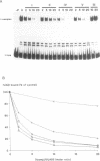
Abstract
The coregulated genes LAC4 and LAC12 encoding beta-galactosidase and lactose permease, respectively, are responsible for the ability of the milk yeast Kluyveromyces lactis to utilise lactose. They are divergently transcribed and separated by an unusually large intergenic region of 2.6 kbp. Mapping of the upstream border of the beta-galactosidase gene (LAC4) promoter by introduction of mutations at the chromosomal locus showed that LAC4 and LAC12 share the same upstream activation sites (UAS). The UASs represent binding sites for the trans-activator LAC9, a K. lactis homologue of GAL4, conforming to the consensus sequence 5'-CGG(N5)A/T(N5)CCG-3'. Two binding sites are located in front of each of the genes at almost symmetrical positions. beta-galactosidase activity measurements as well as quantitation of LAC4 and LAC12 mRNA levels demonstrated that all four sites are required for full induction. LAC4 proximal and a LAC12 proximal sites cooperate in activating transcription of both genes. These sites are more than 1.7 kbp apart and the distal site is located more than 2.3 kbp upstream of the respective start of transcription. Thus, the distance between interacting sites is larger than in any of the well characterised yeast promoters. The contribution to gene activation differs for individual binding sites and correlates with the relative affinity of LAC9 for these sites in vitro suggesting that LAC9 binding is a rate limiting step for LAC promoter function.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beck C. F., Warren R. A. Divergent promoters, a common form of gene organization. Microbiol Rev. 1988 Sep;52(3):318–326. doi: 10.1128/mr.52.3.318-326.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiec J. A., Zhang L., Sasse-Dwight S., Gralla J. D. DNA supercoiling promotes formation of a bent repression loop in lac DNA. J Mol Biol. 1987 Jul 5;196(1):101–111. doi: 10.1016/0022-2836(87)90513-4. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bram R. J., Lue N. F., Kornberg R. D. A GAL family of upstream activating sequences in yeast: roles in both induction and repression of transcription. EMBO J. 1986 Mar;5(3):603–608. doi: 10.1002/j.1460-2075.1986.tb04253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breunig K. D., Dahlems U., Das S., Hollenberg C. P. Analysis of a eukaryotic beta-galactosidase gene: the N-terminal end of the yeast Kluyveromyces lactis protein shows homology to the Escherichia coli lacZ gene product. Nucleic Acids Res. 1984 Mar 12;12(5):2327–2341. doi: 10.1093/nar/12.5.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breunig K. D. Glucose repression of LAC gene expression in yeast is mediated by the transcriptional activator LAC9. Mol Gen Genet. 1989 Apr;216(2-3):422–427. doi: 10.1007/BF00334386. [DOI] [PubMed] [Google Scholar]
- Breunig K. D., Kuger P. Functional homology between the yeast regulatory proteins GAL4 and LAC9: LAC9-mediated transcriptional activation in Kluyveromyces lactis involves protein binding to a regulatory sequence homologous to the GAL4 protein-binding site. Mol Cell Biol. 1987 Dec;7(12):4400–4406. doi: 10.1128/mcb.7.12.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breunig K. D. Multicopy plasmids containing the gene for the transcriptional activator LAC9 are not tolerated by K. lactis cells. Curr Genet. 1989 Feb;15(2):143–148. doi: 10.1007/BF00435461. [DOI] [PubMed] [Google Scholar]
- Carey M., Lin Y. S., Green M. R., Ptashne M. A mechanism for synergistic activation of a mammalian gene by GAL4 derivatives. Nature. 1990 May 24;345(6273):361–364. doi: 10.1038/345361a0. [DOI] [PubMed] [Google Scholar]
- Chang Y. D., Dickson R. C. Primary structure of the lactose permease gene from the yeast Kluyveromyces lactis. Presence of an unusual transcript structure. J Biol Chem. 1988 Nov 15;263(32):16696–16703. [PubMed] [Google Scholar]
- Das S., Breunig K. D., Hollenberg C. P. A positive regulatory element is involved in the induction of the beta-galactosidase gene from Kluyveromyces lactis. EMBO J. 1985 Mar;4(3):793–798. doi: 10.1002/j.1460-2075.1985.tb03699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. C., Barr K. Characterization of lactose transport in Kluyveromyces lactis. J Bacteriol. 1983 Jun;154(3):1245–1251. doi: 10.1128/jb.154.3.1245-1251.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. C., Markin J. S. Molecular cloning and expression in E. coli of a yeast gene coding for beta-galactosidase. Cell. 1978 Sep;15(1):123–130. doi: 10.1016/0092-8674(78)90088-0. [DOI] [PubMed] [Google Scholar]
- Dickson R. C., Markin J. S. Physiological studies of beta-galactosidase induction in Kluyveromyces lactis. J Bacteriol. 1980 Jun;142(3):777–785. doi: 10.1128/jb.142.3.777-785.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficca A. G., Hollenberg C. P. Functional relationship among TATA sequences, gene induction and transcription initiation in the beta-galactosidase, LAC4, gene from Kluyveromyces lactis. Curr Genet. 1989 Apr;15(4):261–269. doi: 10.1007/BF00447041. [DOI] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G., Ptashne M. Negative effect of the transcriptional activator GAL4. Nature. 1988 Aug 25;334(6184):721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- Giniger E., Ptashne M. Cooperative DNA binding of the yeast transcriptional activator GAL4. Proc Natl Acad Sci U S A. 1988 Jan;85(2):382–386. doi: 10.1073/pnas.85.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heus J. J., Zonneveld B. J., Steensma H. Y., Van den Berg J. A. Centromeric DNA of Kluyveromyces lactis. Curr Genet. 1990 Dec;18(6):517–522. doi: 10.1007/BF00327022. [DOI] [PubMed] [Google Scholar]
- Hochschild A., Ptashne M. Cooperative binding of lambda repressors to sites separated by integral turns of the DNA helix. Cell. 1986 Mar 14;44(5):681–687. doi: 10.1016/0092-8674(86)90833-0. [DOI] [PubMed] [Google Scholar]
- Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987 Dec;51(4):458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalnins A., Otto K., Rüther U., Müller-Hill B. Sequence of the lacZ gene of Escherichia coli. EMBO J. 1983;2(4):593–597. doi: 10.1002/j.1460-2075.1983.tb01468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebe R. J., Harriss J. V., Sharp Z. D., Douglas M. G. A general method for polyethylene-glycol-induced genetic transformation of bacteria and yeast. Gene. 1983 Nov;25(2-3):333–341. doi: 10.1016/0378-1119(83)90238-x. [DOI] [PubMed] [Google Scholar]
- Krämer H., Niemöller M., Amouyal M., Revet B., von Wilcken-Bergmann B., Müller-Hill B. lac repressor forms loops with linear DNA carrying two suitably spaced lac operators. EMBO J. 1987 May;6(5):1481–1491. doi: 10.1002/j.1460-2075.1987.tb02390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuger P., Gödecke A., Breunig K. D. A mutation in the Zn-finger of the GAL4 homolog LAC9 results in glucose repression of its target genes. Nucleic Acids Res. 1990 Feb 25;18(4):745–751. doi: 10.1093/nar/18.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lee D. H., Schleif R. F. In vivo DNA loops in araCBAD: size limits and helical repeat. Proc Natl Acad Sci U S A. 1989 Jan;86(2):476–480. doi: 10.1073/pnas.86.2.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardo J. M., Bhairi S. M., Dickson R. C. Identification of upstream activator sequences that regulate induction of the beta-galactosidase gene in Kluyveromyces lactis. Mol Cell Biol. 1987 Dec;7(12):4369–4376. doi: 10.1128/mcb.7.12.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. S., Carey M., Ptashne M., Green M. R. How different eukaryotic transcriptional activators can cooperate promiscuously. Nature. 1990 May 24;345(6273):359–361. doi: 10.1038/345359a0. [DOI] [PubMed] [Google Scholar]
- Mastrangelo I. A., Courey A. J., Wall J. S., Jackson S. P., Hough P. V. DNA looping and Sp1 multimer links: a mechanism for transcriptional synergism and enhancement. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5670–5674. doi: 10.1073/pnas.88.13.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Riley M. I., Hopper J. E., Johnston S. A., Dickson R. C. GAL4 of Saccharomyces cerevisiae activates the lactose-galactose regulon of Kluyveromyces lactis and creates a new phenotype: glucose repression of the regulon. Mol Cell Biol. 1987 Feb;7(2):780–786. doi: 10.1128/mcb.7.2.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Rudolph H., Koenig-Rauseo I., Hinnen A. One-step gene replacement in yeast by cotransformation. Gene. 1985;36(1-2):87–95. doi: 10.1016/0378-1119(85)90072-1. [DOI] [PubMed] [Google Scholar]
- Ruzzi M., Breunig K. D., Ficca A. G., Hollenberg C. P. Positive regulation of the beta-galactosidase gene from Kluyveromyces lactis is mediated by an upstream activation site that shows homology to the GAL upstream activation site of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Mar;7(3):991–997. doi: 10.1128/mcb.7.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron J. M., Jr, Johnston S. A. Analysis of the Kluyveromyces lactis positive regulatory gene LAC9 reveals functional homology to, but sequence divergence from, the Saccharomyces cerevisiae GAL4 gene. Nucleic Acids Res. 1986 Oct 10;14(19):7767–7781. doi: 10.1093/nar/14.19.7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanssens P., Opsomer C., McKeown Y. M., Kramer W., Zabeau M., Fritz H. J. Efficient oligonucleotide-directed construction of mutations in expression vectors by the gapped duplex DNA method using alternating selectable markers. Nucleic Acids Res. 1989 Jun 26;17(12):4441–4454. doi: 10.1093/nar/17.12.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark M. J., Milner J. S. Cloning and analysis of the Kluyveromyces lactis TRP1 gene: a chromosomal locus flanked by genes encoding inorganic pyrophosphatase and histone H3. Yeast. 1989 Jan-Feb;5(1):35–50. doi: 10.1002/yea.320050106. [DOI] [PubMed] [Google Scholar]
- Théveny B., Bailly A., Rauch C., Rauch M., Delain E., Milgrom E. Association of DNA-bound progesterone receptors. Nature. 1987 Sep 3;329(6134):79–81. doi: 10.1038/329079a0. [DOI] [PubMed] [Google Scholar]
- Tschumper G., Carbon J. Sequence of a yeast DNA fragment containing a chromosomal replicator and the TRP1 gene. Gene. 1980 Jul;10(2):157–166. doi: 10.1016/0378-1119(80)90133-x. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Webster T. D., Dickson R. C. The organization and transcription of the galactose gene cluster of Kluyveromyces lactis. Nucleic Acids Res. 1988 Aug 25;16(16):8011–8028. doi: 10.1093/nar/16.16.8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray L. V., Jr, Witte M. M., Dickson R. C., Riley M. I. Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: structural and functional relationships to GAL4 of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Mar;7(3):1111–1121. doi: 10.1128/mcb.7.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]