Scholar]
- 64.Panyukov S, Zhulina EB, Sheiko SS, Randall GC, Brock J, Rubinstein M. Tension amplification in molecular brushes in solutions and on substrates (dagger) J Phys Chem B. 2009;113:3750–3768. doi: 10.1021/jp807671b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anderson M, Haase J, Yeh E, Bloom K. Function and assembly of DNA looping, clustering, and microtubule attachment complexes within a eukaryotic kinetochore. Mol Biol Cell. 2009;20:4131–4139. doi: 10.1091/mbc.E09-05-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]