Abstract
The c-jun gene regulates cellular proliferation and apoptosis via direct regulation of cellular gene expression. Alternative splicing of pre-mRNA increases the diversity of protein functions and alternate splicing events occur in tumors. Here, by targeting the excision of the endogenous c-jun gene within the mouse mammary epithelium, we have identified its selective role as an inhibitor of RNA splicing. Microarray-based assessment of gene expression, on laser capture micro-dissected c-jun−/− mammary epithelium, demonstrated that endogenous c-jun regulates the expression of approximately 50 genes governing RNA splicing. In addition, genome-wide splicing arrays demonstrated that endogenous c-jun regulated the alternate exon of approximately 147 genes, and 18% of these were either alternatively spliced in human tumors or involved in apoptosis. Endogenous c-jun also was shown to reduce splicing activity, which required the c-jun dimerization domain. Together, our findings suggest that c-jun directly attenuates RNA splicing efficiency, which may be of broad biological importance as alternative splicing plays an important role in both cancer development and therapy resistance.
Keywords: Transgenic mice, floxed c-jun, Cre, alternative splicing
Introduction
c-jun is overexpressed in human tumors that display increased cellular proliferation and DNA synthesis while it’s in vivo disruption prevents hepatocarcinoma (1). c-Jun over expression can induce anchorage-independent growth of Rat-1a and human breast MCF-7 cells (2, 3) and can inhibit cellular apoptosis via gene transcription through a p53/p21CIP1 dependent mechanism (1). c-Jun complexes with Jun/Fos and MaF/Nr1 families members (4–6) to form an AP-1 transcription factor complex that can regulate expression of genes that control G1/S phase progression and diverse cellular functions (7).
c-jun mediates oncogenic transformation (8) and cellular apoptosis in a cell and context dependent manner (9–15). c-jun regulates target gene expression that regulates apoptosis, or regulates cell cycle progression in the presence of DNA damage and thereby inducing apoptosis (16). In both scenarios c-jun functions by regulating the gene transcription via cognate DNA-binding sites in the promoter of apoptosis regulatory genes. c-jun function in mammary gland development and growth in vivo is largely unknown as the c-jun−/− mice are embryonic lethal (13). Targeted expression of Cre recombinase in the mammary epithelium of c-junfl/fl mice (MMTV-Cre/c-junfl/fl) demonstrated the loss of c-jun expression in the mammary epithelium correlated with increased apoptosis by tunnel staining (17) although the mechanism by which c-jun inhibits apoptosis remained to be determined. Herein we interrogated the molecular genetic events regulated by endogenous c-jun in mammary epithelial in vivo.
Recent studies have revealed the importance of alternative RNA splicing in cellular apoptosis (18–20), in human disease and cancer (21, 22). Pre-mRNA splicing regulates gene expression in 75% of human genes where as serine/arginine-rich (SR) proteins mediate both constitutive and alternative splicing (23) and act as a switch that controls the conversion of apoptotic genes into pro-apoptotic splice variants (24). Alternative splicing of a subset of genes are enriched in human breast and ovarian cancers. The mechanisms governing alternative splicing in tumors and relative splicing activity are not well understood. The development of a splicing mini-reporter gene to increase splicing activity has been useful in identifying candidate mechanisms.
Herein, microarray analysis performed on laser capture micro-dissected (LCM) mammary epithelial cells from floxed c-jun transgenic mammary epithelium, (devoid of c-jun protein due to Cre-mediated c-jun gene excision), demonstrated that c-jun altered the expression of splicing factors. c-jun directly attenuated splicing activity and regulated target gene splice variants. A significant proportion of these c-jun-dependent alternatively spliced genes were also alternatively spliced in tumors and in apoptosis. Thus, in addition to mediating apoptosis or cell survival via regulating gene transcription, c-jun regulates the mRNA splicing apparatus and splicing of genes involved in apoptosis.
Materials and Methods
Transgenic mice, MEFs, MECs, expression plasmids and reagents
Transgenic animals carrying floxed c-jun alleles, c-junf/f, were previously described (38). All experimental procedures with these mice were approved by the ethics committee of Thomas Jefferson University. Primary mouse mammary epithelial cells (MECs) cultures were prepared by removing the 4th mammary gland from 8 weeks old female mice (39). Production of MEFs and 3T3 and their culture conditions were described in references (39–41). The adenoviruses (Ad-Null and Ad-Cre) used for infecting MEFs or MECs: (42), RSV-c-jun, MSCV-c-jun and their mutants: (43), the splicing double reporter (30) and the c-Fos expression vectors a generous gift of Dr. Tom Curran (44) were previously described.
Immuno-histochemistry, mouse mammary gland whole mounts and western blots
Transgenic animals were euthanized following IACUC allowed procedure. Mammary gland whole mounts were prepared as described in ref: (45).
Laser Capture Microdissection, RNA extraction, RT-PCR, qRT-PCR, Gene Expression Arrays
RNA samples extracted from laser capture micro-dissected mammary epithelial cells from wild-type c-jun+/+ and knockout c-jun−/− bi-transgenic mice mammary glands were processed for Affymetrix 430 2.0 Gene Expression Arrays for ascertaining the differential gene expression of various genes in presence or absence of c-jun expression in mammary epithelial cells.
Analyses of gene splicing using Affymetrix Exon Arrays and Ingenuity Pathway Analyses of alternatively-spliced genes
For genome wide analyses of alternative splicing of various genes, Affymetrix Mouse Exon 1.0 ST arrays were used according to manufacturers prescribed protocols and conditions. Additional details are provided as Supplemental Information.
Results
Somatic excision of c-jun in the mammary gland reduces mammary epithelial cells in vivo
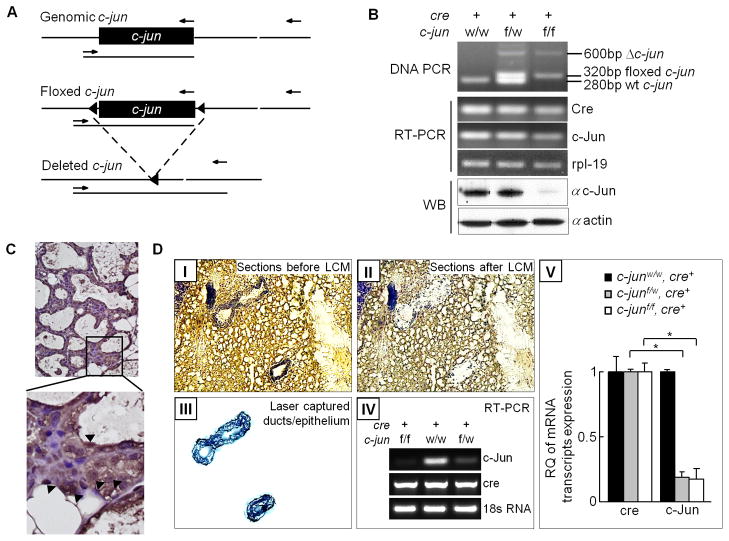
Female progeny, resulting from the crosses between floxed c-jun (c-junf/f) or wild type (c-junw/w) mice with MMTV-Cre transgenic mice expressing mammary specific Cre recombinase were used (Figure 1A–D). Amplification of a 600bp PCR fragment in the mammary gland was indicative of successful c-jun gene excision by Cre recombinase (Figure 1B) and consequent reduction in expression of c-jun mRNA transcripts and proteins (<90%) (Figure 1C). Amplification of residual c-jun mRNA transcripts in whole mammary gland RNA was detected due to the sensitivity of RT-PCR, with resulting amplification from trace amounts of c-jun from other stromal cells, lymphocytes and macrophages. Immunostaining confirmed the expression of Cre-mRNA transcripts and proteins in the mammary glands (Figure 1C). Western blot analysis showed the loss of c-Jun protein on whole mammary gland extracts (Figure 1B). RT-PCR of mammary gland mRNA showed a reduction in c-jun mRNA expression transcripts and proteins (90% reduction) (Figure 1B). LCM was used to select mammary epithelial cells and extract RNA for all subsequent molecular assays (Figures 1D-III). Both RT-PCR and qRT-PCR on LCM RNA confirmed a 10-fold reduction in c-jun expression (Figures 1D-IV, V).
Figure 1. Deletion of the c-jun in the mammary epithelial cells by transgenic mice.
(A) Schematic representation of the genomic c-jun, floxed c-jun allele and deleted c-jun is shown along with primer binding sites (arrows). (B) PCR, RT-PCR and western analyses to validate the c-jun gene excision, expression of Cre recombinase and reduced abundance of c-Jun protein following c-jun excision. (C) Immunohistochemical staining for Cre in bi-transgenic mice mammary gland section (i.e. positive cells indicated by an arrow). (D) Representative laser capture microdissection (LCM) image of a trypan blue stained mammary gland section from transgenic mice. I: Entire section before LCM, II: Section after LCM, III: Green stained micro-dissected mammary epithelial cells on LCM cap, IV: RT-PCR for assessment of c-jun, Cre and housekeeping gene control 18S expression on LCM RNA, V: qRT-PCR based quantification of relative abundance of the Cre-recombinase and c-jun mRNA transcripts following Cre mediated c-jun gene excision.
Endogenous c-jun in mammary epithelial cells inhibits expression of genes governing mRNA splicing
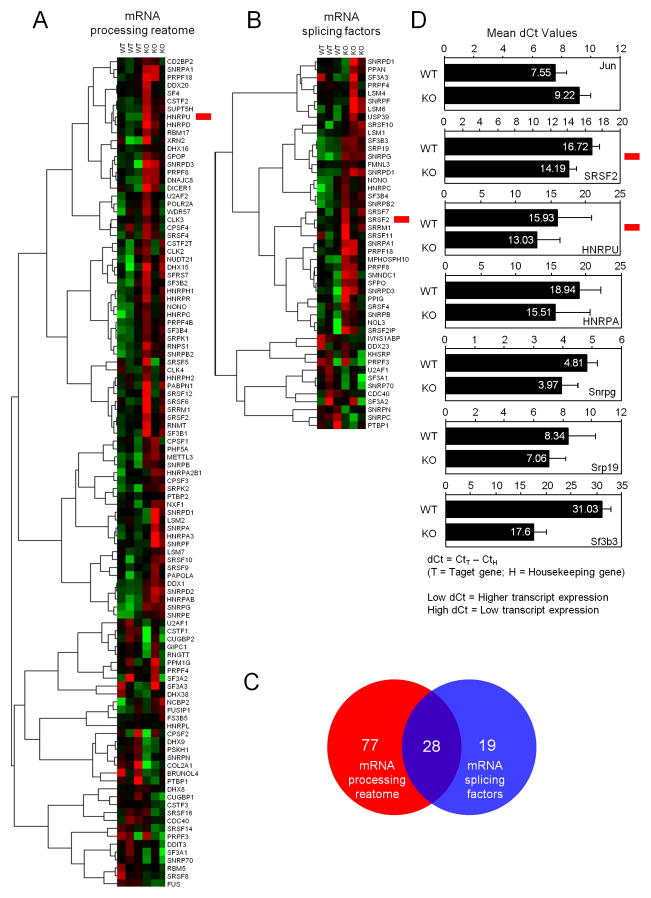
In recent studies of the cellular components of the mammary gland of the bi-transgenic mice, we showed an approximately 50% reduction in area and the number of epithelial cells observed per field in c-jun−/− mice as compared to c-jun+/+ mice (17). Genome-wide transcriptional profiling was performed on RNA obtained by LCM of mammary epithelial cells from MMTV-Cre+/c-junf/f (c-jun−/− or KO) and MMTV-Cre+/c-junw/w (c-jun+/+ or WT) mice. Gene set enrichment analysis by NIH-DAVID, using Gene Ontology terms to characterize the differentially regulated genes by their molecular function and pathway analysis conducted using ASSESS showed c-jun regulated several functional categories (UVA/UVB; RhoGTPases, the G1/S reactome, estrogen signaling, E2F1/E2F1 targets, apoptosis, mRNA processing reactome and mRNA splicing) (17). The finding that the loss of endogenous c-jun correlated with the induction of pathways governing mRNA processing and splicing was novel and warranted further investigation. The enrichment for altered expression of individual genes within the mRNA processing reactome (Figure 2A) and the mRNA splicing factors (Figure 2B), illustrates endogenous c-jun regulates the expression of genes involved in mRNA splicing.
Figure 2. Microarray based gene expression analysis identifies c-jun dependent signaling pathways.
(A). Genome wide expression analysis on LCM RNA from micro dissected mammary epithelium. ASSESS was used to classify the pathways differentially regulated by endogenous c-jun. Red denotes positive enrichment and up-regulation, while green denotes negative enrichment and down-regulation. Tree view analysis of mRNA processing reactome and (B) mRNA splicing factors data showing pathway from six separate transgenic knockout mice (C) Venn Diagram demonstrating overlap between mRNA processing reactome and mRNA splicing factors. (D) Real-time qRT-PCR based validation of Exon Array results. Data displays mean dCt values from 3 WT and 3 KO sample RNAs for each gene target. dCt = CtT -CtH, where T= Target Gene and H=House Keeping Gene, so Low dCt = Higher transcript expression and High dCt = Lower transcript expression. EB= SEM.
Splicing factors include both activators and repressors of splicing (25). The excision of c-jun both induced and repressed activators and repressors of splicing. Members of the SR family (SFR) which encode sequence specific spliceosome factors, basal SnRNA (SNRPB2) and SRRM1 (serine arginine repetitive matrix protein 1) which encode proteins involved in pre- and post-splicing mRNA complexes, were primarily induced but some were repressed in c-jun−/− mammary epithelium. Splicing factors known to function as repressors of splicing (i.e. SNRP70, SNRPN, SNRPC) were mainly inhibited in c-jun−/− cells. Splicing of pre-mRNA occurs in the spliceosome which consists of five ribonucleoprotein subunits. qRT-PCR was therefore conducted to determine the mRNA abundance of these components and the mean date of n=6 for each mRNA is shown as mean dCT values in Figure 2D. The mean dCT value is essentially the reciprocal of fold change. Expression of key mRNA splicing regulatory genes (sfrs2, hnRNPA, hRNPU, snrpg, srp19 and sf3b3) were increased upon c-jun excision (Figure 2D). hnRNP (heterogeneous nucleoprotein) are a family of 20 pre-mRNA binding proteins designated hnRNPA through U. hnRNPU stabilizes specific mRNA by binding their 3′ UTR, and interacts with nucleophosporin to regulate apoptosis (26). Thus, SRSF2 expressed as a reduction in mean dCt values (Figure 2D)(Figure 2B, red tag) was increased in abundance by QT-PCRin c-jun−/− cells.
Endogenous c-jun regulates alternative splicing of genes
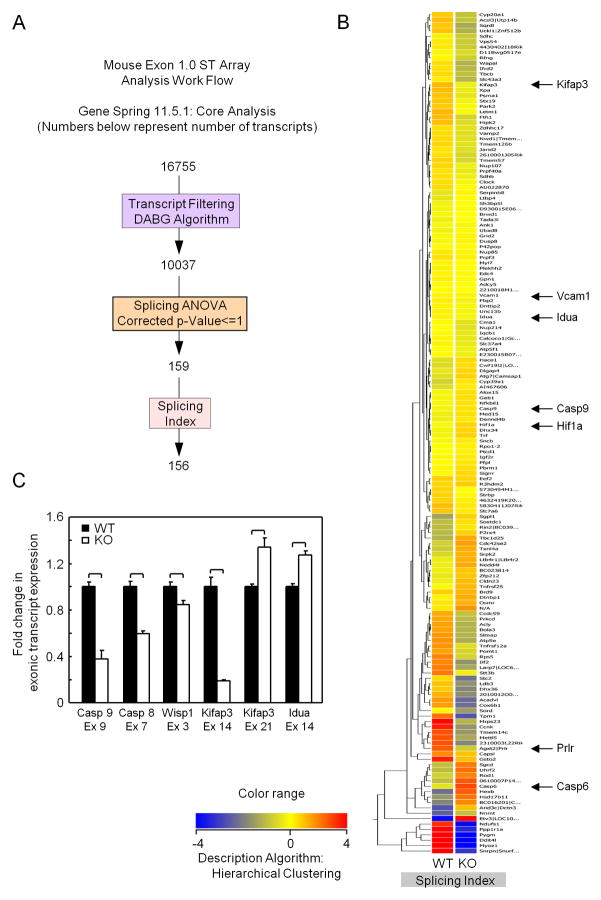
These studies indicate c-jun regulates the abundance of gene products regulating splicing. In order to determine whether alterations in abundance of genes associated with gene splicing of altered gene splicing in vivo, we examined the alternate splice forms of 28,853 gene transcripts represented on the Mouse Exon 1.0 ST chip. Using the Gene Spring “Core Data Analysis”, method and filtering and statistical methods (Figure 3A), we determined the gene splicing index (SI). For the determination of the SI (splicing index) each exonic probesetis normalized by dividing its expression by the gene-level summary value for the entire gene to yield a normalized intensity (NI). The splicing index is then calculated as a-fold change between the two normalized levels. 81 genes were shown to be alternatively spliced in three c-jun+/+ vs three c-jun−/− mice mammary epithelium (Figure 3B). Several of those genes are either known to be involved in apoptosis (Caspase 9, Caspase 6) or have been linked to disease in which apoptosis is a prominent feature (Kifap3 Park2). The alternative splicing of a subset of these genes was validated by qRT-PCR of the affected exons (Figure 3C). In order to display a subset of the analysis of the key individual genes and their alternative splicing the probe sets were indicated on the x-axis and the relative increase or decrease in transcript inclusion or exclusion as a result of cellular mRNA splicing activity is shown as log-10 on the y-axis. The affected gene exons are represented in red below the graphical display of the splicing results (Supplement 1).
Figure 3. Genome-wide splicing array demonstrates endogenous c-jun governs alternative splicing.
Exon inclusion or exclusion (resulting from altered mRNA splicing) were compared in exon arrays using LCM RNA from mammary epithelium of Cre+/c-junf/f (c-jun−/−) versus Cre+/c-junw/w (c-jun+/+). (A) Schematic representation of the alternative splicing analysis on Affymetrix Exon Arrays using Gene spring. (B) Primary Heat Map depicting the splicing index of 81 differentially spliced genes among c-jun WT and KO. Blue lines indicate expression of probe set in WT c-jun (c-jun+/+) and red lines indicates in KO (c-jun−/−) mammary epithelial cell RNA. (C) Graphical representation of exonic inclusion or exclusion of six biologically relevant genes involved in apoptosis showing alternatively spliced exons. Mean transcript exclusion or inclusion data is represented for each of the probe set IDs with 3 samples for each sample type from c-jun+/+ and c-jun−/− animals. *p ≤ 0.05, Error Bars = Standard Deviation.
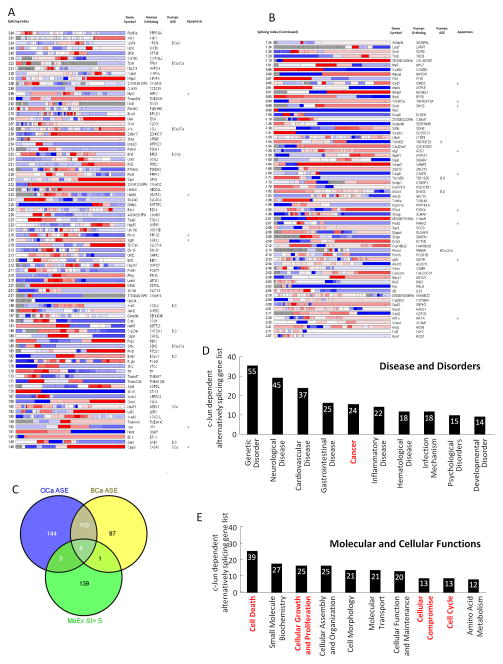
As a complementary interrogation of the genome subjected to alternative splicing by c-jun, comparison was made of c-jun−/− vs c-jun+/+ mammary epithelial cells, and individual exon mapping was conducted using Plotagene. For ease of display exon map, Plotagene displays the structure of the specified gene, coloured according to the expression level of each exon (27). The gene’s average expression is calculated and used to color the overall gene. The genes which are alternatively spliced in a manner dependent upon c-jun and the distribution of the alternately spliced exons, is shown in Figure 4A. Using this approach the population of alternatively spliced genes expanded to include 147 genes. The 147 genes and the alteration in expression of the individual exons determined by the relative abundance of c-jun is shown in Figure 4A. Recent studies have demonstrated active alternative splicing events are changed in breast/ovarian tumors (28). A comparison of these alternatively spliced genes with the c-jun-dependent spliced genes showed an overlap of 8/147 (Fig. 4B, individual genes shown in 4A).
Figure 4. c-jun mediates alternative gene splicing.
(A) A plot gene display of the specific genes colored according to the expression of individual exons for each gene. Average gene expression of the specific exon is shown. The relative splicing index for each gene is shown with the gene symbol human ortholog and the presence of the alternatively spliced gene in human disease either BCa (breast cancer), OCa (ovarian cancer), O (ovarian), B (breast), or in apoptosis from Venables, 2009. (B) Data for human ASE association of alternatively spliced exon is taken from Venables, 2009. (C) Venn diagram representing the overlap between the c-jun dependent alternatively spliced gene sense (MoEx SI>.5) with overlap shown to OCa, ASE and BCa ASE (ovarian cancer alternatively spliced exons or breast cancer alternatively spliced exons). (D) Enrichment for gene function by ingenuity pathway analysis systems (IPA). The relative proportion of genes within the top functional gene networks is shown. (E) Proportion of the top seven genes by molecular and cellular function based on ingenuity pathway analysis.
An advanced bioinformatics analysis was performed to map and classify the patterns of the c-jun-dependent alternatively spliced genes into Functional gene networks and Disease and Disorder networks using the Ingenuity Pathways Analyses System (IPA 7.5). These highest ranked functional networks identified by IPA 7.5 were: (1) Cancer, and Cell Death (Score: 20) (Figure 4D). This finding is consistent with the 14 genes correlating with genes involved in apoptosis (Figure 4A). Approximately 22% of the alternatively spliced genes within “Disease and Disorder” networks were categorized as cancer-related Figure 4D). Approximately 54% of the alternatively spliced genes within the top “Molecular and Cellular Function” networks were functionally linked to cell death or cellular compromise (Figure 4E).
Endogenous c-jun reduces expression of the pro-apoptotic splicing factor SRSF2
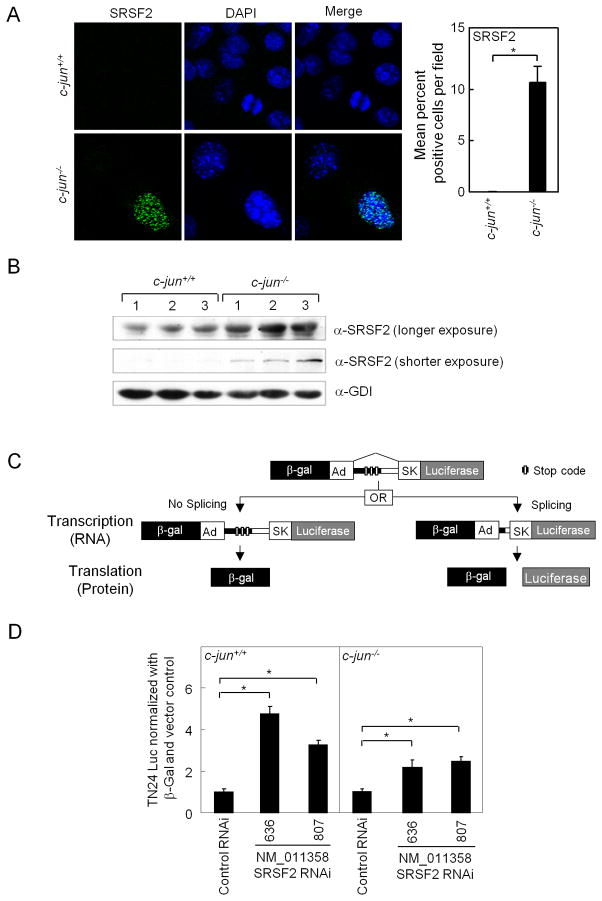
We next examined the relative abundance of an individual splicing factor in c-jun−/− and c-jun+/+ mammary epithelial cells. The SR (serine/arginine family) of RNA binding proteins are essential for multiple steps of spliceosome assembly at both the 5′ and 3′ sites and function in both constitutive and alternative splicing (29). SC35 SRSF2, (the protein product of the Srsf2 gene), is known to promote the alternative splicing of several genes to enhance their proapoptotic phenotype (29). Immunohistochemistry for SRSF2 demonstrated increased abundance in a nuclear speckled distribution in c-jun−/− mammary epithelial breast tumor cells (Figure 5A) and western blot confirmed increased SRSF2 abundance in c-jun−/− MEC (Figure 5B). In order to determine whether the increased abundance of SRSF2 regulated splicing, siRNA to SrSf2 was deployed in conjunction with a synthetic splicing reporter gene (30)(Figure 5C). Two distinct siRNA to Srsf2 enhanced splicing reporter activity ~5-fold (Figure 5D). Consistent with the lower level of Srsf2 in c-jun−/− cells, the induction of splicing activity by Srsf2 siRNA was increased only 2-fold in c-jun+/+ cells. The ~10-fold increase in SRSF2 was abrogated by reintroduction of a c-jun retroviral expression vector, indicating the effect was specific for c-jun. Thus endogenous c-jun inhibits the expression and abundance of SRSF2 and endogenous SRSF2 is capable of regulating activity of a general splicing activity minigene reporter.
Figure 5. Endogenous c-jun determines the relative abundance of the pro-apoptotic splicing factor SRSF2.
(A) Immuno-fluorescence staining for SRSF2 (green nuclear speckles) and nuclear stain (DAPI: blue) was performed on MECs from c-jun+/+ and c-jun−/− and quantification of percentage positivity within 5–10 microscopic fields (B) Western blotting of the SRSF2 in c-jun+/+ and c-jun−/− KO MECs. (C) Schematic representation of the double reporter splicing reporter with two transgenes, the β-galactosidase (β-gal) and luciferase (Luc). Reporter genes are fused in frame by recombinant fragments of the genes encoding adenovirus (Ad). The recombinant fragment contains skeletal muscle isoform (SK) of human tropomyosin. Three in-frame translation stop signals (XXX) are present in the intronic region. (D) Relative splicing activity measured as relative luciferase activity divided by total β-galactosidase (luciferase units × 103) is shown comparing c-jun+/+ and c-jun−/−. Cells were treated either with two different SRSF2 siRNA, or control siRNA. Activity of c-jun−/− and c-jun+/+ were equalized to 1 in the presence of control siRNA to illustrate the effect of SRSF2 siRNA. The assay was conducted in fibroblasts (MEFs) or mammary epithelial cells (MECs) generated from floxed c-jun transgenic mice (c-jun+/+ or c-jun−/−). The data represents mean ± SEM of n>5 separate transfections. *p ≤ 0.05, EB= SEM.
c-jun repression of a splicing minigene reporter requires the c-jun B-ZIP domain
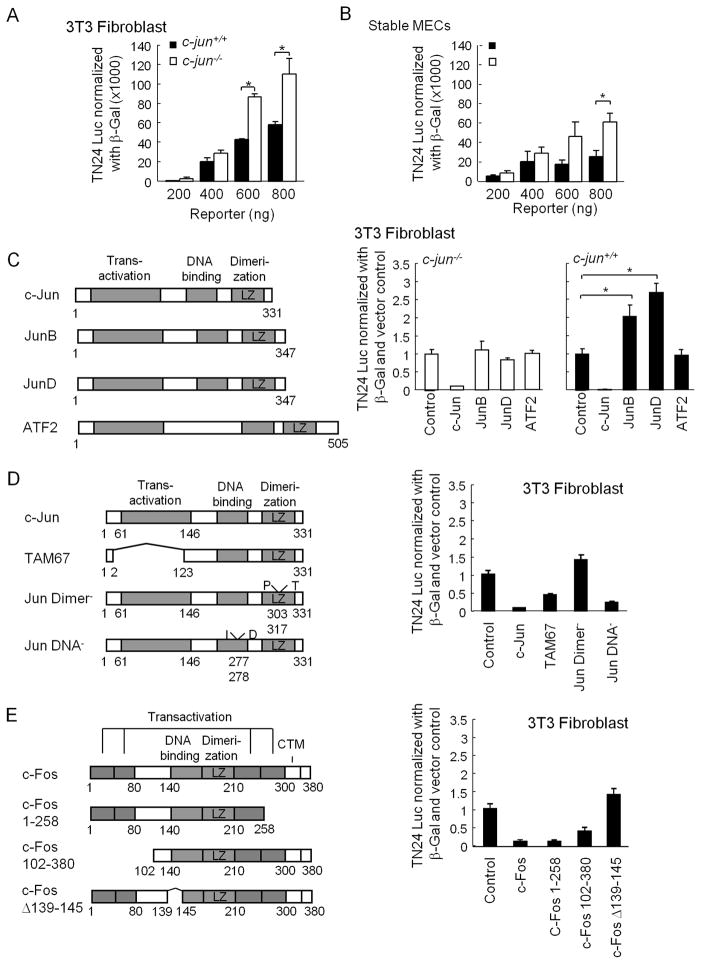
The mRNA gene expression data are consistent with a role for endogenous c-jun as a regulator of genes regulating RNA splicing. However, whether c-jun directly regulated splicing activity remained to be determined. To determine whether c-jun directly repressed splicing, a state-of-the-art splicing reporter gene was deployed (30). Introduction of the β-galactosidase splicing reporter gene system into c-jun−/− versus c-jun+/+, when normalized for transfection efficiency, showed an increase in splicing activity in c-jun−/− cells (Ad-Cre Fig. 6A). Similar results were found using c-jun−/− MEC (MEC from c-junfl/fl + Cre stable line), when compared to c-jun+/+ MEC (c-junfl/fl control) (Fig. 6B). Thus, the relative splicing activity of the splicing activity reporter gene was enhanced in c-jun−/− MEFs and in c-jun−/− MECs indicating endogenous c-jun repressed general splicing activity.
Figure 6. c-jun inhibits splicing activity via the dimerization domain.
(A) The DNT-24 luciferase β-galactosidase dual mini gene reporter system was used to transfect c-jun+/+ with c-jun−/− 3T3 cells. Data has shown as relative luciferase activity c-jun−/− MEC cells were transfected with dual splicing reporter and the data has shown as relative splicing activity. (C) Schematic representation of AP-1 transcription factor plasmids. Relative splicing activity is shown in c-jun−/− cells comparing the control vector with each of the expressed plasmids. Equimolar amounts of AP-1 protein plasmid was used to transfect c-jun−/− with c-jun+/+ 3T3 cells. (D) Schematic representation of c-jun expression plasmids with relative splicing activity shown for each of the co-transfected c-jun mutants in 3T3 cells. (E) Schematic representation of c-Fos mutant expression plasmids with relative splicing activity shown for equimolar amounts of the mutant expression plasmid in 3T3 cells. Data are mean ± SEM n>5 for each transfection.
In order to determine whether c-jun was capable of repressing the splicing reporter gene, c-jun expression vectors were deployed (Fig. 6C). Transduction of c-jun−/− MEFs with a c-jun expression vector repressed splicing reporter activity by 90%. Equimolar amounts of JunB or JunD failed to repress splicing reporter activity. c-jun repressed splicing activity in c-jun+/+ cell. Thus, c-jun is capable of inhibiting splicing when introduced into c-jun−/− or c-jun expressing cells. JunB and JunD enhanced splicing activity in c-jun+/+ cells consistent with the known role of JunB to function as an inhibitor of c-jun activity. In order to identify the domains of c-Jun required for repression, a series of c-Jun mutant expression vectors were used (Fig. 6D). c-Jun repression of splicing activity was reduced 60% by deletion of the N-terminal transactivation/transrepression domain. Mutation by point substitution of the leucine zipper abrogated c-Jun mediated repression, whereas the DNA-binding defective mutant maintained repression. Thus, the leucine zipper domain, but not the DNA-binding domain is required for c-Jun-mediated repression of splicing reporter gene activity. The heterodimeric partner of c-Jun for AP-1-mediated repression, includes the transcription factor c-Fos. Expression of c-Fos repressed splicing reporter activity (Fig. 6E). A c-Fos carboxyl terminal deletion failed to abrogate repression. However, a Δ139-145 mutant of c-Fos, failed to repress splicing reporter activity. Collectively, these studies demonstrate that endogenous c-Jun inhibits splicing activity in both murine mammary epithelial cells and mouse embryo fibroblasts. The repression by c-Jun requires the leucine zipper domain.
Discussion
The current studies provide several independent lines of evidence to support the novel finding that endogenous c-jun regulates gene splicing. First, c-jun regulated a gene expression pathway governing gene splicing. Second c-jun regulated expression of individual mRNA for the genes that regulate mRNA splicing. Third, genome wide splicing analyses showed that c-jun excision altered the splicing pattern of 147 genes, 8 of which are alternatively spliced in cancer and an additional 14 of which are involved in cell death. Fourth, c-Jun directly regulated the activity of a splicing minigene reporter system requiring the B-ZIP domain. Collectively, these studies demonstrate at a genome-wide level, a new role for c-jun in regulating gene splicing.
Here c-jun inhibited the relative abundance of splicing factor including the SRSF2 splicing factor. A subset of alternative splicing events occurs in cancer-related genes and regulates their apoptotic versus pro-apoptotic splice forms. SRSF2 acts as switch between the alternate splicing of apoptotic genes (c-flip, caspase-8, caspase-9, Bcl-X) towards the pro-apoptotic splice variants (24). In our studies of c-jun−/− mice, the E2F1 gene signaling pathway was upregulated (17). In prior studies, the E2F1 upregulated SRSF2 (SC35) in response to DNA damage (24). Thus our studies are consistent with a model in which c-jun may inhibit apoptosis via inhibitors of E2F1 and thereby SRSF2 to inhibit alternative splicing of apoptosis genes.
In the current studies, microarray based genome wide expression profiling revealed that endogenous c-jun altered splicing of several genes regulating cellular apoptosis. In our recent studies, tunnel staining confirmed the increased apoptosis in epithelial cells within c-jun−/− mammary glands and a marked reduction in the number of epithelial cells. c-jun is thought to convey cell type specific pro-or anti-apoptotic functions (16). Thus c-jun conveyed an anti-apoptotic function in the mammary epithelium in vivo consistent with prior studies of hepatocytes and fibroblasts using gene knock down (9, 11, 13, 14, 31). The exon array identified genes that were alternatively spliced in a c-jun dependent manner. The biological pathways in which these alternatively spliced genes participate include cancer and apoptosis.
Several alternatively spliced genes which were regulated by c-jun include anti-apoptotic growth factors and cytokines, such as sostdC1 (sclerostin domain containing 1) which associates with BMPs blocking their binding to their receptors thereby regulating BMP function in cell death. ltbP4 (latent transforming growth factor beta binding protein) Osmr (oncostatin M receptor). Proapoptotic genes that were alternatively spliced in a c-jun dependent manner include caspase9, caspase6, Tnfrsf12A and Park2, (Parkinson’s disease autosomal recessive juvenile 2) which protects neurons against apoptosis. Alternative splicing of caspase 9 has been linked to its apoptotic function (32, 33). SRSF2 for example enhances the pro-apoptotic forms of caspase 9 and herein c-jun attenuated the expression of SRSF2 (24). The current studies showed an enrichment of genes alternatively spliced in tumors amongst the c-jun-mediated alternatively spliced genes. Alternative splicing may contribute to human disease and cancer via different mechanisms (21, 22).
A number of important targets participating in the onset and progression of tumorigenesis are regulated via alternative splicing. However, the molecular mechanisms regulating expression of splicing factors during tumorigenesis are not fully understood (34). Splicing factors and gene splicing are differentially regulated in various tumors (35). c-jun is overexpressed in a human tumors where it promotes cellular proliferation via transcriptional induction of cyclin D1 (7). The current studies raise the possibility that the induction of c-jun expression in tumors may contribute to the altered splicing that occurs during tumorigenesis. The previous models suggest that c-jun regulates cell survival in one or the two ways (i) it alters the transcription of pro- and anti-apoptotic gene products (FasL, Bim, Bcl3) (36) and the relative balance in gene expression determines the apoptotic or survival phenotype; or (ii) c-jun regulates apoptosis via its indirect effects (37), wherein c-jun functions as a homeostatic regulator of cell proliferation. Aberrant control of the cell-cycle by c-jun contributes to apoptosis. The current studies demonstrate endogenous c-jun inhibits mammary epithelial cell apoptosis and alters splicing of genes involved in apoptosis, raising the possibility that c-jun may mediate apoptosis by regulating the splicing efficiency of pro-versus anti-apoptotic genes.
Supplementary Material
Table 1.
Oligonucleotide primers used for PCR genotyping of animals, DNA PCR, RT-PCR, sqRT-PCR and real-time qRT-PCR analyses.
List of primers used for PCR genotyping of animals and real-time quantitative PCR analyses
| Gene | Orientation | Sequence 5′→3′ |
|---|---|---|
| Floxed c-jun | Forward | CTC ATA CCA GTT CGC ACA GGC GGC |
| Reverse | CCG CTA GCA CTC ACG TTG GTA GGC | |
| Reverse | CAG GGC GTT GTG TCA CTG AGC T | |
| Cre | Forward | TGC TCT GTC CGT TTG CCG |
| Reverse | ATC GTG TCC AGA CCA GGC | |
| c-jun | Forward | AGA CGC GTG CCT ACG GCT ACA GTA A |
| Reverse | CGA CGT GAG AAG GTC CGA GTT CTT G | |
| RPL-19 | Forward | CTG AAG GTC AAA GGG AAT GTG |
| Reverse | GGA CAG AGT CTT GAT GAT CTC | |
| Splicing Reporter | Forward | AAC ATC AGC CGC TAC AGT CAA |
| Reverse | ACG TGA TGT TCT CCT CGA TAT | |
| Caspase 9 Exon 9 | Forward | CTA CAC ATG CAG TTG TGG GCG TTT |
| Reverse | GAT GCA TAA TGG CCA GAA CTT GGG | |
| Caspase 8 Exon 7 | Forward | AAA TGG CGG AAC TGT GTG ACT CG |
| Reverse | CCG TGA CTC ACT GTC TTG TTC TCT | |
| Wisp1 Exon 3 | Forward | TAC ACC AAT GGC GAG TCC TTC CAA |
| Reverse | TGG TCT CCT TGC GTC ATC ATC ACA | |
| Kifap3 Exon 14 | Forward | TGC AGC CCA GAT TTC CAG TGA TGA |
| Reverse | ATT GTC AGA TTT GCC AGC GTT CCC | |
| Kifap3 Exon 21 | Forward | ACT AAG TAA GCC TCA AGC AGC CGA |
| Reverse | TAG TAA GGT TCG TCA GGG CGG AAT | |
| Idua Exon 14 | Forward | TGG ACC CTC CCA CAT CAA TCA CTA |
| Reverse | GAG TCA TTT GGG AAG AAG GAA CCT C |
Table 2.
Ingenuity Pathways Analysis (IPA7.5) Results for 81 alternatively spliced genes
| TOP NETWORKS
| |
|---|---|
| Network | Score |
| Cancer, Cell Death, Hematological Disease | 20 |
| Cancer, Developmental Disorder, Embryonic Development | 23 |
| Neurological Disease, Skeletal and Muscular Disorders, Cellular Compromise | 23 |
| Genetic Disorder, Skeletal and Muscular Disorders, Development Disorders | 28 |
| TOP BIO FUNCTIONS
| ||
|---|---|---|
| Disease and Disorders
| ||
| Name | p-Value | Molecules |
| Cancer | 4.11e-03 – 4.57e-02 | 11 |
| Genetic Disorder | 4.11e-03 – 4.83e-02 | 11 |
| Dermatological Diseases and conditions | 4.11e-03 – 4.43e-02 | 10 |
| Skeletal and Muscular Disorders | 3.44e-04 – 4.83e-02 | 9 |
| Cardiovascular Diseases | 4.11e-03 – 4.43e-02 | 7 |
|
| ||
|
Molecular and Cellular Functions
| ||
| Name | p-Value | Molecules |
|
| ||
| Cellular Compromise | 9.40e-04 – 4.49e-02 | 13 |
| Cell Death | 3.44e-04 – 4.83e-02 | 12 |
| Cell Morphology | 4.97e-05 – 1.63e-02 | 8 |
| Protein Degradation | 9.40e-04 – 4.69e-02 | 4 |
| Carbohydrate Metabolism | 1.46e-03 – 4.04e-02 | 3 |
|
| ||
|
Physiological System Development and Function
| ||
| Name | p-Value | Molecules |
|
| ||
| Embryonic Development | 4.11e-03 – 4.04e-02 | 9 |
| Cardiovascular System Development and Function | 4.11e-03 – 3.64e-02 | 5 |
| Digestive System Development and Function | 4.97e-05 – 4.97e-05 | 2 |
| Connective Tissue Development and Function | 4.11e-03 – 4.11e-03 | 1 |
| TOP CANONICAL PATHWAYS
| ||
|---|---|---|
| Name | p-Value | Ratio |
| Glycosaminoglycan Degradation | 3.6e-04 | 3/72 (0.042) |
| Mitochondrial Dysfunction | 1.99e-03 | 4/172 (0.023) |
| Parkinson’s Signaling | 2.17e-03 | 2/17 (0.118) |
| Docosahexaenoic Acid (DHA) Signaling | 9.03e-03 | 2/45 (0.044) |
| Oxidative Phosphorylation | 1.84e-02 | 3/166 (0.018) |
Acknowledgments
This work was supported in part by the following NIH grants R01CA070896, R01CA075503, R01CA107382, R01CA132115, R01CA086072 (R.G.P.) and R01CA120876 (M.P.L.) and Dr. Ralph. This project is funded in part from the Marian C. Falk Medical Research Trust and a grant from the Pennsylvania Department of Health (R.G.P.). The Department specifically disclaims responsibility for analyses, interpretations or conclusions. The Kimmel Cancer Center was supported by the NIH Cancer Center Core Grant P30CA056036 (R.G.P.). We thank Atenssa L. Cheek for the preparation of this manuscript. The authors declare that no conflicts of interest exist.
References
- 1.Eferl R, et al. Liver tumor development. c-Jun antagonizes the proapoptotic activity of p53. Cell. 2003;112(2):181–192. doi: 10.1016/s0092-8674(03)00042-4. [DOI] [PubMed] [Google Scholar]
- 2.Kinoshita I, et al. Identification of cJun-responsive genes in Rat-1a cells using multiple techniques: increased expression of stathmin is necessary for cJun-mediated anchorage-independent growth. Oncogene. 2003;22(18):2710–2722. doi: 10.1038/sj.onc.1206371. [DOI] [PubMed] [Google Scholar]
- 3.Smith LM, et al. cJun overexpression in MCF-7 breast cancer cells produces a tumorigenic, invasive and hormone resistant phenotype. Oncogene. 1999;18(44):6063–6070. doi: 10.1038/sj.onc.1202989. [DOI] [PubMed] [Google Scholar]
- 4.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3(11):859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 5.Karin M, Liu Z, Zandi E. AP-1 function and regulation. Curr Opin Cell Biol. 1997;9(2):240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 6.Pestell RG, Jameson JL. Hormone Action II: transcriptional regulation of endocrine genes by second messenger signalling pathways. In: Weintraub Bruce., editor. Molecular Endocrinology: Basic Concepts and Clinical Correlations. Chapter 5. 1995. pp. 59–76. [Google Scholar]
- 7.Albanese C, et al. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J Biol Chem. 1995;270(40):23589–23597. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- 8.Schuster R, Gerlich WH, Schaefer S. Induction of apoptosis by the transactivating domains of the hepatitis B virus X gene leads to suppression of oncogenic transformation of primary rat embryo fibroblasts. (Translated from eng) Oncogene. 2000;19(9):1173–1180. doi: 10.1038/sj.onc.1203417. (in eng) [DOI] [PubMed] [Google Scholar]
- 9.Wisdom R, Johnson RS, Moore C. c-Jun regulates cell cycle progression and apoptosis by distinct mechanisms. EMBO J. 1999;18(1):188–197. doi: 10.1093/emboj/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bukelskiene V, Baltriukiene D, Stulpinas A, Kalvelyte AV. Study of apoptosis and c-Jun expression in mouse cancer model systems. Biologija Nr. 2003;3:56–59. [Google Scholar]
- 11.Shaulian E, et al. The mammalian UV response: c-Jun induction is required for exit from p53-imposed growth arrest. Cell. 2000;103(6):897–907. doi: 10.1016/s0092-8674(00)00193-8. [DOI] [PubMed] [Google Scholar]
- 12.Kolbus A, et al. c-Jun-dependent CD95-L expression is a rate-limiting step in the induction of apoptosis by alkylating agents. Mol Cell Biol. 2000;20(2):575–582. doi: 10.1128/mcb.20.2.575-582.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eferl R, et al. Functions of c-Jun in liver and heart development. J Cell Biol. 1999;145(5):1049–1061. doi: 10.1083/jcb.145.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivanov VN, et al. Cooperation between STAT3 and c-jun suppresses Fas transcription. Mol Cell. 2001;7(3):517–528. doi: 10.1016/s1097-2765(01)00199-x. [DOI] [PubMed] [Google Scholar]
- 15.Hilberg F, Aguzzi A, Howells N, Wagner EF. c-jun is essential for normal mouse development and hepatogenesis. (Translated from eng) Nature. 1993;365(6442):179–181. doi: 10.1038/365179a0. (in eng) [DOI] [PubMed] [Google Scholar]
- 16.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nature Cell Biol. 2002;4(5):E131–136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 17.Katiyar S, et al. C-jun inhibits mammary apoptosis in vivo. (Translated from eng) Mol Biol Cell. 2010;21(23):4264–4274. doi: 10.1091/mbc.E10-08-0705. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwerk C, Schulze-Osthoff K. Regulation of apoptosis by alternative pre-mRNA splicing. (Translated from eng) Mol Cell. 2005;19(1):1–13. doi: 10.1016/j.molcel.2005.05.026. (in eng) [DOI] [PubMed] [Google Scholar]
- 19.Jiang ZH, Zhang WJ, Rao Y, Wu JY. Regulation of Ich-1 pre-mRNA alternative splicing and apoptosis by mammalian splicing factors. (Translated from eng) Proc Natl Acad Sci U S A. 1998;95(16):9155–9160. doi: 10.1073/pnas.95.16.9155. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X, Wang J, Manley JL. Loss of splicing factor ASF/SF2 induces G2 cell cycle arrest and apoptosis, but inhibits internucleosomal DNA fragmentation. (Translated from eng) Genes Dev. 2005;19(22):2705–2714. doi: 10.1101/gad.1359305. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okumura M, et al. Candidates for tumor-specific alternative splicing. (Translated from eng) Biochem Biophys Res Commun. 2005;334(1):23–29. doi: 10.1016/j.bbrc.2005.06.057. (in eng) [DOI] [PubMed] [Google Scholar]
- 22.Kirschbaum-Slager N, Parmigiani RB, Camargo AA, de Souza SJ. Identification of human exons overexpressed in tumors through the use of genome and expressed sequence data. (Translated from eng) Physiol Genomics. 2005;21(3):423–432. doi: 10.1152/physiolgenomics.00237.2004. (in eng) [DOI] [PubMed] [Google Scholar]
- 23.Sanford JR, Ellis J, Caceres JF. Multiple roles of arginine/serine-rich splicing factors in RNA processing. (Translated from eng) Biochem Soc Trans. 2005;33(Pt 3):443–446. doi: 10.1042/BST0330443. (in eng) [DOI] [PubMed] [Google Scholar]
- 24.Merdzhanova G, et al. E2F1 controls alternative splicing pattern of genes involved in apoptosis through upregulation of the splicing factor SC35. (Translated from eng) Cell Death Differ. 2008;15(12):1815–1823. doi: 10.1038/cdd.2008.135. (in eng) [DOI] [PubMed] [Google Scholar]
- 25.Manley JL, Krainer AR. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). (Translated from eng) Genes Dev. 2010;24(11):1073–1074. doi: 10.1101/gad.1934910. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao Z, et al. B23 acts as a nucleolar stress sensor and promotes cell survival through its dynamic interaction with hnRNPU and hnRNPA1. (Translated from eng) Oncogene. 2010;29(12):1821–1834. doi: 10.1038/onc.2009.473. (in eng) [DOI] [PubMed] [Google Scholar]
- 27.Okoniewski MJ, Miller CJ. Comprehensive analysis of affymetrix exon arrays using BioConductor. (Translated from eng) PLoS Comput Biol. 2008;4(2):e6. doi: 10.1371/journal.pcbi.0040006. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venables JP, et al. Cancer-associated regulation of alternative splicing. (Translated from eng) Nat Struct Mol Biol. 2009;16(6):670–676. doi: 10.1038/nsmb.1608. (in eng) [DOI] [PubMed] [Google Scholar]
- 29.Graveley BR. Sorting out the complexity of SR protein functions. (Translated from eng) Rna. 2000;6(9):1197–1211. doi: 10.1017/s1355838200000960. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nasim MT, Eperon IC. A double-reporter splicing assay for determining splicing efficiency in mammalian cells. (Translated from eng) Nat Protoc. 2006;1(2):1022–1028. doi: 10.1038/nprot.2006.148. (in eng) [DOI] [PubMed] [Google Scholar]
- 31.Hilberg F, Aguzi A, Howells N, Wagner EF. c-jun is essential for normal mouse development and hepatogenesis. Nature. 1993;365:179–181. doi: 10.1038/365179a0. [DOI] [PubMed] [Google Scholar]
- 32.Zheng TS, Hunot S, Kuida K, Flavell RA. Caspase knockouts: matters of life and death. (Translated from eng) Cell Death Differ. 1999;6(11):1043–1053. doi: 10.1038/sj.cdd.4400593. (in eng) [DOI] [PubMed] [Google Scholar]
- 33.Angelastro JM, et al. Characterization of a novel isoform of caspase-9 that inhibits apoptosis. (Translated from eng) J Biol Chem. 2001;276(15):12190–12200. doi: 10.1074/jbc.M009523200. (in eng) [DOI] [PubMed] [Google Scholar]
- 34.Venables JP. Unbalanced alternative splicing and its significance in cancer. (Translated from eng) Bioessays. 2006;28(4):378–386. doi: 10.1002/bies.20390. (in eng) [DOI] [PubMed] [Google Scholar]
- 35.Kirschbaum-Slager N, Lopes GM, Galante PA, Riggins GJ, de Souza SJ. Splicing factors are differentially expressed in tumors. (Translated from eng) Genet Mol Res. 2004;3(4):512–520. (in eng) [PubMed] [Google Scholar]
- 36.Toh WH, Siddique MM, Boominathan L, Lin KW, Sabapathy K. c-Jun regulates the stability and activity of the p53 homologue, p73. (Translated from eng) J Biol Chem. 2004;279(43):44713–44722. doi: 10.1074/jbc.M407672200. (in eng) [DOI] [PubMed] [Google Scholar]
- 37.Stepniak E, et al. c-Jun/AP-1 controls liver regeneration by repressing p53/p21 and p38 MAPK activity. (Translated from eng) Genes Dev. 2006;20(16):2306–2314. doi: 10.1101/gad.390506. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zenz R, et al. c-Jun regulates eyelid closure and skin tumor development through EGFR signaling. Dev Cell. 2003;4(6):879–889. doi: 10.1016/s1534-5807(03)00161-8. [DOI] [PubMed] [Google Scholar]
- 39.Li Z, et al. Cyclin D1 regulates cellular migration through the inhibition of thrombospondin 1 and ROCK signaling. Mol Cell Biol. 2006;26(11):4240–4256. doi: 10.1128/MCB.02124-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Albanese C, et al. Activation of the cyclin D1 gene by the E1A-associated protein p300 through AP-1 inhibits cellular apoptosis. J Biol Chem. 1999;274:34186–34195. doi: 10.1074/jbc.274.48.34186. [DOI] [PubMed] [Google Scholar]
- 41.Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walters RW, et al. Basolateral localization of fiber receptors limits adenovirus infection from the apical surface of airway epithelia. J Biol Chem. 1999;274(15):10219–10226. doi: 10.1074/jbc.274.15.10219. [DOI] [PubMed] [Google Scholar]
- 43.Katiyar S, Jiao X, Wagner E, Lisanti MP, Pestell RG. Somatic excision demonstrates that c-Jun induces cellular migration and invasion through induction of stem cell factor. (Translated from eng) Mol Cell Biol. 2007;27(4):1356–1369. doi: 10.1128/MCB.01061-06. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gius D, et al. Transcriptional activation and repression by Fos are independent functions: the C terminus represses immediate-early gene expression via CArG elements. Mol Cell Biol. 1990;10(8):4243–4255. doi: 10.1128/mcb.10.8.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robinson GW, Hennighausen L. Inhibins and activins regulate mammary epithelial cell differentiation through mesenchymal-epithelial interactions. (Translated from eng) Development. 1997;124(14):2701–2708. doi: 10.1242/dev.124.14.2701. (in eng) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.