Abstract
The proteome of human saliva can be considered as being essentially completed. Diagnostic markers for a number of diseases have been identified among salivary proteins and peptides, taking advantage of saliva as an easy-to-obtain biological fluid. Yet, the majority of disease markers identified so far are serum components and not intrinsic proteins produced by the salivary glands. Furthermore, despite the fact that saliva is essential for protecting the oral integuments and dentition, little progress has been made in finding risk predictors in the salivary proteome for dental caries or periodontal disease. Since salivary proteins, and in particular the attached glycans, play an important role in interactions with the microbial world, the salivary glycoproteome and other post-translational modifications of salivary proteins need to be studied. Risk markers for microbial diseases, including dental caries, are likely to be discovered among the highly glycosylated major protein species in saliva. This review will attempt to raise new ideas and also point to under-researched areas that may hold promise for future applicability in oral diagnostics and prediction of oral disease.
Keywords: bacterial adhesion, dental, glycomics, oral, proteome, saliva, salivary proteins
The proteome of human saliva can be considered as essentially completed. More than 2000 different proteins and peptides have been identified in whole saliva and salivary glandular secretions as a result of the concerted efforts of several groups sponsored over the past few years, through a series of initiatives in the USA by the National Institute of Dental and Craniofacial Research [1,2] and by some European collaboratives [3–6]. Since collection of whole saliva samples, compared with blood, is relatively easy and does not involve invasive methods, this body fluid has recently become a vast treasure trove for identifying biomarkers indicative of disease. Indeed, miniaturized hand-held diagnostic devices are already being developed that promise to allow the future diagnosis of infectious diseases, cancer, heart disease and potentially many other systemic diseases from just a small drop of saliva [7,8]. Yet, the biomarkers identified so far mostly do not belong to the intrinsic components of saliva, but rather are small-molecular-weight inflammatory markers derived from serum that move into saliva by way of diffusion from the surrounding tissues of the oral cavity, most importantly through gingival crevicular fluid [9]. Thus, with regard to these biomarkers, saliva can only be considered as a secondary source of information. In addition, despite the well-known and important function of saliva for protecting the oral tissues, including the mineralized surfaces of teeth, little progress has been made so far in identifying salivary protein markers that could help to predict a predisposition or an increased risk of certain individuals for the two major oral diseases: dental caries and periodontitis [10–12].
It can be assumed that if such markers existed, they may eventually be found among the group of major salivary proteins that are synthesized de novo in the salivary glands. However, even in the dawn of this post-proteomic era, their study is still somewhat elusive due to extremely high variability and heterogeneity, not only in the sequence of their protein backbones but also in various intricate post-translational modifications, potentially including glycosylation, phosphorylation, acetylation, ubiquitination, methylation, deamidation, sulfation and proteolytic processing [13,14]. Among such modifications, glycosylation is particularly important as it adds another huge dimension of complexity to the salivary proteome, namely the ‘glycome’ of saliva. Glycans are arguably the most abundant and structurally diverse class of molecules in nature. Their functional roles and their impact on human disease are now becoming better understood through modern glycomics analyses [15]. The glycans decorating the proteins of saliva are most important for interactions with the microbiota colonizing the mouth and with other infectious and noninfectious microorganisms transiting the oral cavity [16].
This review will only contain a general synopsis of the current state of knowledge about the biological function of the major protein species of saliva, with an emphasis on the glycosylated proteins in the salivary proteome and the ways in which they are thought to interact with the oral microbiome. It will not describe the molecular structures and genetics of singular salivary protein species, as this has already been achieved in previous excellent reviews [3,13,14,17]. It will also not focus on the ongoing and promising attempts to use saliva as a diagnostic medium for systemic disease. The rapidly progressing field of salivary diagnostics has also been previously exhaustively reviewed [3,8,18,19]. This article is rather meant to reignite interest in certain areas of salivary research that have recently fallen somewhat off the wayside of mainstream research. It will further attempt to raise new ideas and also point to under-researched areas that may hold promise for future applicability in oral diagnostics, prediction of risk for oral disease or therapy. As such, it will include a number of unproven hypotheses and leave a lot of open questions at the end.
Most of all, this review will make a pledge to take up the work again on the intrinsic major proteins of saliva, among them mainly the salivary mucins and other high-molecular-weight glycoproteins, and to go back to study their basic biological function by now taking advantage of the rich opportunities that became available through integration of bioinformatics, genetics, proteomics and glycomics – summarized by the fashioned misnomer ‘interactomics’. Even though a lot of work has already been carried out on these intrinsic proteins of saliva in the heyday of salivary biochemistry, mainly in the period from the 1970s to the 1990s (reviewed in [20,21]), there are still many questions remaining to be answered. While the overlaps between saliva and blood plasma proteomes have been investigated [2,22], parallels will also need to be drawn between saliva and other mucous body secretions whose proteomes are currently under study [23–25]. Lastly, a multidisciplinary approach is needed, also including veterinary medicine, to integrate knowledge about the functions of saliva and its proteins in the animal kingdom, most importantly in mammals, and draw comparisons to possible functions in humans. There is reason to hope that further research of these glycoproteins will help to understand individual variations in susceptibility to oral or systemic infectious diseases.
The functions of saliva
“If I try to add up all these observations, I come to the conclusion that we have too few functions and too many components.” Solon Arthur Ellison, 1979 [26].
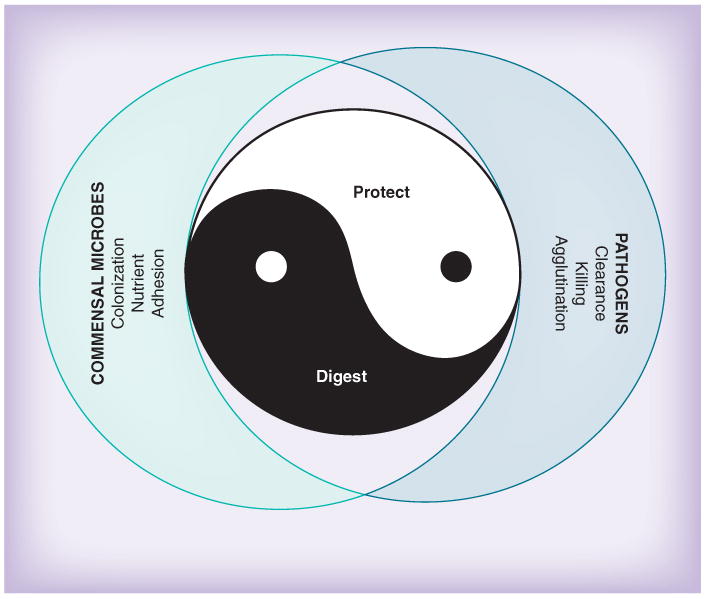
Naturally, the primordial function of saliva is to aid in the preprocessing of food in concert with tooth-mediated mastication [27]. But saliva has other, not so immediately obvious, functions too, which are no less important. Much of what the beneficial functions of saliva are becomes apparent when salivary flow is inhibited or when its composition is altered, such as in patients suffering from dryness of the mouth, sicca syndrome or xerostomia [28,201]. Looking at these patients' symptoms by inference readily delivers a list of the normal physiologic functions of human saliva (Table 1). For the purpose of didactic simplicity, the numerous functions of saliva can be grouped into two major categories, namely digestive and protective functions (Figure 1). Finally, there is yet another function of saliva that is difficult to discern from the other functions, and that is that it interfaces with the outside and inside microbial world, not only the oral microbiota but also systemic pathogens that must traverse the mouth in order to reach their target tissue [16]. One has to be aware, however, that in reality many overlaps exist among those categories and that there are functions that do not easily fit into this simple scheme either. Moreover, a given salivary protein may exert multiple functions. A salivary protein may be taken advantage of by microbes for colonization or nutritional purposes, which may be beneficial to the host as long as it concerns the commensal microflora, but may turn into a Janus-head-like amphi-functionality if pathogens are involved. Lastly, there is most likely functional cooperativity among salivary proteins aided by certain salivary protein species forming complexes with each other [29].
Table 1.
Oral symptoms of patients with oral sicca syndrome or xerostomia.
| Oral symptoms | Missing or altered function | Protein(s) involved |
|---|---|---|
| Dryness of oral mucosa | Lubrication | Mucin MUC5B, mucin MUC7, proline-rich glycoprotein |
| Viscous, sticky, turbid saliva | ||
| Problems with chewing and swallowing | ||
| Problems with speech and singing | ||
| Dry fissured corners of the mouth and lips | ||
|
| ||
| Reduced taste perception (hypogeusia) or bad taste (parageusia) | Not known (hypothesized: transport of taste compounds, presentation to taste receptors) | Carbonic anhydrase VI (synonym: gustin), others not known |
|
| ||
| Red, fissured tongue with atrophic papillae | Not known | Not known |
|
| ||
| Burning tongue syndrome | Not known | Not known |
|
| ||
| Gastroesophageal reflux, reflux esophagitis | Buffering | Carbonic anhydrase VI |
|
| ||
| Predisposition to dental erosion | Buffering, calcium binding, remineralization | Statherin, proline-rich proteins |
|
| ||
| Rampant dental caries, root caries | Antibacterial, balance of oral microflora | Proline-rich proteins, proline-rich glycoprotein, MUC5B, MUC7, S-IgA, other immunoglobulins, amylase, cystatins, histatins, lysozyme, lactoferrin, lactoperoxidase |
|
| ||
| Halitosis | Balance of oral microflora | Not known |
|
| ||
| Mucositis, gingivitis | Antimicrobial, wound healing | Growth factors (EGF, NGF), cytokines, defensins |
|
| ||
| Increased risk for oral fungal (Candida) infection | Antifungal | Histatins, MUC7 |
S-IgA: Secretory IgA
Figure 1.

The ‘Ying and Yang’ of saliva.
A more indirect approach to learn about the functions of saliva is by analogy to other mucous body secretions produced at sites where an interface with the microbial world exists, including nose fluid, tear fluid and the secretions of the genital tract [24,25]. The proteomes of some of these are currently being compiled [23]. Further, an underexplored approach to learn to understand the functions of salivary proteins is to compare salivary proteins among different species and try to relate their expression to the environmental conditions these species have adapted to. Some proteome analyses of saliva from other vertebrates have recently been published [30–33]. All these approaches will be dealt with in more detail below.
Digestive function
Saliva consists of 98% water. Its flow is increased by masticatory activity. This watery solution moistens dry food and further aids in mastication and bolus formation. Highly glycosylated mucous glycoproteins, including the salivary mucins and proline-rich glycoproteins, add lubricity to the food bolus and facilitate swallowing [20,27]. Saliva also contains enzymes, including proteases, lipases and glycohydrolases, which initiate partial digestion of food components. Many of these enzyme activities are not derived from the salivary glands but are of bacterial origin. Some of them add interesting new properties to saliva, such as the digestion of dietary gluten, which is difficult for mammalian proteolytic enzymes to digest [34]. Among the enzymes in saliva, salivary α-amylase is quantitatively by far the most predominant one [35]. Although the role of amylase in digestion of starch seems to be clear-cut, it still remains a matter of debate why there are such immense amounts of amylase in saliva. This becomes even more puzzling because the time to act enzymatically in the mouth during chewing is believed to be too short for amylase to be very effective during mastication. Moreover, after swallowing, most of its enzymatic activity becomes inactivated by the acid environment of the stomach, except for some residual activity remaining in the midst of the food bolus until stomach acid penetrates [36].
From an evolutionary perspective, it is interesting that humans have an approximately threefold higher amylase concentration in saliva than their closest relatives, the great apes. In this regard, it was shown that the increase in salivary amylase was mostly achieved through gene duplication during a relatively short and recent period of evolution. The duplication in copy numbers of the amylase gene corresponds with the consumption of starch and backtraces our evolutionary path from fruit eaters to starch eaters [37]. Therefore, besides pure digestive function, amylase could have helped our ancestors in the search for starch-containing foods by liberating disaccharides and thus making them available for sweet taste perception [38]. One feature that went along with increased starch consumption was stickiness of food remnants to the teeth. Amylase could also be beneficial here by digesting starch residues in areas that are not accessible to the cleansing actions of the tongue and cheeks, particularly the inter-proximal spaces between adjacent teeth. In this respect, salivary amylase may be envisioned as a biochemical toothbrush. So far, no human individuals have been identified who are deficient in salivary amylase; thus, these hypotheses can only be proven once amylase-deficient animal models will become available.
Protective function of saliva
The mouth is the only site in our body where mineralized tissues, namely the enamel crowns of teeth, are exposed to the outside environment. Thus, whereas bone, the other mineralized tissue in our body, is surrounded by a nourishing periosteal tissue and is being constantly remodeled, the crowns of teeth have no such protective tissue once they are erupted. It will be then saliva that takes over this function and, in fact, the salivary glands can be viewed as remote protective organs for our dentition [Oppenheim F, Pers. Comm.]. This is not trivial, as can be seen from the pathologic deterioration of tooth enamel in xerostomic patients [28,201].
Important functions of saliva in this respect are acid neutralization by its buffering systems, as well as acid protection by salivary proteins adsorbed to the enamel surface that form the enamel pellicle [12,39–41]. Saliva is supersaturated with calcium and phosphate and the salivary proline-rich proteins, together with statherin, help to keep these electrolytes in solution and serve as vehicles transferring calcium to the enamel surface for remineralization [21]. There is a balance of de- and re-mineralization that, in concert with the adsorbed pellicle proteins, protects the teeth from decalcification and slows down the abrasive damage caused by attrition during mastication of food [21]. As the electrolyte concentration and buffering capacity of saliva increases with increasing salivary flow [42], it provides a smart system of protection, not only during mastication and consumption of food but also as a reaction to acid taste perception and mechanical irritation of the esophagus preceding regurgitation of acid gastric contents [43].
Saliva also helps keeping the teeth clean by its simple flushing action to remove detritus, and this may be enhanced by the activities of proteolytic and glycolytic enzymes in saliva. Lipids in saliva also help to protect the teeth [44], but little additional evidence of this has been presented in recent years. Considering the important protective functions of saliva, it is not surprising that many studies attempted to identify a correlation between salivary components and caries susceptibility, albeit with limited success thus far [10,12].
The soft integuments of the oral cavity are equally protected by saliva. Saliva moistens the mucosal tissues and aids in wound healing [45,46]. Oral surgeons can attest that the oral cavity is a place where wounds heal at a rapid pace. The wound-healing property of saliva helps to repair microlesions caused by mastication of food and swallowing. Saliva may also help in wound healing of skin lesions. Why else do we reflexly put our finger into our mouth when it has been injured by a cut? Moreover, animals are known to lick their wounds or injured extremities. Interestingly, some indigenous tribes still use saliva as an ingredient for wound dressings.
A number of studies have shown the presence of growth factors in saliva, including EGF, NGF and FGF [46–48]. Saliva was in fact the source of discovery for NGF [49]. More recently, salivary histatins have been found to exert wound-healing properties [45]. Wound-healing activities have been demonstrated in vitro and there are also some in vivo data [46]; however, experiments using defined animal models are still missing. In addition, even though alterations of salivary inflammatory markers occur as a result of periodontal inflammations [50], no convincing correlation has yet been drawn between the composition of salivary proteins and the susceptibility or risk of a given individual for periodontal disease [11,47].
Other functions of saliva
One function that does not fit under the main categories of digestive and protective functions is saliva's help in taste perception [51]. Nevertheless, it may be seen as in some way related to the digestive category because it aids in the selectivity for certain foods and in the avoidance of potentially harmful dietary ingredients. Some of our food preferences, if seen from an evolutionary perspective, may have also affected how saliva is composed in humans. For example, compared with blood plasma or other secretory body fluids, such as tears, saliva has a low salt concentration [42]. Could this be to better enable us and other mammals to taste salt content in our foods? Salt is important for survival, but in our ancestors' diet it was often a sought-after ingredient. Sugar was another highly desirable food ingredient and it has been shown that there is a genetic trait that may explain the cravings for sweet foods [52]. In the same context, as already mentioned, it is hypothesized that salivary α-amylase in our mouth liberates sugars from starch and therefore aids in the detection of starch-rich foods [38].
Saliva may also aid in other modes of taste perception, simply by serving as a diluent for tasty molecules. Thus, xerostomic patients often suffer from a loss of taste acuity [28]. One protein, carbonic anhydrase VI, formerly named gustin, has been associated with taste perception [53,54]. However, it is likely that other salivary proteins exist, which would be worth exploring, that carry and present taste components to the taste buds [51]. Children in particular exhibit an aversion to bitter food ingredients. This may be to avoid potentially harmful polyphenolic compounds in their diet. In this context, it is interesting that the proline-rich proteins in saliva have been shown to protect our digestive system from tannins in our diet by binding and neutralizing these potentially harmful components [32,55]. For these and other overly aversive components, saliva allows simple excretion through spitting. This purgatory process can be frequently observed in infants and children but later in life becomes corrected by cultural restrictions.
Saliva also helps in the maintenance of body hydration by triggering a thirst perception when the mouth becomes dry [28]. Lastly, there are some functions of saliva, mainly in the behavioral, social and sexual domains, which can be observed in animals and may have become virtually lost in humans [56–58]. Mammals use saliva for their fur care, they lick the skin of their litter, or the fur of other group members for social purposes, or leave traces of saliva, presumably containing pheromone-like substances, to mark their territory. From those functions, only rudimentary leftovers may still exist in humans. The only widespread use of saliva among humans in a sociosexual context that is left is kissing, where saliva is allowed to flow freely from one individual's mouth to another [27].
Interactions of saliva with the microbiota
“As a biologist, I find it especially intriguing to consider that the primary habitat on planet Earth for these organisms appears to be the surfaces of human teeth!” Ronald J Gibbons, 1989 [59].
The human oral cavity is the entrance to both the respiratory and digestive tracts. As such, in its interactions with the microbial world, saliva can be seen as a gatekeeper that differentiates between that part of the microbial world permitted to enter the body's interior and to reside as what is called a commensal microflora, and that other part of the microbial world that is un desired. Among the latter are viruses, fungi and pathogenic bacteria (Figure 2). Thus, similar to other body fluids and external secretions, saliva contains a number of antimicrobial active components [25,60]. Fungal growth in the oral cavity is suppressed by the presence of salivary histatins [61,62]. Nevertheless, growth of a physiological commensal oral microflora appears to still be permitted because it is beneficial for maintenance of health in the oral cavity. While the functions of saliva in the interactions with the microbial world can be separated for didactic purposes into anti- and pro-bacterial [16], one has to be aware that great overlap exists between these. Thus, a molecule can serve as an adhesion substrate for one bacterium, but may at the same time act as an agglutinin for another. Such multifunctionality and redundancy of function usually helps to keep a biological system stable, but yet another reason for this complexity can be found in its evolutionary origin [63]. It is assumed that during evolution, a balance has been established in that some bacteria have evolved to bind to salivary proteins in order to colonize oral integuments. Such harmless commensal bacteria are tolerated. Salivary protein glycosylation may have even evolved to foster colonization by certain commensal bacteria that turned out to be beneficial to the oral microenvironment. Other harmful or pathogenic bacteria, viruses or fungi that are not desired will not find adhesion substrata among the salivary proteins but will rather be killed, inactivated or eliminated by agglutination. Some of the salivary agglutinins may function as decoys by presenting host-own oligosaccharide motifs to the invading microbes and, thereby, capture the microorganisms before they can reach their target tissue sites. Thus, it can be hypothesized that salivary proteins may have evolved to keep the good microbes in and the bad ones out.
Figure 2.
Interactions of salivary proteins with the microbiota.
Agglutinins are normally high-molecular-weight proteins or protein complexes that carry extensive glycosylation [16,64]. Agglutinins found in saliva can also be found in other body fluids [25]. Their presumed function is to agglutinate bacteria in their planktonic state and it is assumed that this leads to clearance of unwanted bacteria from the oral cavity through swallowing and subsequent destruction of these microbes in the acidic stomach environment. However, there are some exceptions, as not only agglutinins have evolved to clear undesired bacteria, but bacteria may have evolved in turn to exploit these traps and use them as vehicles instead. Moreover, not all bacteria cleared from the oral cavity are killed in the stomach. A notable exception is Helicobacter pylori, which, by being swallowed, will in fact reach its target tissue for colonization, namely the mucous layer of the stomach epithelium [65]. In addition, by binding highly glycosylated mucins on its surface, it may become better protected against the initial acid assault in the stomach until it can move into the protective mucous layer of the stomach epithelium. It can also be hypothesized that by decorating their outer surface with host-like proteins while traversing the salivary environment in the mouth, gastrointestinal or respiratory pathogens may evade detection by host defense mechanisms.
Saliva as a mediator of microbial adhesion & biofilm formation
All the surfaces lining the oral cavity are normally covered by a thin film of adsorbed proteins, most of which are derived from the surrounding salivary milieu [39–41]. A freshly cleaned tooth will become covered within seconds by this so-called salivary pellicle. Therefore, what a bacterium encounters is not the mineralized tooth surface, carrying the physicochemical properties of hydroxy apatite, but rather a surface coated by salivary proteins that partially mask the original surface properties of the underlying substratum [66]. Thus, it is not surprising that bacteria in the oral cavity have evolved to bind to epitopes on certain salivary proteins in order to adhere to and to be able to colonize these surfaces [67]. Certain salivary proteins also adsorb or bind specifically to the surface of bacteria, where they may mediate adhesion, facilitate bacterial coadhesion or coaggregation [16,67]. Eventually, these processes will help in the build-up of a bacterial biofilm, called dental or oral plaque, which takes several days to mature to full thickness if not removed. Since bacteria that thrive in such biofilms are involved in the pathogenesis of dental caries and periodontal diseases, and since certain systemic pathogens may temporarily or permanently reside within oral biofilms [68–70], it is important to investigate how salivary proteins initiate and modulate bacterial adhesion and subsequent biofilm formation.
Whereas the initial interactions of bacteria with surface-adsorbed salivary proteins are mediated by general physicochemical inter actions, there are a number of well-studied highly specific interactions involving the binding pocket of bacterial adhesins recognizing complementary peptide or oligosaccharide motifs of a given salivary protein, the latter being a lectin–carbohydrate interaction [71]. According to a recent definition, the binding molecule on the bacterium is called an adhesin, the oligosaccharide motif being recognized is called a receptor and the complete protein carrying the receptor motifs is called a ‘counter-receptor’ [72]. The outermost oligosaccharide termini are most important for binding. A well-characterized example is the binding of certain strains of oral viridans-group streptococci to terminal α2–3-linked sialic acids on O-linked glycans decorating salivary glycoproteins, including mucin MUC7 (synonym: MG2) and secretory IgA [73–76]. Interestingly, some bacteria possess enzymes such as sialidases or fucosidases that enable them to gain access to subterminal oligosaccharide motifs for binding. A good example is the binding of oral actinomyces to Galβ1–3GalNAc residues on O-linked glycan chains on MUC7 and secretory IgA1 that requires prior removal of terminal sialic acids by sialidase [73,74,77]. Scenarios can be envisioned in which bacterial strains cooperate in that one strain already attached cleaves off the sugar and thereby aids another strain to gain access to the subterminal sugars. Moreover, it is not only the sequence of oligosaccharides that is important for interaction with bacterial adhesins, but also the type of linkage as well as the substitutions of oligosaccharides by sulfate, acetyl and other chemical groups. Many interactions of bacteria with salivary proteins are mediated by mechanisms other than lectin–carbohydrate recognition. Examples for recognition of salivary peptide motifs by bacterial adhesins or for recognition of bacterial surface components by salivary proteins are the binding of amylase by amylase-binding protein A of Streptococcus gordonii [78], binding of various bacterial species by salivary agglutinin (gp-340, DMBT1) [64,79], as well as the recognition of certain repetitive peptide motifs within salivary proline-rich proteins by complementary adhesins on oral actinomyces and streptococci [73,80–82].
Saliva as a nutritional substrate for oral microbes
Some of the interactions of bacteria with salivary proteins may confer a nutritional benefit to the microbes [83–85]. Thus, the binding of salivary amylase liberates oligosaccharides from starch that can then be metabolized by the bacteria [86]. Moreover, many of the enzymes produced by the bacteria, including fucosidases, sialidases or other glycohydrolases, may enable the micro organisms to forage on salivary glycoproteins. One could even envision attached bacteria grazing on a lawn of surface-adsorbed salivary proteins. Under such a scenario, it may be difficult to discern which biologically active sites serve for adhesion and which for nutritional purposes. It could even be the case that these functions may have evolved from each other. Situations in which bacteria within micro communities cross-feed each other can also be hypothesized. So far, little is known about these processes, but it is certain that saliva plays an important role as a substrate.
Working with saliva as a diagnostic fluid
It has frequently been stated that saliva is the ideal diagnostic fluid because, in contrast to blood, it is easy to obtain by non invasive means [7,19]. Although this is certainly true, one has to keep in mind, however, that salivary flow, composition and protein concentration are quite variable among individuals, and even within individuals are influenced by many confounding parameters, such as time of sampling, flow rate or stress [42]. There are also developmental changes in the composition of salivary proteins that occur during the lifetime of an individual, most notably during infant development [87–89] and in advanced age [90]. Moreover, salivary components are subject to enzymatic degradation in the moment they enter the oral cavity and become exposed to enzymes of host and microbial origin [14]. Thus, reliable diagnostic markers in saliva need to possess a certain robustness in that they are significantly disease-related above and beyond the prevailing physiological variations and fluctuations in salivary composition. Since disease markers with such a high degree of significance have not yet been found in saliva [11], one approach that has been taken recently is multiplexing of several diagnostic markers at the same time [18,48]. This approach is based on the assumption that if one marker alone does not provide a sufficiently significant predictive value, by monitoring several markers at once, statistical power may be gained.
Another approach taken is to learn more about the variability of salivary components within a given individual. This approach accepts the fact that enzymatic degradation takes place and, after careful study of the physiologic variations, even tries to correlate such degradation products such as salivary peptides to certain states of oral disease [4,14]. Some success has been reported in this regard for the prediction of caries risk [91].
A further complication when working with saliva is that certain salivary proteins, including the salivary mucins, form complexes with a number of other salivary proteins [29]. Such protein complexes can also be found organized in macromolecular aggregates or supra-structures such as salivary micelles [92] or the recently discovered exosomes [93,94]. Furthermore, sample collection, sample processing and mode of preservation can all potentially alter the original composition of salivary proteins because degradation takes place, precipitates can form during prolonged storage or after freezing, and certain protein compounds become lost through centrifugation or filtration [95–97].
Despite all these confounding parameters, it has to be kept in mind, however, that the major salivary proteins, which are intrinsically produced by the salivary glands, are genetically determined to a great extent [13]. So far, interindividual variations in quantity or amino acid composition have been found to be of little diagnostic value, with the notable exception of the salivary proline-rich proteins that have been proposed to be related to caries risk [91,98]. Nevertheless, the major saliva-typical protein species that are produced de novo in the salivary glands remain attractive candidates as markers for oral disease. Their abundance in saliva is higher by several orders of magnitude than most biomarkers currently under consideration for diagnostic use (Figure 3). It is not surprising that simple associations of pro-teomic data with clinical disease continue to fail to show significant correlations with disease susceptibility, because most of these major protein species are extensively decorated by post-translational modifications, including phosphorylation, sulfation and glycosylation [14]. For some of these proteins, such as the salivary mucins, their protein component constitutes only a minor part, while most of the molecular mass is due to extensive glycosylation [20]. The oligosaccharides attached to the protein backbone add another dimension of complexity that may be greater than the genetic or proteomic variability by several orders of magnitude [15] and is virtually unexplored in saliva. Since glycosylation is genetically determined to a great degree and, thus, varies from individual to individual, it deserves in-depth structural and functional analysis for individual variations in disease susceptibility. Furthermore, since glycosylation is related to microbial colonization, as has been explained above, it can be hypothesized that functional markers for bacteria-mediated diseases, such as dental caries and periodontal diseases, might be found in there [99,100]. In this respect, it is encouraging that a correlation of caries susceptibility with glycosylation patterns on salivary mucins was reported [99].
Figure 3. Relative abundances of protein and peptide species in human saliva.
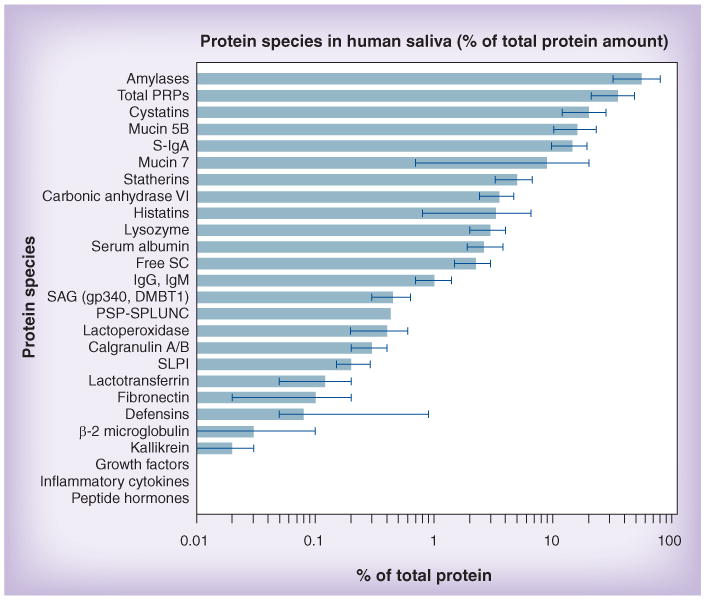
Most biomarkers presently considered for diagnostic use are derived from plasma and are less abundant by several orders of magnitude than the major intrinsic proteins of saliva. The values indicated by bars are good-faith estimates of the author, based on numerous literature sources (too many to cite them all here). The error bars indicate the widely varying measurements reported. For better comparison, concentrations were converted to percentage of total salivary protein. Comprehensive reviews are available [13,20,60,114–116].
PRP: Proline-rich protein; PSP-SPLUNC: Parotid secretory protein-short palate, lung and nasal epithelium clone 2; SAG: Salivary agglutinin; SC: Secretory component; S-IgA: Secretory IgA; SLPI: Secretory leukocyte peptidase inhibitor.
In completing the salivary proteome, great progress has been achieved by employing state-of-the-art shotgun proteomics, including multidimensional protein identification technology (MudPit). Problems with saliva being a complex biological fluid have been mostly overcome by employing various liquid separation techniques, as well as by removing overly abundant protein species such as salivary amylase prior to analysis, thereby increasing the range of lower-abundance proteins detectable [101,102]. By using chemical protein and peptide labeling techniques, such as isotope-coded affinity tagging (ICAT), isotope tags for relative and absolute quantification (iTRAQ), absolute quantification of proteins using internal standards (AQUA) or label-free quantification [103], it has become possible to observe quantitative alterations of the salivary proteome in different physiological or pathological stages [104,105]. These are currently the most valuable assets for the discovery of markers for saliva-based diagnosis.
Considerable advances have also been made in improving 2DE. This technique is still a valuable tool that allows quantitative comparisons and functional probing by a variety of techniques [106]. For the analysis of salivary proteins, one major drawback of 2D PAGE is that high-molecular-weight (>120 kDa) and very low-molecular-weight (<10 kDa) protein species can only be poorly resolved or do not enter the separation at all [6]. Moreover, extremely basic or acidic proteins cannot be separated very well with the current 2DE techniques available to date. However, a great advantage of 2DE is that protein spots can be visualized by a variety of staining methods for general proteins, and further characterized by specific staining methods for phospho-proteins using Pro-Q Diamond®, or glycoproteins using Alcian blue, periodic acid Schiff or Pro-Q-Emerald® [65]. Moreover, quantification of spots and quantitative comparisons are possible through 2D DIGE [106]. Further epitope-specific probing is possible using immunoblotting [107], lectin blotting for the identification of protein- attached glycans [108] or probing for functional activity by far-Western blotting techniques, such as the bacterial overlay technique [65].
The most promising challenge for the coming years will be the exploration of the salivary glycome or glycoproteome. Major advances have been made in improving the tools and techniques used for the characterization of glycans [15]. Novel methods for the study of carbohydrate-based biomarkers, including enrichment of carbohydrate-containing proteins from complex biofluids and subsequent analysis of glycan structures by mass spectrometry or lectin arrays, are now available for the study of salivary glycoproteins [33,108–110]. Thus far, lectin blotting has been used for glycoprofiling of the human salivary proteome [100,108]. It is also likely that some classical biochemical purification methods will become appreciated once again, among them affinity columns using lectins or anticarbohydrate antibodies, as well as other glycoprotein enrichment or pull-down strategies.
Expert commentary: ‘salivary proteome – where is thy function?’
Great progress has been made over the last few years using saliva as a window into the body's interior by measuring protein and peptide biomarkers indicative of systemic malfunction. This has been most successful for the occurrence of inflammatory markers, but was less successful for finding meaningful markers for the early detection or prognosis of dental diseases. It is likely that biomarkers for dental and oral disease will be found among the major groups of intrinsic salivary proteins, as they are known to protect teeth as well as oral integuments, and interact with the oral microbiota. However, much of what we know about the functions of these salivary proteins, we know from the past. Modern proteome analysis has added a vast amount of information that has yet to be sorted out and processed to be able to address functional or clinical relevance. We will only know more once we have gone all the way to complete the salivary phosphoproteome and, most importantly, the salivary glycome or glycoproteome. To achieve this goal, it will become necessary to rediscover classical biochemical purification methods. The likelihood may be great that significant discoveries lie ahead that will help in diagnosis of oral diseases. Will every thing then be completed in salivary research? Of course not. However, to stop at the current stage without continuing what has so enthusiastically been started would be a great mistake.
Five-year view: the salivary ‘interactome’
The hunt for disease biomarker discovery in saliva is currently being pursued with great energy, but little has been achieved in using salivary protein composition for the prediction of risk for infectious diseases in the oral cavity, such as dental caries or periodontitis. More insight is being expected from current efforts to investigate the phosphoproteome [111,112] and other post-translational modifications of salivary proteins. The major challenge will be to decipher the salivary glycome. Recent technological advances, many of which are shared with proteomics, have recently allowed semiquantitative profling of glycans and glyco-proteins [15,110]. Since glycans attached to salivary proteins are recognized by pathogenic as well as commensal microbes, it is an extremely fortunate coincidence that the human oral microbiome is currently being completed too [113,202]. With this at hand, it will become likely that salivary glycoproteomics will lay the ground work for combating oral bacterial diseases, including dental caries and periodontitis.
It is expected that we will gain more knowledge from functional studies. Such studies should not exclusively focus on human saliva but should also include comparisons with animal saliva. To study the biological function of salivary proteins, it will be essential to develop suitable animal models, such as transgenic mice deficient of a certain salivary protein. Foreseeable problems that will need to be solved will occur with such models because both the glycosylation as well as the oral microbiota in such animals will not resemble the situation in humans. When more is understood about the biological functions of salivary proteins and their post-translational modifications, it will become possible to produce better synthetic saliva surrogates that can mimic the important beneficial functions of natural saliva, namely lubrication, remineralization, wound healing and antimicrobial activities. To reach this goal, there is still a long way to go considering the complexity of saliva as a biological fluid, including the structural complexity of its protein components.
Key issues.
The proteome of human saliva is essentially completed.
Promising diagnostic markers for a number of systemic and oral diseases have been identified among salivary proteins and peptides.
Most of the disease markers identified so far are serum components and do not belong to the intrinsic proteins produced in the salivary glands.
Despite the fact that saliva protects the oral integuments and dentition, little progress has been made in finding risk predictors for dental caries or periodontal disease among the salivary proteins.
Functions of salivary proteins in taste perception and wound healing are only beginning to be understood.
Lessons can be learned by studying the role of salivary proteins in other mammalian species.
Salivary proteins and mostly the attached glycans play an important role in interactions with the oral commensal microbial world, as well as with systemic pathogens.
The salivary glycoproteome and other post-translational modifications of salivary proteins need to be studied.
The influences of sample collection, processing and storage need to become better understood.
Risk markers for microbial diseases, including dental caries, are likely to be discovered among the highly glycosylated major protein species, including mucins and agglutinins, in saliva.
To study biological function of salivary proteins, transgenic animal models need to be developed.
A long-term challenge will be the development of artificial saliva surrogates mimicking the functional attributes of natural salivary proteins to help xerostomic patients.
Acknowledgments
The author is grateful for helpful comments from Mira Edgerton, Molakala Reddy and Frank Scannapieco. The reference list is selective and not exhaustive due to space restrictions.
Preparation of this manuscript was supported by grant 5 R01 DE019807–02 (to S Ruhl).
Footnotes
Financial & competing interests disclosure: The author has no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1••.Denny P, Hagen FK, Hardt M, et al. The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions. J Proteome Res. 2008;7(5):1994–2006. doi: 10.1021/pr700764j. A landmark paper reporting the results of a concerted effort of three US research groups to catalogue the salivary proteome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loo JA, Yan W, Ramachandran P, Wong DT. Comparative human salivary and plasma proteomes. J Dent Res. 2010;89(10):1016–1023. doi: 10.1177/0022034510380414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castagnola M, Cabras T, Vitali A, Sanna MT, Messana I. Biotechnological implications of the salivary proteome. Trends Biotechnol. 2011;29(8):409–418. doi: 10.1016/j.tibtech.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Amado F, Lobo MJ, Domingues P, Duarte JA, Vitorino R. Salivary peptidomics. Expert Rev Proteomics. 2010;7(5):709–721. doi: 10.1586/epr.10.48. [DOI] [PubMed] [Google Scholar]
- 5.Ghafouri B, Tagesson C, Lindahl M. Mapping of proteins in human saliva using two-dimensional gel electrophoresis and peptide mass fngerprinting. Proteomics. 2003;3(6):1003–1015. doi: 10.1002/pmic.200300426. [DOI] [PubMed] [Google Scholar]
- 6.Walz A, Stühler K, Wattenberg A, et al. Proteome analysis of glandular parotid and submandibular-sublingual saliva in comparison to whole human saliva by two-dimensional gel electrophoresis. Proteomics. 2006;6(5):1631–1639. doi: 10.1002/pmic.200500125. [DOI] [PubMed] [Google Scholar]
- 7.Spielmann N, Wong DT. Saliva: diagnostics and therapeutic perspectives. Oral Dis. 2011;17(4):345–354. doi: 10.1111/j.1601-0825.2010.01773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8••.Baum BJ, Yates JR, 3rd, Srivastava S, Wong DT, Melvin JE. Scientifc frontiers: emerging technologies for salivary diagnostics. Adv Dent Res. 2011;23(4):360–368. doi: 10.1177/0022034511420433. A comprehensive overview and the introduction to a recent meeting on salivary diagnostics, followed by many other relevant articles on different aspects of salivary diagnostics in the same issue of this journal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inzitari R, Cabras T, Pisano E, et al. HPLC-ESI-MS analysis of oral human fuids reveals that gingival crevicular fluid is the main source of oral thymosins beta(4) and beta(10) J Sep Sci. 2009;32(1):57–63. doi: 10.1002/jssc.200800496. [DOI] [PubMed] [Google Scholar]
- 10.Leone CW, Oppenheim FG. Physical and chemical aspects of saliva as indicators of risk for dental caries in humans. J Dent Educ. 2001;65(10):1054–1062. [PubMed] [Google Scholar]
- 11.Zhang L, Henson BS, Camargo PM, Wong DT. The clinical value of salivary biomarkers for periodontal disease. Periodontol 2000. 2009;51:25–37. doi: 10.1111/j.1600-0757.2009.00315.x. [DOI] [PubMed] [Google Scholar]
- 12.Van Nieuw Amerongen A, Bolscher JG, Veerman EC. Salivary proteins: protective and diagnostic value in cariology? Caries Res. 2004;38(3):247–253. doi: 10.1159/000077762. [DOI] [PubMed] [Google Scholar]
- 13••.Oppenheim FG, Salih E, Siqueira WL, Zhang W, Helmerhorst EJ. Salivary proteome and its genetic polymorphisms. Ann NY Acad Sci. 2007;1098:22–50. doi: 10.1196/annals.1384.030. A detailed overview on the current knowledge about the biochemical structure and genetics of salivary proteins. [DOI] [PubMed] [Google Scholar]
- 14••.Helmerhorst EJ, Oppenheim FG. Saliva: a dynamic proteome. J Dent Res. 2007;86(8):680–693. doi: 10.1177/154405910708600802. An insightful review article describing the variabilities in the salivary proteome due to post-translational modifcations and enzymatic degradation (also see reference [3]) [DOI] [PubMed] [Google Scholar]
- 15••.Hart GW, Copeland RJ. Glycomics hits the big time. Cell. 2010;143(5):672–676. doi: 10.1016/j.cell.2010.11.008. An inspiring perspective on the potential of deciphering glycosylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scannapieco FA. Saliva–bacterium interactions in oral microbial ecology. Crit Rev Oral Biol Med. 1994;5(3–4):203–248. doi: 10.1177/10454411940050030201. [DOI] [PubMed] [Google Scholar]
- 17.Amerongen AV, Veerman EC. Saliva – the defender of the oral cavity. Oral Dis. 2002;8(1):12–22. doi: 10.1034/j.1601-0825.2002.1o816.x. [DOI] [PubMed] [Google Scholar]
- 18.Miller CS, Foley JD, Bailey AL, et al. Current developments in salivary diagnostics. Biomark Med. 2010;4(1):171–189. doi: 10.2217/bmm.09.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, Xiao H, Wong DT. Salivary biomarkers for clinical applications. Mol Diagn Ther. 2009;13(4):245–259. doi: 10.1007/BF03256330. [DOI] [PubMed] [Google Scholar]
- 20.Levine MJ. Salivary macromolecules. A structure/function synopsis. Ann NY Acad Sci. 1993;694:11–16. doi: 10.1111/j.1749-6632.1993.tb18337.x. [DOI] [PubMed] [Google Scholar]
- 21.Lamkin MS, Oppenheim FG. Structural features of salivary function. Crit Rev Oral Biol Med. 1993;4(3–4):251–259. doi: 10.1177/10454411930040030101. [DOI] [PubMed] [Google Scholar]
- 22.Yan W, Apweiler R, Balgley BM, et al. Systematic comparison of the human saliva and plasma proteomes. Proteomics Clin Appl. 2009;3(1):116–134. doi: 10.1002/prca.200800140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li SJ, Peng M, Li H, et al. Sys-BodyFluid: a systematical database for human body fluid proteome research. Nucleic Acids Res. 2009;37(Database issue):D907–D912. doi: 10.1093/nar/gkn849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu S, Loo JA, Wong DT. Human body fuid proteome analysis. Proteomics. 2006;6(23):6326–6353. doi: 10.1002/pmic.200600284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schenkels LC, Veerman EC, Nieuw Amerongen AV. Biochemical composition of human saliva in relation to other mucosal fuids. Crit Rev Oral Biol Med. 1995;6(2):161–175. doi: 10.1177/10454411950060020501. [DOI] [PubMed] [Google Scholar]
- 26.Emmings FG. Oral biology, a dialogue: Solon Arthur Ellison at the State University of New York at Buffalo. J Dent Res. 1999;78(3):725–729. doi: 10.1177/00220345990780030301. [DOI] [PubMed] [Google Scholar]
- 27.Mandel ID. The functions of saliva. J Dent Res. 1987;66 doi: 10.1177/00220345870660S203. Spec No. 623–627. [DOI] [PubMed] [Google Scholar]
- 28.Sreebny LM. Saliva in health and disease: an appraisal and update. Int Dent J. 2000;50(3):140–161. doi: 10.1111/j.1875-595x.2000.tb00554.x. [DOI] [PubMed] [Google Scholar]
- 29.Soares RV, Lin T, Siqueira CC, et al. Salivary micelles: identifcation of complexes containing MG2, sIgA, lactoferrin, amylase, glycosylated proline-rich protein and lysozyme. Arch Oral Biol. 2004;49(5):337–343. doi: 10.1016/j.archoralbio.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 30.Lamy E, Graca G, da Costa G, et al. Changes in mouse whole saliva soluble proteome induced by tannin-enriched diet. Proteome Sci. 2010;8:65. doi: 10.1186/1477-5956-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weldon CL, Mackessy SP. Biological and proteomic analysis of venom from the Puerto Rican Racer (Alsophis portoricensis: Dipsadidae) Toxicon. 2010;55(2–3):558–569. doi: 10.1016/j.toxicon.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 32.Lamy E, da Costa G, Santos R, et al. Sheep and goat saliva proteome analysis: a useful tool for ingestive behavior research? Physiol Behav. 2009;98(4):393–401. doi: 10.1016/j.physbeh.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 33.Ang CS, Binos S, Knight MI, et al. Global survey of the bovine salivary proteome: integrating multidimensional prefractionation, targeted, and glycocapture strategies. J Proteome Res. 2011;10(11):5059–5069. doi: 10.1021/pr200516d. [DOI] [PubMed] [Google Scholar]
- 34.Zamakhchari M, Wei G, Dewhirst F, et al. Identifcation of Rothia bacteria as gluten-degrading natural colonizers of the upper gastro-intestinal tract. PLoS One. 2011;6(9):e24455. doi: 10.1371/journal.pone.0024455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scannapieco FA, Torres G, Levine MJ. Salivary alpha-amylase: role in dental plaque and caries formation. Crit Rev Oral Biol Med. 1993;4(3–4):301–307. doi: 10.1177/10454411930040030701. [DOI] [PubMed] [Google Scholar]
- 36.Fried M, Abramson S, Meyer JH. Passage of salivary amylase through the stomach in humans. Dig Dis Sci. 1987;32(10):1097–1103. doi: 10.1007/BF01300195. [DOI] [PubMed] [Google Scholar]
- 37.Perry GH, Dominy NJ, Claw KG, et al. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 2007;39(10):1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mandel AL, Peyrot des Gachons C, Plank KL, Alarcon S, Breslin PA. Individual differences in AMY1 gene copy number, salivary alpha-amylase levels, and the perception of oral starch. PLoS One. 2010;5(10):e13352. doi: 10.1371/journal.pone.0013352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vitorino R, Calheiros-Lobo MJ, Duarte JA, Domingues PM, Amado FM. Peptide profle of human acquired enamel pellicle using MALDI tandem MS. J Sep Sci. 2008;31(3):523–537. doi: 10.1002/jssc.200700486. [DOI] [PubMed] [Google Scholar]
- 40.Siqueira WL, Zhang W, Helmerhorst EJ, Gygi SP, Oppenheim FG. Identifcation of protein components in in vivo human acquired enamel pellicle using LC-ESI-MS/MS. J Proteome Res. 2007;6(6):2152–2160. doi: 10.1021/pr060580k. [DOI] [PubMed] [Google Scholar]
- 41.Lendenmann U, Grogan J, Oppenheim FG. Saliva and dental pellicle – a review. Adv Dent Res. 2000;14:22–28. doi: 10.1177/08959374000140010301. [DOI] [PubMed] [Google Scholar]
- 42.Dawes C. Salivary fow patterns and the health of hard and soft oral tissues. J Am Dent Assoc. 2008;139(Suppl):18S–24S. doi: 10.14219/jada.archive.2008.0351. [DOI] [PubMed] [Google Scholar]
- 43.Holbrook WP, Furuholm J, Gudmundsson K, Theodors A, Meurman JH. Gastric refux is a signifcant causative factor of tooth erosion. J Dent Res. 2009;88(5):422–426. doi: 10.1177/0022034509336530. [DOI] [PubMed] [Google Scholar]
- 44.Slomiany BL, Murty VL, Slomiany A. Salivary lipids in health and disease. Prog Lipid Res. 1985;24(4):311–324. doi: 10.1016/0163-7827(85)90009-8. [DOI] [PubMed] [Google Scholar]
- 45•.Oudhoff MJ, Blaauboer ME, Nazmi K, et al. The role of salivary histatin and the human cathelicidin LL-37 in wound healing and innate immunity. Biol Chem. 2010;391(5):541–548. doi: 10.1515/BC.2010.057. A recent investigation on the role of saliva in wound healing. [DOI] [PubMed] [Google Scholar]
- 46.Zelles T, Purushotham KR, Macauley SP, Oxford GE, Humphreys-Beher MG. Saliva and growth factors: the fountain of youth resides in us all. J Dent Res. 1995;74(12):1826–1832. doi: 10.1177/00220345950740120301. [DOI] [PubMed] [Google Scholar]
- 47.Ruhl S, Hamberger S, Betz R, et al. Salivary proteins and cytokines in drug-induced gingival overgrowth. J Dent Res. 2004;83(4):322–326. doi: 10.1177/154405910408300410. [DOI] [PubMed] [Google Scholar]
- 48.Blicharz TM, Siqueira WL, Helmerhorst EJ, et al. Fiber-optic microsphere-based antibody array for the analysis of infammatory cytokines in saliva. Anal Chem. 2009;81(6):2106–2114. doi: 10.1021/ac802181j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levi-Montalcini R, Booker B. Excessive growth of the sympathetic ganglia evoked by a protein isolated from mouse salivary glands. Proc Natl Acad Sci USA. 1960;46(3):373–384. doi: 10.1073/pnas.46.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giannobile WV, Beikler T, Kinney JS, et al. Saliva as a diagnostic tool for periodontal disease: current state and future directions. Periodontol 2000. 2009;50:52–64. doi: 10.1111/j.1600-0757.2008.00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mese H, Matsuo R. Salivary secretion, taste and hyposalivation. J Oral Rehabil. 2007;34(10):711–723. doi: 10.1111/j.1365-2842.2007.01794.x. [DOI] [PubMed] [Google Scholar]
- 52.Reed DR, McDaniel AH. The human sweet tooth. BMC Oral Health. 2006;6(Suppl. 1):S17. doi: 10.1186/1472-6831-6-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shatzman AR, Henkin RI. Gustin concentration changes relative to salivary zinc and taste in humans. Proc Natl Acad Sci USA. 1981;78(6):3867–3871. doi: 10.1073/pnas.78.6.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kivelä J, Parkkila S, Parkkila AK, Leinonen J, Rajaniemi H. Salivary carbonic anhydrase isoenzyme VI. J Physiol. 1999;520(Pt 2):315–320. doi: 10.1111/j.1469-7793.1999.t01-1-00315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pascal C, Pate F, Cheynier V, Delsuc MA. Study of the interactions between a proline-rich protein and a favan-3-ol by NMR: residual structures in the natively unfolded protein provides anchorage points for the ligands. Biopolymers. 2009;91(9):745–756. doi: 10.1002/bip.21221. [DOI] [PubMed] [Google Scholar]
- 56.Karn RC, Orth A, Bonhomme F, Boursot P. The complex history of a gene proposed to participate in a sexual isolation mechanism in house mice. Mol Biol Evol. 2002;19(4):462–471. doi: 10.1093/oxfordjournals.molbev.a004102. [DOI] [PubMed] [Google Scholar]
- 57.Smith BA, Block ML. Male saliva cues and female social choice in Mongolian gerbils. Physiol Behav. 1991;50(2):379–384. doi: 10.1016/0031-9384(91)90081-x. [DOI] [PubMed] [Google Scholar]
- 58.Teicher MH, Blass EM. Suckling in newborn rats: eliminated by nipple lavage, reinstated by pup saliva. Science. 1976;193(4251):422–425. doi: 10.1126/science.935878. [DOI] [PubMed] [Google Scholar]
- 59.Gibbons RJ. Bacterial adhesion to oral tissues: a model for infectious diseases. J Dent Res. 1989;68(5):750–760. doi: 10.1177/00220345890680050101. [DOI] [PubMed] [Google Scholar]
- 60•.Gorr SU. Antimicrobial peptides of the oral cavity. Periodontol 2000. 2009;51:152–180. doi: 10.1111/j.1600-0757.2009.00310.x. A recent detailed review on the various antimicrobial peptides in saliva. [DOI] [PubMed] [Google Scholar]
- 61.Jang WS, Bajwa JS, Sun JN, Edgerton M. Salivary histatin 5 internalization by translocation, but not endocytosis, is required for fungicidal activity in Candida albicans. Mol Microbiol. 2010;77(2):354–370. doi: 10.1111/j.1365-2958.2010.07210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oppenheim FG, Xu T, McMillian FM, et al. Histatins, a novel family of histidine-rich proteins in human parotid secretion. Isolation, characterization, primary structure, and fungistatic effects on Candida albicans. J Biol Chem. 1988;263(16):7472–7477. [PubMed] [Google Scholar]
- 63.Varki A. Nothing in glycobiology makes sense, except in the light of evolution. Cell. 2006;126(5):841–845. doi: 10.1016/j.cell.2006.08.022. [DOI] [PubMed] [Google Scholar]
- 64.Leito JT, Ligtenberg AJ, Nazmi K, et al. A common binding motif for various bacteria of the bacteria-binding peptide SRCRP2 of DMBT1/gp-340/salivary agglutinin. Biol Chem. 2008;389(9):1193–1200. doi: 10.1515/BC.2008.135. [DOI] [PubMed] [Google Scholar]
- 65.Walz A, Odenbreit S, Stühler K, et al. Identifcation of glycoprotein receptors within the human salivary proteome for the lectin-like BabA and SabA adhesins of Helicobacter pylori by fuorescence-based 2-D bacterial overlay. Proteomics. 2009;9(6):1582–1592. doi: 10.1002/pmic.200700808. [DOI] [PubMed] [Google Scholar]
- 66.Müller R, Gröger G, Hiller KA, Schmalz G, Ruhl S. Fluorescence-based bacterial overlay method for simultaneous in situ quantifcation of surface-attached bacteria. Appl Environ Microbiol. 2007;73(8):2653–2660. doi: 10.1128/AEM.02884-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nobbs AH, Jenkinson HF, Jakubovics NS. Stick to your gums: mechanisms of oral microbial adherence. J Dent Res. 2011;90(11):1271–1278. doi: 10.1177/0022034511399096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bürgers R, Schneider-Brachert W, Reischl U, et al. Helicobacter pylori in human oral cavity and stomach. Eur J Oral Sci. 2008;116(4):297–304. doi: 10.1111/j.1600-0722.2008.00543.x. [DOI] [PubMed] [Google Scholar]
- 69.Heo SM, Haase EM, Lesse AJ, Gill SR, Scannapieco FA. Genetic relationships between respiratory pathogens isolated from dental plaque and bronchoalveolar lavage fuid from patients in the intensive care unit undergoing mechanical ventilation. Clin Infect Dis. 2008;47(12):1562–1570. doi: 10.1086/593193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Slots J, Slots H. Bacterial and viral pathogens in saliva: disease relationship and infectious risk. Periodontol 2000. 2011;55(1):48–69. doi: 10.1111/j.1600-0757.2010.00361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cisar JO, Takahashi Y, Ruhl S, Donkersloot JA, Sandberg AL. Specifc inhibitors of bacterial adhesion: observations from the study of Gram-positive bacteria that initiate bioflm formation on the tooth surface. Adv Dent Res. 1997;11(1):168–175. doi: 10.1177/08959374970110010801. [DOI] [PubMed] [Google Scholar]
- 72.Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7(4):255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 73.Ruhl S, Sandberg AL, Cisar JO. Salivary receptors for the proline-rich protein-binding and lectin-like adhesins of oral actinomyces and streptococci. J Dent Res. 2004;83(6):505–510. doi: 10.1177/154405910408300614. [DOI] [PubMed] [Google Scholar]
- 74.Ruhl S, Sandberg AL, Cole MF, Cisar JO. Recognition of immunoglobulin A1 by oral actinomyces and streptococcal lectins. Infect Immun. 1996;64(12):5421–5424. doi: 10.1128/iai.64.12.5421-5424.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Murray PA, Prakobphol A, Lee T, Hoover CI, Fisher SJ. Adherence of oral streptococci to salivary glycoproteins. Infect Immun. 1992;60(1):31–38. doi: 10.1128/iai.60.1.31-38.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Takamatsu D, Bensing BA, Prakobphol A, Fisher SJ, Sullam PM. Binding of the streptococcal surface glycoproteins GspB and Hsa to human salivary proteins. Infect Immun. 2006;74(3):1933–1940. doi: 10.1128/IAI.74.3.1933-1940.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mishra A, Devarajan B, Reardon ME, et al. Two autonomous structural modules in the fmbrial shaft adhesin FimA mediate Actinomyces interactions with streptococci and host cells during oral bioflm development. Mol Microbiol. 2011;81(5):1205–1220. doi: 10.1111/j.1365-2958.2011.07745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rogers JD, Palmer RJ, Jr, Kolenbrander PE, Scannapieco FA. Role of Streptococcus gordonii amylase-binding protein A in adhesion to hydroxyapatite, starch metabolism, and bioflm formation. Infect Immun. 2001;69(11):7046–7056. doi: 10.1128/IAI.69.11.7046-7056.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Loimaranta V, Hytönen J, Pulliainen AT, et al. Leucine-rich repeats of bacterial surface proteins serve as common pattern recognition motifs of human scavenger receptor gp340. J Biol Chem. 2009;284(28):18614–18623. doi: 10.1074/jbc.M900581200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gibbons RJ, Hay DI, Schlesinger DH. Delineation of a segment of adsorbed salivary acidic proline-rich proteins which promotes adhesion of Streptococcus gordonii to apatitic surfaces. Infect Immun. 1991;59(9):2948–2954. doi: 10.1128/iai.59.9.2948-2954.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Clark WB, Beem JE, Nesbitt WE, et al. Pellicle receptors for Actinomyces viscosus type 1 fmbriae in vitro. Infect Immun. 1989;57(10):3003–3008. doi: 10.1128/iai.57.10.3003-3008.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu C, Mishra A, Yang J, et al. Dual function of a tip fimbrillin of Actinomyces in fmbrial assembly and receptor binding. J Bacteriol. 2011;193(13):3197–3206. doi: 10.1128/JB.00173-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.De Jong MH, Van der Hoeven JS. The growth of oral bacteria on saliva. J Dent Res. 1987;66(2):498–505. doi: 10.1177/00220345870660021901. [DOI] [PubMed] [Google Scholar]
- 84.Kolenbrander PE. Multispecies communities: interspecies interactions infuence growth on saliva as sole nutritional source. Int J Oral Sci. 2011;3(2):49–54. doi: 10.4248/IJOS11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rohmer L, Hocquet D, Miller SI. Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol. 2011;19(7):341–348. doi: 10.1016/j.tim.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86•.Ragunath C, Manuel SG, Venkataraman V, et al. Probing the role of aromatic residues at the secondary saccharide-binding sites of human salivary alpha-amylase in substrate hydrolysis and bacterial binding. J Mol Biol. 2008;384(5):1232–1248. doi: 10.1016/j.jmb.2008.09.089. A meaningful structural study on the functions of salivary a-amylase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ruhl S, Rayment SA, Schmalz G, Hiller KA, Troxler RF. Proteins in whole saliva during the frst year of infancy. J Dent Res. 2005;84(1):29–34. doi: 10.1177/154405910508400104. [DOI] [PubMed] [Google Scholar]
- 88•.Castagnola M, Inzitari R, Fanali C, et al. The surprising composition of the salivary proteome of preterm human newborn. Mol Cell Proteomics. 2011;10(1) doi: 10.1074/mcp.M110.003467. M110.003467. A recent study on the developmental aspects of salivary proteins (also see reference [87]). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Morzel M, Palicki O, Chabanet C, et al. Saliva electrophoretic protein profles in infants: changes with age and impact of teeth eruption and diet transition. Arch Oral Biol. 2011;56(7):634–642. doi: 10.1016/j.archoralbio.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 90•.Ambatipudi KS, Lu B, Hagen FK, Melvin JE, Yates JR. Quantitative analysis of age specifc variation in the abundance of human female parotid salivary proteins. J Proteome Res. 2009;8(11):5093–5102. doi: 10.1021/pr900478h. A recent study on age-related changes in the salivary proteome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ayad M, Van Wuyckhuyse BC, Minaguchi K, et al. The association of basic proline-rich peptides from human parotid gland secretions with caries experience. J Dent Res. 2000;79(4):976–982. doi: 10.1177/00220345000790041401. [DOI] [PubMed] [Google Scholar]
- 92.Young A, Rykke M, Rölla G. Quantitative and qualitative analyses of human salivary micelle-like globules. Acta Odontol Scand. 1999;57(2):105–110. doi: 10.1080/000163599428995. [DOI] [PubMed] [Google Scholar]
- 93•.Gonzalez-Begne M, Lu B, Han X, et al. Proteomic analysis of human parotid gland exosomes by multidimensional protein identifcation technology (MudPIT) J Proteome Res. 2009;8(3):1304–1314. doi: 10.1021/pr800658c. A proteomic analysis of salivary exosomes (also see reference [94]). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Palanisamy V, Sharma S, Deshpande A, et al. Nanostructural and transcriptomic analyses of human saliva derived exosomes. PLoS One. 2010;5(1):e8577. doi: 10.1371/journal.pone.0008577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ruhl S, Berlenbach P, Langenfelder S, et al. Integrity of proteins in human saliva after sterilization by gamma-irradiation. Appl Environ Microbiol. 2010;77(3):749–755. doi: 10.1128/AEM.01374-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schipper RG, Silletti E, Vingerhoeds MH. Saliva as research material: biochemical, physicochemical and practical aspects. Arch Oral Biol. 2007;52(12):1114–1135. doi: 10.1016/j.archoralbio.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 97.Thomadaki K, Helmerhorst EJ, Tian N, et al. Whole-saliva proteolysis and its impact on salivary diagnostics. J Dent Res. 2011;90(11):1325–1330. doi: 10.1177/0022034511420721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zakhary GM, Clark RM, Bidichandani SI, et al. Acidic proline-rich protein Db and caries in young children. J Dent Res. 2007;86(12):1176–1180. doi: 10.1177/154405910708601207. [DOI] [PubMed] [Google Scholar]
- 99.Denny PC, Denny PA, Takashima J, Galligan J, Navazesh M. A novel caries risk test. Ann NY Acad Sci. 2007;1098:204–215. doi: 10.1196/annals.1384.009. [DOI] [PubMed] [Google Scholar]
- 100.Zehetbauer S, Wojahn T, Hiller KA, Schmalz G, Ruhl S. Resemblance of salivary protein profles between children with early childhood caries and caries-free controls. Eur J Oral Sci. 2009;117(4):369–373. doi: 10.1111/j.1600-0722.2009.00641.x. [DOI] [PubMed] [Google Scholar]
- 101.Krief G, Deutsch O, Gariba S, et al. Improved visualization of low abundance oral fluid proteins after triple depletion of alpha amylase, albumin and IgG. Oral Dis. 2011;17(1):45–52. doi: 10.1111/j.1601-0825.2010.01700.x. [DOI] [PubMed] [Google Scholar]
- 102.Bandhakavi S, Stone MD, Onsongo G, Van Riper SK, Griffn TJ. A dynamic range compression and three-dimensional peptide fractionation analysis platform expands proteome coverage and the diagnostic potential of whole saliva. J Proteome Res. 2009;8(12):5590–5600. doi: 10.1021/pr900675w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem. 2007;389(4):1017–1031. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- 104.Siqueira WL, Dawes C. The salivary proteome: challenges and perspectives. Proteomics Clin Appl. 2011 doi: 10.1002/prca.201100046. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 105.Hardt M, Witkowska HE, Webb S, et al. Assessing the effects of diurnal variation on the composition of human parotid saliva: quantitative analysis of native peptides using iTRAQ reagents. Anal Chem. 2005;77(15):4947–4954. doi: 10.1021/ac050161r. [DOI] [PubMed] [Google Scholar]
- 106.Penque D. Two-dimensional gel electrophoresis and mass spectrometry for biomarker discovery. Proteomics Clin Appl. 2009;3:155–172. doi: 10.1002/prca.200800025. [DOI] [PubMed] [Google Scholar]
- 107.Hu S, Jiang J, Wong DT. Proteomic analysis of saliva: 2D gel electrophoresis, LC-MS/MS, and Western blotting. Methods Mol Biol. 2010;666:31–41. doi: 10.1007/978-1-60761-820-1_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108•.Sondej M, Denny PA, Xie Y, et al. Glycoprofling of the human salivary proteome. Clin Proteomics. 2009;5(1):52–68. doi: 10.1007/s12014-008-9021-0. A pilot study reporting the first characterization of the salivary 2D glycoproteome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Drake PM, Cho W, Li B, et al. Sweetening the pot: adding glycosylation to the biomarker discovery equation. Clin Chem. 2010;56(2):223–236. doi: 10.1373/clinchem.2009.136333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zaia J. Mass spectrometry and glycomics. OMICS. 2010;14(4):401–418. doi: 10.1089/omi.2009.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111•.Stone MD, Chen XB, McGowan T, et al. Large-scale phosphoproteomics analysis of whole saliva reveals a distinct phosphorylation pattern. J Proteome Res. 2011;10(4):1728–1736. doi: 10.1021/pr1010247. A most recent study describing the salivary phosphoproteome (also see reference [112]). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Salih E, Siqueira WL, Helmerhorst EJ, Oppenheim FG. Large-scale phosphoproteome of human whole saliva using disulfde-thiol interchange covalent chromatography and mass spectrometry. Anal Biochem. 2010;407(1):19–33. doi: 10.1016/j.ab.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Peterson J, Garges S, Giovanni M, et al. The NIH human microbiome project. Genome Res. 2009;19(12):2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Messana I, Inzitari R, Fanali C, Cabras T, Castagnola M. Facts and artifacts in proteomics of body fuids. What proteomics of saliva is telling us? J Sep Sci. 2008;31(11):1948–1963. doi: 10.1002/jssc.200800100. [DOI] [PubMed] [Google Scholar]
- 115.Huq NL, Cross KJ, Ung M, et al. A review of the salivary proteome and peptidome and saliva-derived peptide therapeutics. Int J Peptide Res Ther. 2007;13(4):547–564. [Google Scholar]
- 116.Aguirre A, Testa-Weintraub LA, Banderas JA, et al. Sialochemistry: a diagnostic tool? Crit Rev Oral Biol Med. 1993;4(3–4):343–350. doi: 10.1177/10454411930040031201. [DOI] [PubMed] [Google Scholar]
Websites
- 201.NIDCR. Saliva and Salivary Gland Disorders. www.nidcr.nih.gov/OralHealth/Topics/Saliva/
- 202.HOMD. Human Oral Microbiome Database. www.homd.org.


