Abstract
To circumvent limitations of poor antigen presentation and immunogenicity of DNA vaccines that target induction of CD8+ T cell immunity, we have generated single chain MHC I trimers (MHC I SCTs) composed of a single polypeptide chain with a linear composition of antigenic peptide, β2-microglobulin, and heavy chain of a MHC class I molecule connected by flexible linkers. Because of its pre-assembled nature, the SCT presents enhanced expression and presentation of the antigenic peptide/MHC complexes at the cell surface. Furthermore, DNA vaccination with a plasmid DNA encoding an SCT incorporating an immunodominant viral epitope elicited protective CD8+ T cell responses against lethal virus infection. To extend these findings, here we tested the efficacy of SCT DNA vaccines against bacterial infections. In a mouse infection model of Listeria monocytogenes, the SCT DNA vaccine encoding H-2Kd and the immunodominant peptide LLO 91–99 generated functional primary and memory peptide-specific CD8+ T cells that confer partial protection against L. monocytogenes infection. DNA immunization of Kd/LLO91–99 SCTs generated functional memory CD8+ T cells independently of CD4+ T cells, although the expression of cognate or non-cognate CD4+ helper T cell epitopes further enhanced the protective efficacy of SCTs. Our study further demonstrates that the SCT serves as a potent platform for DNA vaccines against various infectious diseases.
Keywords: DNA vaccine, Listeria monocytogenes infection, MHC I single chain trimers, memory CD8+ T cell, CD4+ T cell help
1. Introduction
Vaccination is the most effective method of preventing infectious diseases. Most of the vaccines developed to date have focused on humoral immunity, i.e. production of antibodies for neutralizing viruses and toxins or opsonizing bacteria. However, there are many cases in which the antibody response is not sufficient to protect against pathogens. CD8+ T cells play a major role in controlling infection and disease progression in many infectious diseases. Upon infection, antigen-specific CD8+ T cells are generated and remove infected cells through their cytotoxic activity and/or release of cytokines that inhibit growth of the microbe or impair its ability to survive inside the cell. Ongoing computational and/or experimental approaches have identified antigenic epitopes in a vast number of pathogens and using the identified epitopes to induce CD8+ T cell immune responses has been an important strategy for successful vaccines [1, 2]. However, most immunization approaches with class I binding peptides have failed to induce CD8+ T cell responses strong enough to prevent disease. This failure has been attributed to the lack of CD4+ T cell help and/or difficulty in maintaining a sufficient level of antigen presentation required for CD8+ T cell activation. To circumvent these limitations, we have developed fully assembled MHC molecules that can be expressed as membrane-bound proteins on the cell surface, termed single chain trimers (SCTs) [3, 4]. SCTs are composed of an immunodominant peptide, β2m, and MHC I heavy chain covalently linked by 15–20 amino acid flexible linkers. Because SCTs are expressed as a single polypeptide chain, they do not require peptide processing, or chaperone-assisted peptide loading in the ER. Furthermore, antigen presentation by the SCT bypasses the need to compete with an extensive pool of endogenous peptides for peptide loading.
SCTs are folded properly and T cells respond to SCTs comparably to native peptide/MHC I complexes [4–6]. Various human and mouse class Ia and Ib MHC molecules have been engineered with epitope peptides into SCTs and proven as useful tools to monitor and modulate immune responses [7–10]. The potency of SCT DNA vaccines has been demonstrated in mouse tumor models and virus infection models [11–15]. For example, mice vaccinated with DNA encoding a SCT of an immunodominant CTL epitope of human papilloma virus type 16 (HPV-16) E6 antigen and H-2Kb were protected against a lethal challenge of E6-expressing TC-1 tumor cells [13]. Importantly, SCT-based DNA vaccines appeared more effective at generating CD8+ T cell immunity than subunit or epitope-only DNA vaccines even when targeted to the ER lumen, due to incorporation of a preprocessed and preloaded peptide [13, 14, 16]. Recently, in the study using HLA-A*0201 SCTs in a mouse model of West Nile virus infection, we demonstrated for the first time that SCT DNA vaccination induces protective T cell immunity against virus infection [15]. Furthermore, previous studies have demonstrated the flexibility of the SCT platform by combining it with other strategies to enhance DNA vaccine potency. For example, SCTs were co-expressed with a universal CD4+ T helper cell epitope to stimulate T helper cells that resulted in enhanced CD8+ T cell responses and anti-tumor and anti-virus effects [15, 17]. This suggested that the SCT platform can be incorporated into a composite vaccine targeting multiple aspects of immunity against tumors and viruses and opened the possibility that SCT-based DNA vaccines may be applied to other infectious disease models.
In this study, we extended these findings in the physiologic model system of Listeria monocytogenes infection. Listeria monocytogenes is a gram positive intracellular bacterium that can cause the human disease, listeriosis, particularly in immunocompromised individuals. L. monocytogenes infects a broad range of hosts and mouse models of Listeria infection are often used to study the mammalian immune response to infection [18]. L. monocytogenes infection elicits a robust T cell response that clears the pathogen from infected mice and provides long-lasting immunity while humoral immunity provides only a small contribution to protect mice. CD8+ T cells play a more substantial role than CD4+ T cells in conferring long term protective immunity particularly in the BALB/c mouse model [18–22]. In BALB/c infection, listeriolysin O is one of the most antigenic proteins secreted by L. monocytogenes and the LLO epitope (residues 91–99) restricted by H-2Kd is the most immunodominant as detected by CD8 T cells [23].
The ability to quickly and specifically eliminate recurring infections is a hallmark of immunological memory. Thus, the generation of quality memory CD8+ T cells is an appealing goal for vaccine design against a variety of infectious diseases. While the role of CD4+ T cell help in the generation of CD8+ T cell memory remains controversial, previous studies suggested that the inclusion of a CD4+ T cell epitope in SCT DNA vaccine enhances anti-tumor or -virus effects [15, 17]. However, in these previous studies, it was not reported whether CD4+ T cell responses were actually generated and whether primary or memory CD8+ T cell responses were affected. In this study, we tested whether CD4+ T cell help can improve the efficacy of SCT DNA vaccine by co-expressing CD4+ helper T cell epitopes with the SCT. For this purpose, we compared two known CD4+ helper T cell epitopes, a pan CD4+ T cell epitope and a well-known CD4+ T cell epitope of L. monocytogenes that binds to MHC class II I-Ad, LLO 189–200. Regarding the specificity of CD4+ T cell help required for optimal generation and maintenance of memory CD8+ T cell responses, it has been proposed that both CD4+ and CD8+ T cells are required to be specific for the same antigen [24, 25]. However, recently published data indicate that helper T cell function is not antigen specific and likely results through cytokines [26–30]. The discrepancy between the studies may result from using different experimental systems such as inflammatory or non-inflammatory and/or different precursor frequencies of antigen specific CD8+ T cells. Thus, our study not only examined the role of CD4+ T cell help in SCT-induced memory immune responses, but also compared L. monocytogenes-specific vs. -nonspecific CD4+ T cell epitopes for their ability to help protective CD8+ T cell responses induced by SCT-based DNA vaccines.
We report here the generation of a H-2Kd SCT incorporating the LLO 91–99 epitope that is recognized by peptide-specific CD8+ T cells. DNA vaccination with plasmids encoding Kd/LLO91–99 SCTs by gene gun induced epitope-specific primary and memory CD8+ T cell responses in BALB/c mice and the vaccinated mice showed better control of bacteria upon L. monocytogenes infection. The SCT alone was able to develop functional memory CD8+ T cells. However, providing either cognate or non-cognate help, the presence of CD4+ T cell help enhanced memory CD8+ T cell responses and protective immunity. Therefore, our data suggest that a SCT-based DNA vaccine can elicit potent antibacterial CD8+ T cell immunity and thus broaden the application of SCTs in pathogen diseases.
2. Material and Methods
2.1. Cell lines
Transient transfection was performed on 293T human embryonic kidney cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). LLO 91–99 specific, H-2Kd-restricted T cell hybridoma, 206.15, was a generous gift from Dr. Emil Unanue at Washington University in St. Louis. The hybridoma was maintained in modified DMEM (Invitrogen) supplemented with 10% FBS (HyClone Laboratories, Logan, UT), 1.5mM L-glutamine, 10mM HEPES, 1mM sodium pyruvate, 116mg/L L-arginine, 36mg/L L-asparagine (Sigma, St. Louis, MO) and 100U/mL penicillin/streptomycin (Tissue Culture Support Center, Washington University School of Medicine, St. Louis, MO).
2.2. SCT construct
To generate H-2Kd/LLO91–99 SCT, the H-2Kd SCT with HER2/Neu peptide in pcDNA3.1(−) expression vector was used as a template. Nucleotide oligos encoding LLO 91–99 peptide, GYKDGNEYI, flanked by restriction enzyme sites, AgeI and NheI, were designed in order to replace the existing epitope sequence; 5’-ccggtttgtatgctggctataaagatggcaacgaatatattggaggaggtg-3’ and 5’-ctagcacctcctccaatatattcgttgccatctttatagccagcatacaaa -3’. Oligos were annealed to form a double strand and inserted into the vector cut with AgeI and NheI.
To express SCTs and CD4+ T cell helper epitopes simultaneously, the pIRES expression vector was used and to direct CD4+ T cell helper epitopes to the endosome the invariant chain (Ii) was exploited. The entire sequences encoding SCTs and Ii were cloned from the original plasmids into the pIRES vector using extended PCR and ligation. Using two step overlapping PCR, the CLIP sequence of Ii was replaced with helper epitopes; PADRE (pan T helper epitope), AKFVAAWTLKAAA, or LLO189–200, WNEKYAQAYPNV. For immunization, DNA was prepared using the Plasmid Maxi Prep kit (Qiagen, Valencia, CA) according to the manufacturer's instructions.
2.3. Hybridoma activation assay and ELISA
Activation of the T hybridoma was assessed by measuring IL-2 secreted upon activation. T hybridoma cells were co-cultured with 293T cells transiently transfected with SCTs overnight at 37°C. After incubation, the culture supernatants were taken and the amount of IL-2 was measured by ELISA. The supernatant was added to a 96 well plate pre-coated with anti-IL-2 capture antibody, JES6-1A12 (BioLegend, San Diego, CA), overnight at 4°C and blocked with carbonate/bicarbonate coating buffer. After wash with washing buffer (PBS containing 0.05% Tween-20 (Sigma)), biotin- anti-IL-2 detection antibody, JES6-5H4 (BioLegend), was added to the plates followed by incubation with avidin-HRP (eBiosciences, San Diego, CA). Color was developed by adding TMB substrate (eBiosciences) and the reaction was stopped with 2N sulfuric acid. Plates were read on a plate reader (Bio-Rad, Hercules, CA) at 450nm.
2.4. L. monocytogenes and mouse infection
BALB/c mice were purchased from National Cancer Institute (Frederick, MD), maintained under specific pathogen-free conditions at the Washington University School of Medicine and were handled in accordance with the guidelines set by the Division of Comparative Medicine of Washington University. Listeria monocytogenes strain EGD was stored as frozen glycerol stocks at −80°C. All mice were infected intraperitoneally with various doses of L. monocytogenes EGD strain in pyrogen-free saline. For primary infection, 103 L. monocytogenes were injected. Secondary infection was performed with either 2 × 105 or 5 × 104 L. monocytogenes.
2.5. Flow cytometry
Cell culture supernatant containing SF1-1.1.1, monoclonal anti-H-2Kd antibody, was used to stain Kd/LLO91–99 SCT-expressing cells. The following mAbs were purchased from eBiosciences: anti-CD8 (53-6.7)-FITC, CD4 (GK1.5)-FITC, CD44 (IM7)-APC, CD62L (MEL-14)-APC, CD3 (145-2C11)-APC or -PE, IL-2 (JES6-5H4)-PE, IFNγ (XMG1.2)-PE, rat IgG1 isotype control-PE, and PE-conjugated goat anti-mouse Ig. PE-conjugated Kd/LLO91–99 and control tetramer were obtained from the National Institute of Allergy and Infectious Diseases tetramer facility (Emory University, Atlanta, GA). For intracellular cytokine staining, splenocytes were stimulated in vitro with peptide at 0.1µg/mL in the presence of GolgiPlug (BD Biosciences, San Jose, CA) for 4 hours at 37°C. Cells were washed and incubated with FcR blocking antibody (2.4G2, eBiosciences), stained with anti-CD8α, CD4, CD3, and/or CD62L mAb, and fixed with 1% paraformaldehyde (PFA). After washing twice with buffer containing 0.1% saponin, cells were stained with anti-IFNγ, anti-IL-2 mAb, or isotype control antibody. For tetramer staining, cells were stained with tetramers for 30 min at 4°C, and subsequently anti-CD8α or anti-CD44 mAb was added for an additional 20 min at 4°C. Propidium iodide (PI) was added shortly before flow cytometry to gate out dead cells. Cells were acquired on FACSCalibur and data were analyzed with FlowJo software (Tree Star, Ashland, OR).
2.6. IFNγ ELISpot assay
Spleens were harvested from mice 5 days after the last DNA immunization. After RBC lysis, single cell suspensions were incubated with 1µg/mL LLO 91–99 or control peptides in a PVDF filter plate (Millipore, Billerica, MA) pre-coated with 15µg/mL of anti-IFNγ capture antibody (AN18, Mabtech Inc, Cincinnati, OH). After overnight stimulation, cells were removed and the plate was incubated with biotinylated anti-IFNγ detection antibody (R4-6A2) and subsequently with streptavidin alkaline phosphatase. Spots were developed by adding substrate solution (BCIP/NBT) and counted with an automated ELISpot reader (CTL, Shaker Heights, OH).
2.7. Colony count
L. monocytogenes numbers in the spleen and liver were counted by determining colony forming units (CFU) from tissue homogenates on brain heart infusion agar (BHI) plates using standard procedures. Briefly, spleens or livers were homogenized in buffer containing PBS with 0.05% Triton X-100 (Sigma). Serially diluted tissue homogenates were plated onto BHI plates and incubated for 15–20 hours at 37°C. Colonies on each plate were counted. For all graphs, data were plotted using GraphPad Prism (GraphPad Software, La Jolla, CA). Mann-Whitney U test with two-tailed p-values and 95% confidence intervals was used for all statistical analyses.
3. Results
3.1. H-2Kd SCTs incorporating LLO 91–99 epitope are recognized by peptide-specific CD8+ T cells
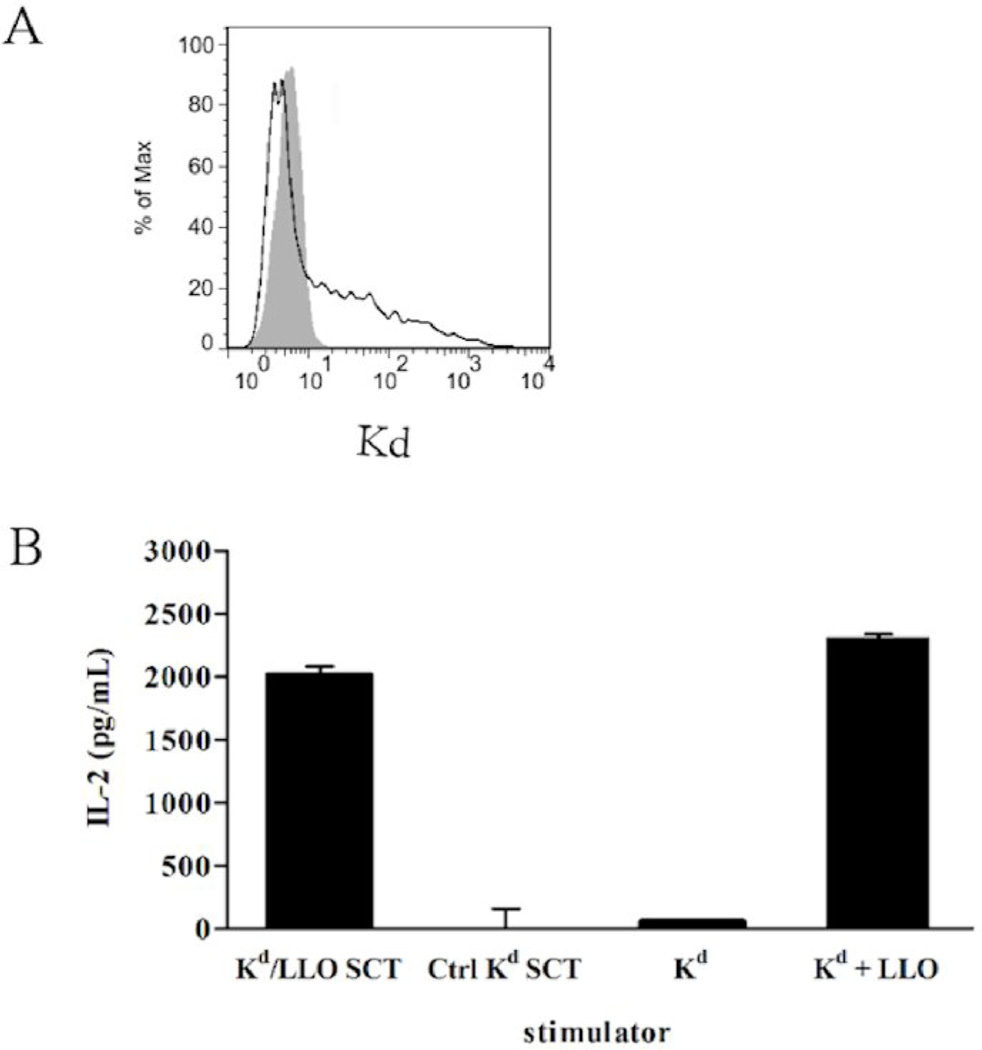
An SCT construct, which consists of the leader sequence of β2m, GYKDGNEYI (LLO 91–99), the first flexible linker of 15 residues (G4S)3, the mature mouse β2m, the second flexible linker of 20 residues (G4S)4, and the heavy chain of H-2Kd was generated in the pcDNA3 expression vector. As a control, the plasmid encoding Kd SCTs with HER2/neu peptide, a breast cancer antigen, in the same vector was used. To check the expression at the cell surface, 293T cells were transiently transfected with plasmids encoding Kd/LLO91–99 SCTs. SCTs were stained with the monoclonal antibody specific for folded Kd, SF1-1.1.1, indicating they were expressed and folded properly on the cell surface after transfection (Fig. 1A). These Kd/LLO91–99 SCT-expressing cells were incubated with CD8+ T hybridoma cells specific for Kd/LLO91–99 complexes for 24 hours to see whether SCTs can activate T cells. T hybridoma cells secreted IL-2 in response to Kd/LLO91–99 SCTs but not to control Kd SCTs (Fig. 1B). These results indicate the successful generation of Kd/LLO91–99 SCTs that are recognized by and activate peptide-specific CD8+ T cells.
Figure 1. Expression and recognition of Kd/LLO91–99 SCTs by CD8+ T cells.
(A) Cell surface expression of Kd/LLO91–99 SCTs. 293T cells were transfected with Kd/LLO91–99 SCT DNA and stained with anti-Kd mAb (SF1-1.1.1, solid line). A shaded histogram shows staining of untransfected cells. (B) Activation of hybridomas specific for LLO 91–99/Kd by Kd/LLO91–99 SCT-expressing cells. Transfected 293T cells were incubated with T hybridomas at the ratios of 1:1 for 24 hours and the amounts of IL-2 in the culture supernatant were measured by ELISA. For controls, P815 cells that express endogenous Kd were incubated with hybridomas with or without 1µg/mL of LLO 91–99 peptide.
3.2. DNA immunization with Kd/LLO91–99 SCTs induces specific CD8+ T cells
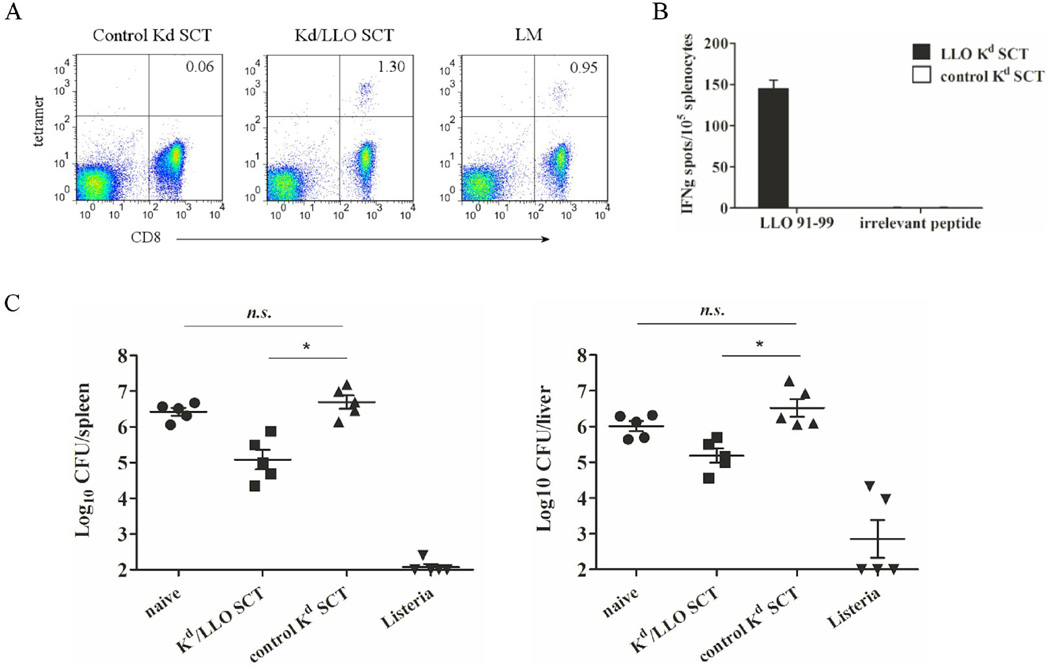
To test the ability of SCT to induce specific CD8+ T cells, BALB/c mice were immunized intradermally with plasmids encoding Kd/LLO91–99 SCTs or control SCTs three times at three day intervals by gene gun. At day 5 after the last immunization, LLO 91–99 specific CD8+ T cells were detected in the spleen from mice immunized with Kd/LLO91–99 SCTs using a tetramer assay (Fig. 2A). In ELISPOT, about the same percentage (~1.3%) of CD8+ T cells produced IFNγ in response to in vitro peptide re-stimulation, but there was no IFNγ response from mice immunized with control SCTs (Fig. 2B). Interestingly, we observed a similar or smaller percentage of Kd/LLO91–99 tetramer positive CD8+ T cells in the spleens from mice infected with a low dose of L. monocytogenes for 7 days (Fig. 2A). Thus, this indicates that Kd/LLO91–99 SCT DNA immunization induces an antigen-specific CD8+ T cell response which is comparable to that induced by natural infection.
Figure 2. SCT DNA immunization induces peptide-specific CD8+ T cells and lowers bacterial burden in L. monocytogenes infection.
Mice were immunized with Kd/LLO91–99 or control Kd SCT DNA three times at 3 day intervals. (A) and (B) At 5 days after the last immunization, splenocytes were harvested. (A) Splenocytes from control (left), Kd/LLO91–99 SCT (middle), or L. monocytogenes (right)-immunized mice were stained with anti-CD8 mAb and LLO 91–99/Kd tetramers. Numbers indicate the percentage of tetramer positive cells among CD3+ CD8+ cells. Representative figures of the flow cytometry data. (B) Splenocytes from each group were stimulated in vitro with LLO 91–99 or irrelevant peptide overnight and IFNγ secretion was measured by ELISPOT. Error bars indicate SE of the experiment. The data presented are from one representative experiment of two performed independently with each group containing 4~5 mice. (C) Immunized mice were infected intraperitoneally with 2×105 of L. monocytogenes at 4 weeks post immunization. For controls, mice were infected with 103 of L. monocytogenes or left uninfected. At 3 days after infection, spleens and livers were harvested and bacteria counts were determined by colony count. Error bars indicate SEM of the experiment. Data were expressed as log10 of colony forming units (CFU). n.s., not significant. *, p<0.05 (Mann-Whitney U test).
3.3. Protection mediated by Kd/LLO91–99 SCT DNA vaccine against L. monocytogenes infection
How a SCT DNA vaccine influences the outcome of L. monocytogenes infection was determined next. At 4 weeks after Kd/LLO91–99 or control SCT DNA vaccination by gene gun as described above, mice were infected intraperitoneally with a high dose of L. monocytogenes (2×105). For controls, naïve mice or mice that had been infected with a low dose of L. monocytogenes (103) for 4 weeks were also infected with a high dose of L. monocytogenes. Mice were sampled for growth of the L. monocytogenes in spleens and livers at 3 days after infection, since, during secondary infection, infected mice usually eliminate L. monocytogenes within 3–4 days [31, 32]. As shown is Fig. 2C, Kd/LLO91–99 SCT DNA vaccinated mice had one log lower bacterial counts (CFU, colony forming units) than control SCT vaccinated mice in both spleen and liver after challenge with L. monocytogenes. Control SCT DNA vaccinated mice had the same bacterial counts as untreated mice while mice immunized with a low dose of L. monocytogenes showed lower bacterial counts than Kd/LLO91–99 SCT DNA vaccinated mice. Thus, this result demonstrates that Kd/LLO91–99 SCT DNA vaccination can induce a protective immune response against L. monocytogenes infection. It should be noted that priming with live and not heat-killed bacteria is required for full protection [33], thus explaining the potency of priming with a low dose of L. monocytogenes. Reduction of bacterial count by one log may not be striking but is a significant level of protection. In the previous studies of DNA vaccines against L. monocytogenes, bacterial count after L. monocytogenes infection was reduced by 1~2 logs by immunization with DNA encoding wild type or mutant LLO protein but only with an adjuvant and/or boost immunizations [34, 35]. In another study which used the codon-optimized DNA sequence of LLO 91–99, DNA immunization decreased bacterial count by about one log [36]. Thus, our result with SCT based vaccination is considerable in that it was done with a single epitope without adjuvants or boost immunizations. It suggests that, relative to the heat-killed bacteria or whole protein, vaccination with the SCT can focus the immune response on an immunodominant CD8+ T cell epitope.
3.4. Memory CD8+ T cell responses after SCT DNA immunization
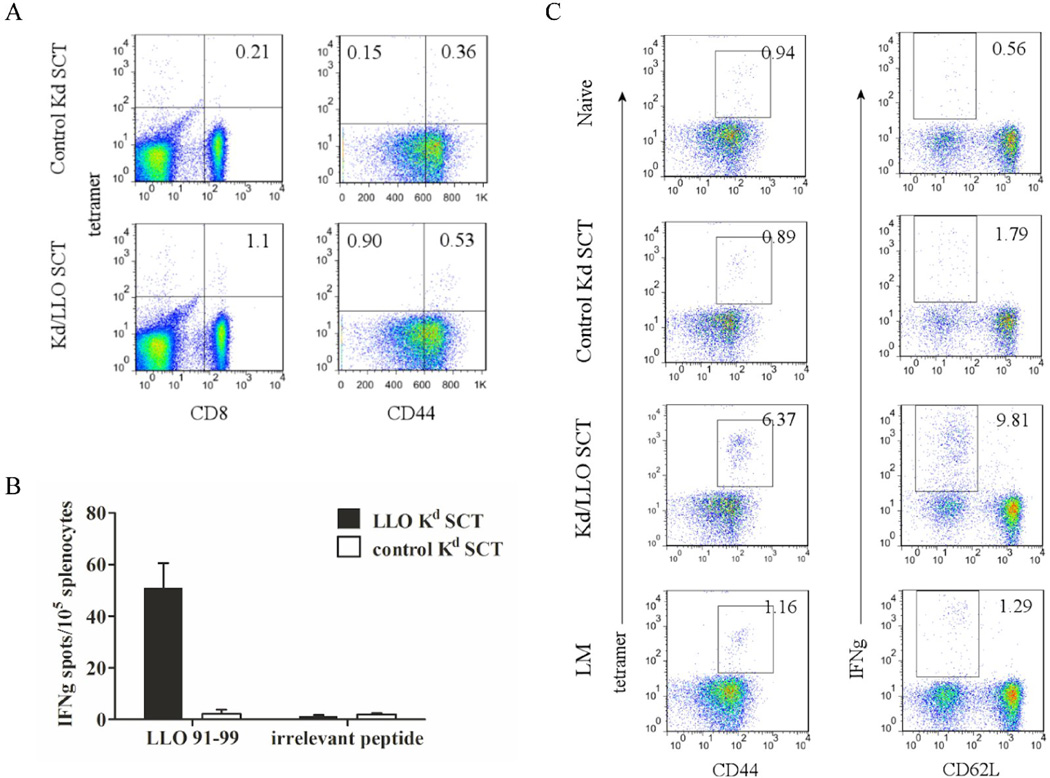
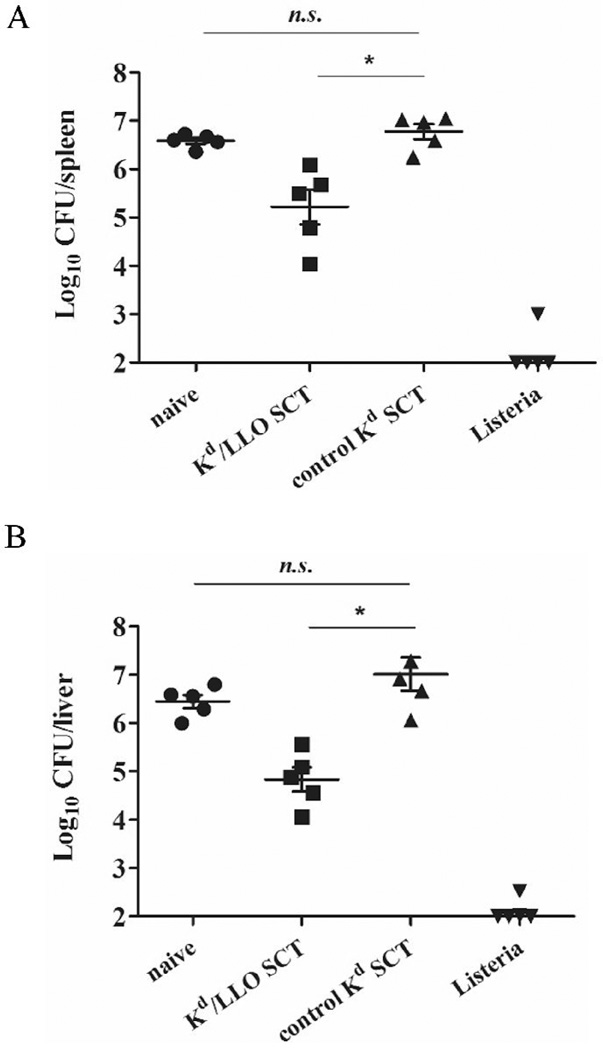
Generation of functional memory responses is a critical issue for vaccines against infection. We showed that mice have protective immunity at 4 weeks after SCT DNA immunization. But antigens may be still expressed at the time of bacteria challenge because it was reported that antigen expression was still detectable at 10 weeks following intradermal DNA injection [37]. Therefore we examined whether memory CD8+ T cells are generated after DNA immunization of SCTs and their functionality at later time points. Mice were immunized with DNA encoding Kd/LLO91–99 or control SCTs as described above and rested for 5 months before L. monocytogenes challenge. Five months after immunization, Kd/LLO91–99 tetramer positive CD8+ T cells were present in the spleen of resting mice immunized with Kd/LLO91–99 SCTs, and IFNγ production was detected after peptide stimulation in vitro (Fig. 3A and B). The CD8+ T cells had memory phenotypes, CD44hi and CD62Llo (Fig. 3A and not shown). To examine the recall response of memory T cells, we infected the vaccinated mice with 1×103 L. monocytogenes and compared CD8+ T cell responses 7 days later between control, Kd/LLO91–99 SCTs-immune, and L. monocytogenes-immune mice. Mice immunized with Kd/LLO91–99 SCTs had 4–6 times more tetramer positive CD8+ T cells that were CD44hi (~6.5%) as well as IFNγ producing CD8+ T cells that were CD62Llo (~10%) than L. monocytogenes- or control SCT -immune mice (Fig. 3C). Consistent with this finding, when mice were infected with 5×104 L. monocytogenes and 3 days later bacteria in the spleen and liver were counted, mice vaccinated with Kd/LLO91–99 SCTs had 10-fold less bacteria than mice immunized with control SCTs in both spleen and liver (Fig. 4). This was a similar result to that from L. monocytogenes challenge at 4 weeks post-immunization described above. Therefore, the results indicate that DNA immunization of Kd/LLO91–99 SCTs generates functional memory CD8+ T cells and provides protection against L. monocytogenes infection.
Figure 3. A SCT DNA vaccine develops memory CD8+ T cells.
Mice were vaccinated with Kd/LLO91–99 SCTs or control Kd SCTs as described above and rested for 5 months before L. monocytogenes challenge. (A) At 5 months post-immunization, splenocytes were stained with anti-CD8, -CD44 mAb and Kd/LLO91–99 tetramers. In the left panels CD3 positive cells are shown and the numbers indicate the percentage of CD3+ CD8+ tetramer+ cells. In the right panels CD3+ CD8+ cells are shown and the numbers indicate the percentage of cells in each quadrant. The flow cytometry data are presented from one representative experiment of three performed independently. (B) Splenocytes from each group were stimulated in vitro with LLO 91–99 or irrelevant peptide overnight and IFNγ secretion was measured by ELISPOT. Error bars indicate SE of the experiment. The data presented are from one representative experiment of two performed independently with each group containing 4~5 mice. (C) Vaccinated mice were challenged with 103 L. monocytogenes. At 7 days post-infection, splenocytes were stained with anti-CD8, -CD44 mAb, and Kd/LLO91–99 tetramers (left) or with anti-CD8, CD62L and -IFNγ mAb after 4 hour stimulation in vitro with LLO or irrelevant peptide in the presence of Golgi-blocking agent (right). CD8 positive cells were shown. Numbers indicate the percentage of cells of the gated population among CD8+ cells. Representative figures of the flow cytometry data.
Figure 4. A SCT DNA vaccine lowers bacterial burden in L. monocytogenes infection.
Mice were vaccinated with Kd/LLO91–99 SCTs or control Kd SCTs as described above and infected with 5 × 104 L. monocytogenes at 5 months post-immunization. 3 days later spleens (left) and livers (right) were harvested and bacteria growth was determined by colony count. Error bars indicate SEM of the experiment. Data were expressed as log10 of colony forming units (CFU). n.s., not significant. *, p<0.05 (Mann-Whitney U test).
3.5. Role of CD4+ helper T cells for the SCT-induced CD8+ T cells
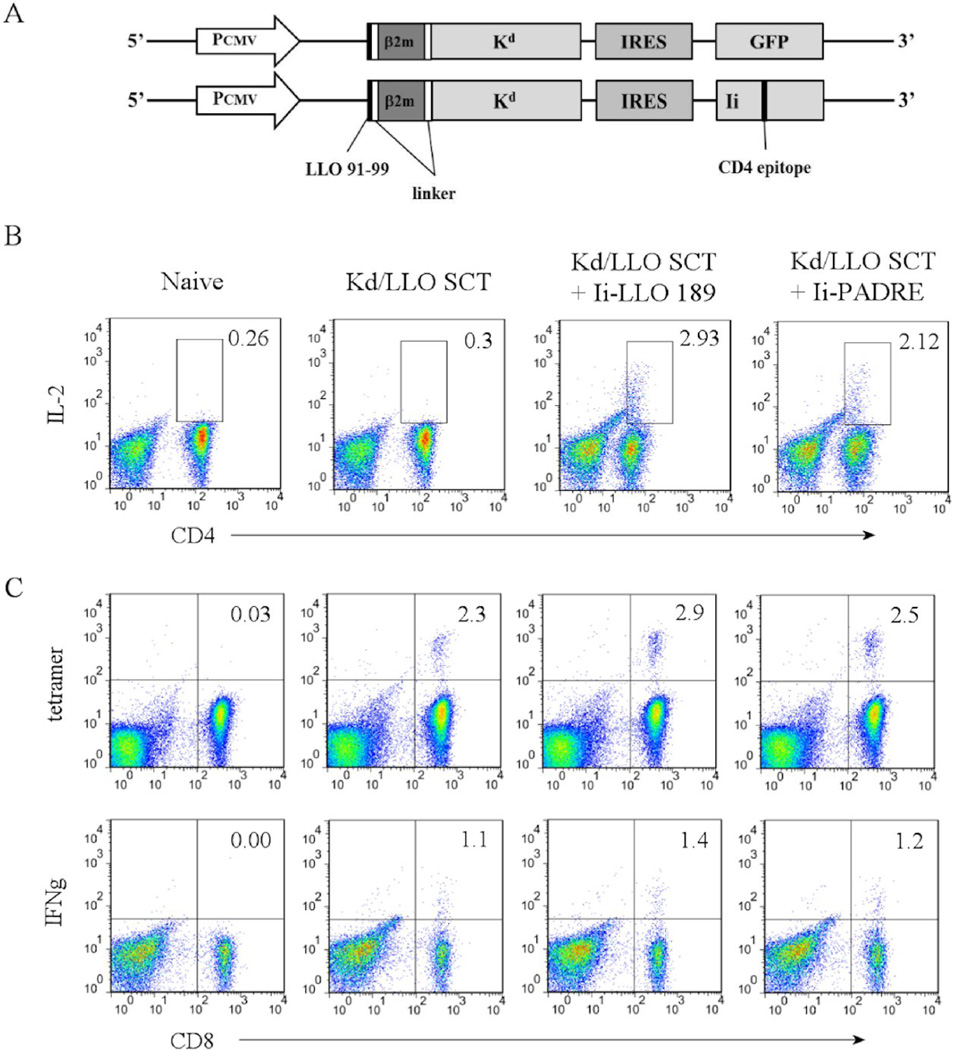
Our results indicated that expression of only CD8+ T cell antigens by SCTs was able to generate functional memory CD8+ T cells. Generally, functional memory CD8+ T cells are known to require the presence of CD4+ helper T cells. Although it is not clear yet how CD4+ T cells help to generate and maintain CD8+ T cell function, it has been reported that memory CD8+ T cell responses are defective when CD4+ T cells are absent during the priming phase [38, 39]. In these studies CD8+ T cells were primed with infectious antigens, which is not the case in our study. It is possible that the CD8+ T cell responses we saw after SCT immunization could be enhanced by the presence of activated CD4+ T cells. To determine whether CD4+ T cells lead to improved SCT-induced CD8+ T cell responses, we designed DNA constructs that expressed CD4+ T cell epitopes with SCTs simultaneously (Fig. 5A). CD4+ T cell epitopes were expressed in the context of an invariant chain (Ii) where a CLIP sequence was replaced with a CD4+ T cell epitope sequence in order for it to be loaded onto MHC II in the appropriate endocytic compartment. Expression of CD4+ T cell epitopes and induction of specific CD4+ T cells using this strategy have been previously reported [40–42]. The CD4+ T cell epitope/Ii was co-expressed with SCTs under the same promoter using an internal ribosomal entry site (IRES). Two different CD4+ helper T cell epitopes were used in the study. One is the peptide LLO 189–200 (one of the dominant CD4+ T cell epitopes of L. monocytogenes that binds to MHC II I-Ad) and the other is the pan CD4+ helper T cell epitope (PADRE). The pan CD4+ T cell epitope (PADRE, AKFVAAWTLKAA), is an engineered peptide that binds to multiple HLA-DR alleles, which also cross-reacts on mouse class II alleles [43]. L. monocytogenes peptide LLO 189–200 (WNEKYAQAYPNV) has been reported to be the most immunodominant CD4+ T cell epitope binding to I-Ad in the BALB/c model of L. monocytogenes infection [44]. Mice were immunized as above and CD4+ and CD8+ T cell responses and bacteria control were compared among groups immunized with SCTs alone, SCTs plus LLO189–200, and SCTs plus PADRE after challenge with L. monocytogenes.
Figure 5. Co-expression of a CD4+ T cell epitope does not affect primary CD8+ T cell responses.
(A) Diagrams of the plasmid DNA encoding Kd/LLO91–99 SCT and a CD4 epitope. PCMV, promoter; IRES, internal ribosomal entry site; Ii, invariant chain. (B) and (C) Mice were immunized with DNA encoding Kd/LLO91–99 SCT only, SCT plus Ii-LLO 189, or SCT plus Ii-PADRE three times at 3 day intervals by gene gun. At 5 days after the last immunization, splenocytes were harvested. (B) Splenocytes were cultured in vitro in the presence of 1µg/mL of LLO 189 or PADRE peptide for 5 days and then stained with anti-CD3, -CD4, and -IL-2 mAb after 4 hour re-stimulation with each peptide in the presence of Golgi-blocking agent. Numbers indicate the percentage of the gated cells of CD3+ cells. (C) Splenocytes were stained with anti-CD8, -CD3 mAb, and Kd/LLO91–99 tetramers (upper) or anti-CD8, -CD3, and -IFNγ mAb after 4 hour stimulation in vitro with LLO 91–99 or irrelevant peptide in the presence of Golgi-blocking agent (lower). Numbers indicate the percentage of the cells in the quadrant among CD8+ cells. CD3+ cells are shown. Representative figures of the flow cytometry data.
First, we confirmed the induction of epitope-specific CD8+ and CD4+ T cells following DNA immunization by measuring IFNγ or IL-2 production after in vitro peptide re-stimulation of splenocytes from the immunized mice at one week post-immunization. Both CD4+ helper T cell epitopes induced a comparable level of CD4+ T cell response, as monitored after in vitro expansion (Fig. 5B). All three groups showed similar CD8+ T cell responses in terms of the number of Kd/LLO91–99 tetramer positive cells as well as IFNγ production (Fig. 5C). These results indicated that the presence of activated CD4+ T cells did not affect the primary CD8+ T cell response.
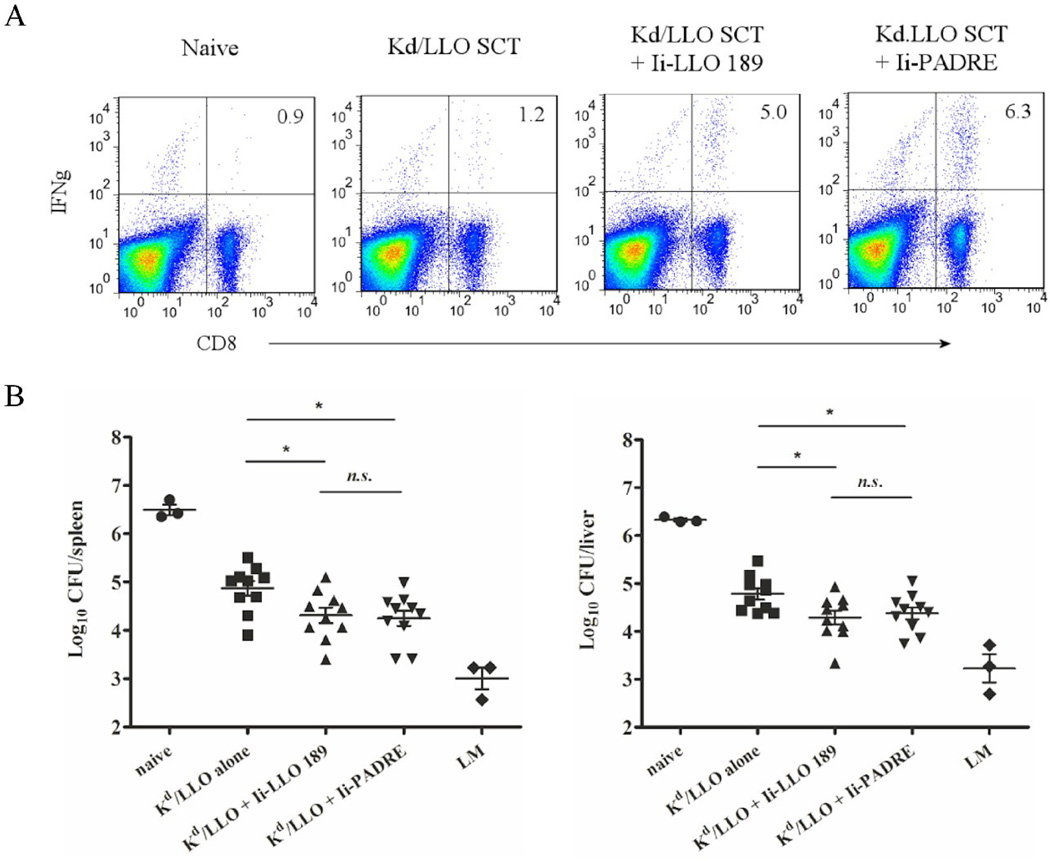
Next, memory CD8+ T cell responses were examined. At 6 weeks post-immunization, about the same number of memory phenotype, CD44hi, CD62Llo and Kd/LLO91–99 tetramer positive CD8+ T cells were present in all three groups and they produced a comparable amount of IFNγ upon in vitro peptide re-stimulation (data not shown). However, after challenge with a low dose of L. monocytogenes, mice that had been vaccinated with SCTs plus LLO189–200 or SCTs plus PADRE showed about 4~5 times more IFNγ-producing CD8+ T cells than mice vaccinated with SCTs alone (Fig. 6A). SCTs plus LLO189–200 and SCTs plus PADRE mounted similar recall responses, indicating that CD4+ T cell help is not necessarily antigen-specific. Finally, the vaccinated mice were challenged with a high dose of L. monocytogenes and 3 days later bacteria in the spleen and liver were counted. Mice that had been vaccinated with SCTs plus LLO189–200 or SCTs plus PADRE showed no significantly different bacterial counts, indicating that they were able to control bacterial infection equally (Fig. 6B). However, mice in both groups had about 10-fold less bacteria than mice vaccinated with SCTs alone. Therefore, DNA immunization of Kd/LLO91–99 SCTs generates functional memory CD8+ T cells and expression of CD4+ helper T cell epitopes can further enhance the protective efficacy of SCTs.
Figure 6. Co-expression of a CD4+ T cell epitope increases memory CD8+ T cell responses and the protective efficacy of a SCT DNA vaccine.
Mice were immunized with DNA encoding Kd/LLO91–99 SCT only, SCT plus Ii-LLO 189, or SCT plus Ii-PADRE as described above. (A) At 6 weeks post-immunization, mice were challenged with 1×103 of L. monocytogenes. 7 days later, splenocytes were stained with anti-CD8, -CD3, and -IFNγ mAb after 4 hour stimulation in vitro with LLO 91–99 or irrelevant peptide in the presence of Golgi-blocking agent. CD3 positive cells are shown. Numbers indicate the percentage of the cells in the quadrant among CD8+ cells. Representative figures of the flow cytometry data. (B) At 6 weeks post-immunization, mice were infected with 5 × 104 L. monocytogenes and 3 days later spleens (left) and livers (right) were harvested and bacteria growth was determined by colony count. Error bars indicate SEM of the experiment. Data were expressed as log10 of colony forming units (CFU). n.s., not significant. *, p<0.05 (Mann-Whitney U test).
4. Discussion
The SCT format has been amenable to different MHC I/peptide complexes, both murine and human, and it is one of the intriguing properties of SCTs [45, 46]. In this study, we successfully generated a SCT with the H-2Kd haplotype for the first time, confirming the broad and potential use of SCTs in clinical applications. Like other SCTs previously reported, Kd/LLO91–99 SCTs were properly folded and expressed at the cell surface after transfection and activated T cells in vitro and primed peptide-specific CD8+ T cells in vivo. This supports the universal application of SCTs and therefore the potential as a DNA vaccine platform.
We demonstrate that Kd/LLO91–99 SCT DNA vaccination provides protection against L. monocytogenes infection. Although it did not achieve the efficacy of vaccination with low dose live bacteria, the SCT DNA vaccine still provided a significant level of protection from infection. The focus of our approach was to optimally target a known immunodominant CD8+ T cell epitope in bacterial infection to the class I antigen presentation pathway and test for protection. It should be noted that heat-killed L. monocytogenes does not provide significant immunological protection [33]. Thus infection with live L. monocytogenes is required for the best protection. Furthermore, there are other CD8+ T cell epitopes in L. monocytogenes besides LLO 91–99 [44] and other arms of the immune system such as neutrophils and macrophages that contribute to L. monocytogenes protection [47, 48]. However, our DNA vaccine using an SCT elicited a more robust response than live L. monocytogenes infection to the immunodominant LLO 91–99 epitope, demonstrating efficient antigen presentation. Furthermore, SCT approaches are likely to benefit from prime-boost strategies or co-expression with different adjuvants such as CpG oligonucleotides (ODN) or IL-12. Indeed, in a previous study, gene gun vaccination with DNA plasmid encoding LLO protein and CpG ODN required a prime/boost inoculation with a 45 day interval to generate significant numbers of Listeria-specific CD8+ T cells [35]. By comparison with our findings, we conclude that SCT approaches can focus early CTL responses to immunodominant CD8+ T cell epitopes.
Our results not only show the efficient priming of naïve peptide-specific CTLs but also indicate that SCT DNA immunization alone, independently of CD4+ T cells, can generate functional memory CD8+ T cells. The presence of CD4+ T cell help improved the recall response and protective efficacy marginally. Generation of functional memory responses is essential to develop successful vaccines against pathogens and considerable effort has been made in developing strategies to induce high frequencies of memory T cells [49]. In the literature, there has been agreement that primary CD8+ T cell responses do not depend on CD4+ T cell help [50, 51], which is consistent with our results. However, the role of CD4+ T cell help for the generation of stable, protective CD8+ T cell memory is still controversial. Our study shows that memory CD8+ T cells were generated without activation of CD4+ T cells, which seems in contrast to the studies supporting a requirement for CD4+ T cell help for the generation of CD8+ T cell memory [26, 38, 39, 51]. However, in those studies CD8+ T cells developed in the absence of CD4+ T cells using MHC class II- or CD4-knockout mice, whereas CD4+ T cells were present during SCT-induced CD8+ T cell generation in our study. Indeed, Bevan et al. demonstrated that effector CD8+ T cells became functionally impaired after transfer into MHC II-deficient recipients, but not into wild-type recipients [26], implying a role of CD4+ T cells in the maintenance of functional memory CD8+ T cells. Our results that co-expression of CD4+ helper T cell epitopes did not affect primary CD8+ T cell responses but increased secondary CD8+ T cell responses and protective efficacy support the conclusion that CD4+ T cells play a role for the maintenance of memory CD8+ T cells. Additionally, the similar extent of augmentation of CD8+ T cell responses by LLO 189–200 and PADRE suggest that CD4+ T cell function is not antigen specific. Our results are consistent with the study by T. C. Wu et al. where co-administration of Ii-PADRE DNA with Kb SCT DNA encoding a tumor epitope further enhanced generation of antigen-specific CD8+ T cells and anti-tumor effects against tumor challenge [17].
The mechanism through which CD4+ T cells provide help for memory CD8+ T cells has not been elucidated. In the cytotoxic T cell response to noninfectious agents such as immunization with protein antigens, activation of CD4+ T cells is important for maturation of antigen presenting cells (APCs) necessary to promote a CD8+ T cell response. CD4+ T cells could function solely via activation of the antigen presenting cells through CD40-CD40L interaction or also via direct CD4-CD8 T cell interaction [51, 52]. Thus the amount of danger signals produced by the immunization seems to be important. Accordingly, in the CTL response to virulent pathogens, the generation of memory CD8+ T cell precursors is CD4+ T cell independent [55, 56]. One of the mechanisms for the immune responses by DNA vaccines is the innate responses directed against the plasmid itself. Because the plasmid is of bacterial origin, its sequences contain CpG oligodeoxynucleotide motifs that bind the pattern recognition receptor Toll-like receptor 9 (TLR9), which is constitutively expressed on dendritic cells, and results in augmentation of the immune response against the antigen encoded by the plasmid [57, 58]. However, the role of TLR9 in DNA vaccines is still inconsistent between studies [59, 60]. Additionally, it has been reported that gene-gun administration of DNA stimulates a type 2 immune response due to the low amount of DNA injected, consequently fewer CpG motifs, whereas a type 1 immune response is critical to clear intracellular bacteria like L. monocytogenes [35, 61, 62]. However, our data show that SCT DNA vaccination using the gene-gun method can provide protection against L. monocytogenes. We speculate that SCT DNA immunization does not largely depend on CD4+ T cell help because the high density expression of epitope/MHC I complexes at the cell surface using a SCT format overcomes the limited adjuvant.
A key to improve future DNA vaccines further is to target the antigen expression to the appropriate APCs. Although, in specific regard to DNA vaccines, the exact mechanism to elicit a T cell immune response is still controversial. Most recent studies highlight the importance of CD8α+ dendritic cells (DCs) for in vivo activation of anti-tumor or anti-viral CD8+ T cells [63–65]. Furthermore this critical role of CD8α+ DCs is thought to reflect a predominant role of cross-presentation for in vivo activation of CD8+ T cells [66]. Indeed there is evidence that cross-presentation plays a key role in antigen presentation after gene gun-delivered DNA vaccines [67, and Lijin Li in preparation]. Thus a future advantage of the SCT format could be its ability to target an appropriate APC, thus, bypassing the need for cross-presentation.
In conclusion, SCT-based DNA vaccines induce pathogen-specific CD8+ T cell responses largely independently of CD4+ T cells and, therefore, can serve as a potent platform for DNA vaccines to develop CD8+ T cell immunity in various disease models including virus and bacterial infection. SCTs should be considered in a composite vaccine targeting multiple aspects of the immune system against a range of infectious diseases.
Highlights.
We tested a MHCI/peptide single chain DNA vaccine against Listeria infection in mice.
This approach was designed to optimize antigen presentation to CD8+ T cells.
Kd/LLOsingle chain DNA vaccine elicited CD8+ T cells and reduced bacterial loads.
Helper T cell responses improved memory but not primary anti-bacterial responses.
Single chain approaches should be considered as part of a composite vaccine.
Acknowledgements
We thank our colleagues Dr. Xiaoli Wang for a help with generation of a DNA construct, Dr. Emil Unanue for advice and reagents for Listeria infection, and Dr. Janet Connolly for critical reading of the manuscript. This work was supported by NIH grant AI55849.
Abbreviations
- SCT
single chain trimer
- PADRE
pan-DR epitope
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Berzofsky JA, Ahlers JD, Belyakov IM. Strategies for designing and optimizing new generation vaccines. Nat Rev Immunol. 2001 Dec;1(3):209–219. doi: 10.1038/35105075. [DOI] [PubMed] [Google Scholar]
- 2.Kutzler MA, Weiner DB. DNA vaccines: ready for prime time? Nat Rev Genet. 2008 Oct;9(10):776–788. doi: 10.1038/nrg2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mottez E, Langlade-Demoyen P, Gournier H, et al. Cells expressing a major histocompatibility complex class I molecule with a single covalently bound peptide are highly immunogenic. J Exp Med. 1995 Feb 1;181(2):493–502. doi: 10.1084/jem.181.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu YY, Netuschil N, Lybarger L, Connolly JM, Hansen TH. Cutting edge: single-chain trimers of MHC class I molecules form stable structures that potently stimulate antigen-specific T cells and B cells. J Immunol. 2002 Apr 1;168(7):3145–3149. doi: 10.4049/jimmunol.168.7.3145. [DOI] [PubMed] [Google Scholar]
- 5.Truscott SM, Lybarger L, Martinko JM, et al. Disulfide bond engineering to trap peptides in the MHC class I binding groove. J Immunol. 2007 May 15;178(10):6280–6289. doi: 10.4049/jimmunol.178.10.6280. [DOI] [PubMed] [Google Scholar]
- 6.Lybarger L, Yu YY, Miley MJ, et al. Enhanced immune presentation of a single-chain major histocompatibility complex class I molecule engineered to optimize linkage of a C-terminally extended peptide. J Biol Chem. 2003 Jul 18;278(29):27105–27111. doi: 10.1074/jbc.M303716200. [DOI] [PubMed] [Google Scholar]
- 7.Truscott SM, Wang X, Lybarger L, et al. Human major histocompatibility complex (MHC) class I molecules with disulfide traps secure disease-related antigenic peptides and exclude competitor peptides. J Biol Chem. 2008 Mar 21;283(12):7480–7490. doi: 10.1074/jbc.M709935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitaksov V, Truscott SM, Lybarger L, Connolly JM, Hansen TH, Fremont DH. Structural engineering of pMHC reagents for T cell vaccines and diagnostics. Chem Biol. 2007 Aug;14(8):909–922. doi: 10.1016/j.chembiol.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen TH, Lybarger L. Exciting applications of single chain trimers of MHC-I molecules. Cancer Immunol Immunother. 2006 Feb;55(2):235–236. doi: 10.1007/s00262-005-0091-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim S, Poursine-Laurent J, Truscott SM, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005 Aug 4;436(7051):709–713. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 11.Cheung YK, Cheng SC, Sin FW, Chan KT, Xie Y. Induction of T-cell response by a DNA vaccine encoding a novel HLA-A*0201 severe acute respiratory syndrome coronavirus epitope. Vaccine. 2007 Aug 10;25(32):6070–6077. doi: 10.1016/j.vaccine.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Li S, Shan M, et al. Hepatitis B virus core antigen epitopes presented by HLA-A2 single-chain trimers induce functional epitope-specific CD8+ T-cell responses in HLA-A2.1/Kb transgenic mice. Immunology. 2007 May;121(1):105–112. doi: 10.1111/j.1365-2567.2007.02543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang CH, Peng S, He L, et al. Cancer immunotherapy using a DNA vaccine encoding a single-chain trimer of MHC class I linked to an HPV-16 E6 immunodominant CTL epitope. Gene Ther. 2005 Aug;12(15):1180–1186. doi: 10.1038/sj.gt.3302519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palmowski MJ, Parker M, Choudhuri K, et al. A single-chain H-2Db molecule presenting an influenza virus nucleoprotein epitope shows enhanced ability at stimulating CD8+ T cell responses in vivo. J Immunol. 2009 Apr 15;182(8):4565–4571. doi: 10.4049/jimmunol.0803893. [DOI] [PubMed] [Google Scholar]
- 15.Kim S, Li L, McMurtrey CP, et al. Single-chain HLA-A2 MHC trimers that incorporate an immundominant peptide elicit protective T cell immunity against lethal West Nile virus infection. J Immunol. 2010 Apr 15;184(8):4423–4430. doi: 10.4049/jimmunol.0903955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L, Herndon JM, Truscott SM, et al. Engineering superior DNA vaccines: MHC class I single chain trimers bypass antigen processing and enhance the immune response to low affinity antigens. Vaccine. 2010 Feb 23;28(8):1911–1918. doi: 10.1016/j.vaccine.2009.10.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hung CF, Tsai YC, He L, Wu TC. DNA vaccines encoding Ii-PADRE generates potent PADRE-specific CD4+ T-cell immune responses and enhances vaccine potency. Mol Ther. 2007 Jun;15(6):1211–1219. doi: 10.1038/sj.mt.6300121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pamer EG. Immune responses to Listeria monocytogenes. Nat Rev Immunol. 2004 Oct;4(10):812–823. doi: 10.1038/nri1461. [DOI] [PubMed] [Google Scholar]
- 19.Zenewicz LA, Shen H. Innate and adaptive immune responses to Listeria monocytogenes: a short overview. Microbes Infect. 2007 Aug;9(10):1208–1215. doi: 10.10110/2/076/j.micinf.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finelli A, Kerksiek KM, Allen SE, et al. MHC class I restricted T cell responses to Listeria monocytogenes, an intracellular bacterial pathogen. Immunol Res. 1999;19(2–3):211–223. doi: 10.1007/BF02786489. [DOI] [PubMed] [Google Scholar]
- 21.Pamer EG. Direct sequence identification and kinetic analysis of an MHC class I-restricted Listeria monocytogenes CTL epitope. J Immunol. 1994 Jan 15;152(2):686–694. [PubMed] [Google Scholar]
- 22.Sijts AJ, Neisig A, Neefjes J, Pamer EG. Two Listeria monocytogenes CTL epitopes are processed from the same antigen with different efficiencies. J Immunol. 1996 Jan 15;156(2):683–692. [PubMed] [Google Scholar]
- 23.Pamer EG, Harty JT, Bevan MJ. Precise prediction of a dominant class I MHC-restricted epitope of Listeria monocytogenes. Nature. 1991 Oct 31;353(6347):852–855. doi: 10.1038/353852a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramsburg EA, Publicover JM, Coppock D, Rose JK. Requirement for CD4 T cell help in maintenance of memory CD8 T cell responses is epitope dependent. J Immunol. 2007 May 15;178(10):6350–6358. doi: 10.4049/jimmunol.178.10.6350. [DOI] [PubMed] [Google Scholar]
- 25.Gao FG, Khammanivong V, Liu WJ, Leggatt GR, Frazer IH, Fernando GJ. Antigen-specific CD4+ T-cell help is required to activate a memory CD8+ T cell to a fully functional tumor killer cell. Cancer Res. 2002 Nov 15;62(22):6438–6441. [PubMed] [Google Scholar]
- 26.Sun JC, Williams MA, Bevan MJ. CD4+ T cells are required for the maintenance, not programming, of memory CD8+ T cells after acute infection. Nat Immunol. 2004 Sep;5(9):927–933. doi: 10.1038/ni1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamilton SE, Jameson SC. The nature of the lymphopenic environment dictates protective function of homeostatic-memory CD8+ T cells. Proc Natl Acad Sci U S A. 2008 Nov 25;105(47):18484–18489. doi: 10.1073/pnas.0806487105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Goer de Herve MG, Cariou A, Simonetta F, Taoufik Y. Heterospecific CD4 help to rescue CD8 T cell killers. J Immunol. 2008 Nov 1;181(9):5974–5980. doi: 10.4049/jimmunol.181.9.5974. [DOI] [PubMed] [Google Scholar]
- 29.Hao S, Yuan J, Xiang J. Nonspecific CD4(+) T cells with uptake of antigen-specific dendritic cell-released exosomes stimulate antigen-specific CD8(+) CTL responses and long-term T cell memory. J Leukoc Biol. 2007 Oct;82(4):829–838. doi: 10.1189/jlb.0407249. [DOI] [PubMed] [Google Scholar]
- 30.Tsukamoto T, Takeda A, Yamamoto T, Yamamoto H, Kawada M, Matano T. Impact of cytotoxic-T-lymphocyte memory induction without virus-specific CD4+ T-Cell help on control of a simian immunodeficiency virus challenge in rhesus macaques. J Virol. 2009 Sep;83(18):9339–9346. doi: 10.1128/JVI.01120-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carrero JA, Calderon B, Unanue ER. Lymphocytes are detrimental during the early innate immune response against Listeria monocytogenes. J Exp Med. 2006 Apr 17;203(4):933–940. doi: 10.1084/jem.20060045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carrero JA, Calderon B, Unanue ER. Type I interferon sensitizes lymphocytes to apoptosis and reduces resistance to Listeria infection. J Exp Med. 2004 Aug 16;200(4):535–540. doi: 10.1084/jem.20040769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lauvau G, Vijh S, Kong P, et al. Priming of memory but not effector CD8 T cells by a killed bacterial vaccine. Science. 2001 Nov 23;294(5547):1735–1739. doi: 10.1126/science.1064571. [DOI] [PubMed] [Google Scholar]
- 34.Cornell KA, Bouwer HG, Hinrichs DJ, Barry RA. Genetic immunization of mice against Listeria monocytogenes using plasmid DNA encoding listeriolysin O. J Immunol. 1999 Jul 1;163(1):322–329. [PubMed] [Google Scholar]
- 35.Fensterle J, Grode L, Hess J, Kaufmann SH. Effective DNA vaccination against listeriosis by prime/boost inoculation with the gene gun. J Immunol. 1999 Oct 15;163(8):4510–4518. [PubMed] [Google Scholar]
- 36.Yoshida A, Nagata T, Uchijima M, Koide Y. Protective CTL response is induced in the absence of CD4+ T cells and IFN-γ by gene gun DNA vaccination with a minigene encoding a CTL epitope of Listeria monocytogenes. Vaccine. 2001 Jul 20;19(30):4297–4306. doi: 10.1016/s0264-410x(01)00146-3. [DOI] [PubMed] [Google Scholar]
- 37.Hovav AH, Panas MW, Rahman S, et al. Duration of antigen expression in vivo following DNA immunization modifies the magnitude, contraction, and secondary responses of CD8+ T lymphocytes. J Immunol. 2007 Nov 15;179(10):6725–6733. doi: 10.4049/jimmunol.179.10.6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun JC, Bevan MJ. Defective CD8 T cell memory following acute infection without CD4 T cell help. Science. 2003 Apr 11;300(5617):339–342. doi: 10.1126/science.1083317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shedlock DJ, Shen H. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science. 2003 Apr 11;300(5617):337–339. doi: 10.1126/science.1082305. [DOI] [PubMed] [Google Scholar]
- 40.van BJ, Schoenberger SP, Verreck F, Amons R, Offringa R, Koning F. Efficient loading of HLA-DR with a T helper epitope by genetic exchange of CLIP. Proc Natl Acad Sci U S A. 1997 Jul 8;94(14):7499–7502. doi: 10.1073/pnas.94.14.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagata T, Aoshi T, Suzuki M, et al. Induction of protective immunity to Listeria monocytogenes by immunization with plasmid DNA expressing a helper T-cell epitope that replaces the class II-associated invariant chain peptide of the invariant chain. Infect Immun. 2002 May;70(5):2676–2680. doi: 10.1128/IAI.70.5.2676-2680.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim D, Monie A, He L, Tsai YC, Hung CF, Wu TC. Role of IL-2 secreted by PADRE-specific CD4+ T cells in enhancing E7-specific CD8+ T-cell immune responses. Gene Ther. 2008 May;15(9):677–687. doi: 10.1038/sj.gt.3303102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alexander J, Sidney J, Southwood S, et al. Development of high potency universal DR-restricted helper epitopes by modification of high affinity DR-blocking peptides. Immunity. 1994 Dec;1(9):751–761. doi: 10.1016/s1074-7613(94)80017-0. [DOI] [PubMed] [Google Scholar]
- 44.Geginat G, Schenk S, Skoberne M, Goebel W, Hof H. A novel approach of direct ex vivo epitope mapping identifies dominant and subdominant CD4 and CD8 T cell epitopes from Listeria monocytogenes. J Immunol. 2001 Feb 1;166(3):1877–1884. doi: 10.4049/jimmunol.166.3.1877. [DOI] [PubMed] [Google Scholar]
- 45.Hansen T, Yu YY, Fremont DH. Preparation of stable single-chain trimers engineered with peptide, beta2 microglobulin, and MHC heavy chain. Chapter 17. Curr Protoc Immunol. 2009 Nov; doi: 10.1002/0471142735.im1705s87. Unit17. [DOI] [PubMed] [Google Scholar]
- 46.Hansen TH, Connolly JM, Gould KG, Fremont DH. Basic and translational applications of engineered MHC class I proteins. Trends Immunol. 2010 Oct;31(10):363–369. doi: 10.1016/j.it.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rogers HW, Unanue ER. Neutrophils are involved in acute, nonspecific resistance to Listeria monocytogenes in mice. Infect Immun. 1993 Dec;61(12):5090–5096. doi: 10.1128/iai.61.12.5090-5096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Czuprynski CJ, Brown JF, Maroushek N, Wagner RD, Steinberg H. Administration of anti-granulocyte mAb RB6-8C5 impairs the resistance of mice to Listeria monocytogenes infection. J Immunol. 1994 Feb 15;152(4):1836–1846. [PubMed] [Google Scholar]
- 49.Sallusto F, Lanzavecchia A, Araki K, Ahmed R. From vaccines to memory and back. Immunity. 2010 Oct 29;33(4):451–463. doi: 10.1016/j.immuni.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bourgeois C, Tanchot C. Mini-review CD4 T cells are required for CD8 T cell memory generation. Eur J Immunol. 2003 Dec;33(12):3225–3231. doi: 10.1002/eji.200324576. [DOI] [PubMed] [Google Scholar]
- 51.Bourgeois C, Rocha B, Tanchot C. A role for CD40 expression on CD8+ T cells in the generation of CD8+ T cell memory. Science. 2002 Sep 20;297(5589):2060–2063. doi: 10.1126/science.1072615. [DOI] [PubMed] [Google Scholar]
- 52.Bennett SR, Carbone FR, Karamalis F, Flavell RA, Miller JF, Heath WR. Help for cytotoxic-T-cell responses is mediated by CD40 signalling. Nature. 1998 Jun 4;393(6684):478–480. doi: 10.1038/30996. [DOI] [PubMed] [Google Scholar]
- 53.Ridge JP, Di RF, Matzinger P. A conditioned dendritic cell can be a temporal bridge between a CD4+ T-helper and a T-killer cell. Nature. 1998 Jun 4;393(6684):474–478. doi: 10.1038/30989. [DOI] [PubMed] [Google Scholar]
- 54.Schoenberger SP, Toes RE, van der Voort EI, Offringa R, Melief CJ. T-cell help for cytotoxic T lymphocytes is mediated by CD40-CD40L interactions. Nature. 1998 Jun 4;393(6684):480–483. doi: 10.1038/31002. [DOI] [PubMed] [Google Scholar]
- 55.Sun JC, Bevan MJ. Cutting edge: long-lived CD8 memory and protective immunity in the absence of CD40 expression on CD8 T cells. J Immunol. 2004 Mar 15;172(6):3385–3389. doi: 10.4049/jimmunol.172.6.3385. [DOI] [PubMed] [Google Scholar]
- 56.Lee BO, Hartson L, Randall TD. CD40-deficient, influenza-specific CD8 memory T cells develop and function normally in a CD40-sufficient environment. J Exp Med. 2003 Dec 1;198(11):1759–1764. doi: 10.1084/jem.20031440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krieg AM. CpG motifs in bacterial DNA and their immune effects. Annu Rev Immunol. 2002;20:709–760. doi: 10.1146/annurev.immunol.20.100301.064842. [DOI] [PubMed] [Google Scholar]
- 58.Bode C, Zhao G, Steinhagen F, Kinjo T, Klinman DM. CpG DNA as a vaccine adjuvant. Expert Rev Vaccines. 2011 Apr;10(4):499–511. doi: 10.1586/erv.10.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Babiuk S, Mookherjee N, Pontarollo R, et al. TLR9−/− and TLR9+/+ mice display similar immune responses to a DNA vaccine. Immunology. 2004 Sep;113(1):114–120. doi: 10.1111/j.1365-2567.2004.01938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rottembourg D, Filippi CM, Bresson D, et al. Essential role for TLR9 in prime but not prime-boost plasmid DNA vaccination to activate dendritic cells and protect from lethal viral infection. J Immunol. 2010 Jun 15;184(12):7100–7107. doi: 10.4049/jimmunol.0803935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feltquate DM, Heaney S, Webster RG, Robinson HL. Different T helper cell types and antibody isotypes generated by saline and gene gun DNA immunization. J Immunol. 1997 Mar 1;158(5):2278–2284. [PubMed] [Google Scholar]
- 62.Strugnell RA, Drew D, Mercieca J, et al. DNA vaccines for bacterial infections. Immunol Cell Biol. 1997 Aug;75(4):364–369. doi: 10.1038/icb.1997.57. [DOI] [PubMed] [Google Scholar]
- 63.Belz GT, Shortman K, Bevan MJ, Heath WR. CD8alpha+ dendritic cells selectively present MHC class I-restricted noncytolytic viral and intracellular bacterial antigens in vivo. J Immunol. 2005 Jul 1;175(1):196–200. doi: 10.4049/jimmunol.175.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hildner K, Edelson BT, Purtha WE, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008 Nov 14;322(5904):1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Allan RS, Smith CM, Belz GT, et al. Epidermal viral immunity induced by CD8alpha+ dendritic cells but not by Langerhans cells. Science. 2003 Sep 26;301(5641):1925–1928. doi: 10.1126/science.1087576. [DOI] [PubMed] [Google Scholar]
- 66.Wakim LM, Bevan MJ. Cross-dressed dendritic cells drive memory CD8+ T-cell activation after viral infection. Nature. 2011 Mar 31;471(7340):629–632. doi: 10.1038/nature09863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lauterbach H, Gruber A, Ried C, Cheminay C, Brocker T. Insufficient APC capacities of dendritic cells in gene gun-mediated DNA vaccination. J Immunol. 2006 Apr 15;176(8):4600–4607. doi: 10.4049/jimmunol.176.8.4600. [DOI] [PubMed] [Google Scholar]