Abstract
Despite a growing number of reports of gene expression analysis from blood-derived RNA sources, there have been few systematic comparisons of various RNA sources in transcriptomic analysis or for biomarker discovery in the context of cardiovascular disease (CVD). As a pilot study of the Systems Approach to Biomarker Research (SABRe) in CVD Initiative, this investigation used Affymetrix Exon arrays to characterize gene expression of three blood-derived RNA sources: lymphoblastoid cell lines (LCL), whole blood using PAXgene tubes (PAX), and peripheral blood mononuclear cells (PBMC). Their performance was compared in relation to identifying transcript associations with sex and CVD risk factors, such as age, high-density lipoprotein, and smoking status, and the differential blood cell count. We also identified a set of exons that vary substantially between participants, but consistently in each RNA source. Such exons are thus stable phenotypes of the participant and may potentially become useful fingerprinting biomarkers. In agreement with previous studies, we found that each of the RNA sources is distinct. Unlike PAX and PBMC, LCL gene expression showed little association with the differential blood count. LCL, however, was able to detect two genes related to smoking status. PAX and PBMC identified Y-chromosome probe sets similarly and slightly better than LCL.
Keywords: microarray, system biology, biomarker discovery, fingerprinting genes, data normalization, X-linked expression, cardiovascular disease
owing to accessibility, practicality, and minimal invasiveness, blood-derived RNA sources, such as lymphoblastoid cell lines (LCL), whole blood cells (PAXgene tubes; PAX), and peripheral blood mononuclear cells (PBMC), have been widely used in gene expression studies for biomarker identification (30, 32) and pathway profiling (10). These RNA sources have been useful for identifying biomarker signatures of lupus (9), cancer (75), and bacterial infection (14). This makes blood-derived RNA valuable even when studying diseases involving remote target tissues (66).
Each of these blood-derived RNA sources is known to have inherent characteristics that will result in a unique gene expression profile (25). PAX samples, derived from whole blood, capture RNA profiles of all cell types in whole blood, including erythrocytes, granulocytes (neutrophils, eosinophils, basophils), lymphocytes, monocytes, and platelets. PBMC samples, derived from a Ficoll-filtered lymphocyte and monocyte subset, are largely devoid of granulocytes, platelets, and reticulocytes. LCL samples, derived from lymphoblastoid cell lines [i.e., B cells infected and immortalized by Epstein-Barr virus (EBV), stored frozen and regrown several years after sample collection], represent RNA from a single cell type. In addition, gene expression differences may also arise from varying RNA isolation protocols and sample handling (6, 25, 50, 80).
Despite a growing number of reports of gene expression analysis from these RNA sources, there have been few systematic comparisons of their suitability for biomarker discovery, especially in the context of cardiovascular disease (CVD). Previous studies (50, 67) have examined gene signature differences among these RNA sources. One study examined the expression profile differences among the sources with respect to age and sex (80) in a spotted-array platform. However, none of these studies has a balanced experimental design that can eliminate certain statistical biases in the analysis.
Therefore, the primary goal of this study, which was undertaken as a feasibility study for the Systems Approach to Biomarker Research (SABRe) in CVD Initiative, was to characterize three blood-derived RNA sources, LCL, PAX, and PBMC, for quantity and quality of RNA, and expression properties using an exon-array platform in a balanced experimental design. The performance of these sources was assessed with regard to identification of differential expression of Y-chromosome probe sets with sex, which is a major CVD risk factor (35). Beyond sex associations, associations of expression with other risk factors, such as age, smoking status, and high-density lipoprotein (HDL) cholesterol level, were also explored. In addition, complete blood counts (CBC) (19) obtained at the time of blood collection allowed tests of association of expression with blood cell proportions.
The balanced experimental design permitted a secondary goal of identifying genes whose expression levels are stable across RNA sources within individuals yet highly variable across the population. Such markers may be useful in fingerprinting the samples for forensic identification or in resolving sample mix-ups, which is a common problem in gene expression studies. Last, we also identified genes that are consistently expressed across multiple RNA sources and across individuals, making them suitable for use as calibration markers.
MATERIALS AND METHODS
Study Samples
The first cohort of the Framingham Heart Study (FHS) included 5,209 men and women between 30 and 60 yr of age who enrolled in 1948 and have undergone biennial examinations (24, 54, 55). In 1971, 5,124 children (spouses of children) of the original cohort were recruited to the Framingham Offspring Study (27). In 2002, 4,095 participants were included in the third generation cohort (71). Blood samples were obtained from 50 consecutive participants from the third generation cohort who attended their second examination cycle clinic visit in January 2009. Immortalized cell lines for these same participants were prepared from samples taken during their initial clinic visit 1, ∼5 years earlier. To investigate for possible sample storage effects, we obtained 24 whole blood samples from the offspring cohort which were sampled in 2005–2006 and stored for 3–4 yr at −80°C prior to RNA preparation. Protocols for participant examinations and collection of genetic materials, including immortalized cell lines, were approved by the Boston University Medical Center Institutional Review Board.
Individual Trait Data
Current smoking status (defined as regularly smoking one or more cigarettes per day during the past year), systolic and diastolic blood pressure (seated, measured twice in the left arm by a physician), total and HDL cholesterol levels, fasting blood glucose level, and body mass index (BMI, weight in kg divided by height in m2) were obtained at the clinic visit. Hypertension was defined a systolic blood pressure of at least 140 mmHg or a diastolic blood pressure of at least 90 mmHg or current use of antihypertensive medication. Diabetes was defined as fasting blood glucose of at least 126 mg/dl or current use of insulin or an oral hypoglycemic medication. CBCs were obtained on samples collected from the third generation at the second examination clinic visit using a Beckman Coulter Counter (Beckman Coulter, Brea, CA).
RNA Isolation and Target Labeling
The three RNA sources collected on each of the 50 consecutive individuals included PAX and PBMC (obtained at the second clinic examination of the third generation cohort), and LCL, obtained at the first clinic examination ∼5 yr earlier.
PAXgene samples.
Blood Specimens (2.5 ml) collected in PAXgene tubes from each participant were incubated at room temperature for 4 h for RNA stabilization and then stored at −80°C. RNA was extracted from whole blood using the PAXgene Blood RNA System Kit following the manufacturer's guidelines (62). In brief, samples were removed from −80°C and incubated at room temperature for 2 h to ensure complete lysis. Following lysis, the tubes were centrifuged for 10 min at 5,000 g, the supernatant was discarded, and 500 μl of RNase-free water was added to the pellet. The tube was vortexed thoroughly to resuspend the pellet, centrifuged for 10 min at 5,000 g, and the entire supernatant was discarded. The pellet was resuspended in 360 μl of buffer BR1 by vortexing and RNA was further purified with on-column DNase digestion. Quality of the purified RNA was verified on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA); RNA concentrations were determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). We amplified 50 ng of total RNA using NuGEN's WT-Ovation Pico RNA Amplification System and labeled it with FL-Ovation cDNA Biotin Module V2 (NuGEN, San Carlos, CA) according to the protocol provided by the supplier.
PBMC samples.
Venous blood (8 ml) from each participant was collected into Vacutainer cell preparation tubes containing sodium citrate and Ficoll (Becton Dickinson, Franklin Lakes, NJ). Purified PBMC suspensions were resuspended in RLT buffer (700–1,000 μl per 107 cells), passed through Qiashredder columns (Qiagen, Valencia, CA), and then stored at −80°C. Total RNA (50 ng) was amplified and labeled using the Affymetrix Whole-Transcript (WT) Sense Target Labeling Protocol without rRNA reduction. Complementary DNA (cDNA) was regenerated through a random-primed reverse transcription using a dNTP mix containing dUTP. The RNA was hydrolyzed with RNase H, and the cDNA was purified. The cDNA was then fragmented by incubation with a mixture of UDG and APE1 with restriction endonucleases and end-labeled via a terminal transferase reaction incorporating a biotinylated dideoxynucleotide.
LCL samples.
Total RNA was extracted from pelleted lymphoblastoid cells of each participant using the Qiagen RNeasy Plus extraction kit according to the manufacturer's protocol (61). The process included a column-based elimination of genomic DNA. Total RNA (50 ng) was amplified and labeled using the Affymetrix Whole-Transcript (WT) Sense Target Labeling Protocol without rRNA reduction. cDNA generation, RNA hydrolysis, fragmentation, and labeling were carried out with the same protocol as described above for PBMC samples.
Microarray Hybridization
We added 5.5 μg of the fragmented, biotinylated cDNA prepared from each of the whole blood, PBMC, and cell line samples to a hybridization cocktail, loaded it on an Affymetrix Human Exon 1.0 ST GeneChip, which contains ∼1.4 million probe sets in total, and hybridized it for 16 h at 45°C and 60 rpm (3). Following hybridization, the array was washed and stained according to the manufacturer's protocol. The stained array was scanned at 532 nm using an Affymetrix GeneChip Scanner 3000, generating CEL files for each array. Aside from 10 samples (4 LCL, 3 PAX, 3 PBMC) with insufficient RNA, all samples were chipped in two batches.
Expression Data Analysis
We applied the robust multichip average (RMA) method (34) to normalize expression values for the remaining 140 samples using the Affymetrix Power Tool (APT) (4) version 1.12.0. We used the following metrics (2) to determine the quality of the hybridized samples: all_probeset_mean, all_probeset_rle_mean, pm_mean, and pos_vs_neg_auc. Two LCL and five PBMC samples failed on these metrics, and were excluded, along with the other samples from the same individuals. After inspecting Y-chromosome probe sets for agreement across the samples from each individual, we additionally removed two PAX and two PBMC samples due to apparent mislabeling. This left 35 individuals with satisfactory results for all three sample types, giving 105 samples. We repeated probe set-level RMA normalization on these samples, retaining only core-level, RefSeq-annotated probe sets, giving 287,329 probe sets in all, representing 18,282 distinct genes. We also performed transcript cluster-level normalization at the “core level,” giving 17,330 RefSeq-annotated genes. The gene counts and annotations are based on Affymetrix NetAffx release 31 (1).
Quality control.
Having discarded samples from participants 21, 23, and 35 due to insufficient RNA, we normalized the CEL files of the remaining 140 chipable samples with the RMA method using the APT software in three runs, one per RNA source. The quality control parameters of these samples are shown in Supplementary Table S13.1 Since participant 45 did not yield sufficient RNA in its LCL sample, its PAX and PBMC samples were discarded. PBMC samples of participants 2 and 8 and PAX samples of participants 43 and 44 were found mislabeled by the inspection of Y chromosome probe sets. These four samples were discarded. LCL sample of participant 18 was identical to that of participant 17, and LCL sample of participant 42 was identical to that of participant 43. Only LCL sample from participant 18 could be restored. We select only participants with samples having all probe set RLE mean of at most 0.75. This step removed samples from participants 37, 38, 40, 41, 46, and 47. We renormalized the 105 CEL files altogether from the remaining 35 patients.
Postnormalization methods.
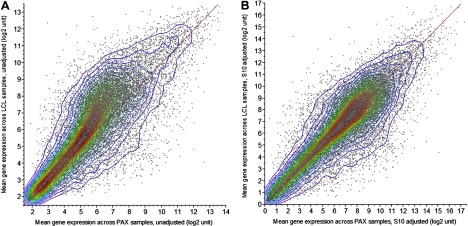
To address an apparent systematic bias in gene expression values between PAX results and either PBMC or LCL (Fig. 1A), likely arising from the differences in labeling protocol, we further normalized the data with the S10 postnormalization procedure (52), a variance-stabilizing and quantile-normalizing transform. In addition to S10, we also considered quantile postnormalization (QPN), which is a quantile-normalization transform. We will choose the transform that minimizing variance across participants.
Fig. 1.
Comparison of robust multichip average (RMA)-normalized gene expression mean values across 35 samples for lymphoblastoid cell lines (LCL) vs. PAXgene tubes (PAX). A: before S10 postnormalization; B: after. RMA normalization alone failed to normalize gene expression quantiles between LCL and PAX samples. This skew also appears in peripheral blood mononuclear cells (PBMC) vs. PAX. Diagonal lines are identity lines.
Using QPN, we computed the mean value of each probe set per RNA source, yielding three sets of mean values. We chose PBMC as the reference distribution because its mean values correlate well with those of LCL and PAX. Such selection is aimed to minimize drastic quantile correction. After the mean value of each gene for LCL and PAX was quantile-normalized against that of PBMC, its individual expression values were shifted by the difference between the original and normalized mean values.
For S10, we computed the anti-log of the RMA expression values, calculated the normal quantiles, then computed mean and standard deviations across samples, and then fit a spline to the standard deviation as a function of the mean. A variance-stabilizing transform function is computed from this smooth function, and then applied to the data. Finally, the log base 2 was computed on the normalized data.
After postnormalization, the QPN-transformed mean densities were identical to that of PBMC, while those S10-transformed were diagonally aligned (Fig. 1B). Using two-way ANOVA with RNA source and participant as fixed factors, we determined to use S10 because it minimizes variance across participants while normalizing the quantiles.
Statistical Methods
All statistical analyses were performed both at the exon/probe-set level and at the gene/transcript cluster level using R (63) version 2.11.1 or JMP 9 (SAS, Cary, NC). The MSCL Analyst's Toolbox (freely available at http://abs.cit.nih.gov/MSCLtoolbox/) was used for initial exploratory analyses and feature discovery. We discarded 92,157 probe sets where the intensity of 52 or fewer (≤50%) of the 105 samples was significantly above background [i.e., with detected-above-background (DABG) P values ≤ 0.05], leaving 195,172 for subsequent analysis. DABG filtering was applied to exon-level data and not to gene-level data. In all cases, we calculated the false discovery rates (FDR) with Benjamini and Hochberg's method (11).
To determine the separation of expression patterns across cell types, we performed principal component analysis (PCA) on all 105 arrays on DABG-filtered exon-level data. The PCA was performed using the “prcomp” function (76) of R on centered, but unscaled data.
To determine differentially expressed genes between each pair of RNA sources, we used a two-way ANOVA with fixed factors for sample type (n = 3) and participant (n = 35). We counted probe sets and transcript clusters where mean expression differences among RNA sources were declared significant based on the sample type F-statistic (FDR ≤ 0.05). Comparison of expression between pairs of RNA sources used a post hoc t-test statistic with the same FDR threshold. To identify genes that were uniquely overexpressed in each RNA source compared with the other two sources, we computed the minimum fold-change for each paired comparison and required this to be greater than eightfold.
Stable fingerprinting genes/exons are those with expression levels strongly related to participant, irrespective of RNA source. Using the same two-way ANOVA, we selected such genes/exons with a significant participant effect at FDR ≤ 0.05. A subset of the most significant exons having participant-effect standard deviations of at least twofold change were clustered using Ward's method (53) on their expression level, after subtracting the sample-type effect. Conversely, housekeeping or calibration genes were selected as those with the smallest variation across sample type or across participant. We selected such genes with 1) P value for sample type >0.2, 2) P value for participant >0.2, 3) standard deviation of the within-participant effects less than twofold change, and 4) mean expression level greater than background threshold (4 RMA units).
Transcript profile associations with age, sex, and selected CVD risk factors.
To discover genes that are expressed differentially in men vs. women, we performed a two-sample t-test with unequal variance assumption for each RNA source. We required the exons and genes to pass the FDR ≤ 0.05 threshold.
For trait-based biomarker discovery, we regressed the RMA expression of each gene for each RNA source against the trait, adjusting for age and sex, using the linear mixed-effects model implemented in the R package “lmer” (59). Owing to small sample size, we relaxed the multiple-testing penalty by setting a P value cutoff of 0.05 and selecting only genes with a significant association in more than one RNA source. For confirmatory testing of previously identified biomarkers using our data, we regressed the RMA expression of each exon against the trait, adjusting for age and sex using P value ≤ 0.05.
Differential blood count analysis.
Because blood is a complex tissue made up of varying proportions of several cell types, each with a distinct expression profile, expression of some genes would be expected to vary proportionally to these components. To find such genes, we used a multiple regression model with factors for the absolute count per μl of each measured component: red blood cells, platelets, neutrophils, lymphocytes, monocytes, eosinophils, and basophils. We collected the significance levels (P values) for each factor and performed FDR adjustment as above. We also relaxed the FDR cutoff to 0.2, since few results were obtained at lower levels. The test was repeated for each RNA source.
Gene ontology analysis.
We performed gene ontology (GO) (7) enrichment analysis of the differentially expressed genes between each pair of RNA sources based on exon-level or gene-level data using GOrilla (26). This method determines whether the number of differentially expressed genes having a particular GO assignment is significantly higher than would be expected by chance, given the total number of genes, the total number having that assignment, and the number of differentially expressed genes, overall. We removed unannotated genes and required the remaining genes to pass 1) an FDR ≤ 0.05 threshold and 2) at least a fourfold difference in expression. We ran GOrilla using genes in the Affymetrix NetAffx core-level annotation version 31 for the Human Exon 1.0 ST GeneChip as the background set of genes.
RESULTS
RNA Source Comparison
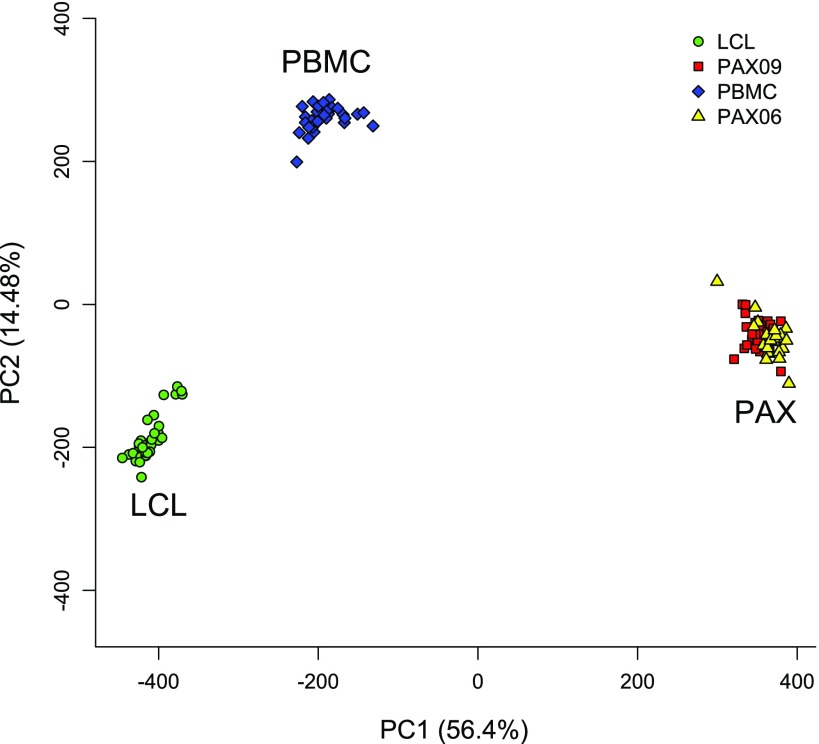
The clinical characteristics of the study sample are provided in Table 1. PCA revealed striking differences in expression patterns of the three RNA sources (Fig. 2). The first two principal components, attributable to RNA source differences, accounted for 70.88% of overall variance in expression. The PCA plot of the 24 older PAX samples coincide with the newer PAX samples. Therefore, the striking differences among the three RNA sources are much larger than any possible sample storage or aging effects.
Table 1.
Characteristics of the study participants
| Characteristic | Value |
|---|---|
| Age, yr | 51 ± 7.3 (27–59) |
| Sex | 21 males/14 females |
| Body mass index, kg/m2 | 30.2 ± 5.8 (20.5–42.0) |
| Systolic blood pressure, mmHg | 120.5 ± 13.5 (96–153) |
| Diastolic blood pressure, mmHg | 76.9 ± 7.5 (59–91) |
| Total cholesterol, mg/dl | 202.7 ± 39.1 (145–323) |
| High density lipoprotein, mg/dl | 56.6 ± 18.1 (27–112) |
| Fasting blood glucose, mg/dl | 96.6 ± 7.6 (81–108) |
| Smoking status | 6 males/2 females |
| Hypertension status | 4 males/3 females |
| Lipid medication use | 5 males/4 females |
| White blood cell count (× 103/μl) | 6.22 ± 1.48 (3.5–9.7) |
| Red blood cell count (× 106/μl) | 4.53 ± 0.47 (3.72–5.47) |
| Hemoglobin, g/dl | 14.06 ± 1.46 (11.3–16.8) |
| Hematocrit, % | 40.90 ± 4.25 (32.6–49.0) |
| Mean corpuscular volume, fl | 90.37 ± 3.86 (83.4–100.2) |
| Mean corpuscular hemoglobin, pg | 31.07 ± 1.94 (27.3–37.3) |
| Mean corpuscular hemoglobin concentration, g/dl | 34.37 ± 1.19 (32.7–39.3) |
| Red blood cell distribution width, % | 12.46 ± 0.60 (11.4–14.1) |
| Platelet count | 232.85 ± 63.32 (48–379) |
| Mean platelet volume, fl | 8.65 ± 0.90 (6.8–10.9) |
| Neutrophil, % | 57.4% ± 8.17 (35.3–72.2) |
| Lymphocyte, % | 30.39% ± 7.48 (19.2–50.8) |
| Monocyte, % | 8.07% ± 2.32 (5.2–14.9) |
| Eosinophil, % | 3.23% ± 1.70 (1.2–8.6) |
| Basophil, % | 0.87% ± 0.36 (0.3–1.7) |
| Neutrophil count (× 103/μl) | 3.63 ± 1.22 (1.3–6.9) |
| Lymphocyte count (× 103/μl) | 1.83 ± 0.42 (1.1–3.2) |
| Monocyte count (× 103/μl) | 0.50 ± 0.16 (0.2–0.8) |
| Eosinophil count (× 103/μl) | 0.21 ± 0.13 (0.1–0.7) |
| Basophil count (× 103/μl) | 0.05 ± 0.05 (0.0–0.1) |
Values are means ± SD (minimum–maximum); n =35.
Fig. 2.
Principal component plot of the 3 RNA sources at the exon level. The RNA source difference explains 70.88% of total variation. The stored samples coincide with the newer ones, indicating a lack of storage effect. PAX06 indicates PAXgene samples assayed in 2005–2006, while PAX09 those assayed in 2009.
About 90% of probe sets (176,641 of the 195,172 expressed above background) were found to differ across RNA sources (FDR ≤ 0.05). Even when an exceedingly low FDR cutoff (≤1×10−8) was set, more than half the exon probe sets (105,709) differed significantly across RNA sources (Table 2). A similar percentage showed expression differences at the gene level. Most of these expression differences were seen in the PAX vs. LCL and PAX vs. PBMC comparisons. Genes that are uniquely overexpressed by ≥8-fold in each RNA source compared with the other two sources were ranked by level of overexpression, and the topmost are presented in Tables 3–5. The corresponding tables based on exon-level analysis are given in Supplementary Tables S1–S3.
Table 2.
Number of probe sets and transcripts with expression differences among RNA sources
| Exon Level | Gene Level | |
|---|---|---|
| Differed among 3 RNA sources | 105,709 (14,811)* | 10,253 |
| PAX vs. LCL | 97,219 (14,728) | 9,323 |
| LCL vs. PBMC | 51,922 (7,427) | 5,708 |
| PBMC vs. PAX | 86,829 (14,480) | 8,169 |
| Expressed ≥ 8-fold over other 2 RNA sources | ||
| PAX | 4,859 (2,512) | 119 |
| LCL | 2,436 (495) | 188 |
| PBMC | 883 (341) | 19 |
| Expressed ≥ 4-fold over other 2 RNA sources | ||
| PAX | 12,970 (5,339) | 336 |
| LCL | 6,312 (1,173) | 426 |
| PBMC | 2,859 (831) | 113 |
Results based on S10 postnormalization, application of detected-above-background filtering and significance at false discovery rate (FDR) ≤10−8 .
Values in parentheses are the number of genes that include the detected probe sets.
PAX, PAXgene tubes; LCL, lymphoblastoid cell lines; PBMC, peripheral blood mononuclear cells.
Table 3.
Genes overexpressed in PAX
| Ranka | Transcript Cluster ID | Gene Symbol | Chr. | Description | Mean PAXb | PAX/LCLc | PAX/PBMCc | Min. FCd |
|---|---|---|---|---|---|---|---|---|
| 1 | 2787958 | GYPB | 4 | glycophorin B (MNS blood group)*(13) | 10.5 | 319 | 218 | 218 |
| 2 | 2907173 | HCRP1 | 6 | hepatocellular carcinoma-related HCRP1 | 11.1 | 95 | 102 | 95 |
| 3 | 2648677 | MME | 3 | membrane metallo-endopeptidase | 10.2 | 97 | 65 | 65 |
| 4 | 3453732 | TUBA1B | 12 | tubulin, alpha 1b | 9.1 | 57 | 174 | 57 |
| 5 | 4009849 | ALAS2 | X | aminolevulinate, delta-, synthase 2*(23) | 12.4 | 118 | 56 | 56 |
| 6 | 3037100 | RSPH10B | 7 | radial spoke head 10 homolog B (Chlamydomonas) | 7.9 | 60 | 56 | 56 |
| 7 | 3996598 | NCRNA00204 | X | nonprotein coding RNA 204 | 8.2 | 136 | 48 | 48 |
| 8 | 2765935 | GAFA3 | 4 | FGF-2 activity-associated protein 3 | 9.6 | 48 | 47 | 47 |
| 9 | 3906007 | PRO0628 | 20 | uncharacterized protein PRO0628-like | 7.7 | 46 | 49 | 46 |
| 10 | 4010152 | LOC442454 | X | ubiquinol-cytochrome c reductase binding protein pseudogene | 7.5 | 59 | 45 | 45 |
| 11 | 3679643 | C16orf72 | 16 | chromosome 16 open reading frame 72 | 9.7 | 46 | 42 | 42 |
| 12 | 3617458 | GOLGA8A | 15 | golgin A8 family, member A | 8.5 | 41 | 47 | 41 |
| 13 | 3489673 | KCNRG | 13 | potassium channel regulator | 10.8 | 63 | 41 | 41 |
| 14 | 3399623 | THYN1 | 11 | thymocyte nuclear protein 1 | 12.9 | 53 | 39 | 39 |
| 15 | 3421118 | RAP1B | 12 | RAP1B, member of RAS oncogene family | 10.5 | 195 | 37 | 37 |
| 16 | 2375338 | OCR1 | 1 | ovarian cancer-related protein 1 | 9.1 | 67 | 36 | 36 |
| 17 | 2325877 | RHD | 1 | Rh blood group, D antigen*(8) | 6.1 | 45 | 36 | 36 |
| 18 | 4037708 | MIR1974 | M | microRNA 1974 | 13.9 | 48 | 35 | 35 |
| 19 | 3886765 | PI3 | 20 | peptidase inhibitor 3, skin-derived | 10.9 | 35 | 36 | 35 |
| 20 | 3416483 | HNRNPA1 | 12 | heterogeneous nuclear ribonucleoprotein A1 | 10.8 | 33 | 47 | 33 |
| 21 | 3090006 | SLC25A37 | 8 | solute carrier family 25, member 37 | 13.9 | 41 | 31 | 31 |
| 22 | 3498476 | LOC100132099 | 13 | FRSS1829 | 11.0 | 72 | 31 | 31 |
| 23 | 2701294 | TMEM14E | 3 | transmembrane protein 14E | 9.7 | 41 | 29 | 29 |
| 24 | 3823304 | CYP4F3 | 19 | cytochrome P450, family 4, subfamily F, polypeptide 3 | 7.8 | 32 | 29 | 29 |
| 25 | 3360401 | HBB | 11 | hemoglobin, beta*(31) | 15.1 | 1651 | 27 | 27 |
| 26 | 2923270 | PLN | 6 | phospholamban | 8.0 | 26 | 25 | 25 |
| 27 | 2527580 | CXCR2 | 2 | chemokine (C-X-C motif) receptor 2†(44) | 12.0 | 131 | 24 | 24 |
| 28 | 3830484 | FFAR2 | 19 | free fatty acid receptor 2 | 9.1 | 33 | 23 | 23 |
| 29 | 3918696 | SON | 21 | SON DNA binding protein | 13.8 | 21 | 26 | 21 |
| 30 | 3920850 | KCNJ15 | 21 | potassium inwardly-rectifying channel, subfamily J, member 15 | 11.0 | 66 | 19 | 19 |
| Genes known to be expressed in erythrocytes or neutrophils | ||||||||
| 36 | 2496907 | IL1R2 | 2 | interleukin 1 receptor, type II†(15) | 10.2 | 26 | 17 | 17 |
| 47 | 3657253 | AHSP | 16 | alpha hemoglobin stabilizing protein* | 9.0 | 26 | 16 | 16 |
| 52 | 3907190 | SLPI | 20 | secretory leukocyte peptidase inhibitor†(37) | 7.5 | 25 | 14 | 14 |
| 58 | 3475782 | GPR109A | 12 | G protein-coupled receptor 109A†(43) | 10.3 | 18 | 13 | 13 |
| 76 | 3759006 | SLC4A1 | 17 | solute carrier family 4, anion exchanger, member 1 (erythrocyte membrane protein band 3, Diego blood group) *(74) | 10.1 | 16 | 11 | 11 |
| 85 | 3621029 | EPB42 | 15 | erythrocyte membrane protein band 4.2*(69) | 8.9 | 13 | 10 | 10 |
| 98 | 3360417 | HBD | 11 | hemoglobin, delta*(31) | 7.4 | 18 | 10 | 10 |
| Genes known to be expressed in erythrocytes or neutrophils but with fold change < 8-fold | ||||||||
| 160 | 3217077 | HEMGN | 9 | hemogen*(46) | 8.8 | 33 | 6 | 6 |
| 182 | 3533435 | PNN | 14 | pinin, desmosome associated protein | 11.0 | 6 | 9 | 6 |
| 267 | 2731381 | CXCL1 | 4 | chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) †(77) | 7.7 | 6 | 5 | 5 |
| 306 | 3388751 | MMP8 | 11 | matrix metallopeptidase 8 (neutrophil collagenase) †(42) | 4.4 | 6 | 4 | 4 |
| 612 | 2327677 | EPB41 | 1 | erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) *(22) | 11.9 | 11 | 3 | 3 |
| 840 | 3089102 | EPB49 | 8 | erythrocyte membrane protein band 4.9 (dematin) *(65) | 10.1 | 13 | 2 | 2 |
Ranked according to minimum fold change (FC).
RMA units, log2scale.
FC ratio of gene expression between PAX and LCL or between PAX and PBMC.
Minimum FC.
Expressed in erythrocytes (literature reference given within parentheses).
Expressed in neutrophils (literature reference).
Table 4.
Genes overexpressed in LCL
| Ranka | Transcript Cluster ID | Gene Symbol | Chr. | Description | Mean LCLb | LCL/PAXc | LCL/PBMCc | Min. FCd |
|---|---|---|---|---|---|---|---|---|
| 1 | 3248289 | CDK1 | 10 | cyclin-dependent kinase 1¶ | 8.4 | 111 | 68 | 68 |
| 2 | 3595979 | CCNB2 | 15 | cyclin B2¶ | 9.5 | 100 | 65 | 65 |
| 3 | 3662687 | CCL22 | 16 | chemokine (C-C motif) ligand 22 | 13.7 | 63 | 93 | 63 |
| 4 | 2333136 | CDC20 | 1 | cell division cycle 20 homolog (S. cerevisiae)¶ | 11.5 | 207 | 58 | 58 |
| 5 | 3129149 | PBK | 8 | PDZ binding kinase¶ | 7.8 | 77 | 58 | 58 |
| 6 | 3041816 | DFNA5 | 7 | deafness, autosomal dominant 5 | 9.4 | 59 | 54 | 54 |
| 7 | 2742935 | HSPA4L | 4 | heat shock 70 kDa protein 4-like | 8.3 | 69 | 53 | 53 |
| 8 | 3565663 | DLGAP5 | 14 | discs, large (Drosophila) homolog-associated protein 5¶ | 9.3 | 68 | 49 | 49 |
| 9 | 3756193 | TOP2A | 17 | topoisomerase (DNA) II alpha 170 kDa¶ | 10.0 | 75 | 49 | 49 |
| 10 | 2946225 | HIST1H2BB | 6 | histone cluster 1, H2bb | 7.3 | 115 | 47 | 47 |
| 11 | 3629103 | KIAA0101 | 15 | KIAA0101 | 9.5 | 118 | 46 | 46 |
| 12 | 3258168 | KIF11 | 10 | kinesin family member 11¶ | 8.4 | 74 | 46 | 46 |
| 13 | 3589697 | BUB1B | 15 | budding uninhibited by benzimidazoles 1 homolog beta (yeast) ¶ | 9.3 | 55 | 45 | 45 |
| 14 | 3331903 | FAM111B | 11 | family with sequence similarity 111, member B | 10.0 | 72 | 43 | 43 |
| 15 | 3443206 | AICDA | 12 | activation-induced cytidine deaminase | 9.9 | 49 | 42 | 42 |
| 16 | 2378937 | DTL | 1 | denticleless homolog (Drosophila) | 10.9 | 55 | 41 | 41 |
| 17 | 3040518 | MACC1 | 7 | metastasis associated in colon cancer 1 | 9.4 | 42 | 40 | 40 |
| 18 | 3788049 | SKA1 | 18 | spindle and kinetochore associated complex subunit 1¶ | 6.9 | 54 | 39 | 39 |
| 19 | 2585933 | SPC25 | 2 | SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae)¶ | 6.9 | 48 | 38 | 38 |
| 20 | 3881443 | TPX2 | 20 | TPX2, microtubule-associated, homolog (Xenopus laevis)¶ | 10.5 | 46 | 38 | 38 |
| 21 | 2914777 | TTK | 6 | TTK protein kinase¶ | 7.6 | 57 | 38 | 38 |
| 22 | 2838656 | HMMR | 5 | hyaluronan-mediated motility receptor (RHAMM) | 7.5 | 40 | 38 | 38 |
| 23 | 2830638 | KIF20A | 5 | kinesin family member 20A¶ | 9.4 | 49 | 37 | 37 |
| 24 | 3160658 | SLC1A1 | 9 | solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 | 8.5 | 37 | 38 | 37 |
| 25 | 2417528 | DEPDC1 | 1 | DEP domain containing 1 | 7.5 | 55 | 36 | 36 |
| 26 | 3258444 | CEP55 | 10 | centrosomal protein 55 kDa¶ | 8.6 | 58 | 36 | 36 |
| 27 | 3260586 | SCD | 10 | stearoyl-CoA desaturase (delta-9-desaturase) | 11.3 | 44 | 36 | 36 |
| 28 | 3354799 | CHEK1 | 11 | CHK1 checkpoint homolog (S. pombe)¶ | 8.9 | 64 | 35 | 35 |
| 29 | 3648391 | TNFRSF17 | 16 | tumor necrosis factor receptor superfamily, member 17 | 9.3 | 33 | 33 | 33 |
| 30 | 2720251 | NCAPG | 4 | nonSMC condensin I complex, subunit G¶ | 8.9 | 37 | 33 | 33 |
| 31 | 3720896 | CDC6 | 17 | cell division cycle 6 homolog (S. cerevisiae)¶ | 8.3 | 31 | 34 | 31 |
| Known EBV-inducible genes | ||||||||
| 32 | 3817380 | EBI3 | 19 | Epstein-Barr virus induced 3*(45) | 11.0 | 46 | 30 | 30 |
| 112 | 2377283 | CR2 | 1 | complement component (3d/Epstein Barr virus) receptor 2 *(12) | 9.0 | 27 | 12 | 12 |
| 117 | 3848492 | FCER2 | 19 | Fc fragment of IgE, low affinity II, receptor for (CD23) *(12) | 12.3 | 20 | 12 | 12 |
| Known EBV-inducible genes, significant but with fold change <8.0 | ||||||||
| 302 | 3332403 | MS4A1 | 11 | membrane-spanning 4-domains, subfamily A, member 1*(12) | 12.2 | 14 | 6 | 6 |
| 308 | 2438892 | FCRL5 | 1 | Fc receptor-like 5*(51) | 8.0 | 5 | 6 | 5 |
| 319 | 3063685 | MCM7 | 7 | minichromosome maintenance complex component 7*(38) | 11.1 | 6 | 5 | 5 |
| 422 | 2402459 | STMN1 | 1 | stathmin 1*(20, 38) | 10.5 | 4 | 6 | 4 |
| 438 | 3677752 | TRAP1 | 16 | TNF receptor-associated protein 1*(38) | 10.7 | 7 | 4 | 4 |
| 359 | 2440327 | SLAMF1 | 1 | signaling lymphocytic activation molecule family member 1* | 9.5 | 5 | 5 | 5 |
| 561 | 2901913 | TUBB | 6 | tubulin, beta*(38) | 13.5 | 4 | 3 | 3 |
| 770 | 3259253 | ENTPD1 | 10 | ectonucleoside triphosphate diphosphohydrolase 1*(12) | 11.8 | 3 | 4 | 3 |
| 778 | 2317317 | TP73 | 1 | tumor protein p73*(18) | 8.7 | 4 | 3 | 3 |
| 912 | 2320683 | TNFRSF8 | 1 | tumor necrosis factor receptor superfamily, member 8*(12) | 9.5 | 2 | 2 | 2 |
| 977 | 3820443 | ICAM1 | 19 | intercellular adhesion molecule 1*(68) | 10.4 | 7 | 2 | 2 |
| 1064 | 2592268 | STAT1 | 2 | signal transducer and activator of transcription 1, 91 kDa *(49) | 12.3 | 2 | 2 | 2 |
| 1216 | 2526759 | ATIC | 2 | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase*(38) | 11.1 | 2 | 3 | 2 |
| 1323 | 2877508 | HSPA9 | 5 | heat shock 70 kDa protein 9 (mortalin)*(38) | 11.0 | 2 | 2 | 2 |
| 2389 | 3743906 | TP53 | 17 | tumor protein p53*(41) | 11.4 | 7 | 2 | 2 |
Ranked according to minimum FC.
RMA units, log2scale.
FC ratio of gene expression between LCL and PAX or between LCL and PBMC.
Minimum FC.
Epstein Barr-Virus-inducible genes (literature reference given within parentheses).
Cell-cycle related genes by Gene Ontology (GO).
Table 5.
Genes overexpressed in PBMC
| Ranka | Transcript Cluster ID | Gene Symbol | Chr. | Description | Mean PBMCb | PBMC/LCLc | PBMC/PAXc | Min. FCd |
|---|---|---|---|---|---|---|---|---|
| 1 | 3012978 | GNG11 | 7 | guanine nucleotide binding protein (G protein), gamma 11 | 9.9 | 35 | 41 | 35 |
| 2 | 2701081 | P2RY12 | 3 | purinergic receptor P2Y, G-protein coupled, 12¶ | 7.7 | 21 | 18 | 18 |
| 3 | 3589458 | THBS1 | 15 | thrombospondin 1¶ | 10.8 | 48 | 16 | 16 |
| 4 | 2761837 | FGFBP2 | 4 | fibroblast growth factor binding protein 2 | 9.5 | 33 | 15 | 15 |
| 5 | 3535780 | PTGER2 | 14 | prostaglandin E receptor 2 (subtype EP2), 53 kDa | 10.1 | 17 | 14 | 14 |
| 6 | 3724545 | ITGB3 | 17 | integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61)¶ | 11.7 | 14 | 12 | 12 |
| 7 | 3891342 | TUBB1 | 20 | tubulin, beta 1 | 12.7 | 246 | 12 | 12 |
| 8 | 2773972 | CXCL11 | 4 | chemokine (C-X-C motif) ligand 11 | 5.6 | 10 | 11 | 10 |
| 9 | 3841506 | LAIR2 | 19 | leukocyte-associated immunoglobulin-like receptor 2 | 7.3 | 29 | 10 | 10 |
| 10 | 3866831 | CABP5 | 19 | calcium binding protein 5 | 5.4 | 14 | 9 | 9 |
| 11 | 2443417 | SELP | 1 | selectin P (granule membrane protein 140 kDa, antigen CD62)¶ | 9.2 | 29 | 9 | 9 |
| 12 | 2987544 | LFNG | 7 | LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase | 10.1 | 9 | 11 | 9 |
| 13 | 3729052 | YPEL2 | 17 | yippee-like 2 (Drosophila) | 8.2 | 10 | 9 | 9 |
| 14 | 3417842 | LRP1 | 12 | low density lipoprotein receptor-related protein 1 | 10.6 | 13 | 8 | 8 |
| 15 | 3904508 | SLA2 | 20 | Src-like-adaptor 2 | 10.4 | 19 | 8 | 8 |
| 16 | 2783596 | PDE5A | 4 | phosphodiesterase 5A, cGMP-specific¶ | 7.8 | 28 | 8 | 8 |
| 17 | 3579114 | BCL11B | 14 | B-cell CLL/lymphoma 11B (zinc finger protein) | 10.0 | 8 | 10 | 8 |
| 18 | 2902609 | C6orf25 | 6 | chromosome 6 open reading frame 25 | 12.3 | 30 | 8 | 8 |
| 19 | 3188111 | PTGS1 | 9 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | 11.4 | 16 | 8 | 8 |
Ranked according to minimum FC.
bRMA units, log2scale.
FC ratio of gene expression between PBMC and LCL or between PBMC and PAX.
Minimum FC.
Platelet-related genes by GO.
Seven red blood cell-related genes were overexpressed in PAX compared with the other two RNA sources (Table 3) including the top-ranked GYPB (glycophorin B) gene. Hemoglobin and hemoglobin-related genes, including HBB, HBD, ALAS2, RHD, and AHSP are seen in high-ranking positions. Many known erythrocyte-related genes, such as HEMGN, EPB42, EPB49, and SLC4A1, were also significantly higher in PAX, but were seen above the eightfold cutoff only in the exon-level analysis. These observations clearly result from the fact that PAX samples are derived from whole blood, which comprises predominantly erythrocytes and reticulocytes as well as white blood cells. Genes associated with neutrophils such as SLPI were also overexpressed in PAX compared with LCL or PBMC (Table 3). Most of these genes were also moderately expressed in PBMC but markedly lower in LCL expression. The top GO (7) categories (see Table 6) for genes most highly expressed in PAX are related to RNA splicing and processing.
Table 6.
GO categories for overexpressed genes in each RNA source
| GO ID | GO Category | −log10(P)a | Significant Genes, nb | Genes in Category, n |
|---|---|---|---|---|
| PAX | ||||
| GO:0008380 | RNA splicing | 6 | 17 | 259 |
| GO:0006397 | mRNA processing | 5 | 19 | 335 |
| GO:0000375 | RNA splicing, via transesterification reactions | 5 | 13 | 166 |
| GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile | 5 | 12 | 160 |
| GO:0000398 | nuclear mRNA splicing, via spliceosome | 5 | 12 | 160 |
| GO:0016071 | mRNA metabolic process | 4 | 21 | 462 |
| GO:0006396 | RNA processing | 3 | 22 | 560 |
| LCL | ||||
| GO:0022402 | cell cycle process | 76 | 130 | 723 |
| GO:0022403 | cell cycle phase | 69 | 103 | 462 |
| GO:0007049 | cell cycle | 69 | 119 | 665 |
| GO:0000278 | mitotic cell cycle | 47 | 69 | 293 |
| GO:0051301 | cell division | 40 | 62 | 278 |
| GO:0010564 | regulation of cell cycle process | 37 | 62 | 319 |
| GO:0000087 | M phase of mitotic cell cycle | 35 | 37 | 90 |
| GO:0000279 | M phase | 35 | 37 | 92 |
| GO:0000236 | mitotic prometaphase | 29 | 32 | 82 |
| GO:0006996 | organelle organization | 29 | 110 | 1356 |
| GO:0071156 | regulation of cell cycle arrest | 29 | 45 | 208 |
| GO:0051726 | regulation of cell cycle | 28 | 70 | 585 |
| GO:0000075 | cell cycle checkpoint | 27 | 42 | 192 |
| GO:0000280 | nuclear division | 26 | 40 | 175 |
| GO:0007067 | mitosis | 26 | 40 | 175 |
| GO:0048285 | organelle fission | 26 | 41 | 186 |
| PBMC | ||||
| GO:0030168 | platelet activation | 13 | 17 | 224 |
| GO:0001775 | cell activation | 12 | 22 | 455 |
| GO:0002576 | platelet degranulation | 10 | 10 | 76 |
| GO:0050817 | coagulation | 10 | 19 | 434 |
| GO:0007596 | blood coagulation | 10 | 19 | 434 |
| GO:0007599 | hemostasis | 10 | 19 | 438 |
| GO:0050878 | regulation of body fluid levels | 9 | 19 | 499 |
| GO:0002376 | immune system process | 8 | 25 | 922 |
| GO:0006955 | immune response | 8 | 19 | 545 |
| GO:0050896 | response to stimulus | 7 | 64 | 5133 |
| GO:0006887 | exocytosis | 7 | 10 | 156 |
| GO:0007165 | signal transduction | 7 | 44 | 2942 |
| GO:0006952 | defense response | 6 | 17 | 620 |
| GO:0046903 | secretion | 5 | 13 | 391 |
| GO:0051716 | cellular response to stimulus | 5 | 46 | 3515 |
Determined by GOrilla (26).
Overexpressed by at least 4-fold compared with other 2 RNA sources and met the FDR ≤0.05 threshold on gene-level analysis.
The top 32 genes specific to LCL (Table 4) are rich in cell cycle-related genes. For example, CDK1 (cyclin-dependent kinase 1) and CCNB2 (cyclin B2) are 68- and 65-fold overexpressed in LCL compared with the other two RNA sources. Top GO categories for genes most highly expressed in LCL (see Table 6) are related to cell cycle and mitosis, which are indicative of a cell line undergoing rapid cell division. Several genes known to be induced by EBV were also overexpressed in LCL. Of these, EBI3 (Epstein-Barr induced 3) was the most highly differentially expressed. Fifteen other known EBV-induced genes show significant overexpression of two- to sixfold. In general, exon-level analysis was more sensitive than gene-level analysis in identifying such genes (Supplementary Table S2). Comparison of LCL with PAX expression appeared to be generally more sensitive to EBV-induced differences than the comparison with PBMC (e.g., CR2, Table 4).
PBMC overexpression (Table 5) was seen in many genes known to be platelet specific, or involved in coagulation. For example, P2RY12, THBS1, ITGB3, PTGS1 were abundantly expressed in PBMC vs. PAX and PBMC vs. LCL. Evidently, the inclusion of platelets within the PBMC fraction is sufficient to for allow detection of these genes (64). The top GO categories (Table 6) for genes most highly expressed in PBMC are related to immune response, platelet activation, and blood coagulation reflecting the primary presence of lymphocytes and monocytes, and some platelets, in this sample type.
Analysis of Differential Blood Count Data
We were able to identify the associations of numerous genes with individual blood elements in the differential blood count (Table 7). In PAX samples, most of the genes with positive associations were associated with neutrophil or lymphocyte counts, while in PBMC, the genes were generally associated with lymphocyte (36, 70, 73) and monocyte counts, as would be expected based on the cell-type composition of these sources. Some of the neutrophil-associated genes, such as SLPI and IL1R2, are also reported in Table 3. As before, the exon-level analysis often detected more genes than did the gene-level analysis. GO analysis of these genes showed overrepresentation in the categories of immune system regulation, lymphocyte, leukocyte, and T-cell activation (Supplementary Tables S4–S6). In contrast, no genes were associated with the differential blood count in RNA from LCL. This may be due to the single cell type represented in LCL or because the LCL samples were derived from whole blood obtained 5 yr prior to the differential blood counts whereas PAX and PBMC were drawn at the same time as the blood counts were performed.
Table 7.
Number of genes with positive association* with differential blood count, for each RNA source at the gene and exon level
| Gene Level |
Exon Levela |
|||||
|---|---|---|---|---|---|---|
| LCL | PAX | PBMC | LCL | PAX | PBMC | |
| Red blood cell count | 0 | 0 | 3 | 0 | 0 | 15 (14) |
| Neutrophil count | 0 | 354 | 0 | 0 | 1,178 (475) | 0 |
| Lymphocyte count | 0 | 636 | 131 | 0 | 354 (259) | 458 (241) |
| Monocyte count | 0 | 2 | 154 | 0 | 1 (1) | 321 (200) |
| Eosinophil count | 0 | 2 | 0 | 0 | 0 | 0 |
| Basophil count | 0 | 0 | 0 | 0 | 0 | 0 |
| Platelet count | 0 | 0 | 0 | 0 | 0 | 1 (1) |
Significant at FDR ≤0.2 level, counting only genes with positive association.
Number of exons (number of genes).
Identification of Genes Associated With Major CVD Risk Factors
Sex.
Not surprisingly, a search for biomarkers of sex in our study yielded many Y-chromosome genes (Table 8). All three sample types identified 128 exons within nine distinct genes residing on the Y-chromosome at FDR ≤ 0.05 level. An additional 28 exons on 11 Y-chromosome genes were detected in one or more of the RNA sources, with PAX able to detect 16 exons, PBMC 15 exons, and LCL 9 exons at this FDR level. Only 14 exons of two X-linked genes [KDM5C, KDM6A, lysine (K)-specific demethylase 5C and 6A] were differentially expressed in women vs. men in all three RNA sources. However, 142 exons in 22 genes (Table 8) showed differential expression in at least one source. Several of these genes are obvious homologs to their Y-linked counterparts (DDX3X, EIF1AX, NLGN4X, PRKX, RPS4X, ZFX). Interestingly, in LCL samples more X-linked overexpression in women was detected, with 99 additional exons beyond those detected in all three sources, compared with 28 for PAX and 25 for PBMC. The key gene responsible for X inactivation (XIST), which is ordinarily highly overexpressed in women, is less overexpressed in women in LCL samples (Supplementary Table S7), compared with PAX or PBMC (female to male fold-change of 25 in LCL, 140 in PAX, and 52 in PBMC). Furthermore, we observed that XIST expression in LCL in women is significantly correlated with 339 of 648 X-linked genes (FDR ≤ 0.2 genome wide). The majority (206) are negatively correlated with X-linked expression, further supporting the idea that XIST-mediated X inactivation is substantially and variably disrupted by EBV infection/transformation and/or culture conditions of the LCL samples.
Table 8.
X- and Y-linked exons detected as expression biomarkers of sex using 3 RNA sources
| Number of Exons Detected by |
Number of Additional Exons Detected with: |
||||||
|---|---|---|---|---|---|---|---|
| Gene Symbol | Description | Total Exons | Any of 3 | All 3 | LCL | PAX | PBMC |
| Y-chromosome | |||||||
| CYorf15B | chromosome Y open reading frame 15B | 18 | 13 | 9 | 0 | 4 | 2 |
| DDX3Y | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked | 22 | 20 | 16 | 4 | 0 | 3 |
| EIF1AY | eukaryotic translation initiation factor 1A, Y-linked | 9 | 8 | 7 | 1 | 0 | 1 |
| NLGN4Y | neuroligin 4, Y-linked | 6 | 1 | 0 | 1 | 0 | 0 |
| PRKY | protein kinase, Y-linked | 16 | 7 | 4 | 1 | 2 | 1 |
| PRY | PTPN13-like, Y-linked | 22 | 1 | 0 | 0 | 1 | 0 |
| RPS4Y1 | ribosomal protein S4, Y-linked 1 | 12 | 12 | 12 | 0 | 0 | 0 |
| RPS4Y2 | ribosomal protein S4, Y-linked 2 | 7 | 4 | 2 | 0 | 1 | 1 |
| TMSB4Y | thymosin beta 4, Y-linked | 6 | 2 | 0 | 0 | 1 | 2 |
| USP9Y | Ubiquitin-specific peptidase 9, Y-linked | 53 | 40 | 38 | 1 | 2 | 0 |
| UTY | ubiquitously transcribed tetratricopeptide repeat gene, Y-linked | 52 | 42 | 35 | 0 | 5 | 4 |
| ZFY | zinc finger protein, Y-linked | 10 | 6 | 5 | 1 | 0 | 1 |
| Total | 233 | 156 | 128 | 9 | 16 | 15 | |
| X-chromosome | |||||||
| DDX3X | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked | 28 | 6 | 0 | 3 | 2 | 2 |
| EIF1AX | eukaryotic translation initiation factor 1A, X-linked | 15 | 6 | 0 | 5 | 2 | 0 |
| EIF2S3 | eukaryotic translation initiation factor 2, subunit 3 gamma, 52 kDa | 15 | 4 | 0 | 4 | 1 | 0 |
| HDHD1A | haloacid dehalogenase-like hydrolase domain containing 1A | 7 | 4 | 0 | 4 | 1 | 0 |
| KDM5C | lysine (K)-specific demethylase 5C | 37 | 27 | 5 | 20 | 2 | 5 |
| KDM6A | lysine (K)-specific demethylase 6A | 42 | 29 | 9 | 17 | 2 | 6 |
| NLGN4X | neuroligin 4, X-linked | 27 | 2 | 0 | 2 | 0 | 0 |
| PLCXD1 | phosphatidylinositol-specific phospholipase C, X domain containing 1 | 26 | 1 | 0 | 0 | 0 | 1 |
| PNPLA4 | patatin-like phospholipase domain containing 4 | 8 | 5 | 0 | 5 | 0 | 0 |
| PRKX | protein kinase, X-linked | 18 | 6 | 0 | 2 | 3 | 3 |
| RPS4X | ribosomal protein S4, X-linked | 11 | 8 | 0 | 8 | 1 | 0 |
| SEPT6 | septin 6 | 16 | 2 | 0 | 0 | 0 | 2 |
| SMC1A | structural maintenance of chromosomes 1A | 32 | 12 | 0 | 12 | 2 | 0 |
| STS | steroid sulfatase (microsomal), isozyme S | 24 | 8 | 0 | 8 | 0 | 0 |
| TCEANC | transcription elongation factor A (SII) N-terminal and central domain containing | 8 | 1 | 0 | 0 | 1 | 0 |
| TIMM8A | translocase of inner mitochondrial membrane 8 homolog A (yeast) | 4 | 1 | 0 | 0 | 1 | 0 |
| TXLNG | taxilin gamma | 14 | 4 | 0 | 0 | 2 | 2 |
| VSIG4 | V-set and immunoglobulin domain containing 4 | 14 | 3 | 0 | 0 | 3 | 0 |
| XPNPEP2 | X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound | 28 | 2 | 0 | 0 | 0 | 2 |
| ZFX | zinc finger protein, X-linked | 13 | 6 | 0 | 6 | 4 | 0 |
| ZRSR2 | zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 2 | 20 | 4 | 0 | 2 | 1 | 2 |
| ZXDB | zinc finger, X-linked, duplicated B | 12 | 1 | 0 | 1 | 0 | 0 |
| Total | 419 | 142 | 14 | 99 | 28 | 25 | |
There were 90 autosomal exons (74 genes) associated with sex at FDR ≤ 0.05, none was significant in more than one RNA source (Table 9). PBMC identified exons from 52 genes, while LCL and PAX identified 19 genes each.
Table 9.
Autosomal genes associated with sex
| Transcript cluster ID | Gene Symbol | Chr. | Description | Effect LCL* | Effect PAX* | Effect PBMC* |
|---|---|---|---|---|---|---|
| 3631397 | UACA | 15 | uveal autoantigen with coiled-coil domains and ankyrin repeats | 0.43 | 0.29 | 0.14 |
| 2880361 | JAKMIP2 | 5 | janus kinase and microtubule interacting protein 2 | 0.25 | 0.60 | 0.69 |
| 3712675 | RAI1 | 17 | retinoic acid induced 1 | −0.22 | −0.10 | 0.13 |
| 3373946 | TIMM10 | 11 | translocase of inner mitochondrial membrane 10 homolog (yeast) | 0.17 | 0.15 | 0.36 |
| 3725602 | ABI3 | 17 | ABI family, member 3 | −0.16 | 0.03 | 0.40 |
| 2439101 | FCRL1 | 1 | Fc receptor-like 1 | −0.12 | −0.58 | −0.81 |
| 2893109 | LOC100129033 | 6 | QIQN5815 | −0.10 | −0.58 | 0.07 |
| 3857811 | C19orf12 | 19 | chromosome 19 open reading frame 12 | 0.08 | 0.20 | 0.30 |
| 3223687 | PHF19 | 9 | PHD finger protein 19 | 0.04 | 0.03 | 0.31 |
| 3264621 | TCF7L2 | 10 | transcription factor 7-like 2 (T-cell specific, HMG-box) | −0.04 | 0.22 | 0.61 |
| 3417184 | SUOX | 12 | sulfite oxidase | −0.04 | 0.06 | 0.35 |
| 3543935 | COQ6 | 14 | coenzyme Q6 homolog, monooxygenase (S. cerevisiae) | −0.04 | −0.26 | 0.06 |
| 2607055 | PASK | 2 | PAS domain containing serine/threonine kinase | 0.04 | −0.18 | −0.21 |
| 3870990 | GP6 | 19 | glycoprotein VI (platelet) | 0.04 | −0.36 | −0.24 |
| 3534866 | MGAT2 | 14 | mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase | 0.02 | −0.04 | 0.26 |
| 3940992 | ASPHD2 | 22 | aspartate beta-hydroxylase domain containing 2 | 0.01 | 0.15 | 0.40 |
In log2 RMA units. Positive effects are highly expressed in males. Partial list, genes are significant in at least 1 RNA source, FDR ≤0.2.
Smoking.
Several probe sets in the CHRNA3 (cholinergic receptor, nicotinic, alpha 3) gene were downregulated in smokers in all three RNA sources in our study, though not significantly (P < 0.10). PAX and PBMC samples showed a stronger tendency toward downregulation (P = 0.06) on probe set ID 3634334. Variants of CHRNA3 have been associated with smoking behavior and susceptibility to lung cancer (5). Genetic variants in ALDH2 (aldehyde dehydrogenase 2) have been studied extensively in relation to smoking and lung cancer risk (57). In our study, LCL samples detected 1.36- to 2.84-fold lower expression of this gene in smokers, with 14 of 16 probe sets having P values < 0.05. Average expression of all 16 exons differed significantly in LCL samples (P = 0.004) was borderline for PBMC (P = 0.054) but did not differ for PAX (P = 0.247).
Age.
The relatively narrow age range of the participants hindered biomarker detection for age. Nevertheless, five genes were associated with age (P < 0.05) for each of the three RNA sources (Supplementary Table S8). One of them, TP53, has been associated with senescence (28). The magnitude of expression differences was small with only three out of five genes having the same directional difference in all three RNA sources.
HDL cholesterol levels.
Since the small sample size hindered discovery of gene expression signatures of HDL cholesterol, we sought to confirm previously observed associations with HDL. Four such genes were seen to be associated in PAX at P < 0.05, four were associated in PBMC and none in LCL (Supplementary Table S9). Two genes, FADS1 (fatty acid desaturase 1) and LDLR (low-density lipoprotein receptor), were associated with HDL levels in both PAX and PBMC, with small but consistent inverse associations of higher expression with lower HDL. These genes are known to influence circulating lipid levels and risk of coronary artery disease (78). The remaining CVD risk factors listed in Table 1, including BMI, total cholesterol, and blood pressure, were analyzed but did not reveal any significant association with gene expression.
Robust and Consistent Markers
“Fingerprinting” genes.
We identified a number of exons that strongly distinguished individual participants, irrespective of RNA source. These fingerprinting exons have robust expression levels (i.e., their relative expression is independent of RNA source) and may allow for identification of individuals within a large study sample.
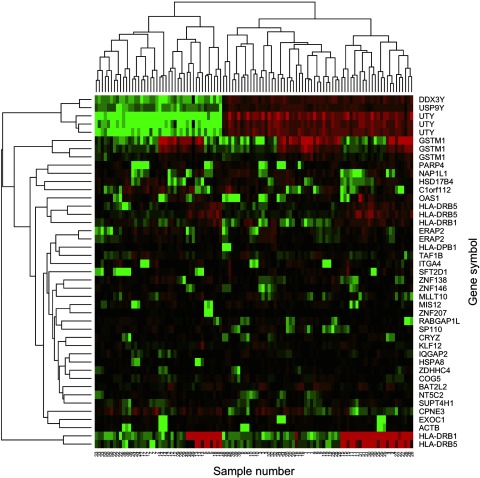
We selected 423 such exons drawn from 247 distinct genes having statistical significance (Table 10, Supplementary Table S10). Among the top results were several histocompatibility antigen genes (HLA-DRB1, HLA-DRB5, HLA-DPB1, HLA-B, HLA-DQA2, HLA-DQB2). HLA genes code for antigenic surface proteins used by the immune system to recognize “self” and thus are highly specific to an individual's ancestry. These genes have been suggested as biomarkers for autoimmune diseases (56, 60). These 423 selected exons were able to cluster the three samples from each participant perfectly. Indeed, a subset of only 38 autosomal exons exhibiting the largest F-ratio for participant effects together with five exons on the X- and Y-chromosomes were sufficient to cluster the participants perfectly (Fig. 3). Of note, these fingerprinting markers include one exon of the β-actin gene (ACTB), commonly used as a calibration standard or housekeeping gene. This ACTB exon exhibits a strongly bimodal expression pattern (Fig. 4), possibly due to the influence of an underlying or associated SNP. A similar bimodal pattern is also seen in other probe sets (Fig. 5), such as exons of genes GSTM1, HLA-DRB1, and OAS1. OAS1, which encodes a protein vital to immune response to viral infection, is associated with multiple diseases (40) and contains common functional variation that strongly affects exon inclusion (58). In the case of GSTM1, the bimodal pattern is evident in eight consecutive probe sets covering seven distinct exons, suggesting a true pattern of bimodal expression or extensive splice variation, rather than the direct influence of a single SNP. GSTM1 is an important drug and xenobiotic metabolizing enzyme that is known to exhibit common copy number variation that likely contributes to the observed bimodal pattern of expression (33). The complete list of fingerprinting exons is given in Supplementary Table S10.
Table 10.
Partial list of exons with FDR ≤0.05 for the participant effect and SD across participant of at least 2-fold for this effect
| Probe Set IDa | Gene Symbol | Chr. | Description | F(subject)b | Det. Exonsc | Total Exons, nd |
|---|---|---|---|---|---|---|
| 4030178 | DDX3Y | Y | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked | 85.2 | 18 | 22 |
| 4035087 | UTY | Y | ubiquitously transcribed tetratricopeptide repeat gene, Y-linked | 74.4 | 33 | 52 |
| 4030146 | USP9Y | Y | ubiquitin-specific peptidase 9, Y-linked | 73.5 | 34 | 53 |
| 4028553 | RPS4Y1 | Y | ribosomal protein S4, Y-linked 1 | 46.4 | 12 | 12 |
| 3764386 | SUPT4H1 | 17 | suppressor of Ty 4 homolog 1 (S. cerevisiae) | 46.0 | 1 | 11 |
| 4048279 | HLA-DRB1 | 6 | major histocompatibility complex, class II, DR beta 1 | 45.9 | 4 | 8 |
| 2350995 | GSTM1 | 1 | glutathione S-transferase mu 1 | 44.4 | 3 | 14 |
| 3717652 | ZNF207 | 17 | zinc finger protein 207 | 43.7 | 1 | 24 |
| 3505812 | PARP4 | 13 | poly (ADP-ribose) polymerase family, member 4 | 41.3 | 1 | 43 |
| 4031141 | EIF1AY | Y | eukaryotic translation initiation factor 1A, Y-linked | 41.2 | 7 | 9 |
| 2825746 | HSD17B4 | 5 | hydroxysteroid (17-beta) dehydrogenase 4 | 38.9 | 1 | 26 |
| 4028588 | ZFY | Y | zinc finger protein, Y-linked | 35.4 | 5 | 10 |
| 3988474 | DOCK11 | X | dedicator of cytokinesis 11 | 34.7 | 1 | 57 |
| 3036926 | ACTB | 7 | actin, beta | 34.7 | 1 | 11 |
| 3432446 | OAS1 | 12 | 2′,5′-oligoadenylate synthetase 1, 40/46 kDa | 31.5 | 1 | 15 |
| 2367199 | BAT2L2 | 1 | HLA-B associated transcript 2-like 2 | 31.3 | 1 | 48 |
| 3304629 | NT5C2 | 10 | 5′-nucleotidase, cytosolic II | 28.6 | 1 | 22 |
| 2984580 | SFT2D1 | 6 | SFT2 domain containing 1 | 28.4 | 1 | 9 |
| 3462877 | NAP1L1 | 12 | nucleosome assembly protein 1-like 1 | 27.9 | 1 | 22 |
| 4028462 | CD99 | Y | CD99 molecule | 25.5 | 1 | 27 |
| 4048249 | HLA-DRB5 | 6 | major histocompatibility complex, class II, DR beta 5 | 25.2 | 5 | 11 |
| 2727952 | EXOC1 | 4 | exocyst complex component 1 | 24.7 | 1 | 25 |
| 3831276 | ZNF146 | 19 | zinc finger protein 146 | 24.5 | 1 | 8 |
| 4025365 | IDS | X | iduronate 2-sulfatase | 23.7 | 1 | 20 |
| 2469139 | TAF1B | 2 | TATA box binding protein (TBP)-associated factor, RNA polymerase I, B, 63 kDa | 23.5 | 1 | 17 |
| 3067144 | COG5 | 7 | component of oligomeric golgi complex 5 | 23.4 | 1 | 33 |
| 2366603 | C1orf112 | 1 | SCY1-like 3 (S. cerevisiae) | 23.1 | 1 | 36 |
| 2903428 | HLA-DPB1 | 6 | major histocompatibility complex, class II, DP beta 1 | 23.1 | 1 | 8 |
| 2367974 | RABGAP1L | 1 | RAB GTPase-activating protein 1-like | 21.6 | 1 | 47 |
| 2418460 | CRYZ | 1 | crystallin, zeta (quinone reductase) | 21.3 | 1 | 15 |
| 3105938 | CPNE3 | 8 | copine III | 20.2 | 1 | 22 |
| 2989124 | ZDHHC4 | 7 | zinc finger, DHHC-type containing 4 | 19.4 | 1 | 13 |
| 2603075 | SP110 | 2 | SP110 nuclear body protein | 19.4 | 1 | 24 |
| 3975522 | KDM6A | X | lysine (K)-specific demethylase 6A | 19.1 | 1 | 42 |
| 2821406 | ERAP2 | 5 | endoplasmic reticulum aminopeptidase 2 | 18.8 | 22 | 27 |
| 3004680 | ZNF138 | 7 | zinc finger protein 138 | 18.7 | 1 | 15 |
| 3395427 | HSPA8 | 11 | heat shock 70 kDa protein 8 | 18.5 | 1 | 17 |
| 3238248 | MLLT10 | 10 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 | 17.7 | 1 | 38 |
| 4015713 | BTK | X | Bruton agammaglobulinemia tyrosine kinase | 17.5 | 1 | 24 |
| 2518349 | ITGA4 | 2 | integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) | 17.4 | 1 | 40 |
| 3707765 | MIS12 | 17 | MIS12, MIND kinetochore complex component, homolog (S. pombe) | 16.5 | 1 | 5 |
| 2816364 | IQGAP2 | 5 | IQ motif containing GTPase-activating protein 2 | 16.5 | 1 | 45 |
| 3517836 | KLF12 | 13 | Kruppel-like factor 12 | 16.1 | 1 | 23 |
| 2542747 | LAPTM4A | 2 | lysosomal protein transmembrane 4 alpha | 15.8 | 1 | 10 |
| 2948952 | HLA-B | 6 | major histocompatibility complex, class I, B | 15.6 | 1 | 12 |
| 2351023 | GSTM5 | 1 | glutathione S-transferase mu 5 | 15.5 | 1 | 11 |
| 3056088 | BAZ1B | 7 | bromodomain adjacent to zinc finger domain, 1B | 15.3 | 1 | 39 |
| 3385778 | CTSC | 11 | cathepsin C | 15.2 | 1 | 15 |
| 3932139 | PSMG1 | 21 | proteasome (prosome, macropain) assembly chaperone 1 | 14.9 | 1 | 11 |
| 2961826 | PHIP | 6 | pleckstrin homology domain-interacting protein | 14.9 | 1 | 48 |
| 3879393 | PLK1S1 | 20 | polo-like kinase 1 substrate 1 | 14.4 | 1 | 19 |
| 2723770 | TBC1D1 | 4 | TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 | 14.4 | 1 | 32 |
| 3576822 | TRIP11 | 14 | thyroid hormone receptor interactor 11 | 14.3 | 1 | 30 |
| 2903265 | HLA-DQA2 | 6 | major histocompatibility complex, class II, DQ alpha 2 | 14.3 | 1 | 5 |
| 3996335 | RPL10 | X | ribosomal protein L10 | 14.2 | 1 | 14 |
| 3485880 | EXOSC8 | 13 | exosome component 8 | 14.2 | 1 | 12 |
| 3584495 | SNRPN | 15 | small nuclear ribonucleoprotein polypeptide N | 14.0 | 1 | 23 |
| 4031106 | CYorf15B | Y | chromosome Y open reading frame 15B | 13.6 | 10 | 18 |
| 3425122 | C12orf29 | 12 | chromosome 12 open reading frame 29 | 13.5 | 1 | 11 |
| 2530554 | MFF | 2 | mitochondrial fission factor | 13.2 | 1 | 14 |
| 3243742 | BMS1 | 10 | BMS1 homolog, ribosome assembly protein (yeast) | 13.2 | 1 | 31 |
| 3169339 | ALDH1B1 | 9 | aldehyde dehydrogenase 1 family, member B1 | 13.1 | 1 | 10 |
| 2739191 | CCDC109B | 4 | coiled-coil domain containing 109B | 13.0 | 1 | 12 |
| 2571102 | ANAPC1 | 2 | anaphase-promoting complex subunit 1 | 12.8 | 1 | 70 |
| 3046682 | TRGV5 | 7 | TCR gamma alternate reading frame protein | 12.7 | 1 | 1 |
| 2446619 | STX6 | 1 | syntaxin 6 | 12.7 | 1 | 11 |
| 4031175 | RPS4Y2 | Y | ribosomal protein S4, Y-linked 2 | 12.7 | 2 | 7 |
| 3458101 | NACA | 12 | nascent polypeptide-associated complex alpha subunit | 12.0 | 1 | 10 |
| 2350940 | GSTM4 | 1 | glutathione S-transferase mu 4 | 11.9 | 1 | 12 |
| 2676049 | WDR82 | 3 | WD repeat domain 82 | 11.6 | 1 | 12 |
| 3907879 | ELMO2 | 20 | engulfment and cell motility 2 | 11.4 | 1 | 27 |
| 3759912 | LRRC37A4 | 17 | leucine-rich repeat containing 37, member A4 (pseudogene) | 11.3 | 3 | 18 |
| 3315556 | PSMD13 | 11 | proteasome (prosome, macropain) 26S subunit, nonATPase, 13 | 11.2 | 1 | 16 |
| 2369585 | SOAT1 | 1 | sterol O-acyltransferase 1 | 11.2 | 1 | 21 |
| 2492088 | KDM3A | 2 | lysine (K)-specific demethylase 3A | 10.8 | 1 | 35 |
| 3003206 | CCT6A | 7 | chaperonin-containing TCP1, subunit 6A (zeta 1) | 10.7 | 1 | 23 |
| 2821249 | CAST | 5 | calpastatin | 10.7 | 1 | 42 |
| 3641887 | LINS1 | 15 | lines homolog 1 (Drosophila) | 10.3 | 1 | 16 |
| 3971880 | EIF2S3 | X | eukaryotic translation initiation factor 2, subunit 3 gamma, 52 kDa | 10.3 | 1 | 15 |
| 3850437 | KRI1 | 19 | KRI1 homolog (S. cerevisiae) | 10.1 | 1 | 24 |
| 3908171 | ZMYND8 | 20 | zinc finger, MYND-type containing 8 | 10.0 | 1 | 33 |
| 4029193 | PRKY | Y | protein kinase, Y-linked | 9.7 | 3 | 16 |
| 2501343 | LOC654433 | 2 | hypothetical LOC654433 | 9.6 | 1 | 8 |
| 3487448 | DNAJC15 | 13 | DnaJ (Hsp40) homolog, subfamily C, member 15 | 9.4 | 1 | 8 |
| 3462702 | KRR1 | 12 | KRR1, small subunit (SSU) processome component, homolog (yeast) | 9.2 | 1 | 17 |
| 3742635 | C17orf87 | 17 | chromosome 17 open reading frame 87 | 9.0 | 1 | 6 |
| 3140703 | STAU2 | 8 | staufen, RNA binding protein, homolog 2 (Drosophila) | 9.0 | 1 | 29 |
ID of the probe set of the gene with the highest F statistics.
F statistics of the top probe set of the gene.
Number of exons of the gene with ≥2 F-statistics.
Total number of exons of the gene in the annotation. The probe sets are sorted by the F-score of the participant.
Fig. 3.
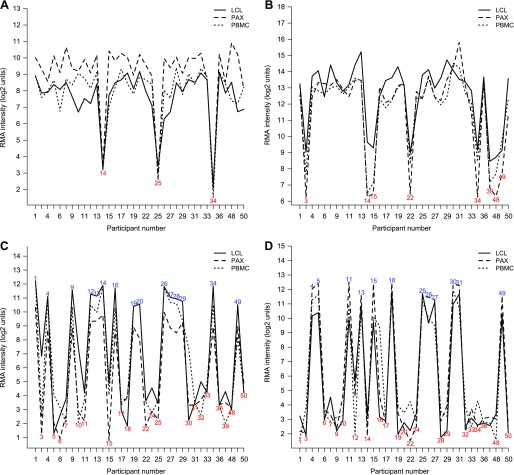
Heat map of expression values of the top “fingerprinting” probe sets. These probe sets have the largest F-values for participant effects (Table 10, Supplementary Table S10) and are clustered with Ward's hierarchical clustering. The first 5 are Y-linked. Colors indicate expression values after subtracting the mean within RNA source, with red having high values. The participants are clustered perfectly in groups of 3, 1 for each RNA source indicating that perfect self-identification is possible from expression data, even across different RNA sources. Participant number indicated at bottom.
Fig. 4.
Expression profile of gene β-actin (ACTB) on chromosome 7p22. ACTB has 15 known exons and 14 known RefSeq transcripts. According to the Alternative Splicing Database (72), this gene is known to have 10 splice variants. A: gene level profile; B: 1 of 11 RefSeq-core probe sets, 3036926. Since the bimodal distribution is seen at the gene level, it is not likely to be solely the result of splice variation.
Fig. 5.
Expression profile of some fingerprinting probe sets. A: EXOC1 probe set 2727952, B: OAS1 probe set 3432446, C: GSTM1 probe set 2350993, and D: HLA-DRB1 probe set 4048279. These probe sets show strong, participant-specific variation in expression consistently in all 3 sample types and may reflect genetically determined variation in expression levels (e.g., functional SNPs, CNVs, imprinting, LOH) or variation in mRNA sequence in each participant (e.g., SNP) compared with the Affymetrix probe sequence.
Stable “calibration” genes.
Conversely, we also searched for genes expressed above background (>4.0 in log2 RMA scale) and that had nonsignificant expression changes (<2.0-fold change, P value >0.2) across RNA sources and across participants. These genes would be valuable for batch corrections, meta-analysis across RNA sources or platforms, and for calibrating expression levels of transcripts of other genes (17). We found 139 genes meeting these criteria (Supplementary Table S11). Most are well-known and well-annotated protein coding genes. Many are known to be expressed in whole blood. Some of the most stable genes were CLCN6, TEAD3, ART5, COX6A2, SIRT5, ACTL6B, GPR50, GPR32, and RAB8B. Although these may not commonly be used as housekeeping genes, they are likely to be quite stable as calibration standards in future analyses using this platform. At the exon level, we found 1,544 exons representing 1,355 genes that passed similar selection criteria. Of these exons 25, representing 22 distinct genes, were common to the set selected at the gene level, including CLCN6, CSNK1G3, FAM48A, and RAB8B.
DISCUSSION
Each of the three RNA sources bears distinct characteristics, evident by the clear separation in the first two principal components (Fig. 2) and the finding that most genes were differentially expressed among the different sources (Table 2). Since most genes are expressed differentially across the RNA sources, their associations with each of the traits we studied are also different, warranting careful selection of the RNA source in a gene expression experiment. For the gene expression signature of sex, all three RNA sources yielded a large common subset of Y-chromosome genes strongly linked to sex. LCL samples were able to detect expression differences in X-chromosome genes between men and women, but this may be due to reversal of X-chromosome inactivation during EBV infection, cell immortalization, and culture. PBMC were better able to detect sex-linked autosomal genes than the other two RNA sources, although apparently none of the detected genes were also detected in prior studies (39), suggesting that our observation may be unique to our sample.
As cultured cells, LCL samples are less likely than PAX or PBMC samples to reflect in vivo expression changes. For example, LCL did not detect association between lymphocyte-related genes and lymphocyte differential counts. These findings, together with the perturbation of expression attributable to the EBV transformation process itself, suggest that LCL may be of limited value in identifying expression signatures of many health related traits. Prior work has shown limitations in the use of expression signatures in LCL due to their ex vivo status (16, 21). However, the ability of LCL to detect downregulation of ALDH2 in smokers suggests that epigenetic influences conditioned by the environment may still be encoded in LCL expression profiles.
It is important to note that a proportion of the differences observed between PAXgene and the other two sample types may be due to differences in preparation kits. As noted in materials and methods, PAX require a distinct preparation kit from that used for PBMC and LCL. However, by focusing on the minimal difference observed between each type vs. the other two (see Table 3), we attempted to report differences most likely attributable to underlying biological differences rather than simply due to technical sources. For example, the comparison of LCL with PBMC (which use the same preparation kit) shows very large differences for genes involved in cell-cycle pathways, as might be expected in transformed LCL cells.
Our study has several important advantages over prior studies. A balanced study design with three blood-derived RNA sources from each of 35 participants allows investigation of biomarkers and source-invariant genes to be undertaken more thoroughly. Indeed, few population-based expression studies include replicate samples in as many participants as are included here. This study includes multiple samples from the same individual, separated in time by as much as 5 yr. Expression patterns that persist across these samples are more likely to represent true stable phenotypes of the individual, than are those based on single, one-time measurements. Genes and exons showing variation in expression across the population, yet remaining consistent within the individual over years are likely to be enriched in useful expression biomarkers of risk factors or disease, compared with other genes. Furthermore, such genes and exons may be more likely to be associated with genetic factors (such as expression single nucleotide polymorphisms), than are genes having greater within-individual variation.
We showed that some of genes or exons showing variation in expression across our study sample can be used to distinguish individuals, suggesting that microarray expression data alone provide a personally identifiable fingerprint. In our study, only a tiny fraction of all exons distinguished individuals perfectly. This finding may prompt consideration of the identifiability of individuals within public microarray databases and whether safeguards are needed to protect their privacy. Conversely, we also provided result on stable and robust markers that may help researchers to calibrate their gene expression results. Calibration has been one of the major issues in gene expression analysis. We showed that conventional calibration genes, such as ACTB, may not be reliable.
We believe fingerprinting genes are useful in two contexts. First, in quality control of high-throughput assays, the identity of samples is sometimes questioned. Estimates of sample mix-ups often range up to 18% (79). If left unaddressed, this can introduce errors in the analysis and may possibly lead to the weakened or incorrect conclusion (47). Indeed some mix-ups were detected in the current study by aligning predicted sex based on Y-chromosome expression with that recorded in the database for the subject. When multiple samples from the same individuals are assayed, analysis of fingerprinting gene expression levels can be used to further identify mislabeled samples by clustering of such genes. The second context would be in searching for eQTLs (expression quantitative trait loci). A quantitative trait should be tightly coupled to the genome and recognizable regardless of when or in what tissue the gene expression level is measured. The set of fingerprinting genes are here shown to be stable within individual (in the small number of tissues tested) and over time (since the LCL cells were derived from an earlier blood draw, compared with the PAX and PBMC samples) and are thus good candidate quantitative traits. In searching for eQTLs, i.e., loci in the genome associated with quantitative traits, the fingerprinting genes, should be an excellent place to start. It has previously been noted that some genes are expressed in a bimodal fashion in the population (e.g., ACTB) and that a disproportionately large number of such genes have associations to disease (48). Many of our fingerprinting genes appear to express bimodally. Thus, it is reasonable to hypothesize that our fingerprinting genes might also contain a large fraction of genes (e.g., the HLA genes) related to disease or disease propensities.
Our study considered only blood-derived RNA sources, because this is one source likely to be widely available in a large population-based study. Although a desired tissue, such as brain in stroke patients, may be inaccessible, one can sometimes use blood as a surrogate, provided the relevant transcripts are similarly expressed in blood and brain. In certain situations (e.g., angioplasty, heart transplant, or coronary artery bypass graft surgery), it may be possible to obtain paired blood and heart tissue samples, from which the relevant transcripts expressed similarly in both can be determined. Accumulating such information will ultimately make blood-derived expression data in population-based studies more valuable in the future.
A larger sample size would have improved our power for biomarker discovery. The relatively narrow age range in this study likely prevented detection of extensive associations with age. In addition, analysis of many complex traits influenced by multiple genes each having modest effects (29) will require larger sample sizes. Larger sample size (or combining results of many studies) would have the additional benefit of further characterizing the measurement platform. The Affymetrix Exon array has ∼1.4 million probe sets, of which only about one-fourth were analyzed here. These probe sets were used because they correspond to well-annotated transcripts and have good performance characteristics. Many of the remaining probe sets have unknown performance characteristics or correspond to unannotated regions of the genome or to weakly annotated genes. Pooling experience from the growing number of published results on this platform will allow us to more sharply focus on the better-performing probe sets, while the general improvement of genome annotation will make other probe sets more useful in the future.
Although our pilot study was small and not intended for biomarker discovery, we were able to confirm associations of expression with lipid levels in two previously implicated genes, FADS1 and LDLR. While the observed effects were small, the magnitude, direction, and significance were consistent in PAX and PBMC samples, but not in LCL. This, again, suggests that LCL samples are less appropriate for detecting signatures related to health-related traits. The ability of even a small study to confirm associations with these well-established lipid-controlling genes lends optimism that more associations would be detected in a larger study, using either PAX or PBMC. Based on the results of this pilot, the larger, population-based SABRe in CVD Initiative will be using PAX as its RNA source and the Affymetrix Exon array platform. Completion of data collection is anticipated in late 2011.
GRANTS
The National Heart, Lung, and Blood Institute's (NHLBI's) FHS is supported by National Institutes of Health Grant NO1-HC-25195. The SABRe CVD Initiative is funded by the Division of Intramural Research, NHLBI, Bethesda, MD.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
D. L and P. J. M. designed, directed, and supervised the experiment. D. L. was responsible for funding of the project. R. J. and P. J. M. drafted the manuscript. P. J. M., D. L., A. D. J., and C. J. O. revised and edited the manuscript. R. J. and P. J. M. performed the statistical analysis. J. J. B. performed S10 normalization of the data. N. R., P. L., and K. A. W. collected the data. All authors have read and approved the final version of the manuscript.
Supplementary Material
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Affymetrix. Transcript assignment for NetAffx(TM) Annotations [online]. http://www.affymetrix.com/support/technical/byproduct.affx?product=huexon-st, 2006.
- 2.Affymetrix. Quality Assessment of Exon and Gene Arrays, 2007. [Google Scholar]
- 3.Affymetrix. GeneChip Whole Transcript (WT) Sense Target Labeling Assay Manual [online]. http://www.affymetrix.com/support/downloads/manuals/wt_sensetarget_label_manual.pdf.
- 4.Affymetrix. Affymetrix Power Tools [online]. http://www.affymetrix.com/partners_programs/programs/developer/tools/powertools.affx.
- 5.Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, Sullivan K, Matakidou A, Wang Y, Mills G, Doheny K, Tsai YY, Chen WV, Shete S, Spitz MR, Houlston RS. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet 40: 616–622, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asare AL, Kolchinsky SA, Gao Z, Wang R, Raddassi K, Bourcier K, Seyfert-Margolis V. Differential gene expression profiles are dependent upon method of peripheral blood collection and RNA isolation. BMC Genomics 9: 474, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avent ND, Reid ME. The Rh blood group system: a review. Blood 95: 375–387, 2000. [PubMed] [Google Scholar]
- 9.Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, Shark KB, Grande WJ, Hughes KM, Kapur V, Gregersen PK, Behrens TW. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci USA 100: 2610–2615, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barr TL, Conley Y, Ding J, Dillman A, Warach S, Singleton A, Matarin M. Genomic biomarkers and cellular pathways of ischemic stroke by RNA gene expression profiling. Neurology 75: 1009–1014, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B 57: 289–300, 1995. [Google Scholar]
- 12.Birkenbach M, Josefsen K, Yalamanchili R, Lenoir G, Kieff E. Epstein-Barr virus-induced genes: first lymphocyte-specific G protein-coupled peptide receptors. J Virol 67: 2209–2220, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blumenfeld OO, Huang CH. Molecular genetics of the glycophorin gene family, the antigens for MNSs blood groups: multiple gene rearrangements and modulation of splice site usage result in extensive diversification. Hum Mutat 6: 199–209, 1995. [DOI] [PubMed] [Google Scholar]
- 14.Boldrick JC, Alizadeh AA, Diehn M, Dudoit S, Liu CL, Belcher CE, Botstein D, Staudt LM, Brown PO, Relman DA. Stereotyped and specific gene expression programs in human innate immune responses to bacteria. Proc Natl Acad Sci USA 99: 972–977, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bourke E, Cassetti A, Villa A, Fadlon E, Colotta F, Mantovani A. IL-1 beta scavenging by the type II IL-1 decoy receptor in human neutrophils. J Immunol 170: 5999–6005, 2003. [DOI] [PubMed] [Google Scholar]
- 16.Cain CE, Blekhman R, Marioni JC, Gilad Y. Gene expression differences among primates are associated with changes in a histone epigenetic modification. Genetics 187: 1225–1234, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caradec J, Sirab N, Keumeugni C, Moutereau S, Chimingqi M, Matar C, Revaud D, Bah M, Manivet P, Conti M, Loric S. “Desperate house genes”: the dramatic example of hypoxia. Br J Cancer 102: 1037–1043, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casabonne D, Reina O, Benavente Y, Becker N, Maynadié M, Foretová L, Cocco P, González-Neira A, Nieters A, Boffetta P, Middeldorp JM, de Sanjose S. Single nucleotide polymorphisms of matrix metalloproteinase 9 (MMP9) and tumor protein 73 (TP73) interact with Epstein-Barr virus in chronic lymphocytic leukemia: results from the European case-control study EpiLymph. Haematologica 96: 323–327, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.CDC. Understanding Your Complete Blood Count [online]. http://www.cc.nih.gov/ccc/patient_education/pepubs/cbc97.pdf, 2008.
- 20.Chen PW, Lin SJ, Tsai SC, Lin JH, Chen MR, Wang JT, Lee CP, Tsai CH. Regulation of microtubule dynamics through phosphorylation on stathmin by Epstein-Barr virus kinase BGLF4. J Biol Chem 285: 10053–10063, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choy E, Yelensky R, Bonakdar S, Plenge RM, Saxena R, De Jager PL, Shaw SY, Wolfish CS, Slavik JM, Cotsapas C, Rivas M, Dermitzakis ET, Cahir-McFarland E, Kieff E, Hafler D, Daly MJ, Altshuler D. Genetic analysis of human traits in vitro: drug response and gene expression in lymphoblastoid cell lines. PLoS Genet 4: e1000287, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conboy JG. Structure, function, and molecular genetics of erythroid membrane skeletal protein 4.1 in normal and abnormal red blood cells. Semin Hematol 30: 58–73, 1993. [PubMed] [Google Scholar]
- 23.Cox TC, Sadlon TJ, Schwarz QP, Matthews CS, Wise PD, Cox LL, Bottomley SS, May BK. The major splice variant of human 5-aminolevulinate synthase-2 contributes significantly to erythroid heme biosynthesis. Int J Biochem Cell Biol 36: 281–295, 2004. [DOI] [PubMed] [Google Scholar]
- 24.Dawber TR, Meadors GF, Moore FE., Jr Epidemiological approaches to heart disease: the Framingham Study. Am J Public Health Nations Health 41: 279–281, 1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Debey S, Schoenbeck U, Hellmich M, Gathof BS, Pillai R, Zander T, Schultze JL. Comparison of different isolation techniques prior gene expression profiling of blood derived cells: impact on physiological responses, on overall expression and the role of different cell types. Pharmacogenomics J 4: 193–207, 2004. [DOI] [PubMed] [Google Scholar]
- 26.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feinleib M, Kannel WB, Garrison RJ, McNamara PM, Castelli WP. The Framingham Offspring Study. Prev Med 4: 518–525, 1975. [DOI] [PubMed] [Google Scholar]
- 28.Fujita K, Horikawa I, Mondal AM, Jenkins LMM, Appella E, Vojtesek B, Bourdon JC, Lane DP, Harris CC. Positive feedback between p53 and TRF2 during telomere-damage signalling and cellular senescence. Nat Cell Biol 12: 1205–1212, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Govindaraju DR, Larson MG, Yin X, Benjamin EJ, Rao MB, Vasan RS. Association between SNP heterozygosity and quantitative traits in the Framingham Heart Study. Ann Hum Genet 73: 465–473, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grünblatt E, Bartl J, Zehetmayer S, Ringel TM, Bauer P, Riederer P, Jacob CP. Gene expression as peripheral biomarkers for sporadic Alzheimer's disease. J Alzheimers Dis 16: 627–634, 2009. [DOI] [PubMed] [Google Scholar]
- 31.Higgs DR, Vickers MA, Wilkie AO, Pretorius IM, Jarman AP, Weatherall DJ. A review of the molecular genetics of the human alpha-globin gene cluster. Blood 73: 1081–1104, 1989. [PubMed] [Google Scholar]
- 32.Hindle AK, Edwards C, McCaffrey T, Fu SW, Brody F. Reactivation of adiponectin expression in obese patients after bariatric surgery. Surg Endosc 24: 1367–1373, 2010. [DOI] [PubMed] [Google Scholar]
- 33.Huang RS, Chen P, Wisel S, Duan S, Zhang W, Cook EH, Das S, Cox NJ, Dolan ME. Population-specific GSTM1 copy number variation. Hum Mol Genet 18: 366–372, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264, 2003. [DOI] [PubMed] [Google Scholar]
- 35.Isensee J, Witt H, Pregla R, Hetzer R, Regitz-Zagrosek V, Noppinger PR. Sexually dimorphic gene expression in the heart of mice and men. J Mol Med 86: 61–74, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iyoda T, Ushida M, Kimura Y, Minamino K, Hayuka A, Yokohata S, Ehara H, Inaba K. Invariant NKT cell anergy is induced by a strong TCR-mediated signal plus co-stimulation. Int Immunol 22: 905–913, 2010. [DOI] [PubMed] [Google Scholar]
- 37.Jacobsen LC, Sørensen OE, Cowland JB, Borregaard N, Theilgaard-Mönch K. The secretory leukocyte protease inhibitor (SLPI) and the secondary granule protein lactoferrin are synthesized in myelocytes, colocalize in subcellular fractions of neutrophils, and are coreleased by activated neutrophils. J Leukoc Biol 83: 1155–1164, 2008. [DOI] [PubMed] [Google Scholar]
- 38.Jeon JP, Kim JW, Park B, Nam HY, Shim SM, Lee MH, Han BG. Identification of tumor necrosis factor signaling-related proteins during Epstein-Barr virus-induced B cell transformation. Acta Virol 52: 151–159, 2008. [PubMed] [Google Scholar]
- 39.Jison ML, Munson PJ, Barb JJ, Suffredini AF, Talwar S, Logun C, Raghavachari N, Beigel JH, Shelhamer JH, Danner RL, Gladwin MT. Blood mononuclear cell gene expression profiles characterize the oxidant, hemolytic, and inflammatory stress of sickle cell disease. Blood 104: 270–280, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson AD, O'Donnell CJ. An open access database of genome-wide association results. BMC Med Genet 10: 6, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kashuba E, Yurchenko M, Yenamandra SP, Snopok B, Szekely L, Bercovich B, Ciechanover A, Klein G. Epstein-Barr virus-encoded EBNA-5 forms trimolecular protein complexes with MDM2 and p53 and inhibits the transactivating function of p53. Int J Cancer 128: 817–825, 2011. [DOI] [PubMed] [Google Scholar]
- 42.Khanna-Gupta A, Zibello T, Idone V, Sun H, Lekstrom-Himes J, Berliner N. Human neutrophil collagenase expression is C/EBP-dependent during myeloid development. Exp Hematol 33: 42–52, 2005. [DOI] [PubMed] [Google Scholar]
- 43.Kostylina G, Simon D, Fey MF, Yousefi S, Simon HU. Neutrophil apoptosis mediated by nicotinic acid receptors (GPR109A). Cell Death Differ 15: 134–142, 2008. [DOI] [PubMed] [Google Scholar]
- 44.Lane HC, Anand AR, Ganju RK. Cbl and Akt regulate CXCL8-induced and CXCR1- and CXCR2-mediated chemotaxis. Int Immunol 18: 1315–1325, 2006. [DOI] [PubMed] [Google Scholar]
- 45.Larousserie F, Bardel E, Coulomb L'Herminé A, Canioni D, Brousse N, Kastelein RA, Devergne O. Variable expression of Epstein-Barr virus-induced gene 3 during normal B-cell differentiation and among B-cell lymphomas. J Pathol 209: 360–368, 2006. [DOI] [PubMed] [Google Scholar]
- 46.Li CY, Zhan YQ, Xu CW, Xu WX, Wang SY, Lv J, Zhou Y, Yue PB, Chen B, Yang XM. EDAG regulates the proliferation and differentiation of hematopoietic cells and resists cell apoptosis through the activation of nuclear factor-kappa B. Cell Death Differ 11: 1299–1308, 2004. [DOI] [PubMed] [Google Scholar]
- 47.Malossini A, Blanzieri E, Ng RT. Assessment of SVM reliability of microarrays data analysis. 14th Dutch-Belgian Conference of Machine Learning. WP05–03, 2005. [Google Scholar]
- 48.Mason CC, Hanson RL, Ossowski V, Bian L, Baier LJ, Krakoff J, Bogardus C. Bimodal distribution of RNA expression levels in human skeletal muscle tissue. BMC Genomics 12: 98, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McLaren JE, Zuo J, Grimstead J, Poghosyan Z, Bell AI, Rowe M, Brennan P. STAT1 contributes to the maintenance of the latency III viral programme observed in Epstein-Barr virus-transformed B cells and their recognition by CD8+ T cells. J Gen Virol 90: 2239–2250, 2009. [DOI] [PubMed] [Google Scholar]
- 50.Min JL, Barrett A, Watts T, Pettersson FH, Lockstone HE, Lindgren CM, Taylor JM, Allen M, Zondervan KT, McCarthy MI. Variability of gene expression profiles in human blood and lymphoblastoid cell lines. BMC Genomics 11: 96, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mohan J, Dement-Brown J, Maier S, Ise T, Kempkes B, Tolnay M. Epstein-Barr virus nuclear antigen 2 induces FcRH5 expression through CBF1. Blood 107: 4433–4439, 2006. [DOI] [PubMed] [Google Scholar]
- 52.Munson PJ. A consistency test for determining the significance of gene expression changes on replicate samples and two convenient variance-stabilizing transformations [online]. GeneLogic Workshop on Low Level Analysis of Affymetrix GeneChip Data. http://stat-www.berkeley.edu/users/terry/zarray/Affy/GL_Workshop/genelogic2001.html.
- 53.Murtagh F. Multidimensional Clustering Algorithms. Würzburg: Physica-Verlag, 1985. [Google Scholar]
- 54.O'Donnell CJ, Elosua R. Cardiovascular risk factors. Insights from Framingham Heart Study. Rev Esp Cardiol 61: 299–310, 2008. [PubMed] [Google Scholar]
- 55.Oppenheimer GM. Becoming the Framingham Study 1947–1950. Am J Public Health 95: 602–610, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Palikhe NS, Kim JH, Park HS. Biomarkers predicting isocyanate-induced asthma. Allergy Asthma Immunol Res 3: 21–26, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park JY, Matsuo K, Suzuki T, Ito H, Hosono S, Kawase T, Watanabe M, Oze I, Hida T, Yatabe Y, Mitsudomi T, Takezaki T, Tajima K, Tanaka H. Impact of smoking on lung cancer risk is stronger in those with the homozygous aldehyde dehydrogenase 2 null allele in a Japanese population. Carcinogenesis 31: 660–665, 2010. [DOI] [PubMed] [Google Scholar]
- 58.Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E, Veyrieras JB, Stephens M, Gilad Y, Pritchard JK. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 464: 768–772, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pinheiro J, Bates D. Mixed-effects Models in S and S-PLUS. New York: Springer, 2009. [Google Scholar]
- 60.Provan S, Angel K, Semb AG, Atar D, Kvien TK. NT-proBNP predicts mortality in patients with rheumatoid arthritis: results from 10-year follow-up of the EURIDISS study. Ann Rheum Dis 69: 1946–1950, 2010. [DOI] [PubMed] [Google Scholar]
- 61.QIAGEN. RNeasy Plus Handbook [online]. http://www.qiagen.com/literature/render.aspx?id=103686.
- 62.QIAGEN. PAXgene Blood RNA Kit Handbook Version 2 [online]. http://www.qiagen.com/literature/render.aspx?id=104458.
- 63.R Development Core Team. R: A Language and Environment for Statistical Computing [online]. http://www.R-project.org.
- 64.Raghavachari N, Xu X, Harris A, Villagra J, Logun C, Barb J, Solomon MA, Suffredini AF, Danner RL, Kato G, Munson PJ, Morris SM, Jr, Gladwin MT. Amplified expression profiling of platelet transcriptome reveals changes in arginine metabolic pathways in patients with sickle cell disease. Circulation 115: 1551–1562, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rana AP, Ruff P, Maalouf GJ, Speicher DW, Chishti AH. Cloning of human erythroid dematin reveals another member of the villin family. Proc Natl Acad Sci USA 90: 6651–6655, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rockett JC, Burczynski ME, Fornace AJ, Herrmann PC, Krawetz SA, Dix DJ. Surrogate tissue analysis: monitoring toxicant exposure and health status of inaccessible tissues through the analysis of accessible tissues and cells. Toxicol Appl Pharmacol 194: 189–199, 2004. [DOI] [PubMed] [Google Scholar]
- 67.Rollins B, Martin MV, Morgan L, Vawter MP. Analysis of whole genome biomarker expression in blood and brain. Am J Med Genet B Neuropsychiatr Genet 153B: 919–936, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rowe M, Lear AL, Croom-Carter D, Davies AH, Rickinson AB. Three pathways of Epstein-Barr virus gene activation from EBNA1-positive latency in B lymphocytes. J Virol 66: 122–131, 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rybicki AC, Musto S, Schwartz RS. Identification of a band-3 binding site near the N-terminus of erythrocyte membrane protein 4.2. Biochem J 309: 677–681, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schaniel C, Rolink AG, Melchers F. Attractions and migrations of lymphoid cells in the organization of humoral immune responses. Adv Immunol 78: 111–168, 2001. [DOI] [PubMed] [Google Scholar]
- 71.Splansky GL, Corey D, Yang Q, Atwood LD, Cupples LA, Benjamin EJ, D'Agostino RB, Fox CS, Larson MG, Murabito JM, O'Donnell CJ, Vasan RS, Wolf PA, Levy D. The third generation cohort of the National Heart, Lung, and Blood Institute's Framingham Heart Study: design, recruitment, and initial examination. Am J Epidemiol 165: 1328–1335, 2007. [DOI] [PubMed] [Google Scholar]
- 72.Stamm S, Riethoven JJ, Le Texier V, Gopalakrishnan C, Kumanduri V, Tang Y, Barbosa-Morais NL, Thanaraj TA. ASD: a bioinformatics resource on alternative splicing. Nucleic Acids Res 34: D46–D55, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stanietsky N, Mandelboim O. Paired NK cell receptors controlling NK cytotoxicity. FEBS Lett 584: 4895–4900, 2010. [DOI] [PubMed] [Google Scholar]
- 74.Tanner MJ. Molecular and cellular biology of the erythrocyte anion exchanger (AE1). Semin Hematol 30: 34–57, 1993. [PubMed] [Google Scholar]
- 75.Twine NC, Stover JA, Marshall B, Dukart G, Hidalgo M, Stadler W, Logan T, Dutcher J, Hudes G, Dorner AJ, Slonim DK, Trepicchio WL, Burczynski ME. Disease-associated expression profiles in peripheral blood mononuclear cells from patients with advanced renal cell carcinoma. Cancer Res 63: 6069–6075, 2003. [PubMed] [Google Scholar]
- 76.Venables WN, Ripley BD. Modern Applied Statistics With S (4th ed.). New York: Springer, 2002. [Google Scholar]
- 77.Wallace AE, Sales KJ, Catalano RD, Anderson RA, Williams ARW, Wilson MR, Schwarze J, Wang H, Rossi AG, Jabbour HN. Prostaglandin F2alpha-F-prostanoid receptor signaling promotes neutrophil chemotaxis via chemokine (C-X-C motif) ligand 1 in endometrial adenocarcinoma. Cancer Res 69: 5726–5733, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Waterworth DM, Ricketts SL, Song K, Chen L, Zhao JH, Ripatti S, Aulchenko YS, Zhang W, Yuan X, Lim N, Luan J, Ashford S, Wheeler E, Young EH, Hadley D, Thompson JR, Braund PS, Johnson T, Struchalin M, Surakka I, Luben R, Khaw KT, Rodwell SA, Loos RJF, Boekholdt SM, Inouye M, Deloukas P, Elliott P, Schlessinger D, Sanna S, Scuteri A, Jackson A, Mohlke KL, Tuomilehto J, Roberts R, Stewart A, Kesäniemi YA, Mahley RW, Grundy SM, McArdle W, Cardon L, Waeber G, Vollenweider P, Chambers JC, Boehnke M, Abecasis GR, Salomaa V, Järvelin MR, Ruokonen A, Barroso I, Epstein SE, Hakonarson HH, Rader DJ, Reilly MP, Witteman JCM, Hall AS, Samani NJ, Strachan DP, Barter P, van Duijn CM, Kooner JS, Peltonen L, Wareham NJ, McPherson R, Mooser V, Sandhu MS. Genetic variants influencing circulating lipid levels and risk of coronary artery disease. Arterioscler Thromb Vasc Biol 30: 2264–2276, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.West M, Blanchette C, Dressman H, Huang E, Ishida S, Spang R, Zuzan H, Olson JA, Jr, Marks JR, Nevins JR. Predicting the clinical status of human breast cancer by using gene expression profiles. Proc Natl Acad Sci USA 98: 11462–11467, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Whitney AR, Diehn M, Popper SJ, Alizadeh AA, Boldrick JC, Relman DA, Brown PO. Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci USA 100: 1896–1901, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.