Abstract
Podosomes are highly dynamic actin-rich adhesion structures in cells of myeloid lineage and some transformed cells. Unlike transformed mesenchymal cell types, podosomes are the sole adhesion structure in macrophage and thus mediate all contact with adhesion substrate, including movement through complex tissues for immune surveillance. The existence of podosomes in inflammatory macrophages and transformed cell types suggest an important role in tissue invasion. The proteome, assembly, and maintenance of podosomes are emerging, but remain incompletely defined. Previously, we reported a formin homology sequence and actin assembly activity in association with macrophage β-3 integrin. In this study we demonstrate by quantitative RT-PCR and Western blotting that the formin FRL1 is specifically upregulated during monocyte differentiation to macrophages. We show that the formin FRL1 localizes to the actin-rich cores of primary macrophage podosomes. FRL1 co-precipitates with beta-3 integrin and both fixed and live cell fluorescence microscopy show that endogenous and overexpressed FRL1 selectively localize to macrophage podosomes. Targeted disruption of FRL1 by siRNA results in reduced cell adhesion and disruption of podosome dynamics. Our data suggest that FRL1 is responsible for modifying actin at the macrophage podosome and may be involved in actin cytoskeleton dynamics during adhesion and migration within tissues.
Keywords: FRL1, FMNL1, podosome, integrin, cell adhesion
Introduction
Differentiation from circulating monocytes endows macrophages with unique capabililties, including flattened morphology, phagocytic activity, receptivity to chemotactic signaling, and the ability to migrate through complex extracellular matrices (ECM). Macrophages interact with the ECM through podosomes, an integrin-based adhesion structure unique to invasive cells, particularly those of myelocytic lineage. Podosomes are dynamic, punctate, actin-rich structures that form perpendicular to the ventral cell surface and project into the substratum [Trotter, 1981]. On rigid structures, podosomes form stalagmite-like structures. They are composed of a filamentous actin core surrounded by a concentric ring of proteins including paxillin, talin, and vinculin [Marchisio et al., 1984; Marchisio et al., 1988; Pfaff and Jurdic, 2001; vid-Pfeuty and Singer, 1980] in the fashion of a bullseye. Integrins of the β2 and β3 families provide adhesion and localize to the outer ring and core, respectively [Marchisio et al., 1988; Pfaff and Jurdic, 2001]. Notably, the integrin αvβ3 is specifically upregulated during monocyte differentiation to macrophages in culture [DeNichilo and Burns, 1993] and is a major podosome constituent. The promiscuity of αvβ3 ligand specificity and its prominence in podosomes likely contributes to successful migration in complex tissues. Despite their prominence, it remains undetermined how the actin structures within podosomes are generated, maintained or deconstructed.
Several classes of proteins capable of nucleating and polymerizing actin filaments have been identified: the Arp2/3 complex, the formins, Spire, and Cobl. The activity of at least three of these classes of proteins is subject to Rho-family GTPase regulation, a prominent downstream signal of integrin ligation, including αvβ3 [Chhabra and Higgs, 2007; Wellington et al., 1999; Gao and Blystone, 2009]. The most well-studied of the actin polymerizing proteins, the Arp2/3 complex, has been shown to localize to the macrophage podosome, specifically at the ends of the filamentous actin core [Linder et al., 2000]. However, our own studies have shown that Arp2/3 localization to leukocyte β3 adhesions is transient [Chandhoke et al., 2004]. We have demonstrated that after integrin clustering, a purified complex of proteins associated with the αvβ3 integrin is capable of nucleating and polymerizing actin in vitro, yet devoid of detectable Arp2/3. The same study demonstrated that a purified formin inhibitory domain can inhibit actin assembly by the purified complex [Butler et al., 2006].
Of the three known classes of actin nucleating proteins, formins are the broadest subset and have been demonstrated capable of nucleating, polymerizing, bundling, and severing actin filaments in vitro [Harris et al., 2004; Harris et al., 2006; Pruyne et al., 2002]. To date, there are 15 known mammalian formins, grouped into seven families [Higgs and Peterson, 2005]. Formins nucleate actin filaments in vitro and remain bound to the barbed end, generating unbranched actin filaments through a processive capping mechanism [Otomo et al., 2005, Romero et al., 2004]. At least one of these families, the diaphanous formins, is autoinhibited through an interaction between the N-terminal DID (Diaphanous Inhibitory Domain) and C-terminal DAD (Diaphanous Activating Domain). Active RhoGTPase disrupts the inhibitory interaction between DID and DAD domains by binding the GBD (GTPase binding domain), which partially overlaps the DID domain. Similar autoregulation has been demonstrated for FRL1 and INF2, and likely applies to all members of the Dia, FRL, and DAAM groups, based on sequence homology [Chhabra and Higgs, 2006; Higgs and Peterson, 2006, Seth et al., 2006]. FRL1 (also referred to as FRLα or FMNL1) and mDia2 have been demonstrated capable of bundling preexisting actin filaments [Harris et al., 2006] and FRL1 has been shown to sever filaments in vitro [Harris et al., 2004]. While the biochemical features have been well characterized in vitro, it is not clear what function these capabilities serve in vivo.
Recent reports have begun to elucidate the role of FRL1 in leukocytes. FRL1 has been shown to be upregulated in T cells of patients with non-Hodgkins lymphoma, although the function of the protein within these cells is unclear [Favaro et al., 2006]. Han et al. have recently shown that the gamma isoform of FRL1 (FRL1γ) is involved in regulating membrane integrity. This isoform contains an additional C-terminal sequence lacking in the α and β isoforms and may be involved in localizing the protein to the cell membrane in response to myristoylation [Han et al., 2009]. Gomez et al. demonstrated that FRL1 is involved in positioning the centrosome during cell mediated cytotoxicity in leukocytes [Gomez et al., 2007].
In this study, we show that upon differentiation, human peripheral blood monocyte-derived macrophages (PBMC) increase expression of FRL1 six-fold. In macrophages, FRL1 associates selectively with β3, but not β2 integrin. Correspondingly, FRL1 localizes with β3 to the actin core in podosomes. We show that both endogenous and overexpressed FRL1 colocalize spatially and temporally with actin and β3 integrin in podosome cores. We demonstrate that loss of FRL1 in primary macrophage leads to a significant decrease in cell adhesion and podosome number; average podosome size, in contrast, is increased. This is the first study that demonstrates localization of a formin to podosomes in primary cells; it suggests an in vivo role for our previous observations describing an association between β3 integrin, formins and actin polymerization.
Materials & Methods
Cells and Materials
ATCC THP-1 cells were cultured in RMPI-1640/10% FCS with 1mM pyruvate, adding fresh media every 2-3 days. Podosomes in THP-1 cells were elicited in media containing 160nM phorbol myristate acid for three days before use [Tsuchiya et al., 1982]. Meg01 and H2K cells were obtained from ATCC (Manassas, VA). RPMI-1640, PBS, HBSS, Opti-MEM and Lipofectamine 2000 were purchased from Invitrogen (Grand Island, NY). FCS was purchased from Tissue Culture Biologicals (Tulare, CA). Mouse monoclonal anti-FRL1 (2369E4a) and goat polyclonal anti-FRL1 antibody (P-17) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). These antibodies are directed against an internal region of FRL1 and are therefore do not distinguish between isoforms. The anti-β2 monoclonal IB4, and anti-β3 monoclonal 7G2, 1A2, and AP3 were gifts from Eric. J. Brown, Genentech. Mouse monoclonal anti-actin, anti-vinculin clone hVin, and anti-talin clone 8D4 were purchased from Sigma Aldrich (St. Louis, MO), anti-paxillin clone 349 from BD Biosciences (Franklin Lakes, NJ), and FITC labeled donkey anti-mouse and anti-rabbit secondary antibodies from Jackson ImmunoResearch Laboratories, Inc (West Grove, PA). Mouse monoclonal anti-myc antibody 9E10 was ordered from the Developmental Studies Hybrodoma Bank (Iowa City, Iowa). Mouse monoclonal anti-Xpress recognizes the eight amino acid sequence “Asp-Leu-Tyr-Asp-Asp-Asp-Asp-Lys” and was ordered from Invitrogen (St. Louis, MO). Rhodamine phalloidin was purchased from Cytoskeleton (Denver, CO). The GM-CSF used is available commercially as Leukine from Berlex (Seattle, WA). GTPase constructs were gifts from Dr. Nicholas Deakin, SUNY Upstate Medical University.
Monocyte Purification and Culture
Monocytes were isolated from fresh, citrated peripheral blood with a 30 minute dextran sedimentation followed by density gradient centrifugation using Ficoll-Paque (Amersham Pharmacia Biotech AB, Uppsala, Sweden). The mononuclear layer was then washed once with HBSS and resuspended in RPMI-1640/20% FCS. To induce differentiation, 125pg/mL GM-CSF was added to the media. Cells were allowed to differentiate at 37°C and 5% CO2.
Macrophage Transfection
After eight days in culture, macrophages were transfected with Lipofectamine 2000. For each well of a 24 well plate, 0.5μL of lipofectamine 2000 was complexed with 1μg of DNA encoding GFP-FRL1, GFP-actin, or GFP-β3 in 100μL of Opti-MEM for 20 minutes prior addition to culture media. Cells were then cultured for 48 hours before use.
Fusion Constructs
The pEGFP-actin vector encoding GFP-actin was purchased from Clontech (Mountain View, CA). The pEGFP-β3 vector encoding GFP-β3 was previously described in [Chandhoke et al., 2004]. The pEGFP-FRL1γ vector was generated from pCMV-His-FMNL1 which was a gift from Dr. Angela Krackhardt [Schuster et al., 2007]. The FMNL1 (FRL1γ) cDNA was inserted into pcDNA-GFP-C1, previously generated using the existing GFP ORF from pEGFP-C1 (Clontech) and inserting into pcDNA3.1(+) (Invitrogen, St. Louis, MO), yielding an amino-terminal GFP fusion with FRL1. Xpress-FRL1 was generated by inserting the FRL1γ cDNA into pcDNA3.1/His C vector, which includes the eight amino acid Xpress tag N-terminal to the MCS (Invitrogen, St. Louis, MO).
RT-PCR
Three independent mRNA samples were generated for each cell type utilized. These samples were analyzed with a real-time quantitative reverse transcriptase (RT) polymerase chain reaction (qRT–PCR). For the RT reaction, 250 ng total RNA (25 μL in water) from each RNA preparation was incubated with 250 pmoles of an oligo (dT)24 primer at 70°C for 10 min. Then, 3.5 μL 10 × PCR Gold buffer (Applied Biosystems, Foster City, CA, USA), 15 μL of 25 mM MgCl2 (Applied Biosystems), 2.0 μL of 25 mM dNTP, 0.50 μL RNase inhibitor (40 U/mL; Ambion, Austin, TX, USA), and 0.63 μL SuperScript II reverse transcriptase (200 U/mL; Invitrogen, Carlsbad, CA, USA) were added. The reaction was incubated at 25°C for 10 min, 48°C for 30 min, 95°C for 5 min. Subsequently, samples were diluted five-fold with nuclease-free water. For quantification of transcript differences, 1.0 μL of the RT reaction from each of the samples was evaluated in triplicate PCR reactions for each gene of interest on 96-well plates. Each 25 μL PCR reaction contained 1 × TaqMan Universal PCR Master Mix (Applied Biosystems), 0.20 μM of each custom-designed, formin-specific forward and reverse primer as listed above, 0.25 μL of SYBRGreen dye (Invitrogen) and 1.0 μL of diluted RT. Amplification in the absence of template failed to produce any signal that would have occurred due to primer dimerization and extension. End point melt-curve analysis confirmed the presence of single amplicons in each reaction well. Average cycles to threshold for each cell/formin combination was compared to controls quantifying ribosomal RNA levels and expressed as a percentage of that constant.
PCR Oligonucleotides
For both non-quantitative PCR and qRT-PCR, oligonucleotides employed were as follows: For human Dia1, 5′-GCTTGTGGCTGAGGACCTCTCCC-3′ and 5′-GATCATAGACTCAGTCAGAACAGC-3′ generated a 316bp product. For human Dia2, 5′-GATCAGACCTCATGAAATGACTG-3′ and 5′-CTGAATCATAGACTCTGCCAACCG-3′ generated 307bp product. For Dia3, 5′-GGTCAAAGATTGAACCCACAG-3′ and 5′-GGTTCTGAATTAAAGCCTCACTCAGC-3′ generated a 324bp product. For human FHOD1, 5′-CGTGACGTGAAGCTGGCTGGGGG-3′ and 5′-GCTCTTCCTCCGTGGGCATCATGG-3′ generated a 343bp product. For FRL1, 5′-GCACTGAAACCCAGCCAGATCACC-3′ and 5′-GGAAGTCCAGGCCCAGAGCCTGC-3′ generated a 301bp product. For human FMN1, 5′-GCTGAAGAAGGGGGCTACCGC-3′ and 5′-GGAGAGTGGGAGTGGCCTTCG-3′ generated a 179bp product.
Isoform specific oligonucleotides were as follows: For FRL1α and FRL1γ, 5′-GCCTACAAGAAAGCTGAGCAGGAGGTGG-3′ and 5′-GCATCTCTTCTCCCAGGCTGGCC-′3 generated a 309bp product for FRL1α and a 483bp product for FRL1γ. For FRL1β, 5′-GCATCTCTTCTCCCAGGCTGGCC-3′ and 5′-CGAGAGGTCGGAGGTGACCTGCAGTGGG-′3 generated a 324bp product.
Immunoprecipitation
For integrin immunoprecipitation, monocytes were differentiated on tissue culture plates for 10 days with 125pg/mL GM-CSF. To prepare lysates, cells were washed twice with HBSS with 1mM MgCl2, 1mM CaCl2, then scraped off using ice cold lysis buffer (50mM Tris, pH 7.6, 150mM NaCl, 5mM MgCl2, 1% NP-40, 1mM PMSF, 10ug/mL leupeptin, 10ug/mL aprotinin) with 50 μM Na3VO4. Immunopreciptation was performed as described in [Gao et al., 2005] using 10ug/mL IB4 for β2 or a 50:50 mixture of 7G2 and 1A2 tissue culture supernatant for β3.
FRL1 siRNA
Macrophages were differentiated for seven days as described above before transfection with siRNA. The siRNA oligomers s2227 and s2228 (Ambion, Inc, Austin, TX) target exons 21 and 23 of FMNL1, respectively, both of which are part of the FH2 domain of FRL1. These siRNAs target regions that are common to all three currently known isoforms of FRL1: α, β, and γ. The siRNA oligomer s4093 (Ambion, Inc, Austin, TX) targets exon 11 of hDia1. siRNA oligomers were complexed with INTERFERin (PolyPlus Transfection, New York, NY) per manufacturer’s instructions. Cells were treated at a final concentration of 400nM siRNA for 48 hours. THP-1 cells were differentiated with 160mM PMA before being treated with 400nM siRNA. RAW cells were allowed to adhere for 24 hours before treatment with 200nM siRNA. Forty eight hours after siRNA treatment, cells were harvested for biochemical or morphologic analysis. Macrophage adhesion was determined following siRNA treatment by manual counting of adherent cells and reported as the average of cells per cm2.
Microscopy
PBMCs were differentiated for 10 days with 125pg/mL GM-CSF on 12mm glass coverslips (Fisher Scientific, Pittsburg, PA) in 24 well cell culture plates (Corning Incorporated, Corning, NY). Cells were washed twice with PBS before fixing with 3.7% formaldehyde (Fisher Scientific, Fair Lawn, NJ) in PBS at 4° C for one hour, followed by permeabilization with ice cold 0.0005% NP-40 Alternative (BMD Biosciences Inc, La Jolla, CA) in PBS for 12 seconds. Primary antibody was diluted in PBS with 5% goat serum and 0.1% Triton X-100 and incubated with the cells for 30 minutes at 37° C. Primary antibodies were diluted as follows: 1:100 anti-FRL1; 1:400 anti-vinculin, 1:250 anti-paxillin, 1:200 anti-talin, 1.5ug/mL 1B4 and 1.5ug/mL AP3. After two washes with PBS, either 1:100 FITC labeled donkey anti-mouse or 1:100 FITC labeled donkey anti-rabbit secondary antibody and 1:3500 rhodamine phalloidin were added in PBS for 30 minutes at 37° C. After five final washes with PBS, coverslips were overturned onto o-phenylenediamine (Sigma Aldrich, St Louis, MO) in glycerol (Fisher Scientific, Pittsburg, PA). Confocal movies were digitally captured with a Nikon Ti microscope fitted with a Perkin Elmer spinning disk laser running Volocity 4.4 software. Fixed cell epifluorescence images were captured using a Nikon Eclipse E800 fluorescent microscope (Nikon, Melville, NY) equipped with a Hamamatsu ORCA-ER digital camera (Bridgewater, NJ) running Simple PCI 5.3.1 software. Images were then processed with Adobe Photoshop CS software.
Podosome Quantitation
Volocity Quantitation (Perkin Elmer, Waltham, MA) software image recognition module was used to count podosomes in still frames from the GFP-actin movie (Supplemental Movie 2). Podosomes were identified as greater than 10% of maximum intensity, within the outline of the body of the cell, and greater than 0.2μm2. Identified objects were then screened for two centers of intensity using the “separate touching objects” tool, and newly identified objects smaller than 0.15 μm2 were excluded. This program was then applied to still frames from three movies of cells expressing each construct. Still images from one movie for each cell type are shown in Figure 3B. For fixed cell microscopy, objects were identified as greater than 50% of maximum intensity.
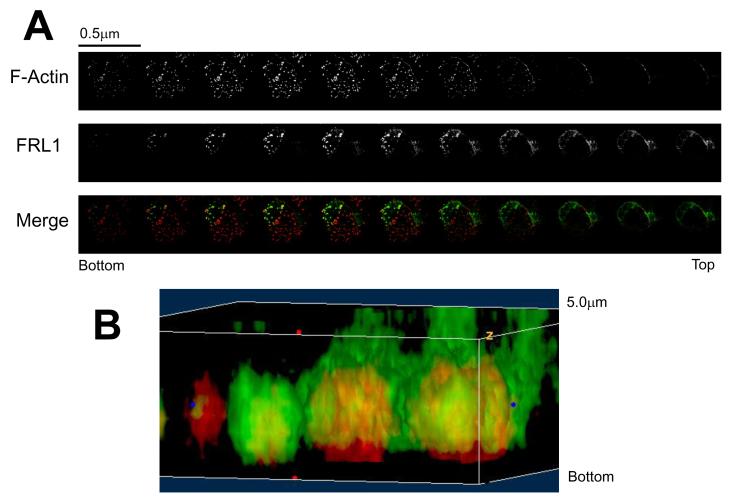
Figure 3. FRL1 Localizes above the Podosome Tip.
A. Z-axis kymograph composed of confocal microscopy images of a primary macrophage. Cells were fixed, permeabilized, and stained for FRL1 and actin using goat anti-FRL1 followed by FITC tagged anti-goat secondary antibody and rhodamine phalloidin, respectively. Progressive sections are incremented by 0.5μm. B. Three-dimensional reconstruction of podosomes stained for FRL1 and actin. FRL1 has been stained green using anti-FRL1 antibody followed by FITC-tagged secondary antibody while actin has been stained red with rhodamine phalloidin.
Results
FRL1 is upregulated during monocyte differentiation to macrophages
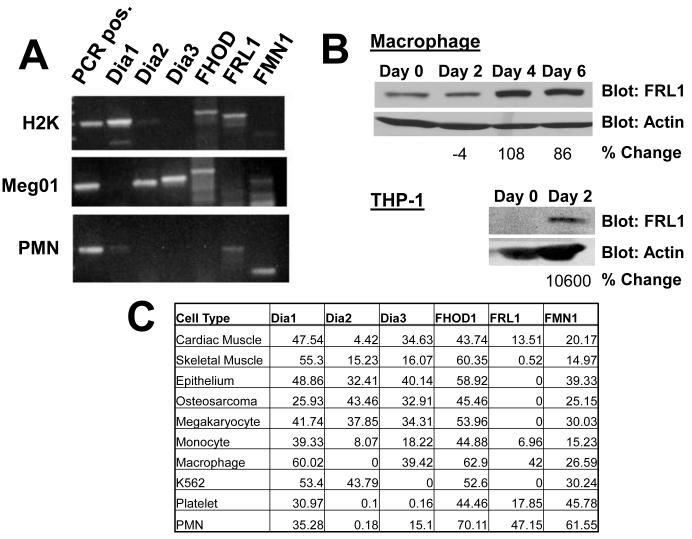
We have reported that a purified complex of proteins associated with the β3 integrin is capable of generating unbranched actin filaments in vitro [Butler et al., 2006]. Sequence efforts indicated the presence of a formin family member in this complex which is potentially responsible for actin engagement of the integrin complex. Since at least 15 mammalian formins have been described, we used RT-PCR to quantitate mRNA for formins in monocytes and differentiated macrophages, and in comparison with other cell types. PCR oligomers were first tested on mRNA isolated from H2K epithelial cells, Meg01 megakaryocytes, and primary neutrophils (PMNs), among others, to verify that each pair of oligomers produced a unique PCR product (Figure 1A). PCR oligomers were then used quantitatively on mRNA isolated from primary monocytes and macrophages differentiated in the presence of GM-CSF. While five of the six formins we examined showed a modest increase in mRNA levels, the formin FRL1 showed a more prominent six-fold increase in mRNA levels normalized to static levels of 28S RNA controls after 5 days of differentiation (Figure 1C). By non-quantitative PCR, we determined that differentiated macrophages express a mixture of FRL1α and FRL1γ, but no FRL1β. We were unable to design oligomers to quantitatively determine the relative expression of the isoforms. In order to verify that protein levels increased along with mRNA, we blotted for FRL1 in both monocyte and macrophage lysates. A correlating pattern of increased FRL1 protein expression was observed (Figure 1B) following differentiation. In the representative experiment, fully differentiated macrophages showed a 108% increase in FRL1 expression compared to freshly isolated monocytes.These experiments were repeated on THP-1 cells and on THP-1 cells induced to differentiate into a macrophage phenotype (Figures 1B and 1C). In THP-1 cells, the mRNA levels increased even more dramatically—26-fold—and subsequent Western blotting showed that FRL1 was detectible in THP-1 lysate only after differentiation, increasing 10600% in the representative experiment. Interestingly, FRL1 was the least diversely expressed formin tested in varied tissues. Further, we find that most cell types examined expressed mRNA for multiple formins simultaneously.
Figure 1. FRL1 Expression is Upregulated in Macrophages.
A. RT-PCR performed on mRNA of H2K epithelial cells, Meg01 megakaryocyte cells, and polymorphonuclear cells (PMN) in order to verify that each primer pair, listed in Methods, produces a unique fragment of the predicted size, and that each formin primer pair produces a product in at least one cell type. B. Western blot performed on cell lysates of control (Control) and differentiated (Diff) primary macrophages and THP-1 cells. Cell differentiation and sample preparation are described in Methods. FRL1 levels are significantly increased in differentiated macrophages and THP-1 cells. C. Quantitative RT-PCR, reporting relative percent expression of the listed formins in various cell types, compared to static ribosome transcript.
FRL1 localizes to macrophage podosomes
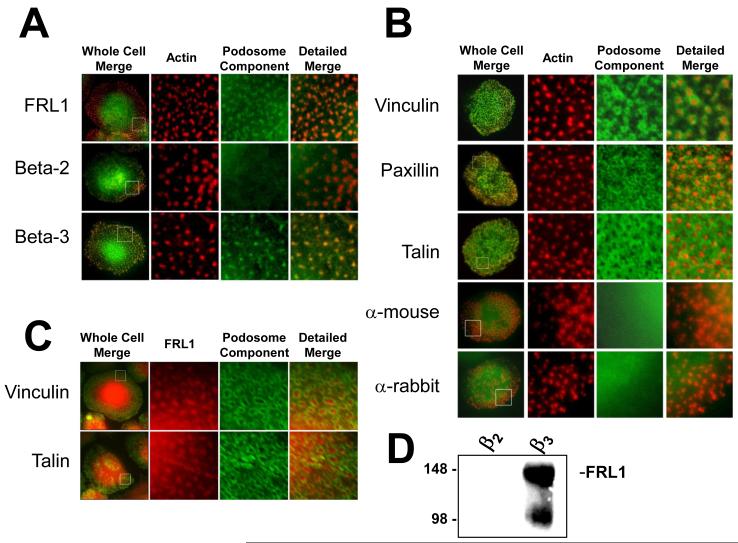
β3 integrin is known to be upregulated during macrophage differentiation in GM-CSF [DeNichilo and Burns, 1993] and FRL1 displayed a similar pattern of expression. Since we previously reported actin assembly activity in association with purified macrophage β3 integrin complexes, we examined whether FRL1 localized to β3 integrin adhesions in macrophages. We find that FRL1 and β3 integrin co-localize to punctate actin structures known as podosomes (Figure 2A). The dense accumulation of actin within podosomes cores mandates low concentrations of rhodamine phalloidin to avoid overexposure. Figure 2B shows macrophages stained for vinculin, paxillin, and talin, known podosomes markers [Kopp and Linder, 2005], as well as macrophages stained with secondary antibodies alone. Vinculin forms intense rings around the actin-rich podosome core, while paxillin forms rings of puncta, and talin forms a modified sheet throughout the cell but is absent from actin cores. All three podosome markers are excluded from the actin-rich core, in contrast to FRL1 and β3 integrin, but define the FRL1 localization to the podosome. To further verify that FRL1 localizes to the podosome core, we co-stained cells for FRL1 and either vinculin or talin. In both cases, FRL1 localized to the core of the podosome, while vinculin and talin circumscribed the core and defined the podosome ring (Figure 2C).
Figure 2. FRL1 Localizes to Podosomes in Differentiated Macrophages.
A. Fluorescence microscopy images of macrophages stained for FRL1 and integrins. The boxed areas in the first column are enlarged in the second, third and fourth columns, with the second column showing only rhodamine phalloidin staining of actin, the third showing only FITC staining, and the fourth showing a merged image of the second and third columns. FRL1 is visualized using a rabbit primary antibody followed by an FITC-tagged anti-rabbit secondary antibody. Integrins beta-2 and beta-3 are visualized using monoclonal mouse primary antibody followed by FITC-tagged anti-mouse secondary antibody. Boxed regions are 10μm by 10μm. B. The first column in each row shows a representative macrophage stained with rhodamine phalloidin and either no primary antibody, hVin α-vinculin, 349 α-paxillin, or 8D4 α-talin followed by FITC tagged anti-mouse or FITC tagged anti-rabbit. The boxed areas in the first column are enlarged in the second, third and fourth columns, with the second column showing only rhodamine phalloidin staining of actin, the third showing only FITC staining, and the fourth showing both. The anti-mouse and anti-rabbit controls show normal podosomes and demonstrate that the rhodamine phalloidin stain does not bleed into the green channel. Vinculin staining forms a concentric ring around the actin-rich podosome and is excluded from the core. Paxillin staining appears to surround the podosome and is excluded from the core but appears as punctute points surrounding the core rather than a continuous ring. Talin staining forms a modified sheet throughout the cell but is excluded from the actin-rich core. Boxed regions are 10μm by 10μm. C. The first column in each row shows a representative macrophage stained with goat anti-FRL1 antibody followed by TRITC-tagged anti-goat antibody and either 349 α-paxillin or 8D4 α-talin followed by FITC tagged anti-mouse or FITC tagged anti-rabbit. The boxed areas in the first column are enlarged in the second, third and fourth columns, with the second column showing only FRL1 staining, the third showing only FITC staining, and the fourth showing a merged image of the second and third columns. Boxed regions are 10μm by 10μm. D. Western blot of proteins immunoprecipitated from differentiated primary macrophage lysates using anti-β2 IB4 monoclonal antibody and anti-β3 7G2 and 1A2 monoclonal antibodies as described in Methods. IPs were separated by SDS-PAGE and blotted for FRL1. FRL1 co-immunoprecipitates with β3 integrin but not with β2 integrin.
Immunoprecipitates of macrophage β3 and β2 integrin were also blotted for FRL1, and show selective association of FRL1 with β3 (Figure 2D). The Western blot also shows the presence of a lower molecular weight band at around 100kDa. We believe that this is a currently unidentified isoform of FRL1. Two bands are seen, the lower of which appears to be an undefined post-translational modification or alternate splice product. The intense upper band at 140kDa represents the full-length transcript and is the primary species seen in macrophages.
In addition to epiflourescence microscopy, we examined FRL1 localization using three dimension confocal microscopy. Figure 3A shows z-slices of a macrophage stained for FRL1 and F-actin, separated by 0.5μm. The slices are reconstructed into a three dimensional image in Figure 3B. This image shows that FRL1 localizes within and slightly outside of the podosome core but does not extend as ventrally as the actin core does. Rather, it forms a cloud surrounding the dorsal aspect of the actin core.
GFP and Xpress-Tagged FRL1 localizes to podosomes
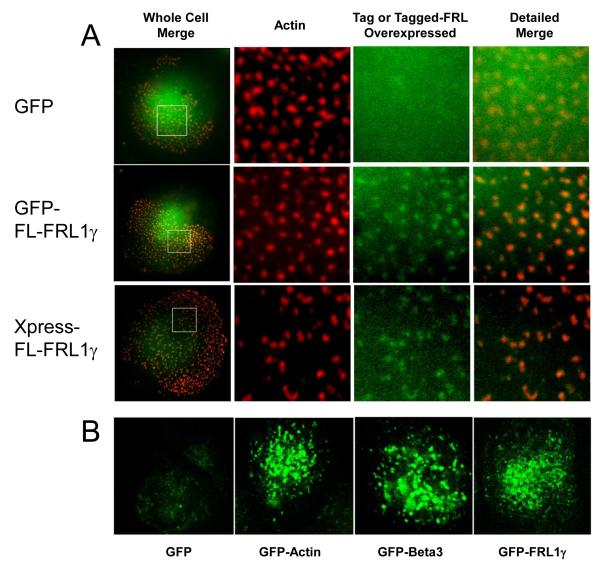
Once we established that endogenous FRL1 localizes to podosomes by antibody staining, we generated and expressed constructs encoding GFP-full-length FRL1γ and Xpress-full-length-FRL1γ. Figure 4A shows that both constructs localized to podosome cores in the same manner as the endogenous FRL1, while GFP alone remained diffusely distributed in the cytoplasm. Macrophages expressing GFP-tagged FRL1γ were examined, along with GFP-actin and GFP-β3 using live confocal microscopy (Supplemental Movies 1, 2, and 3, respectively). Figure 4B shows representative single frames from each of these movies. All three movies show the formation of punctae of GFP-tagged protein with a half life of approximately 1-2 minutes. No such concentrations of protein were observed with GFP alone (Supplemental Movie 4).
Figure 4. GFP- and Xpress-tagged FRL1 localize to podosomes.
A. Fixed cell microscopy of macrophages expressing GFP alone, GFP-FRL1, and Xpress-FRL1. Cells were fixed, permeabilized, and stained with rhodamine phalloidin to visualize actin. Cells transfected with Xpress-FRL were stained with monoclonal anti-Xpress antibody followed by FITC tagged anti-mouse secondary antibody. The boxed area from the images in the first column are enlarged in the second, third and fourth. The second column shows rhodamine phalloidin staining, the third column either GFP or FITC, and the fourth column a merged image of the second and third columns. Boxed regions are 10μm by 10μm. B. Confocal live cell microscopy images of macrophages expressing GFP alone or GFP tagged actin, beta-3, or FRL1.
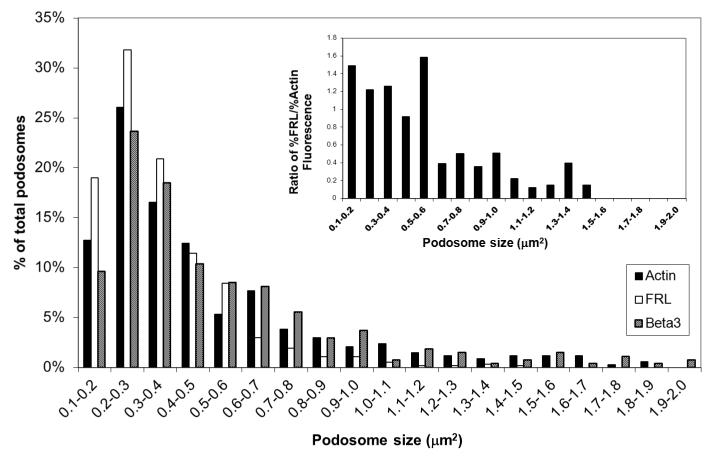
Podosomes visualized by GFP-actin, -β3 integrin, and -FRL1γ were identified using an object recognition feature in Volocity software and were quantified for average number, size, and distribution. GFP-actin, GFP-β3, and GFP-FRL1γ showed an average of 113, 136, and 190 podosomes per cell with an average podosome size of 0.53, 0.57, and 0.35 μm2, respectively. Figure 5 shows the distribution by size of the podosomes in each cell type. All three podosome markers identify podosomes with a median diameter of 0.6-0.9μm and ranges from 0.3-1.6μm in agreement with Linder [Linder, 2008]. Interestingly, the GFP-FRL1γ podosomes have a lower average size and higher average number per cell than either GFP-actin or GFP-β3. Their size distribution is also biased toward the lower end. The inset in Figure 5 shows the ratio of percent of total podosomes of FRL1 to percent of total podosomes of actin for the given ranges. The data indicates that below 0.6μm2 there is a greater predominance of podosomes with GFP-FRL1γ. This, along with the smaller average size of podosomes marked by GFP-FRL1γ, suggests that FRL1 concentrates toward the center of the podosome core, and may indicate their early presence in newly forming podosomes.
Figure 5. Distribution of Podosome Size.
The chart shows the size distribution in 0.1μm2 intervals of podosomes identified by GFP-actin, GFP-FRL, and GFP-β3 fluorescence as a percentage of total podosomes per cell. The inset chart shows the ratio of the percent of total podosomes of FRL to the percent of total podosomes of actin for the given ranges as determined by the presence or absence of GFP in the podosome for each construct. There is a dramatic decrease in ratios from the 0.1-0.6 μm2 range to the 0.6-1.5 μm2 range, indicating a preferred distribution of FRL in smaller podosomes.
RhoGTPase activity modulates FRL1 podosomal distribution
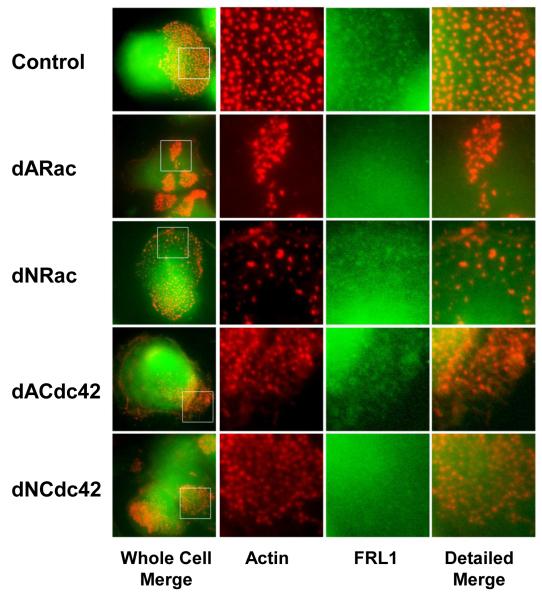
GTPases regulate of a number of actin modifying proteins, most notably the Arp2/3 complex through WASp and the formin family of proteins. Previous studies have indicated that FRL1 activation and localization is regulated by either Rac1 or Cdc42 [Seth et al., 2006, Yayoshi-Yamamoto et al., 2000]. We examined the effects of Rac1 and Cdc42 on the size and distribution of podosomes and as well as localization of FRL1 to podosomes in primary human macrophages. Cells were transfected with dominant active and dominant negative mutants of each GTPase. Figure 6 shows that dominant active Rac1 altered the distribution of podosomes into small clusters throughout the cells without any concentrations of FRL1 while dominant negative Rac1 reduced the concentration of podosomes but did not affect localization of FRL1. Overexpression of dominant active Cdc42 resulted in a reduction in the number of podosomes and an increase in lamellipodia, but did not alter localization of FRL1 to podosomes. Overexpression of the dominant negative mutant of Cdc42 resulted in altered podosome patterns and also a loss of FRL1 localization at macrophage podosomes. When we quantified the average podosomes per cell (Supplemental Figure 1B), we found that altering the GTPase balance by overexpressing any of the GTPase constructs resulted in a decrease in the average number of podosomes as reflected in the representative images in Figure 6. The average size of the podosomes still present in these cells was not dramatically altered (Supplemental Figure 1C). Overall, constructs excluding dominant negative Rac1 and dominant active Cdc42 resulted in a loss of FRL1 localization at the macrophage podosome. We were unable to quantify podosomes using antibody staining for FRL1 due to high background staining. However, the images clearly show differences in localization of FRL1 to the podosome. A previous study has demonstrated that active Cdc42 relieves autoinhibition of FRL1 and induces membrane localization [Seth et al., 2006]. The presence of FRL1 in podosomes in cells expressing dominant active Cdc42 but not in cells expressing dominant negative Cdc42 suggests that FRL1 localization to podosomes is likely positively regulated by Cdc42. The opposite effect of Rac1 construct activity further supports this, as Rac1 and Cdc42 levels are inversely regulated. Our own studies have shown that the predominant active GTPase evolves as the β3 adhesion site matures [Gao et al., 2005]. Within the first ten minutes after contact between the integrin and ligand, Cdc42 is the dominant active GTPase after which it becomes progressively inactivated. Active Rac1 levels continuously increase and overwhelm active Cdc42 around 30 minutes after contact with the ligand. Our results suggest that Cdc42 is required for localization of FRL1 while Rac1 inhibits localization. Taken together, these results suggest that Cdc42 is involved in recruiting FRL1 to macrophage podosomes while Rac1 negatively regulates FRL1 localization but not podosome formation.
Figure 6. The Effects of Rho GTPases on FRL1 Localization to Podosomes.
Fixed cell microscopy of macrophages transfected with a vector encoding dominant active Rac (dARac), dominant negative Rac (dNRac), dominant active Cdc42 (dACdc42), and dominant negative Cdc42 (dNCdc42). Cells were fixed, permeabilized, and stained with rhodamine phalloidin to visualize actin and anti-FRL1 followed by a FITC tagged anti-goat secondary antibody. The boxed area from the images in the first column are enlarged in the second, third and fourth. The second column shows rhodamine phalloidin staining, the third column FITC, and the fourth column a merged image. Boxed regions are 10μm by 10μm.
Loss of FRL1 results in decreased macrophage adhesion
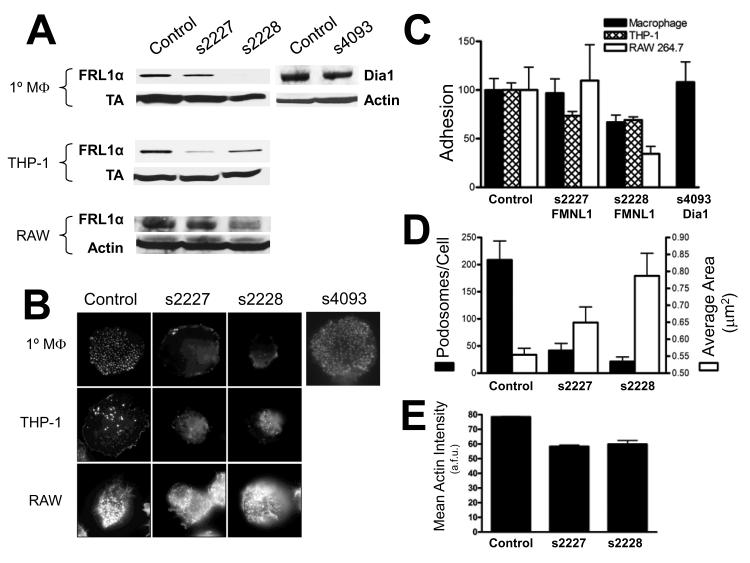
In order to explore the functional role of FRL1 in macrophage podosomes, we treated primary macrophages with siRNA targeting two different exons of FRL1. Figure 7A shows Western blots of lysates from cells in which FRL1 levels were reduced with siRNA. In primary macrophages, s2227 reduced FRL1 levels slightly while s2228 reduced FRL1 to nearly undetectable levels. In both cases we found that adhesion of primary macrophages was reduced as compared to controls and correlated to the levels of FRL1 as shown by Western blotting (Figure 7C). The s2227 oligomer did not significantly reduce FRL1 expression and caused a similarly insignificant 3% reduction (p=0.870) in adherent cells. The s2228 oligomer, however, reduced FRL1 levels by an average of 89% and led to a correspondingly significant 33% (p=0.045) reduction in the number of adherent cells. Depletion of Dia1 with an s4093 oligomer resulted in a 45% decrease in Dia1 expression but no significant decrease in cell adhesion (p=0.743). We verified our observations in two transformed macrophage cells lines; THP-1 cells are an undifferentiated human monocyte cell line that can be differentiated to macrophages with PMA while RAW 264.7 cells are a fully differentiated mouse macrophage cell line. PMA differentiation of THP-1 cells induces the formation of podosomes [Akimov and Belkin, 2001]. We found that s2228 significantly reduced adhesion of both cell types; Western blotting showed a reduction of 25% of endogenous FRL1 in THP-1 cells and 79% in RAW 264.7 cells. The depletion of FRL1 resulted in a 31% reduction (p=0.001) in adherent THP-1 cells after treatment with s2228 and a 65% reduction (p=0.029) in RAW 264.7 cells. While treatment with the s2227 oligomer resulted in a 59% decrease in endogenous FRL1 expression in THP-1 cells and a 33% reduction in RAW 264.7 cells, only the THP-1 cells showed a significant 26.5% decrease (p=.006) in adhesion when FRL1 was depleted with this oligomer.
Figure 7. siRNA Targeting of FRL1 Reduces Macrophage Adhesion and Podosome Number.
A. Western blots of lysates from differentiated macrophages, THP-1 cells, and RAW 264.7 cells treated with control siRNA, s2227 siRNA, s2228, or s4093 siRNA as described in Methods. Lysates were separated on SDS-PAGE gels and blotted for FRL1 with mouse monoclonal anti-FRL1 antibody followed by anti-mouse HRP tagged antibody or for Dia1 with goat polyclonal anti-Dia1 antibody followed by anti-goat HRP tagged antibody. Lysates were also blotted for either transaldolase or actin as controls. B. Fixed cell microscopy of cell treated with siRNA targeting formins as described for A. Cells were fixed, permeabilized, and stained with rhodamine phalloidin to visualize actin. Images are 40μm by 40μm. C. Adhesion of macrophages following FRL1 reduction by siRNA. Graph depicting average cell numbers in one square centimeter in 24 well plates after treatment with siRNA to reduce expression of formins as described for A. Macrophage counts are shown as solid bars, THP-1 cell counts are shown as open bars, and RAW 264.7 cell counts are shown as cross-hatched bars. Results are reported as mean +/− SEM. D. Podosome number and area in macrophages following FRL1 reduction by siRNA. Graph depicting average podosome counts and average individual podosome area in primary macrophages treated with siRNA. Podosome count data is shown as solid bars while average podosome size data is shown as open bars. Results are reported as mean +/− SEM. E. Actin Intensity in Podosomes. Graph depicting average actin intensity per podosome in primary macrophages treated with siRNA. Mean Actin Intensity is shown as solid bars. Results are reported as mean +/− SEM.
When we examined the remaining adherent cells microscopically, we found that the size and number of podosomes was altered from normal (Figures 7C and 7D). Using the method described above for calculating podosome size and number, but with an intensity threshold of 50%, we calculated the average number of podosomes and area per podosome from fixed cell images stained with rhodamine phalloidin. Using this method, we found that macrophages transfected with control siRNA had an average of 208.7 podosomes per cell with an average area of 0.55μm2 per podosome. Cells treated with the s2227 oligomer had an average of 41.7 podosomes per cell with an average area of 0.65 μm2 per podosome, corresponding to an 80% decrease (p=0.011) and a 17% increase, respectively. Cells treated with the s2228 oligomer had an average of 21.3 podosomes per cell with an average area of 0.79 μm2 per podosome, corresponding to a 90% decrease (p=0.006) and a 42% increase, respectively. Together these data show that reduction of FRL1 corresponds with loss of macrophage adhesion and a progressive decrease in podosome number, but an increase in podosome size.
The increase in podosome size prompted us to examine whether the amount of actin within each podosome had changed. When we examined the mean concentration of actin within each podosome, defined by the mean intensity of rhodamine phalloidin staining, we found that both FRL1 targeting siRNA oligomers decreased mean actin concentration (58.4 a.f.u. for s2227 and 59.9 a.f.u. for s2228) compared to controls (78.5 a.f.u.) (Figure 7E). This effect was significant for both oligomers (p<0.0001).
We approximated the average total actin within individual podosomes by multiplying the mean of the area of individual podosomes by the mean fluorescence intensity of rhodamine phalloidin staining in individual podosomes. Control cells had an average total actin fluorescence per podosome of 43.5 units, while cells treated with the s2227 oligomer had an average total actin per podosome of 37.9 units and cells treated with the s2228 oligomer had an average total actin per podosome of 47.1 units. (Supplemental Figure 1B). These data indicate that there is not a significant reduction in total actin per podosome when FRL1 levels are reduced. However, since the average number of podosomes per cell is reduced, the total adhesion contact area is significantly decreased when FRL1 levels are reduced. We approximated total adhesion contact area by multiplying total actin per podosome by the number of podosomes per cell. Control cells had a total adhesion contact area of 9078 units. while macrophages treated with the s2227 oligomer had a total adhesion contact area of 1580 units, and macrophages treated with the s2228 olgimer had a total adhesion contact area of 1005 units (Supplemental Figure 1B). In summary, while loss of FRL1 results in larger podosomes, the total actin within podosomes does not change. In addition, while the total area per individual podosomes increases, the significant decrease in the number of podosomes per cell results in a dramatically reduced total adhesion surface area, resulting in a loss of adhesion.
Discussion
A great deal of work has been done in elucidating the biochemistry of formins. It is now clear that individual formins are capable of nucleating, elongating, severing, and bundling actin filaments. Some recent studies have begun to describe the functions of formins in actin-based structures within cells. However, little attention has been given to the expression patterns of formins within cells. With the array of formins currently known, it is likely that expression patterns vary significantly between cell types. We examined a broad range of cell types and found significant variation in expression levels, with each of 15 cell types expressing 3-5 of the six formins examined (Figure 1C). Such variations in expression likely play a significant role in the phenotypes observed in different cells, particularly in the types of actin-based structures that a given cell generates.
The differentiation of monocytes to macrophages is one of the most dramatic changes seen in eukaryotic cells. Macrophages gain the ability to diapedese through the endothelium, migrate through inflammatory tissues in response to chemotactic signals, phagocytose pathogens, and present processed antigens to lymphocytic cells. All of these phenotypic traits require the generation of new actin filaments. While Arp2/3 has been shown to be present in many of these structures, our previous studies have indicated that actin nucleation at β3 integrin adhesion sites may be formin dependent. However, it was not clear which of these formins were present at the β3 adhesion site, nor which formins are expressed endogenously in macrophages. We examined the expression and localization of formins in both undifferentiated monocytes and differentiated macrophages.
We found that following differentiation with GM-CSF in culture for five days, cells increase expression of Dia1, Dia3, FHOD1, FRL1, and FMN1 mRNA by 52.6%, 116.4%, 40.2%, 503.5%, 74.6% respectively, while decreasing Dia2 expression to zero. Such drastic changes in formin expression likely account for many of the phenotypic changes observed during macrophage differentiation. A change in the subset of formins expressed in a cell may shift the phenotype produced in a cell in response to activation of a particular GTPase. A cell expressing one particular formin regulated by a given GTPase may produce entirely different actin-based structures than another expressing a different formin regulated by the same GTPase. For example, Moreau et al. recently indicated that expression of a constituitively active mutant of Cdc42 in vascular endothelial cells yields structures consistent with podosomes [Moreau et al., 2006]. In our own experiments with K562 cells, we have found that a Cdc42 dominant phenotype produces extensive filopodia [Gao et al., 2005]. It is possible that these two cell types express different Cdc42 regulated formins. Thus, variations in formin expression may account for differences in phenotypes produced in response to activation of a given GTPase between cell types. The most striking shift in expression during macrophage differentiation occurs in FRL1. We found that FRL1 was not expressed in epithelial cells, megakaryocytes (data not shown), or undifferentiated monocytes, but is expressed in differentiated macrophages. These results agree with Yayoshi-Yamamoto et al.’s findings [Yayoshi-Yamamoto et al., 2000]. FRL1 has been shown to be capable of nucleating, elongating, bundling, and severing actin filaments in an autoregulated, GTPase dependent manner [Harris et al., 2004; Seth et al., 2006]. The GTPase responsible for autoregulation, however, is still unclear. In contrast to what has been observed in the case of Dia1, two studies have already indicated that FRL1 is not regulated by Rho. Yayoshi-Yamamoto et al. showed that mouse FRL1 associates with GTPase bound Rac1 in COS7 cells [Yayoshi-Yamamoto et al., 2000]. Seth et al. indicated that GMPPNP loaded Cdc42 relieved autoinhibition of FRL1 and induced localization of FRL1 to the membrane in RAW macrophages [Seth et al., 2006]. It is likely that the regulation of murine FRL1 is identical to that of human FRL1, as 94% of the first 450 amino acids, including the DID and GBD, are identical. Yayoshi-Yamamoti et al. described two isoforms of FRL1, α and β, isolated from mouse brain and spleen respectively [Yayoshi-Yamamoto et al., 2000]. The β form they described contains an alternatively spliced 38 amino acid N-terminus, which replaces the 35 amino acid N-terminus found on the α and γ isoforms. A third isoform, FRLγ, has also been recently described, although it was not examined for GTPase regulation [Han et al., 2009]. We find that macrophages express both the α and γ, but not β isoforms (data not shown). In these studies, we introduced FRL1 cDNA encoding γ, the longest possible transcript.
One previous study indicated that FRL1 may play a role in Fc receptor mediated phagocytosis [Seth et al., 2006]. Here we found that FRL1 is upregulated in parallel with β3 integrin, associates with activated β3 integrin and localizes to the podosome. Podosomes are actin rich structures approximately 0.5 μm in diameter and 0.5 μm in height that are observed in differentiated macrophages [Marchisio et al, 1988]. They have also been shown to be rich in vinculin, paxillin, talin, β2, and β3. A role for FRL1 in both podosomes and phagosomes is not unreasonable. Both functions have been shown to be Cdc42 driven [Seth et al., 2006; Tatin et al., 2006; Trotter, 1981] and occur in mature macrophages exclusive of undifferentiated monocytes. Our own results using GTPase constructs demonstrate that dominant active Cdc42 leaves podosomes and FRL1 localization to podosomes intact while dominant negative Cdc42 eliminates both podosomes and FRL1 localization.
A number of previous studies have indicated that the Arp2/3 complex localizes to podosomes and is required for podosome formation. Arp2/3 has been placed at the tips of podosomes by fluorescent microscopy [Calle et al., 2006]. Additionally, inhibition of Arp2/3 by CA constructs, which act competitively to sequester active WASp, appears to abolish podosome formation [Linder et al., 2006]. While Arp2/3 appears to play a significant role in actin turnover in podosomes, its presence does not rule out a role for FRL1 in regulating actin filaments in podosomes. Our own studies have indicated that Arp2/3 is apparent at early β3 adhesion contacts but its presence is transient [Chandhoke et al., 2004]. The specific role of either of these proteins in nucleating actin is not clear. It may also be possible that FRL1 functions in bundling actin fibers generated by Arp2/3. Treating macrophages with siRNA targeting FRL1 showed that loss of FRL1 results in significantly reduced adhesion. Fluorescent microscopy also showed significant loss of podosomes and an increase in the size of the podosomes. In contrast, loss of Dia1 left adhesion and podosomes unchanged. When we quantified the total actin per podosome when FRL1 levels are reduced by siRNA, we found that the total actin does not change significantly. While podosome size increases with reduced FRL1, the total adhesion contact area decreases, since the number of podosomes is reduced. This loss of adhesion contact surface area likely results in the loss of adhesion that we observed. A connection between macrophage adhesion and FRL1 was previously demonstrated by Yayoshi-Yamamoto et al [Yayoshi-Yamamoto et al., 2000]. In their study, they showed that macrophages transfected with the FH3 domain of FRL, which has since been redefined as the GBD/DID domains, significantly reduced adhesion of macrophages. Their results suggest that the presence of excess DID domain inhibits FRL1 and reduced macrophage adhesion. However, more recent studies have shown that DID domains can act in a trans fashion and therefore may be inhibiting formins other than FRL1. Our study shows that specific knockdown of FRL1 reduced macrophage adhesion and disrupted normal podosome structure.
Based on three dimensional reconstructions of the podosomes using confocal fluorescent microscopy images, FRL1 appears to localize slightly above the actin rich podosome structure (Figure 3B). Placing this in the context of a recent study by Luxenburg et al, FRL1 appears to interact primarily with the network of radial actin filaments surrounding the podosomes. Destaing et al. showed that the rate of turnover of actin within podosomes is 2-3 times the rate of turnover of actin in the surrounding network. Such a difference in actin turnover rates may be accounted for by the presence of two different actin nucleating proteins working within the podosome. Another possibility is that FRL1 is not generating new actin filaments but rather is bundling or severing previously formed filaments.
The severing function is of particular interest, as Destaing et al proposed a model in which actin filaments are polymerized at the base of the podosome and randomly severed throughout its length [Destaing et al., 2003]. Our quantitative analysis of the effects of siRNA targeting FRL1 on podosomes strongly suggests that FRL1 is directly involved in stabilizing podosomes during their life cycle. Loss of FRL1 led to a dramatic decrease in podosome number and a significant increase in average podosome size. Such an effect could be explained by the bundling function of FRL1. The fact that total actin within podosomes does not change, but podosome size increases when FRL1 levels decrease further supports a role for FRL1 in bundling filaments within podosomes. Podosomes are highly dynamic structures with a half-life of 2-12 minutes, after which they can either dissolve or fuse with other podosomes. They can form de novo or by fission of existing podosomes [Kopp and Linder, 2005]. The loss of FRL1 as an actin bundling protein within podosomes would lead to a dispersion of the actin fibers within the structure, and a weakening of the adhesion contact, leading to a loss of cell adhesion, as we observed. Determining which activity of FRL1 is important in podosomes will be a future challenge. The same regions of the protein appear to be involved in all four known activities, and it is not yet clear whether severing or bundling are under the same GTPase controlled autoregulation that actin assembly appears to be.
In this report, we demonstrate for the first time that a formin localizes to the macrophage podosome and is critical both for macrophage adhesion and for podosome stability. In light of our previous study, it is likely that FRL1 plays a role in modifying the actin filaments at the β3 adhesion site [Butler et al., 2006]. However, it is not clear what role it plays in this modification. Given the multitiude of functions described for FRL1 in vitro, identifying the specific role of FRL1 at the podosome will require a method to isolate or eliminate individual functions in vivo. Additionally, the connection between β3 integrin and FRL1 requires clarification. While the two protein co-immunoprecipitate, it is not clear which, if any, components of the β3 signaling pathway are required for FRL1 localization or activation. In our previous studies, we found that Vav1 is activated downstream of β3 integrin and functions to activate Rho [Gao et al., 2005]. Since FRL1 contains a GTPase binding domain, it is possible that this pathway directly regulates FRL1. This interaction will best be clarified by targeting individual components of the β3 signaling pathway along with individual domains of the FRL1 protein. Clarifying the function and regulation of FRL1 at the podosome will be of great interest because of its potentially critical role in the extravasation of macrophages.
Supplementary Material
Acknowledgements
The authors would like thank Dr. Angela Krackhardt (Helmholtz Zentrum Munchen, Munich, Germany) for generously sharing her pCMV-His-FMNL1 vector, Nicholas Deakin (SUNY Upstate Medical University, Syracuse, NY) for sharing his GTPase constructs and his assistance with confocal microscopy, and Matthew P. Williams (Washington University, St. Louis) for his technical assistance. This work is supported by NIH grant R01DK79884 (to S. Blystone).
Reference List
- Akimov SS, Belkin AM. Cell surface tissue transglutaminase is involved in adhesion and migration of monocytic cells on fibronectin. Blood. 2001;98:1567–76. doi: 10.1182/blood.v98.5.1567. [DOI] [PubMed] [Google Scholar]
- Butler B, Gao C, Mersich AT, Blystone SD. Purified integrin adhesion complexes exhibit actin-polymerization activity. Curr Biol. 2006;16:242–251. doi: 10.1016/j.cub.2005.12.033. [DOI] [PubMed] [Google Scholar]
- Calle Y, Burns S, Thrasher AJ, Jones GE. The leukocyte podosome. Eur J Cell Biol. 2006;85:151–157. doi: 10.1016/j.ejcb.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Chandhoke SK, Williams M, Schaefer E, Zorn L, Blystone SD. Beta 3 integrin phosphorylation is essential for Arp3 organization into leukocyte alpha V beta 3-vitronectin adhesion contacts. J Cell Sci. 2004;117:1431–1441. doi: 10.1242/jcs.00987. [DOI] [PubMed] [Google Scholar]
- Chhabra ES, Higgs HN. INF2 Is a WASP homology 2 motif-containing formin that severs actin filaments and accelerates both polymerization and depolymerization. J Biol Chem. 2006;281:26754–26767. doi: 10.1074/jbc.M604666200. [DOI] [PubMed] [Google Scholar]
- Chhabra ES, Higgs HN. The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol. 2007;9:1110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- Copeland SJ, Green BJ, Burchat S, Papalia GA, Banner D, Copeland JW. The DID-DAD interaction is able to mediate heterodimerization between mDia1 and mDia2. J Biol Chem. 2007;282:30120–30. doi: 10.1074/jbc.M703834200. [DOI] [PubMed] [Google Scholar]
- De Nichilo MO, Burns GF. Granulocyte-macrophage and macrophage colony-stimulating factors differentially regulate alpha v integrin expression on cultured human macrophages. Proc Natl Acad Sci USA. 1993;90:2517–2521. doi: 10.1073/pnas.90.6.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destaing O, Saltel F, Geminard JC, Jurdic P, Bard F. Podosomes display actin turnover and dynamic self-organization in osteoclasts expressing actin-green fluorescent protein. Mol Biol Cell. 2003;14:407–416. doi: 10.1091/mbc.E02-07-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro PM, Traina F, Vassallo J, Brousset P, Delsol G, Costa FF, Saad ST. High expression of FMNL1 protein in T non-Hodgkin’s Lymphoma. Leuk.Res. 2006;30:735–8. doi: 10.1016/j.leukres.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Gao C, Blystone SD. A Pyk2-Vav1 complex is recruited to beta3-adhesion sites to initiate Rho activation. Biochem.J. 2009;420:49–56. doi: 10.1042/BJ20090037. [DOI] [PubMed] [Google Scholar]
- Gao C, Schaefer E, Lakkis M, Blystone SD. Beta3 tyrosine phosphorylation and alphavbeta3-mediated adhesion are required for Vav1 association and Rho activation in leukocytes. J Biol Chem. 2005;280:15422–15429. doi: 10.1074/jbc.M414457200. [DOI] [PubMed] [Google Scholar]
- Gomez TS, Kumar K, Medeiros RB, Shimizu Y, Leibson PJ, Billadeau DD. Formins regulate the actin-related protein 2/3 complex-independent polarization of the centrosome to the immunological synapse. Immunity. 2007;25:177–90. doi: 10.1016/j.immuni.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Eppinger E, Schuster IG, Weigand LU, Liang X, Kremmer E, Peschel C, Krackhardt AM. Formin-like 1 (FMNL1) is regulated by N-terminal myristoylation and induces polarized membrane blebbing. J.Biol.Chem. 2009;284:33409–17. doi: 10.1074/jbc.M109.060699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris ES, Li F, Higgs HN. The mouse formin, FRLalpha, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments. J.Biol.Chem. 2004;279:20076–20087. doi: 10.1074/jbc.M312718200. [DOI] [PubMed] [Google Scholar]
- Harris ES, Rouiller I, Hanein D, Higgs HN. Mechanistic differences in actin bundling activity of two mammalian formins, FRL1 and mDia2. J Biol Chem. 2006;281:14383–14392. doi: 10.1074/jbc.M510923200. [DOI] [PubMed] [Google Scholar]
- Higgs HN, Peterson KJ. Phylogenetic analysis of the formin homology 2 domain. Mol Biol Cell. 2005;16:1–13. doi: 10.1091/mbc.E04-07-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp P, Linder S. Podosomes at a glance. J Cell Sci. 2005;18:2079–82. doi: 10.1242/jcs.02390. [DOI] [PubMed] [Google Scholar]
- Linder S, Nelson D, Weiss M, Aepfelbacher M. Wiskott-Aldrich syndrome protein regulates podosomes in primary human macrophages. Proc Natl Acad Sci USA. 1999;96:9648–9653. doi: 10.1073/pnas.96.17.9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder S, Higgs H, Hufner K, Schwarz K, Pannicke U, Aepfelbacher M. The polarization defect of Wiskott-Aldrich syndrome macrophages is linked to dislocalization of the Arp2/3 complex. J Immunol. 2000;165:221–225. doi: 10.4049/jimmunol.165.1.221. [DOI] [PubMed] [Google Scholar]
- Linder S. The matrix corroded: podosomes and invadopodia in extracellular matrix degradation. Trends in Cell Biology. 2006;17:107–117. doi: 10.1016/j.tcb.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Marchisio PC, Cirillo D, Naldini L, Primavera MV, Teti A, Zambonin-Zallone A. Cell-substratum interaction of cultured avian osteoclasts is mediated by specific adhesion structures. J Cell Biol. 1984;99:1696–1705. doi: 10.1083/jcb.99.5.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchisio PC, Bergui L, Corbascio GC, Cremona O, D’Urso N, Schena M, Tesio L, Caligaris-Cappio F. Vinculin, talin, and integrins are localized at specific adhesion sites of malignant B lymphocytes. Blood. 1988;72:830–833. [PubMed] [Google Scholar]
- Moreau V, Tatin F, Varon C, Anies G, Savona-Baron C, Genot E. Cdc42-driven podosome formation in endothelial cells. Eur J Cell Biol. 2006;85:319–325. doi: 10.1016/j.ejcb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MK. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433:488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- Pfaff M, Jurdic P. Podosomes in osteoclast-like cells: structural analysis and cooperative roles of paxillin, proline-rich tyrosine kinase 2 (Pyk2) and integrin alphaVbeta3. J Cell Sci. 2001;114:2775–2786. doi: 10.1242/jcs.114.15.2775. [DOI] [PubMed] [Google Scholar]
- Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A, Boone C. Role of formins in actin assembly: nucleation and barbed-end association. Science. 2002;297:612–615. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- Romero S, Le CC, Didry D, Egile C, Pantaloni D, Carlier MF. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119:419–429. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- Seth A, Otomo C, Rosen MK. Autoinhibition regulates cellular localization and actin assembly activity of the diaphanous-related formins FRLalpha and mDia1. J Cell Biol. 2006;174:701–713. doi: 10.1083/jcb.200605006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster IG, Busch DH, Eppinger E, Kremmer E, Milosevic S, Hennard C, Kuttler C, Ellwart JW, Frankenberger B, Nossner E, Salat C, Bogner C, Borkhardt A, Kolb HJ, Krackhardt AM. Allorestricted T cells with specificity for the FMNL1-derived peptide PP2 have potent antitumor activity against hematologic and other malignancies. Blood. 2007;110:2931–2939. doi: 10.1182/blood-2006-11-058750. [DOI] [PubMed] [Google Scholar]
- Tatin F, Varon C, Genot E, Moreau V. A signalling cascade involving PKC, Src and Cdc42 regulates podosome assembly in cultured endothelial cells in response to phorbol ester. J Cell Sci. 2006;119:769–781. doi: 10.1242/jcs.02787. [DOI] [PubMed] [Google Scholar]
- Tominaga T, Sahai E, Chardin P, McCormick F, Courtneidge SA, Alberts AS. Diaphanous-related formins bridge Rho GTPase and Src tyrosine kinase signaling. Mol Cell. 2000;5:13–25. doi: 10.1016/s1097-2765(00)80399-8. [DOI] [PubMed] [Google Scholar]
- Trotter JA. The organization of actin in spreading macrophages. The actin-cytoskeleton of peritoneal macrophages is linked to the substratum via transmembrane connections. Exp Cell Res. 1981;132:235–248. doi: 10.1016/0014-4827(81)90099-9. [DOI] [PubMed] [Google Scholar]
- Tsuchiya S, Kobayashi Y, Goto Y, Okumura H, Nakae S, Konno T, Tada K. Induction of maturation in cultured human monocytic leukemia cells by a phorbol diester. Cancer Res. 1982;42:1530–1536. [PubMed] [Google Scholar]
- vid-Pfeuty T, Singer SJ. Altered distributions of the cytoskeletal proteins vinculin and alpha-actinin in cultured fibroblasts transformed by Rous sarcoma virus. Proc Natl Acad Sci USA. 1980;77:6687–6691. doi: 10.1073/pnas.77.11.6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellington A, Emmons S, James B, Calley J, Grover M, Tolias P, Manseau L. Spire contains actin binding domains and is related to ascidian posterior end mark-5. Development. 1999;126:5267–5274. doi: 10.1242/dev.126.23.5267. [DOI] [PubMed] [Google Scholar]
- Yayoshi-Yamamoto S, Taniuchi I, Watanabe T. FRL, a novel formin-related protein, binds to Rac and regulates cell motility and survival of macrophages. Mol Cell Biol. 2000;20:6872–6881. doi: 10.1128/mcb.20.18.6872-6881.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.