Abstract
Feeding mice with protease inhibitor (PI) leads to increased endogenous cholecystokinin (CCK) release and results in pancreatic growth. This adaptive response requires calcineurin (CN)-NFAT and AKT-mTOR pathways, but the genes involved, the dynamics of their expression, and other regulatory pathways remain unknown. Here, we examined the early (1–8 h) transcriptional program that underlies pancreatic growth. We found 314 upregulated and 219 downregulated genes with diverse temporal and functional profiles. Several new identifications include the following: stress response genes Gdf15 and Txnip, metabolic mediators Pitpnc1 and Hmges2, as well as components of growth factor response Fgf21, Atf3, and Egr1. The genes fell into seven self-organizing clusters, each with a distinct pattern of expression; a representative gene within each of the upregulated clusters (Egr1, Gadd45b, Rgs2, and Serpinb1a) was validated by qRT-PCR. Genes up at any point throughout the time course and CN-dependent genes were subjected to further bioinformatics-based networking and promoter analysis, yielding STATs as potential transcriptional regulators. As shown by PCR, qPCR, and Western blots, the active phospho-form of STAT3 and the Jak-STAT feedback inhibitor Socs2 were both increased throughout early pancreatic growth. Moreover, immunohistochemistry showed a CCK-dependent and acinar cell-specific increase in nuclear localization of p-STAT3, with >75% nuclear occupancy in PI-fed mice vs. <0.1% in controls. Thus, the study identified novel genes likely to be important for CCK-driven pancreatic growth, characterized and biologically validated the dynamic pattern of their expression and investigated STAT-Socs signaling as a new player in this trophic response.
Keywords: cholecystokinin, pancreas, regeneration, signal transducer and activator of transcription
the adult pancreas is typically static in size and exhibits little cellular turnover. The otherwise quiescent pancreas, however, can increase in size in response to exogenous stimuli (5, 31); hyperphagia, increased dietary protein, or injury can all promote pancreatic growth (20). The best-studied mediators of mitogenesis and hypertrophy of exocrine pancreas are gastrointestinal hormones, particularly cholecystokinin (CCK). Exogenous administration of CCK stimulates exocrine pancreatic mitogenesis and growth (33, 38, 44) and leads to increased acinar cell division in culture (21, 23). In vivo, increased endogenous CCK induced by feeding chow containing protease inhibitors (PI) also leads to robust growth, more than doubling the size of the rodent pancreas relative to body weight (27, 41). Furthermore, this effect is abrogated in both CCK (41) and CCK-A receptor-deficient mice (36).
Work on CCK-driven pancreatic growth in vivo has thus far focused on the following three pathways: mitogen-activated protein kinases ERKs and JNKs (8), nutritional sensors AKT and mammalian target of rapamycin (mTOR) (3), as well as the calcineurin-nuclear factor of activated T-cells (CN/NFAT) cascade (41). First, elevated CCK-induced by administration of protease inhibitor (PI)-containing chow has been shown to activate ERK, JNK, and their downstream mediators, including activated protein-1 (AP-1) (8). Second, CCK also activates the atypical kinase mTOR, which leads to phosphorylation of initiation factor 4E binding protein and protein S6 kinase, as well as resultant increase in protein synthesis (1). In addition, rapamycin, a well-known inhibitor of mTOR, blocks CCK-induced pancreatic growth and acinar cell division (3). Finally, CCK activates CN and its downstream transcriptional effector nuclear NFATs, both in vivo and in isolated acini (10). CN is necessary for pancreatic growth, as administration of its inhibitors FK506 and cyclosporin A leads to near complete ablation of this response (41). Likewise, acinar cell-specific overexpression of Rcan1 (regulator of calcineurin 1), an endogenous feedback inhibitor of CN, is also sufficient to block this response (9). A limited, single time point gene expression array analysis carried out as part of the above-mentioned study, however, also suggested that adaptive growth of the pancreas is a complex response involving multiple other pathways.
Classically characterized as mediators of cytokine signaling, the components of the JAK-STAT pathway have now been shown to play a critical role in gastrointestinal homeostasis (6, 15). Disregulation of JAK-STAT may be an important component of inflammatory bowel disease (37). IL-6 receptor and leukemia inhibitory factor, both of which signal via Jak-STAT, are key in liver regeneration following partial hepatectomy (29). Furthermore, deletion of suppressor of cytokine signaling 2 (SOCS2) leads to disinhibition of Jak-STAT and resultant 30–40% increase of lean body weight (28). In the pancreas, the role of Jak-STAT signaling has been examined almost exclusively in the islets of Langerhans. STAT5 and Socs3, for instance, have been shown to regulate pancreatic β-cell mass and proliferation (18). Activation of Jak-STATs in the exocrine compartment and the potential role of this pathway in GI hormone-mediated pancreatic growth, however, remain unexplored.
Several large-scale profiling studies to characterize pancreatitis and pancreatic cancer have been performed (14, 22), but analogous study of adaptive, hormonally driven pancreatic growth has yet to be completed. A better understanding of pancreatic growth and regeneration should inform the development of clinical applications aimed at manipulation of digestion and metabolism. Insights from work on adaptive growth mediated by GI hormones may also impact the efforts to manipulate exocrine fate for therapies aimed at pancreatic cancer, pancreatitis, or even diabetes. Recent advances surrounding the plasticity of acinar cells further highlight this topic (13, 30).
The objective of this study was to perform rigorous computational and experimental analysis of the genetic program that underlies the early, 1–8 h time course of CCK-mediated pancreatic growth. This work led us to identify and evaluate Jak-STAT signaling as a novel and potentially important pathway activated in the course of this hormonally driven trophic response.
METHODS
Materials.
Synthetic PI camostat, also known as FOY-305, was provided by Ono Pharmaceuticals (Osaka, Japan). TaqMan reverse transcription reagents and expand high-fidelity PCR system were from Roche (Basel, Switzerland). Trizol and PCR primers were obtained from Invitrogen (Carlsbad, CA) and RNAlater from Ambion (Austin, TX). Antibodies were obtained as follows: phospho-Tyr705 STAT3 (#D3A7) from Cell Signaling (Boston, MA), total STAT3 (#61018) from BD Transduction Laboratories, Socs2 (Ab3692) from Abcam (Cambridge, MA), Lamin A/C (sc-20681) from Santa Cruz Biotechnology (Santa Cruz, CA), and cyclophilin A (07-313) from Upstate Biotechnology (Lake Placid, NY). Secondary antibodies and enhanced chemiluminescence (ECL) reagents came from Amersham Pharmacia (Piscataway, NJ). Precast gels, nitrocellulose membrane, and SDS-PAGE standard markers were from Bio-Rad (Hercules, CA). CCK-8 peptide was from Research Plus (Bayonne, NJ) and MiniComplete protease inhibitor cocktail from Roche. All other chemical reagents were obtained from Sigma (St. Louis, MO).
Animals and treatment.
Male ICR mice (Harlan Sprague-Dawley, Indianapolis, IN) weighing 25–30 g were used for all experiments. The mice were housed at 22–24°C on a 12 h light/dark cycle with free access to water and a standard, pellet chow (5001 Rodent Diet; PMI Nutrition International, St. Louis, MO). Prior to initiating any study, we first acclimated animals to the powder form of the chow for at least 2 days. The mice were then fasted overnight, refed with control powder chow or powder chow containing 0.1% PI for 1–8 h, and then sacrificed. All studies were approved by the University of Michigan committee on Use and Care of Animals.
RNA isolation, RT, and PCR.
RNA isolation, cDNA synthesis, and PCR followed the same procedures as previously described (9). Briefly, total RNA was isolated from pancreas using TRIzol and RNeasy spin columns (Qiagen, Valencia, CA). RNA quality was evaluated by agarose gel electrophoresis and nanodrop spectrophotometer (Thermo Scientific, Wilmington, DE). We reverse transcribed 1 μg of total RNA using TaqMAN reagents with random hexamers as primers. PCR and qPCR were carried out using reagents from Expand High Fidelity PCR system in an Eppendorf Epgradient Mastercycler and realPlex, respectively (Hamburg, Germany). The primers were designed with Primer3 ver4.0 based on gene sequences retrieved from GenBank NCBI.
Microarray and bioinformatics.
RNA was isolated as above, with cDNA synthesis, hybridization, and fluorescence signal optimization carried out on Affymetrix 430A gene chips at University of Michigan microarray core according to manufacturer's protocols. The data sets were initially compared by ANOVA with P values of the groups subject to F-test. False discovery rate (FDR) and associated q-values were then derived, as previously described (39).
Pertinent details can be obtained at: http://www.gurda.net/PMID_2210007/, and the complete set of data has also been deposited in Gene Expression Omnibus at http://www.ncbi.nlm.nih.gov/geo/. Bioinformatics analysis was done in an analogous fashion to our previously published work (9). Briefly, we performed and periodically updated a batch annotation of all probe sets using Netaffx software, available online through Affymetrix (Santa Clara, CA). We computed measures for FDR and defined an appropriate cutoff (q < 0.04–0.06) using open-source bioconductor package in R. Differentially regulated genes were then more extensively annotated and analyzed using Gene Ontology Tree Machine (GOTM) software (45), per authors' instructions. Further pathway and disease-directed, time point-specific analysis was also carried out using Ingenuity software, per authors' instructions. Network association analysis was performed as previously described (42), excluding oncomine database but otherwise at default settings. To analyze transcriptional regulation, we used the default 600 bp region of the promoter upstream of the start site, obtained using automatic retrieval tools on Genomatix software suite (2); sequence identification of TRANSFAC-based transcription factor binding site motifs were then carried out using Region Miner tool within Genomatix, per manufacturers' instructions.
Lysate preparation and Western blotting.
To generate whole tissue protein lysate, pancreatic tissue was homogenized with a polytron homogenizer in an ice-cold phosphate-buffered saline with MiniComplete protease inhibitor and then briefly sonicated; debris was removed by centrifugation, and supernatant was saved for further analysis. Protein concentrations were determined using protein assay reagent (Bio-Rad, Hercules, CA). Aliquots of the lysates were mixed with 4× SDS stop solution and boiled for 5 min. They were then loaded onto precast SDS-polyacrylamide gels (Bio-Rad), followed by electrophoresis, transfer onto nitrocellulose membranes, blocking, and incubation overnight with primary antibody at 4°C. The membranes were then washed, incubated with the appropriate secondary antibody, and visualized by ECL chemiluminescence using Alpha Ease FC8900 imaging system (Alpha Innotech, San Leandro, CA).
Immunohistochemistry.
Cryostat sections (6 μm thick) of snap-frozen pancreas were picked up on glass slides and fixed for 10 min in 100% methanol at −20°C. Immunofluorescence localization was performed following methods described previously (8). Rabbit monoclonal anti-P-STAT3 (#D3A7) antibody from Cell Signaling was diluted 1:800; secondary antibody was Alexa 594 diluted 1:200. DAPI (4′6-diamidino-2-phenylindole) was added to the mounting medium to counterstain nuclei. Digital images were taken with an Olympus BX-51 microscope and processed using Photoshop CS2 software (Adobe System, Mountain View, CA). Quantitation of nuclear signal was carried out using Metamorph software (Olympus, Japan), with mean and standard deviation derived from automated counts of eight ×40 fields per animal. Results were calculated as the ratio of pSTAT3-positive to DAPI-positive nuclei.
Statistics.
Statistical treatment of array and sequence data was performed as described above. For other experimental work, multiple comparisons were performed by one-way ANOVA followed by Dunnett's posttest, whereas two-way comparisons were performed via two-tailed Student's t-test; these were carried out on Graphpad Prism software (Hearne Scientific, Auckland, New Zealand).
RESULTS
Defining the genetic program of CCK-mediated pancreatic growth.
Expression profiling was performed on PI-fed mice, an established model of hormonally driven pancreatic growth (41); feeding was synchronized by an overnight fast and fasted animals served as controls. The pancreases of all animals appeared grossly and histologically unremarkable. We utilized Affymetrix 430A GeneChips and surveyed 22,665 probe sets, representing 13,850 unique genes and expressed sequence tags (ESTs). To focus on the early and more likely primary events, pancreatic mRNA was obtained from mice fed PI-containing chow for 1, 2, 4, 8 h as well as the fasted littermates. The material was sampled individually from four animals per group, for a total of 20 chips, with the data showing close intragroup correlation. Employing strict selection criteria (q value <0.04–0.06 and relative fold change ≥3), we found large number of differentially regulated genes, 533 in total; of these, 314 were up and 219 down vs. fasting (Table 1; the entire dataset can be found in Supplementary Table S2).1 Overall, almost all differentially expressed genes were upregulated at 1 and 2 h time points, whereas the distribution of upregulated vs. downregulated genes at the 4 and 8 h points was more even (Table 1). We also performed an overlay of the data at the 2 h time point with other previously published, array-generated sets of genes, including our prior work focused on CN-NFAT signaling at the 2 h time point of PI feeding (9), pancreatitis (14), and pancreatic cancer (22). We found a large degree of overlap with our past data sets (54% and 45%), a more moderate overlap with pancreatitis (12%), and no overlap with pancreatic adenocarcinoma (0%) (Supplementary Table S1). Lastly, we annotated and listed the top 5 differentially regulated genes at each time point (Table 2). Examples of these include the following: stress response genes Gdf15 and Txnip, regulators of cell metabolism Pitpnc1 and Hmges, as well as growth factor response genes Fgf21, Atf3, and Egr-1. Another category includes feedback inhibitors, such as Rgs2, Socs2, Socs3, and Sel1l, as well as previously documented Rcan1, which together may act as a brake for adaptive growth. Most of these genes are previously unknown in context of hormonally driven pancreatic growth and comprise a set of candidate mediators of this adaptive response.
Table 1.
Expression analysis of early (1–8 h) time course of PI feeding
| 1 h | 2 h | 4 h | 8 h | Any Point | |
|---|---|---|---|---|---|
| Up ↑ | 43 | 74 | 140 | 233 | 314 |
| Down ↓ | 2 | 5 | 132 | 136 | 219 |
Summary data of the number of genes significantly up or down (≤ 3-fold, false discovery rate >0.04–0.06) with protease inhibitor (PI) feeding vs. fasted controls at 1, 2, 4, and 8 h and at any point along the entire 1–8 h time course of early pancreatic growth.
Table 2.
Genes differentially regulated in PI-refed vs. fasted mice
| GenBank Accession | Gene Symbol | Product | Functional Annotation | PI vs. Fast (fold up) | |
|---|---|---|---|---|---|
| ↑ 1 h | NM_019466.1 | Rcan1 | regulator of calcineurin 1 | calcium signaling, phosphatase regulator, muscle development | 164 |
| NM_020013.1 | Fgf21 | fibroblast growth factor 2 | growth factor, extracellular space | 32.1 | |
| NM_010415.1 | HbEGF | heparin binding EGF-like GF | angiogenesis, cell migration, proliferation | 27.8 | |
| AY061760.1 | Nfil3 | nuclear factor, interleukin 3, regulated | transcriptional regulator, rhythmic process | 22.1 | |
| BC019946.1 | Atf3 | activating transcription factor 3 | transcriptional regulator, positive regulation of cell proliferation | 20.4 | |
| ↑ 2 h | BB241535 | Socs3 | suppressor of cytokine signaling 3 | JAK-STAT. negative regulator of signaling, organ regeneration | 43.6 |
| BG067321 | Rgs2 | regulator of G protein signaling 2 | cell cycle, negative regulator of signaling | 22.2 | |
| BB049138 | Pvr | poliovirus receptor | cell-cell adhesion, cell migration | 21.6 | |
| NM_011819.1 | Gdf15 | growth differentiation factor 15 | catabolism, stress response | 20.4 | |
| NM_008416.1 | Junb | Jun-B oncogene | vasculogenesis, transcription, blood vessel development | 19.2 | |
| Repeat: Rcan1, FGF21 | 71.7, 20.0 | ||||
| ↑ 4 h | AF356876.1 | Acat2 | acetyl-Coenzyme acyltransferase 2 | metabolic process | 36.9 |
| AI323528 | Gadd45b | growth arrest and DNA-damageinducible 45 beta | cell cycle, negative regulator of signaling | 18.0 | |
| NM_007913.1 | Egr1 | early growth response 1 | activation of MAPKK activity, cell differentiation | 16.4 | |
| BC028271.1 | Pitpnc1 | phosphatidylinositol transfer protein | transports, signal transduction+ | 16.4 | |
| BF318536 | Tcrb-J | T-cell receptor beta, joining | immune response | 15.1 | |
| Repeat: Rcan1 | 32.7 | ||||
| ↑ 8 h | BB535494 | Nedd9 | neural precursor cell expressed, downregulated gene 9 | cell cycle, regulation of growth | 33.2 |
| BB479063 | Ldb3 | LIM domain binding 3 | 31.6 | ||
| NM_009789.1 | S100 g | S100 calcium binding protein G | calcium-mediated signaling | 24.2 | |
| NM_018792.1 | Hils1 | histone H1-like protein in spermatids 1 | transcription, organismal development | 23.3 | |
| AA589629 | Slc6a6 | solute carrier family 6, member 6 | beta-alanine transport, taurine transport | 23.1 | |
| Repeat: Rcan1, Acat2, Pitpnc1 | 51.2, 35.6, 32.8 | ||||
| ↓ 1 h | BG076140 | Sesn1 | sestrin 1 | cell cycle arrest | 0.17 |
| NM_023719.1 | Txnip | thioredoxin interacting protein | transcriptional regulator, response to oxidative stress, cell cycle | 0.21 | |
| ↓ 2 h | AK005023.1 | Sel1l | sel-1 suppressor of lin-12-like (C. elegans) | Notch signaling pathway | 0.14 |
| BF687395 | Aass | aminoadipatesemialdehyde synthase | metabolic process, oxidation reduction | 0.24 | |
| NM_013873.1 | Sult4a1 | sulfotransferase family 4A, member 1 | lipid metabolism, steroid metabolism | 0.27 | |
| BM934224 | Cyb5b | cytochrome b5 type B | transport, electron transport chain | 0.28 | |
| Repeat: Sesn1 | 0.17 | ||||
| ↓ 4 h | BC014714.1 | Hmgcs2 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 | response to nutrient, response to peptide hormone stimulus | 0.01 |
| BC018220.1 | Dym | dymeclin | 0.03 | ||
| NM_016772.1 | Ech1 | enoyl coenzyme A hydratase 1 | lipid metabolic process+ | 0.05 | |
| BB703414 | Higd1c | HIG1 domain family, 1C | metabolic process | 0.06 | |
| BC019410.1 | Abhd14b | abhydrolase domain containing 14b | 0.08 | ||
| ↓ 8 h | AI196411 | Gsta3 | glutathione S-transferase, alpha 3 | metabolic process | 0.03 |
| BM227770 | Camk2 g | CAM-dependent protein kinase II gamma | G1/S transition of mitotic cell cycle, protein amino acid phosphorylation | 0.08 | |
| AK019346.1 | Dgcr6 | DiGeorge syndrome critical region gene 6 | 0.11 | ||
| AV171622 | Mettl7a1 | methyltransferas like 7A1 | metabolic process | 0.11 | |
| NM_009752.1 | Glb1 | galactosidase, beta 1 | carbohydrate metabolic process | 0.11 | |
| Repeat: Ech1, Sesn1, Abhd14b, Dym | 0.05, 0.07, 0.08, 0.08 |
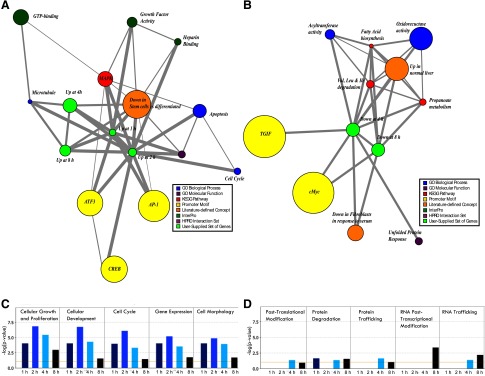
To define common characteristics and patterns among differentially expressed genes, we performed association network analysis using Molecular Concept Mapping (MCM) as well as automated functional annotation via Ingenuity software suite and GOTM. The analyses confirmed previous observations and generated some unexpected findings. Among MCM associations, upregulated genes were linked to concepts including: the MAPK pathway and its downstream transcriptional effectors AP-1 and ATF3, cell cycle and apoptosis, growth factor and heparin binding activity, as well as genes down in stem cells vs. differentiated counterparts (Fig. 1A); downregulated genes were linked to concepts such as: branched amino acid and fatty acid metabolism, oxidoreductase and unfolded protein response, serum response, and transcription factor motifs for TGIF and cMyc (Fig. 1B). Ingenuity-based analysis showed two temporally distinct programs: genes altered at 1–4 h were enriched for functional categories of signaling necessary for initiation of growth (top 5, Fig. 1C), whereas those at 4–8 h were enriched among categories centered on metabolism and mobilization of intracellular machinery during early stages of growth (top 5, Fig. 1D). The complete list and measures of statistical significance for all significantly enriched functional categories are shown in Supplementary Table S3; similar results were observed even when less stringent criteria (≥2-fold change, q ≤ 0.1) were applied (Supplementary Fig. S1, C and D). An independent GOTM-based analysis led to analogous findings. Upregulated genes, again the majority at 1–4 h, showed significant enrichment for functional categories related to early signaling events such as those mediating cell migration and angiogenesis, whereas downregulated genes, mainly at 4–8 h, were enriched for genes responsible for oxidation/reduction and metabolism (Supplementary Fig. S2).
Fig. 1.
A, B: molecular concept mapping (MCM): graphical representation of statistically derived association networks between genes of interest and color-coded molecular concepts (see color legend). Size of a node reflects number of genes in the set and the thickness of the line relates to odds ratio of the association. A: associations with gene sets up at 1,2,4 and 8 h of protease inhibitor (PI) feeding (green nodes). B: associations with gene sets down at 4 and 8 h of PI feeding (green nodes). Not enough genes were down at 1 or 2 h to conduct an analysis. C, D: a compiled Ingenuity cell and molecular function categorization analysis for 1 h, 2 h, 4 h and 8 h of PI feeding. C: top 5 functional categories altered at 1–4 h. D: top 5 functional categories altered at 4–8 h.
Transcriptional dynamics underlying initiation of pancreatic growth.
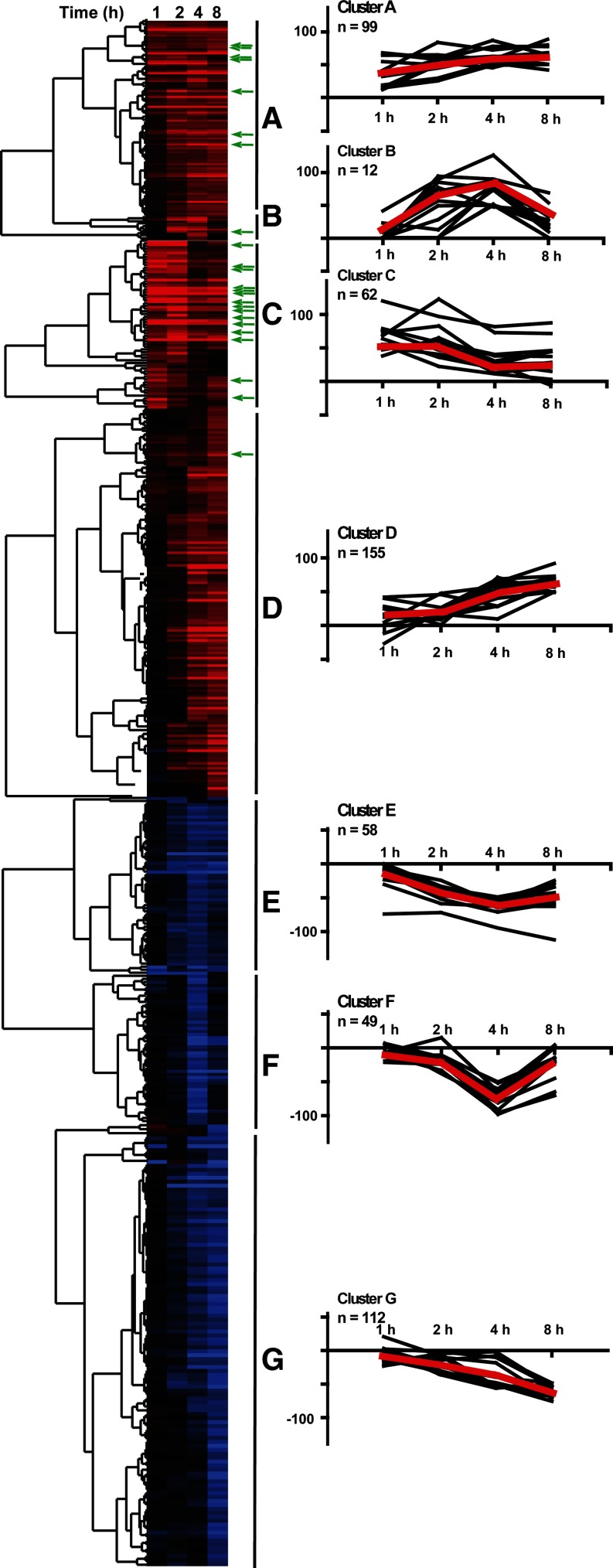
The work described thus far treated differentially expressed genes at each time point as independent “snapshots” in time. To examine the dynamic interrelationships between genes in context of the entire 1–8 h time course, we carried out hierarchical clustering. The genes significantly altered at any point of early pancreatic growth organized into seven distinct groups (Fig. 2, clusters A–G), each with a clear temporal pattern: peak at 2–8 h (cluster A), up at 4 h and/or 8 h (cluster B), peak at 1–2 h (cluster C), up at 4–8 h (cluster D), down at 2–8 h (cluster E), down at 4 h (cluster F) and decreasing, with maximal trough at 8 h (cluster G). For illustrative purposes, we also graphed the expression profiles of 12 random genes per cluster (black) and mean fold changes for the entire cluster (red). In addition, we performed an overlay of these data with CN-dependent/FK506-sensitive genes (green arrows) obtained from our earlier work (9); the CN-dependent genes were concentrated largely in cluster C with 25% overlap vs. 8% in cluster B and 6% in cluster A.
Fig. 2.
Left: clustering analysis of all genes (n = 533, corresponding to 608 probes) significantly altered at least once in the 1–8 h course of pancreatic growth (up or down at any point). Gene cluster into 7 groups with temporally distinct patterns (A–G). A: peak at 2–8 h; B: up at 4 h and/or 8 h; C: peak at 1–2 h; D: up at 4–8 h; E: decreased at 2–8 h; F: down at 4 h; G: slow decrease with max trough at 8 h. Green arrows point to 38 calcineurin (CN)-dependent (FK506-inhibited) genes. Right: graphical representation of 12 random genes within a cluster (black lines) and averaged time course of all genes within a cluster (red line) shown as fold induction (log 10 scale) at 1–8 h vs. fasted controls.
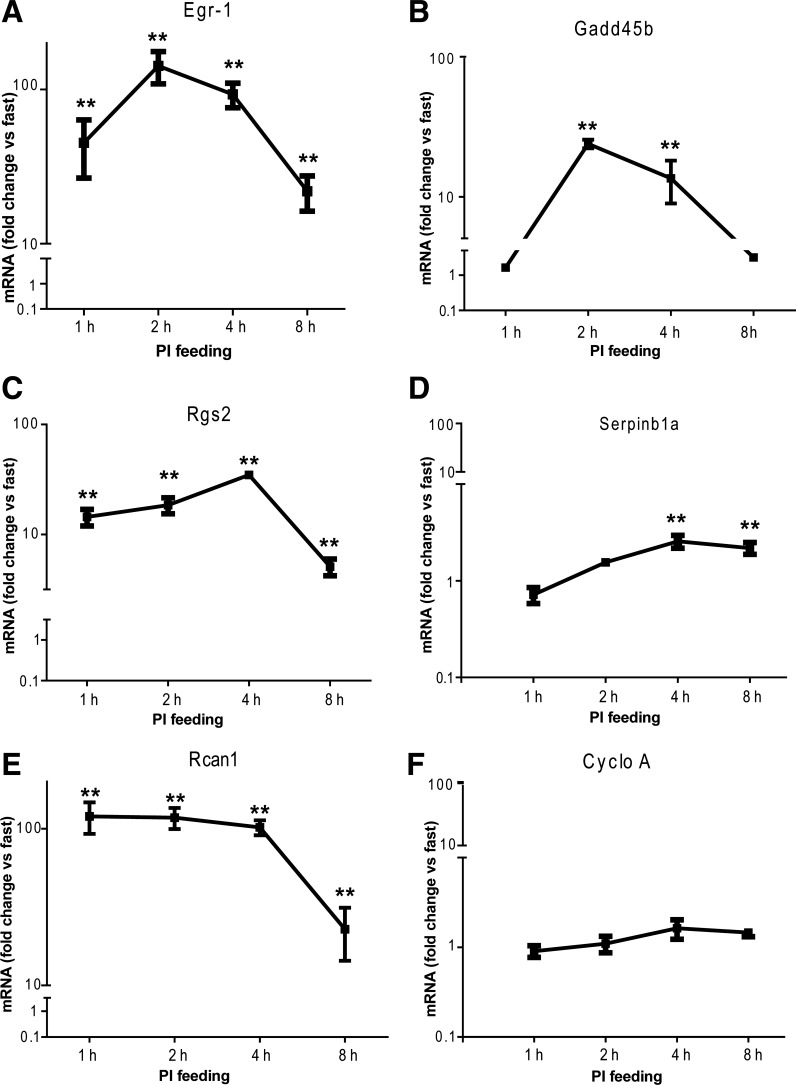
Next, we used qRT-PCR to experimentally validate a representative gene in each of the upregulated clusters: Egr-1 for cluster A, Gadd45b for cluster B, Rgs2 for cluster C, and Serpinb1a for cluster D (Fig. 3, A–D). We also examined the time course of two control genes: Rcan1, a previously characterized and robustly induced gene (Fig. 3E), and cyclophilin A, a gene with a static level of expression throughout early pancreatic growth (Fig. 3F). Aside from the expected null-result for cyclophilin A, all of the genes we examined produced a significant (P ≤ 0.01) increase in expression vs. fasting, with fold changes similar to those observed in microarray data.
Fig. 3.
RT-qPCR assays for a representative gene from each upregulated cluster: Egr1 (A), Gadd45b (B), Rgs2 (C), and Serpinb1a (D). Rcan1 (E) and cyclophilin (CycloA) (F) were shown to be upregulated and unchanged, respectively; n = 4–7, **P < 0.01.
Identifying regulatory mechanisms and Jak-STAT as a novel activated pathway.
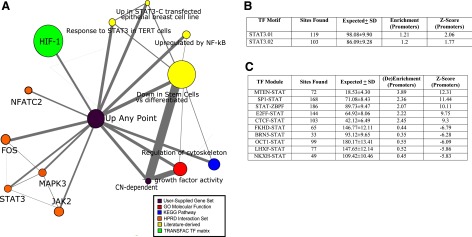
The dynamic genetic programs that drive physiological responses like pancreatic growth are orchestrated by complex networks, which in turn converge upon two main gateways: control of expression and posttranscriptional feedback (35). We investigated genes upregulated at any point in the course of our 1–8 h profile of pancreatic growth (≥3-fold induction vs. fasting and q ≤ 0.04–0.06), yielding a set of 314 genes. In addition, we also used a previously defined set of 51 probes, representing 38 unique CN-dependent genes (10). Analyzing the two groups in conjunction via MCM, we showed associations with growth factor activity, cytoskeletal organization and apoptosis, NF-κB and Jak-STAT signaling, as well as two literature-derived concepts related to differentiation (Fig. 4A). Search for STAT transcription motifs among promoters of genes significantly induced at any point along the 1–8 h course of PI feeding demonstrated a small, but significant enrichment among STAT3 sites (Fig. 4B). An analogous strategy was carried out to search for STAT-containing transcription factor complexes or “modules,” yielding 41 enriched and 61 de-enriched modules vs. expected rates in genome-wide promoter sequence. Top 5 among both categories, sorted by z-score, are shown (Fig. 4C).
Fig. 4.
Two sets of genes analyzed using bioinformatics-based tools: 1) 314 genes up at any point of the early 1–8 h course of pancreatic growth and 2) 38 CN-dependent genes, corresponding to green arrows in Fig. 2. A: MCM association network; B: regionminer (Genomatix) transcription factor motif sites found vs expected with enrichment and Z-score. C: analogous analysis for transcription factor modules, with top 5 enriched and de-enriched modules by Z-score.
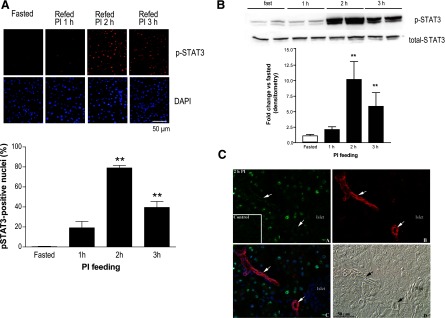
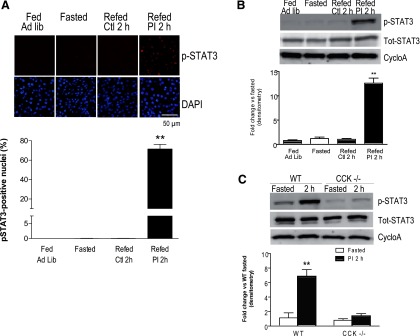
Informed by these findings, we proceeded to experimentally test activation of STAT signaling in the course of hormonally driven pancreatic growth. The active, phospho-Tyr705 form of STAT3, showed a strong nuclear signal in the pancreases of PI-fed mice, but no staining in fasted controls; the staining showed an increase at 1 h, peaked at over 75% nuclear occupancy at 2 h, and decreased at 3 h (Fig. 5A). These changes were paralleled by Western blot data with p-STAT3 signal increasing to a peak at 2–3 h, with ∼10-fold change vs. fasted controls; no change was observed for total STAT3 (Fig. 5B). To examine subcellular localization of p-STAT3 and cell-type specific changes, we overlaid multiple immunohistochemical markers at the peak 2 h time point and showed that STAT3 activation occurred specifically in acinar cells, with no increase in nuclear p-STAT3 in either islets or ducts (Fig. 5C). Moreover, the increase in p-STAT3 occurred only in the setting of elevated endogenous CCK induced by PI feeding; in the two control treatments known to have much less effect on endogenous CCK (mice fed ad libitum, or those fasted and refed control chow) the levels of p-STAT3 were no different than in the baseline fasted mice. This was evidenced both by immunohistochemistry (Fig. 6A) and Western blots (Fig. 6B), with quantitation of the former experiments showing over 75% nuclear p-STAT3 in PI-fed mice and <0.1% nuclear p-STAT3 in the remaining groups (Fig. 6A). We also showed the increase in p-STAT3 requires CCK, as the effect was abolished in CCK-deficient mice (Fig. 6C).
Fig. 5.
STAT3 is activated early in the course of cholecystokinin (CCK)-mediated growth of the exocrine compartment of the pancreas. A: immunohistochemistry for P-Tyr705 STAT3 and 4′6-diamidino-2-phenylindole (DAPI) nuclear stain in fasted mice or mice fasted and then refed PI-containing chow for 1, 2, or 3 h. Representative ×40 field (top) and quantitation of percent nuclear localization of P-STAT3 (bottom), with 3 mice per group (n = 3, **P < 0.01). B: representative Western blot (top) with summary of densitometry data below of p-Tyr705 STAT3 and total STAT3; 4 independent experiments (n = 4, **P < 0.01). C: immunohistochemistry for p-Tyr 705 STAT3 in exocrine pancreas of fasted (inset) and 2 h PI-fed mice (top left, A); immunohistochemistry for a ductal marker CK19 with arrows used for emphasis of duct-specific labeling (top right, B); overlay of p-Tyr705 STAT3 (green), ductal marker CK19 (red), and nuclear label DAPI (blue), showing the activation and nuclear localization of p-STAT3 occurs specifically in acinar cells, but not in islets or ducts (bottom left, C); corresponding Nomarski image (bottom right, D).
Fig. 6.
Activation of STAT3 requires an increase in endogenous CCK induced by PI feeding. A: immunohistochemistry for P-Tyr705 STAT3 and staining for DAPI in exocrine pancreas for the following conditions: fasting, ad libitum feeding, and fasting with refeeding of either control chow or chow containing PI. Representative ×40 field (top) and quantitation of percent nuclear localization of P-STAT3 (bottom), with 4 mice per group (n = 4, **P < 0.01). B: representative Western blot (top) and summary data of P-Tyr705 STAT3, total STAT3, and CycloA (control) for same conditions as listed in A; 4 mice per group (n = 4, **P < 0.01). C: representative Western blot (top) and summary data for wild-type (WT) and CCK-deficient (CCK−/−) mice that were fasted or fasted and then refed PI-containing chow; 3 mice per group (n = 3, **P < 0.01).
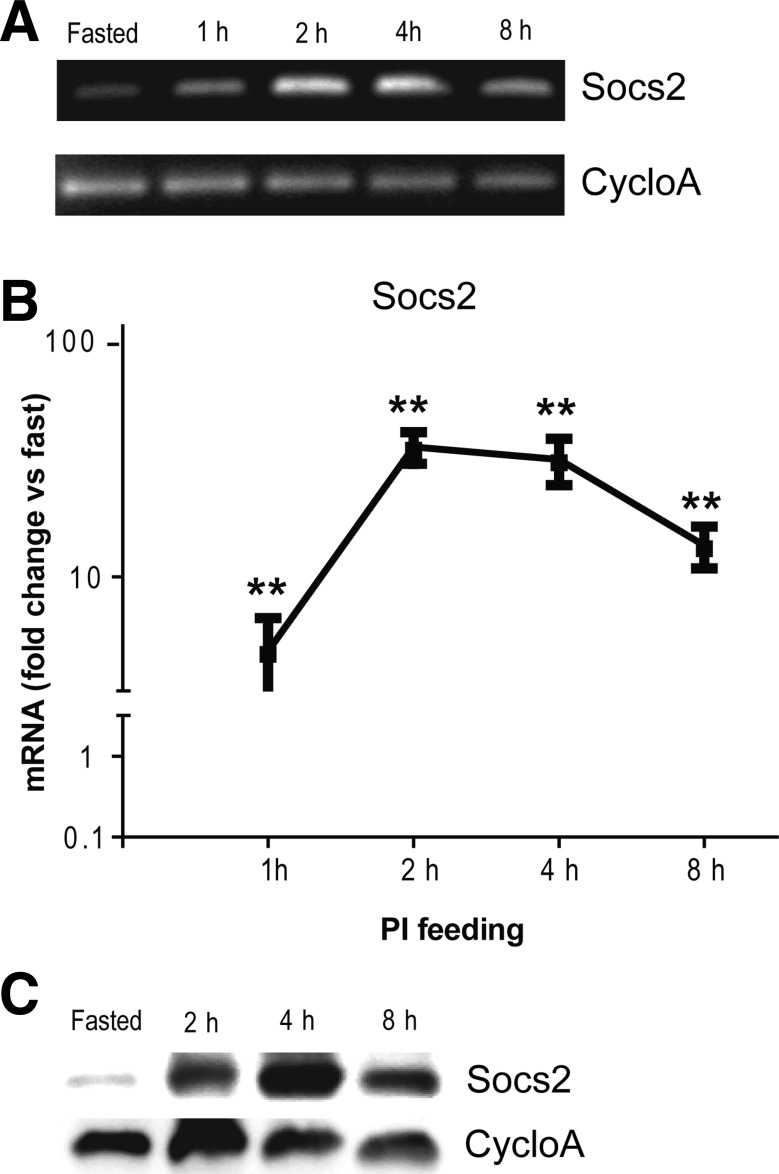
Socs2, which serves as a negative feedback regulator of Jak-STAT, is a transcriptional target of STATs. Consequently, we used Socs2 expression as readout of Jak-STAT activation. Using end-point RT-PCR and then quantitative, real-time RT-PCR, we showed that Socs2 mRNA increases in context of early pancreatic growth with a peak at 2–4 h (Fig. 7, A and B). Socs2 protein expression likewise increased, though with a slightly delayed peak at 2–8 h (Fig. 7C). With long-term PI feeding, Socs2 protein remained elevated up to 48 h, the last of the 12 h intervals we tested (data not shown). We also studied Socs3 and phospho-STAT5 in less detail; we found an increase in Socs3 mRNA by qRT-PCR and increased protein expression of both Socs3 and STAT5 by Western blotting (data not shown).
Fig. 7.
Socs2, a feedback regulator of Jak-STAT signaling, is activated in the early course of CCK-mediated pancreatic growth. A: RT-PCR for Socs2 expression during 1–8 h course of PI feeding; static gene CycloA served as control. B: quantitative RT-PCR for Socs2 expression during early course of PI feeding; values normalized to fasting controls (n = 4–7, P < 0.01). C: representative Western blot for protein expression of Socs2 during early course of PI feeding; CycloA as control.
DISCUSSION
The pancreas has been shown to be an adaptable organ with regenerative potential to recover from inflammation or resection, as well as robust growth in response to high protein diet, hyperphagia, pregnancy, and lactation. The molecular mechanisms that govern metabolic and trophic adaptation of the pancreas, however, are poorly understood with only a handful of signals characterized thus far (43). Past efforts to decipher molecular underpinnings of adaptive, hormonally driven pancreatic growth were carried out by candidate-driven approaches and centered almost exclusively on the MAPK cascade (8), AKT-mTOR (3), and CN-NFAT (10). More recently, our work demonstrated the power of unbiased global expression profiling with identification of Rcan1, an endogenous feedback inhibitor of CN-NFAT; acinar cell-specific overexpression of this protein was sufficient to completely block pancreatic growth (9).
Here, we employed an unbiased, global approach. We identified a large, new set of 533 genes (314 up and 219 down) significantly altered throughout early 1–8 h course of CCK-driven pancreatic growth. We found that fewer genes are upregulated at the earliest 1–2 h time points and almost none are repressed. The initial preponderance of upregulated, or so called “immediate early response” genes, has been observed in other systems modeling biological adaptation (19, 26). The complete list of differentially regulated genes contained many intriguing candidates and should prove to be fertile ground for further hypothesis-driven research (Supplementary Table S2). In particular, we noted a number of genes involved in growth factor and extracellular signaling (FGF21 and HbEGF), transcriptional regulation (Atf3, Egr1, and Jun), as well as feedback inhibition (Rcan1, Socs3, and Rgs2). Several upregulated and downregulated genes, like Nedd9, Gadd45b, and Sesn1, were involved in regulation of the cell cycle; others, including Acat2, Slc6a6, Ech1, and Gsta3, were involved in metabolism.
A statistical, bioinformatics-driven analysis of differentially expressed genes yielded further insights. Association networks produced by molecular concept mapping (MCM) confirmed the likely importance of MAPK signaling, transcription and growth factors, cell cycle, and small G proteins as components of the early trophic response. We also analyzed genes representing repressed or downregulated transcripts as well as those altered predominantly at later time points (4–8 h); in these groups, most associations relate to protein modification, trafficking, and metabolism, suggesting that intracellular reorganization and metabolic reprogramming may be an important priming step for growth. Another intriguing observation was that our network associations favored differentiated vs. stem cell/progenitor profile (Fig. 1, A and B, and 4A), while analysis of the promoters of genes (data not shown) and modules (Fig. 4C) showed de-enrichment for transcription factors that may play a role in determination/maintenance of embryonic stem cell fate (40). This paralleled our earlier work showing that increased BrdU incorporation in context of CCK-driven pancreatic growth took place in mature-appearing acinar cells with strong staining for amylase (9). In contrast, there is evidence that biological processes in CCK-driven acinar cell division in vitro (23), pancreatic regeneration following pancreatitis (13), or unrestrained cellular proliferation of pancreatic cancer (30) may all involve either progenitor cells or dedifferentiation from an acinar cell profile. Thus, there may be two different paradigms of growth: division of mature acinar cells or progenitors/de-differentiated cells.
The sets of differentially expressed genes can be studied as independent entities at each time point; here, however, we also took advantage of the opportunity to analyze temporal dynamics of gene expression. We performed hierarchical clustering and thereby identified and characterized seven distinct groups of genes, each with an independent pattern of expression (Fig. 2, A–G). We went on to validate expression for a gene in each of the four upregulated clusters by qRT-PCR. These were chosen, in part, based on subjectively relevant biological function; Egr1 had been shown to be a key regulator of gene expression in acute pancreatitis (14), Gadd45b as an important DNA repair and cell cycle regulator in pancreatic cancer (12), Rgs2 as feedback regulator of G protein receptors and perhaps CCK1 receptor (16), and Serpinb1a as a cytoprotective gene in pancreatic growth following partial pancreatectomy in exedin-4-treated mice (4). Any of these genes may be a centerpiece for future work, particularly comparative studies of pancreatic development, adaptation and pathogenesis.
Coordinately expressed genes are more likely to share common regulatory frameworks and much of the work in the genomics community has been focused on regulation of transcription. Here, we performed two bioinformatics-based analyses. First, we overlaid the previously identified 38 CN-dependent genes onto the cluster of 314 genes upregulated at any point in the 1–8 h course of pancreatic growth. The majority fall in cluster C with a peak at 1–2 h, corresponding to activation kinetics of CN-driven NFAT transcription factors. Second, we used 314 genes up at any point of early pancreatic growth in conjunction with the 38 CN-dependent genes to perform further MCM network associations as well as a Genomatix-based search for STAT transcription factor motifs and modules. We identified an enrichment for Jak-STAT signaling, particularly STAT3, pointing to this pathway as a potential new player in hormonally driven pancreatic growth.
Jak-STAT pathway is a well-known regulator of the immune response, and more recently it has also been shown to play a role in metabolism and growth (7). STAT activation drives the expression of many genes, including SOCS proteins, which act as endogenous feedback inhibitors that terminate the STAT signal. In the pancreas, Jak2-STAT3 pathway is activated in pancreatic cancer (7) and regulates angiogenesis and metastatic potential (7), whereas inhibition of the pathway suppresses malignant proliferation (17). Also of interest, cross talk between Jak-STAT and CN-NFAT signaling has recently been uncovered in the context of cardiac hypertrophy (24). Here, we showed an acinar cell specific and CCK-dependent increase in the active, phospho-form of STAT3 in the initiation of hormonally-driven pancreatic growth. Moreover, we showed an increase in mRNA and protein levels of Socs2, a target of STAT signaling. By predicting and then documenting Jak-STAT activation, we demonstrated the hypothesis-generating power of large-scale profiling in physiological models of adaptation. Whether Jak-STAT signal is activated directly or as a result of paracrine effects remains to be seen. With this work, STAT3-Socs2 joins CN-NFAT-Rcan1 as potential molecular switch in CCK-mediated, adaptive growth of the pancreas.
Recent advances have underlined the inherent plasticity of cell fate in the pancreas, bringing renewed promise for diagnostic and therapeutic tools useful in pancreatitis, pancreatic cancer, and diabetes. The expression profile of hormonally mediated pancreatic growth could prove useful in efforts to untangle the constraints of differentiation, particularly because in contrast to other models, cell division induced by PI feeding appears to take place in mature acinar cells, not progenitors or dedifferentiated cells. In terms of Jak-STAT signaling, activation of pathways formerly thought to be functionally restricted to inflammation has now shown to be critical in adaptation, including growth, metabolic reprogramming and adipogenesis (11, 32). This concept is not without precedent in the exocrine pancreas. We have shown that NFATs, known to be involved in T-cell differentiation and cytokine production (25), are activated in the course of CCK-mediated pancreatic growth (10). Furthermore, a key transcriptional effector of microbial and cytokine-induced signaling, NF-κB, has now been shown to be activated in pancreatitis and may be an important determinant of disease severity (34). Our work here, particularly the identification of STAT3-Socs2 activation in context of adaptive pancreatic growth, should prove a good point of departure for continued research in this area.
In summary, we have carefully dissected the genetic program that underlies early events in hormonally mediated pancreatic growth. We identified and experimentally validated novel growth-related genes, analyzed their interrelationships, and thereby uncovered new functional, temporal, and regulatory frameworks. This work led us to computationally identify and experimentally evaluate STAT-Socs signaling as a novel player in adaptive, hormonally driven pancreatic growth.
GRANTS
The research was supported by National Institutes of Health Grants DK-59578 to J. A. Williams and P30 DK-34933 (Michigan Gastrointestinal Peptide Center). G. T. Gurda was supported by Systems and Integrative Biology Training Grant (T32 GM-008322) and the Medical Scientist Training Program.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
ACKNOWLEDGMENTS
We thank Bradley Nelson for help with immunohistochemistry, Jessica Schwartz laboratory for advice and reagents, Arul Chinnayian laboratory for assistance with bioinformatics, and Dr. Linda Samuelson for critical reading of this manuscript.
Current addresses: G. T. Gurda, Dept. of Pathology, Johns Hopkins Hospital, 600 N. Wolfe St./Carnegie 489, Baltimore, MD 21287; L. Guo, Joslin Diabetes Center, 1 Joslin Pl., Boston, MA 02215.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1. Bragado MJ, Groblewski GE, Williams JA. Regulation of protein synthesis by cholecystokinin in rat pancreatic acini involves PHAS-I and the p70 S6 kinase pathway. Gastroenterology 115: 733–742, 1998. [DOI] [PubMed] [Google Scholar]
- 2. Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21: 2933–2942, 2005. [DOI] [PubMed] [Google Scholar]
- 3. Crozier SJ, Sans MD, Guo L, D'Alecy LG, Williams JA. Activation of the mTOR signalling pathway is required for pancreatic growth in protease-inhibitor-fed mice. J Physiol 573: 775–786, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. De Leon DD, Farzad C, Crutchlow MF, Brestelli J, Tobias J, Kaestner KH, Stoffers DA. Identification of transcriptional targets during pancreatic growth after partial pancreatectomy and exendin-4 treatment. Physiol Genomics 24: 133–143, 2006. [DOI] [PubMed] [Google Scholar]
- 5. Elsasser H-P, Adler G, Kern HF. Replication and regeneration of the pancreas. In The Pancreas: Biology, Pathobiology, and Disease, edited by Vay Liang W. New York: Raven, 1993 p. 75–86. [Google Scholar]
- 6. Gao B. Cytokines, STATs and liver disease. Cell Mol Immunol 2: 92–100, 2005. [PubMed] [Google Scholar]
- 7. Greten FR, Weber CK, Greten TF, Schneider G, Wagner M, Adler G, Schmid RM. Stat3 and NF-kappaB activation prevents apoptosis in pancreatic carcinogenesis. Gastroenterology 123: 2052–2063, 2002. [DOI] [PubMed] [Google Scholar]
- 8. Guo L, Sans MD, Gurda GT, Lee SH, Ernst SA, Williams JA. Induction of early response genes in trypsin inhibitor-induced pancreatic growth. Am J Physiol Gastrointest Liver Physiol 292: G667–G677, 2007. [DOI] [PubMed] [Google Scholar]
- 9. Gurda GT, Crozier SJ, Ji B, Ernst SA, Logsdon CD, Rothermel BA, Williams JA. Regulator of calcineurin 1 controls growth plasticity of adult pancreas. Gastroenterology 139: 609–619, 619 e601–606, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gurda GT, Guo L, Lee SH, Molkentin JD, Williams JA. Cholecystokinin activates pancreatic calcineurin-NFAT signaling in vitro and in vivo. Mol Biol Cell 19: 198–206, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Herrington J, Smit LS, Schwartz J, Carter-Su C. The role of STAT proteins in growth hormone signaling. Oncogene 19: 2585–2597, 2000. [DOI] [PubMed] [Google Scholar]
- 12. Hildesheim J, Bulavin DV, Anver MR, Alvord WG, Hollander MC, Vardanian L, Fornace AJ., Jr Gadd45a protects against UV irradiation-induced skin tumors, and promotes apoptosis and stress signaling via MAPK and p53. Cancer Res 62: 7305–7315, 2002. [PubMed] [Google Scholar]
- 13. Jensen JN, Cameron E, Garay MV, Starkey TW, Gianani R, Jensen J. Recapitulation of elements of embryonic development in adult mouse pancreatic regeneration. Gastroenterology 128: 728–741, 2005. [DOI] [PubMed] [Google Scholar]
- 14. Ji B, Chen XQ, Misek DE, Kuick R, Hanash S, Ernst S, Najarian R, Logsdon CD. Pancreatic gene expression during the initiation of acute pancreatitis: identification of EGR-1 as a key regulator. Physiol Genomics 14: 59–72, 2003. [DOI] [PubMed] [Google Scholar]
- 15. Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137: 1343–1355, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Le Page SL, Bi Y, Williams JA. CCK-A receptor activates RhoA through G alpha 12/13 in NIH3T3 cells. Am J Physiol Cell Physiol 285: C1197–C1206, 2003. [DOI] [PubMed] [Google Scholar]
- 17. Lin L, Hutzen B, Zuo M, Ball S, Deangelis S, Foust E, Pandit B, Ihnat MA, Shenoy SS, Kulp S, Li PK, Li C, Fuchs J, Lin J. Novel STAT3 phosphorylation inhibitors exhibit potent growth-suppressive activity in pancreatic and breast cancer cells. Cancer Res 70: 2445–2454, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lindberg K, Ronn SG, Tornehave D, Richter H, Hansen JA, Romer J, Jackerott M, Billestrup N. Regulation of pancreatic beta-cell mass and proliferation by SOCS-3. J Mol Endocrinol 35: 231–243, 2005. [DOI] [PubMed] [Google Scholar]
- 19. Locker J, Tian J, Carver R, Concas D, Cossu C, Ledda-Columbano GM, Columbano A. A common set of immediate-early response genes in liver regeneration and hyperplasia. Hepatology 38: 314–325, 2003. [DOI] [PubMed] [Google Scholar]
- 20. Logsdon CD. Role of cholecystokinin in physiologic and pathophysiologic growth of the pancreas. In Gastrointestinal Endocrinology, edited by Greely GH., Jr Totowa, NJ: Humana, 1999, p. 393–422. [Google Scholar]
- 21. Logsdon CD. Stimulation of pancreatic acinar cell growth by CCK, epidermal growth factor, and insulin in vitro. Am J Physiol Gastrointest Liver Physiol 251: G487–G494, 1986. [DOI] [PubMed] [Google Scholar]
- 22. Logsdon CD, Simeone DM, Binkley C, Arumugam T, Greenson JK, Giordano TJ, Misek DE, Kuick R, Hanash S. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res 63: 2649–2657, 2003. [PubMed] [Google Scholar]
- 23. Logsdon CD, Williams JA. Pancreatic acinar cells in monolayer culture: direct trophic effects of caerulein in vitro. Am J Physiol Gastrointest Liver Physiol 250: G440–G447, 1986. [DOI] [PubMed] [Google Scholar]
- 24. Manukyan I, Galatioto J, Mascareno E, Bhaduri S, Siddiqui MA. Cross-talk between calcineurin/NFAT and Jak/STAT signalling induces cardioprotective alphaB-crystallin gene expression in response to hypertrophic stimuli. J Cell Mol Med 14: 1707–1716, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Masuda ES, Imamura R, Amasaki Y, Arai K, Arai N. Signalling into the T-cell nucleus: NFAT regulation. Cell Signal 10: 599–611, 1998. [DOI] [PubMed] [Google Scholar]
- 26. Mayer H, Bilban M, Kurtev V, Gruber F, Wagner O, Binder BR, de Martin R. Deciphering regulatory patterns of inflammatory gene expression from interleukin-1-stimulated human endothelial cells. Arterioscler Thromb Vasc Biol 24: 1192–1198, 2004. [DOI] [PubMed] [Google Scholar]
- 27. Melmed RN, Bouchier IA. Effects of trypsin inhibitors on the acinar cells of rat pancreas. Gut 9: 729, 1968. [PubMed] [Google Scholar]
- 28. Metcalf D, Greenhalgh CJ, Viney E, Willson TA, Starr R, Nicola NA, Hilton DJ, Alexander WS. Gigantism in mice lacking suppressor of cytokine signalling-2. Nature 405: 1069–1073, 2000. [DOI] [PubMed] [Google Scholar]
- 29. Michalopoulos GK. Liver regeneration after partial hepatectomy: critical analysis of mechanistic dilemmas. Am J Pathol 176: 2–13, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morris JP, 4th, Cano DA, Sekine S, Wang SC, Hebrok M. Beta-catenin blocks Kras-dependent reprogramming of acini into pancreatic cancer precursor lesions in mice. J Clin Invest 120: 508–520, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morisset J. Hormonal control of pancreatic growth during fetal, neonatal and adult life. Adv Med Sci 53: 99–118, 2008. [DOI] [PubMed] [Google Scholar]
- 32. Nanbu-Wakao R, Morikawa Y, Matsumura I, Masuho Y, Muramatsu MA, Senba E, Wakao H. Stimulation of 3T3–L1 adipogenesis by signal transducer and activator of transcription 5. Mol Endocrinol 16: 1565–1576, 2002. [DOI] [PubMed] [Google Scholar]
- 33. Niederau C, Liddle RA, Williams JA, Grendell JH. Pancreatic growth: interaction of exogenous cholecystokinin, a protease inhibitor, and a cholecystokinin receptor antagonist in mice. Gut 28: 63–69, 1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rakonczay Z, Hegyi P, Takacs T, McCarroll J, Saluja AK. The role of NF-kappa B activation in the pathogenesis of acute pancreatitis. Gut 57: 259–267, 2008. [DOI] [PubMed] [Google Scholar]
- 35. Ray JC, Igoshin OA. Adaptable functionality of transcriptional feedback in bacterial two-component systems. PLoS Comput Biol 6: e1000676, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sato N, Suzuki S, Kanai S, Ohta M, Jimi A, Noda T, Takiguchi S, Funakoshi A, Miyasaka K. Different effects of oral administration of synthetic trypsin inhibitor on the pancreas between cholecystokinin-A receptor gene knockout mice and wild type mice. Jpn J Pharmacol 89: 290–295, 2002. [DOI] [PubMed] [Google Scholar]
- 37. Schreiber S, Rosenstiel P, Hampe J, Nikolaus S, Groessner B, Schottelius A, Kuhbacher T, Hamling J, Folsch UR, Seegert D. Activation of signal transducer and activator of transcription (STAT) 1 in human chronic inflammatory bowel disease. Gut 51: 379–385, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Solomon TE, Petersen H, Elashoff J, Grossman MI. Interaction of caerulein and secretin on pancreatic size and composition in rat. Am J Physiol Endocrinol Metab Gastrointest Physiol 235: E714–E719, 1978. [DOI] [PubMed] [Google Scholar]
- 39. Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sun Y, Li H, Liu Y, Mattson MP, Rao MS, Zhan M. Evolutionarily conserved transcriptional co-expression guiding embryonic stem cell differentiation. PLoS One 3: e3406, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tashiro M, Samuelson LC, Liddle RA, Williams JA. Calcineurin mediates pancreatic growth in protease inhibitor-treated mice. Am J Physiol Gastrointest Liver Physiol 286: G784–G790, 2004. [DOI] [PubMed] [Google Scholar]
- 42. Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, Kalyana-Sundaram S, Wei JT, Rubin MA, Pienta KJ, Shah RB, Chinnaiyan AM. Integrative molecular concept modeling of prostate cancer progression. Nat Genet 39: 41–51, 2007. [DOI] [PubMed] [Google Scholar]
- 43. Williams JA. Intracellular signaling mechanisms activated by cholecystokinin-regulating synthesis and secretion of digestive enzymes in pancreatic acinar cells. Annu Rev Physiol 63: 77–97, 2001. [DOI] [PubMed] [Google Scholar]
- 44. Yamamoto M, Otani M, Jia DM, Fukumitsu KI, Yoshikawa H, Akiyama T, Otsuki M. Differential mechanism and site of action of CCK on the pancreatic secretion and growth in rats. Am J Physiol Gastrointest Liver Physiol 285: G681–G687, 2003. [DOI] [PubMed] [Google Scholar]
- 45. Zhang B, Schmoyer D, Kirov S, Snoddy J. GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinformatics 5: 16, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]