Abstract
Fourier transform tandem mass spectrometry (MS/MS) provides high mass accuracy, high sensitivity, and analytical versatility and has therefore emerged as an indispensable tool for structural elucidation of biomolecules. Glycosylation is one of the most common posttranslational modifications, occurring in ~50% of proteins. However, due to the structural diversity of carbohydrates, arising from non-template driven biosynthesis, achievement of detailed structural insight is highly challenging. This review briefly discusses carbohydrate sample preparation and ionization methods, and highlights recent developments in alternative high-resolution MS/MS strategies, including infrared multiphoton dissociation (IRMPD), electron capture dissociation (ECD), and electron detachment dissociation (EDD), for carbohydrates with a focus on glycans and proteoglycans from mammalian glycoproteins.
Keywords: Tandem mass spectrometry, glycosylation, oligosaccharides, infrared multiphoton dissociation (IRMPD), electron capture dissociation (ECD), electron detachment dissociation (EDD)
INTRODUCTION
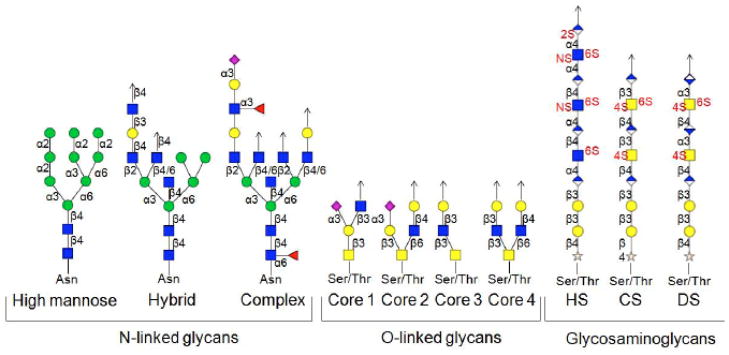
Glycosylation is one of the most prevalent posttranslational modifications (PTMs) and a variety of different types of glycans are also frequently attached to lipids. Glycans participate in many important biological processes, including protein folding and assembly, cell adhesion, cell signaling, immune response, molecular trafficking, and cancer metastasis [1–5]. Although the biological significance of glycosylation is well established, the structural diversity arising from complex nontemplate-based biosynthesis, makes glycan structural characterization a highly challenging task. Glycosylation encompasses, e.g., N-linked glycans, O-linked glycans, and the glycosaminoglycan (GAG) family of polysaccharides. As shown in Fig. (1) [6], N-glycans are branched carbohydrates containing a pentasaccharide core, normally attached to asparagine residues of proteins in an Asn-Xxx-Ser (or Thr) amino acid consensus sequence with Xxx ≠ proline. O-glycans are branched and linked with either serine or threonine but without a consensus amino acid sequence. GAGs are also attached to serine or threonine, but they are linear carbohydrates and often highly sulfated [7–8].
Fig. (1).
Representative structures of N-linked glycans, O-linked glycans, and glycosaminoglycans [6].
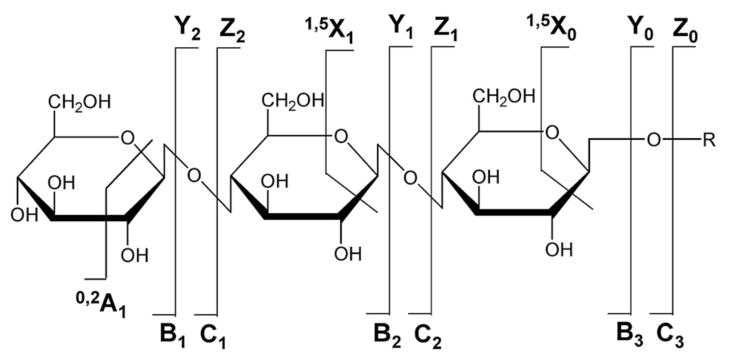
In order to fully understand the key roles glycans play in various cellular processes, knowledge of only the sequence of these compounds is far from sufficient. The linkage, degree of branching, and stereochemistry should also be determined. Traditionally, exoglycosidase digestion, nuclear magnetic resonance (NMR) spectroscopy, X-ray crystallography, and gas chromatography/mass spectrometry (GC/MS) have all been used for structural characterization of carbohydrates [9]. However, such methods do not provide the required sensitivity for glycoproteomics. Modern mass spectrometry is an important tool for structural analysis of carbohydrates, offering high sensitivity, high precision, and analytical versatility. Accurate masses obtained from high-resolution tandem mass spectrometry (MS/MS or MSn) provide reproducible and reliable information for glycan structural characterization. There are two types of bond cleavages in MS/MS, including glycosidic cleavages (B-, C-, Y-, Z-type) and cross-ring cleavages (A- and X-type) as shown in Fig. (2) [10]. Glycosidic cleavages involve bond rupture between monosaccharides and provide information regarding monosaccharide composition. Cross-ring cleavages, which occur across carbohydrate rings, are particularly helpful in determining linkage type.
Fig. (2).
Nomenclature for tandem mass spectrometric product ions of glycans [10].
This review begins with a brief description of ionization techniques and sample preparation methods, and then highlights recent developments in Fourier transform tandem mass spectrometry for carbohydrate analysis with an emphasis on glycans and proteoglycans from mammalian glycoproteins. MSn in low resolution ion traps, including collision activated dissociation (CAD) [11–13] and negative electron transfer dissociation (NETD) [14–15], and ultraviolet photodissociation (UVPD) in both ion traps and time-of-flight (TOF) instruments [16–18] are also powerful current strategies for oligosaccharide characterization. The resolution and mass accuracy of TOF analyzers are rapidly improving, however, Fourier transform mass analyzers are still superior in performance. Due to our focus on Fourier transform tandem mass spectrometric methods, other techniques are not covered. For more comprehensive reviews of carbohydrate analysis by MS we refer to previous literature [7–9, 19–22].
CARBOHYDRATE IONIZATION METHODS
Matrix-assisted laser desorption/ionization (MALDI) and electrospray ionization (ESI) are the two major modern ionization methods for carbohydrates. MALDI involves co-crystallization of analytes and an organic matrix which acts as a chromophore for laser irradiation. The MALDI mechanism is still under debate, but it has been demonstrated that the choice of matrix plays a crucial role in obtaining strong signals from carbohydrates [19]. MALDI demonstrates high sensitivity, relatively high tolerance to contamination, and predominantly produces singly charged ions, such as (M + H)+ and (M + Na)+ ions in positive-ion mode. However, acidic (sialylated, sulfated, or phosphorylated) carbohydrates may undergo fragmentation in positive-ion mode during ionization, both in-source and post-source [23–24]. In negative-ion mode, MALDI mostly produces (M - H)− ions.
ESI is a soft ionization method and particularly suitable to use with high-performance liquid chromatography (HPLC). Depending on sample composition, ESI produces both singly and multiply charged ions, which makes ESI more compatible with various MS/MS strategies than MALDI. It is believed that the sensitivity of ESI for carbohydrates is limited by their hydrophilicity [25]: there are three major steps in the formation of gas-phase ions during the ESI process: (a) formation of charged analyte-containing droplets; (b) disruption of charged droplets due to solvent evaporation and Coulomb repulsion; (c) formation of gas-phase ions from secondary droplets [26]. When the initial droplets are formed, hydrophobic analytes (such as most proteins and peptides) are distributed preferentially on the droplet surfaces, whereas hydrophilic analytes (such as oligosaccharides or heavily glycosylated proteins and peptides) are located inside the droplets. Consequently, ion signal from hydrophilic compounds is suppressed in the ESI process, resulting in low sensitivity [25]. Permethylated carbohydrates and carbohydrates derivatized with hydrophobic tags thus demonstrate enhanced sensitivity, up to 1,000 fold [21, 27–28]. Nano electrospray ionization (nano-ESI) shows significantly improved sensitivity and higher tolerance to salts and other contaminants compared to conventional ESI due to decreased initial droplet sizes [25, 29]. Nano-ESI also facilitates glycan structural analysis with extremely low sample consumption and has been widely utilized with nano-scale LC [30–33].
SAMPLE PREPARATION METHODS
a) Enzymatic and Chemical Release Methods
For enzymatic release of N-linked glycans, peptide-N-glycosidase F (PNGase F) is most commonly used. This enzyme cleaves the amide bond between asparagine residues and the glycan. PNGase F releases most N-linked glycans, except for those containing an α1→3 fucose linkage at the reducing end N-acetyl glucosamine (GlcNAc) [34]. In such cases, peptide-N-glycosidase A (PNGase A) can be used as an alternative. Endoglycosidase-H (Endo-H) is another popular enzyme for N-glycan release. This enzyme cleaves the glycan between the two GlcNAcs at the chitobiose core of high-mannose and hybrid glycans [35]. However, Endo-H derived N-glycans do not provide information regarding core fucosylation, because the reducing end GlcNAc remains bound to the protein. There is currently no universal enzyme for releasing O-glycans. Endo-α-N-acetylgalactosaminidase (O-glycanase) cleaves exclusively at serine/threonine-glycan bonds, but it is only active for core-1 type O-glycans [36]. Therefore, chemical release methods are more commonly used for releasing O-glycans.
Hydrazinolysis is a commonly used chemical method [37–39], which has the advantage of releasing both N- and O-linked oligosaccharides, but also frequently introduces side reactions, such as loss of N- and O-acetyl and glycolyl groups under harsh reaction conditions [20]. Reductive β-elimination with sodium hydroxide and borohydride is also a popular method to chemically release O-glycans. When using strong base such as sodium hydroxide, non-selective release of other Ser/Thr PTMs and glycan degradation (peeling) are often observed [40–41]. Therefore, usage of mild bases such as ammonia or dimethylamine [40] has been explored. Recently, a novel O-glycan release method combining complete enzyme degradation with chemical release during the process of solid-phase permethylation was introduced [42]. Glycans attached to serine and threonine are effectively cleaved, but those attached to arginine are not, rendering this strategy highly specific.
b) Derivatization Methods
Permethylation of glycans prior to mass spectrometric analysis is a widely used method. When glycans are permethylated, they become significantly more hydrophobic, which has been reported to increase glycan sensitivity in MS with both ESI and MALDI [19, 43]. Moreover, permethylation enables structural analysis of sialylated glycans in positive-ion mode by stabilizing the labile sialic acid in MALDI [44–45]. Traditionally, permethylation is based on the reaction of glycans with iodomethane and sodium hydroxide prepared in dimethyl sulfoxide (DMSO) [46]. More recently, Novotny, Mechref and co-workers have developed high-throughput solid-phase permethylation of glycans prior to MS [47–48] by utilizing fused-silica capillaries or spin columns packed with sodium hydroxide powders or beads and also demonstrated that sequential double-permethylation can be used for improved structural characterization of sulfated glycans, including localization of sulfate groups [49].
Reductive amination is another widely used derivatization method. 2-aminobenzoic acid (2-AA), 2-aminobenzamide (2-AB) and 2-aminopyridine (2-AP) are some of the commonly used reagents [18–19, 50–52]. Reductive amination serves multiple purposes: first, by introducing a chromophore to the reducing end of glycans, UV or fluorescence detection becomes possible in HPLC. Second, because the introduced chromophore is hydrophobic, the retention in reverse phase HPLC improves at the same time that MS sensitivity increases. However, in order to ensure high yield of labeled carbohydrates, excess of labeling reagent is used, which requires a clean-up step. More detailed information on carbohydrate derivatization can be found in previous reviews [41, 53–54].
In addition to protonated/deprotonated carbohydrates, alkali, alkaline earth, and transition metals are widely used for ionization of glycans prior to MS/MS analysis [55–61]. Metal cations can stabilize labile acidic groups, e.g. sulfate groups [62–63]. It has also been demonstrated that metal-adducted glycans result in more cross-ring cleavages than their protonated counterparts in CAD [55, 58, 61]. Another advantage of metal adduction is elimination of rearrangement reactions during fragmentation. Tandem mass spectrometry of reductively aminated glycans and glycopeptides in their protonated form is known to result in fucose and hexose rearrangements, but this phenomenon is not observed for their sodium-adducted counterparts [52, 64–67]. Therefore, one should be aware of the possibility of rearrangements when assigning MS/MS spectra of protonated glycans and glycoconjugates.
Compared to metal adducts, anionic adducts are not as frequently used in mass spectrometry. Harvey explored a variety of anions in negative-ion mode ESI and reported that nitrate adducts yielded the most satisfactory spectra of N-linked glycans with little in-source fragmentation and high signal abundance [68]. CAD of nitrate- and chloride-adducted N-glycans resulted in abundant A- and C-type product ions, whereas bromide and iodide adducts generated few fragments. Cole and co-workers examined chloride adducts in both negative-ion MALDI and ESI, and found unique structurally-informative product ions [69–71]. For example, the abundance ratio of Cl−-adducted/non-Cl−-adducted product ions in CAD spectra was utilized to differentiate anomeric configurations of disaccharides [70].
HIGH RESOLUTION MASS ANALYZERS
The tremendous complexity of biological samples continues to push the technical limitations of analytical instrumentation. Fourier transform ion cyclotron resonance (FT-ICR) [72] and orbitrap mass analyzers [73–75] provide the highest resolving power of current mass spectrometers and such high-end instruments are utilized in a number of areas, including biomolecular identification, characterization of PTMs, quantification in proteomics, glycomics, and metabolomics, as well as petroleum characterization. In addition to the superior resolution, FT-ICR and orbitrap instruments also provide ultrahigh mass accuracy [76–77]. Makarov et al. used automatic gain control (AGC) in an LTQ orbitrap mass spectrometer to control the number of ions in each fill over several orders of magnitude of analyte concentration [78]. With this technique, mass accuracy of <5 ppm was achieved with >95% probability at a dynamic range >5000, representing at least an order of magnitude higher than typical values for TOF instruments. Both FT-ICR and orbitrap mass analyzers are able to achieve <5 ppm mass error for externally calibrated spectra and <2 ppm for internally calibrated spectra [76].
Unlike mass analyzers that measure ion deflection (electric/magnetic sectors), stability of ion trajectories (quadrupole mass analyzer, quadrupole ion trap), or time of ion transit (TOF), FT-ICR and orbitrap mass analyzers are based on detection of induced image current in the time domain followed by Fourier transformation to yield a frequency domain spectrum, which is then converted to an m/z spectrum through the ion cyclotron equation (equation (1)) for FT-ICR mass analyzers [72] and through equation (2) for orbitrap mass analyzers [74]:
| (1) |
in which ωc is the ion cyclotron frequency, z is an integer, e is the elementary charge, B is the magnetic field strength, and m is the ion mass.
| (2) |
in which ωa is the ion axial oscillation frequency inside the orbitrap and k is the field curvature of the applied electric field. This principle inherently allows high resolution because resolution is proportional to the ion observation time and this time can be rather long (several seconds) because FT-ICR and orbitrap mass analyzers operate at ultrahigh vacuum [76]. A major difference between these two high resolution mass analyzers is that the FT-ICR requires a highly homogenous magnetic field of several Tesla whereas the orbitrap is purely electrostatic.
Sufficiently accurate mass measurements allow unique identification of biomolecular elemental composition. For example, the mass difference between potassium and sodium is 16 Da, which is also the mass difference between hexose (Hex, e.g., mannose or glucose) and deoxyhexose (dHex, e.g., fucose or rhamnose), and between N-glycolyl neuraminic acid (NeuGc) and N-acetyl neuraminic acid (NeuAc). With a conventional ion trap mass analyzer, it is impossible to distinguish, e.g., (Hex + Na − H) (mass 184.03029) and (dHex + K − H) (mass 184.00931) [79], whereas high resolution mass analyzers demonstrate outstanding capability for such applications [80]. In addition, high resolution mass spectrometers combined with isotopic labeling of glycans is a powerful tool for glycan quantification. For example, Orlando and co-workers reported a novel permethylation method using 13CH3I and 12CH2DI for comparative glycomics, introducing a mass difference of 0.002922 Da at each permethylation site.
Gas-phase ion-electron reactions, such as electron capture dissociation (ECD) [81], electron detachment dissociation (EDD) [82], and electron transfer dissociation (ETD) [83] have received increasing attention as alternative MS/MS strategies to CAD and infrared multiphoton dissociation (IRMPD), particularly for structural characterization of PTMs (see discussion below). ECD and EDD are best implemented in FT-ICR mass spectrometers, although ECD is also available in three-dimensional quadrupole ion traps [84–86] and digital ion traps [87–88]. The superiority of the FT-ICR for ECD and EDD is due to the required trapping of both electrons and polycations or anions, respectively, which is challenging in ion traps due to their low m/z cut-off [73]. In addition, ECD and EDD often generate complex MS/MS spectra, including mixtures of radical and even-electron species. Therefore, high resolution mass analysis is essential for unambiguously assigning product ions. ETD was first introduced in a radio-frequency linear quadrupole ion trap instrument [83], however, analogous to ECD and similar to EDD, ETD generates complex product ion spectra that are challenging to interpret without high resolving power and high mass accuracy [89–91]. ETD is commercially available on both FT-ICR and orbitrap instruments.
MS/MS TECHNIQUES IN HIGH RESOLUTION MASS SPECTROMETRY
a) Collision Activated Dissociation (CAD)
CAD is the most widely utilized fragmentation technique for structural analysis of carbohydrates. Low-energy CAD, which involves multiple inelastic collisions with neutral molecules or atoms, typically produces glycosidic cleavages [21]. Cross-ring cleavages are less likely to be formed because higher energy is required to break two bonds. In positive-ion mode of neutral oligosaccharides, B- and Y-type ions are commonly observed, whereas in negative ion mode, cross-ring and abundant C-type fragments are often generated [68, 92–94]. For sialylated oligosaccharides, negative-ion CAD is dramatically different compared to non-sialylated glycans and higher energy is required for dissociation [95]. High-energy CAD provides more cross-ring cleavages, such as 1,5X- and 3,5A-type ions in both positive and negative ion mode [50, 96–98] compared to low-energy CAD. In addition to providing extensive structural information regarding glycan sequence and branching, high-energy CAD is capable of differentiating structural isomers [97, 99]. For example, the N-glycan Man7 released from ribonuclease B has three structural isomers. High-energy CAD of Man7 generated several characteristic product ions for each isomer, such as 0,4A3 and 3,5A3, and the intensity ratios of these species matched with the ratios determined by NMR spectroscopy [97]. However, high-energy CAD is currently mainly available in MALDI tandem time-of-flight (TOF/TOF) instruments, which limits its applicability. Further, high-energy CAD involves significant ion scattering which reduces sensitivity.
In Fourier transform mass spectrometry, low-energy CAD can be performed external to the mass analyzer, either in a linear quadrupole ion trap (for orbitrap and FT-ICR instruments), or in a hexapole collision cell (for FT-ICR). The FT-ICR mass analyzer also allows CAD inside the ICR cell by adding collision gas. In the latter case, two main variants of CAD can be performed: in on-resonance CAD, an excitation frequency equal to the ICR frequency of a precursor ion is applied while collision gas is pulsed into the ICR cell [22]. In this approach, product ions are formed off-axis, which limits the practicality of further fragmentation stages and also limits resolving power [100]. In sustained off-resonance irradiation (SORI)-CAD, the excitation frequency is slightly offset from the precursor ions’ cyclotron frequency. Consequently, ions are repeatedly excited to a relatively small cyclotron radius and then come back to the center of the ICR cell, thereby removing the shortcomings of on-resonance CAD. “In cell” CAD and CAD in an external linear ion trap both have the advantage of allowing multiple stages of tandem mass spectrometry, MSn where n>2.
In SORI-CAD, precursor ions undergo multiple low-energy collisions with added background gas and are continuously vibrationally excited in a manner analogous to IRMPD [101] (see below). Solouki et al. reported that SORI-CAD MSn (n>2) of permethylated oligosaccharides provided more extensive fragmentation compared to conventional CAD in a triple quadrupole mass spectrometer [102]. SORI-CAD has also been applied to protonated carrageenan sulfated oligosaccharides for determining the positions of sulfated residues [103], to deprotonated monosaccharides for determining phosphate locations [104], and to alkali metal-adducted disaccharides for studying the influence of metal ions on fragmentation behavior [105]. The latter work showed that, in SORI-CAD, the dissociation thresholds of oligosaccharide glycosidic cleavages depend on the size of alkali metals, while activation barriers for cross-ring cleavages are independent of alkali metal identity, but dependent on linkage type [105].
With the orbitrap mass analyzer, in addition to CAD in the linear ion trap (ion trap-type CAD, which favors low energy fragmentation pathways [106–107]), CAD can be accomplished within the C-trap used to focus ions prior to injection into the orbitrap. This mode of CAD, termed high-energy C-trap dissociation (HCD) [108] yields similar MS/MS spectra to “beam-type” CAD in an external multi-pole. For example, for peptides, immonium ions are observed in addition to b- and y-type ions from backbone cleavage. Such ions, which cannot be observed in ion trap-type CAD due to the low m/z cut-off of such devices, can aid the identification of modified amino acids. The absence of a low m/z cut-off in HCD also allows quantification of, e.g., phosphopeptides with iTRAQ labels [109–110]. Such labels have low molecular weight and are therefore difficult to observe in ion trap-type CAD. HCD in orbitrap mass spectrometers thus shows great potential for both qualitative and quantitative analysis with excellent mass accuracy, resolution, and sensitivity. The absence of a low m/z cut-off is also beneficial in carbohydrate analysis because it allows detection of monosaccharide “marker” ions.
b) Infrared Multiphoton Dissociation (IRMPD)
IRMPD is a valuable tool in FT-ICR and other ion trap instruments because of its ability to readily yield secondary fragmentation and thereby provide higher fragmentation efficiency compared to CAD [22]. IRMPD has been applied to various kinds of biomolecules, such as proteins, peptides, carbohydrates, and oligonucleotides [111]. Typically, a 10.6 μm CO2 laser is used for infrared irradiation, but tunable CO2 laser IRMPD was recently implemented [112–114]. The latter approach was able to differentiate anomers of lithium-adducted methyl-glucopyranoside by comparing relative product ion abundances as function of laser wavelength [113]. Differentiation of lithium-tagged glucose-containing disaccharides was previously reported by Polfer et al. with 7–11 μm photons from a free electron laser [115]. That work was able to assign both linkage position and sugar anomeric configuration.
During IR activation, multiple photons are absorbed and the corresponding energy is redistributed over all precursor ion vibrational modes [113]. Therefore, multiple fragmentation events may take place. One of the major advantages of IRMPD in FT-ICR MS is that no collision gas needs to be introduced to the ICR cell [116]. In addition, IRMPD provides on-axis fragmentation, thus there is no loss of resolution.
Lebrilla and co-workers have compared IRMPD fragmentation patterns to those from CAD of model oligosaccharides, N-linked glycans, and O-linked glycans ionized with alkali metals in FT-ICR MS [24, 59, 117–118]. Similar to CAD, IRMPD favors low energy dissociation pathways, thus mostly glycosidic cleavages but also some cross-ring cleavages are observed in IRMPD spectra. However, contrary to CAD, IRMPD fragmentation efficiency increases with increasing glycan size: for large O-glycans, single-stage IRMPD resulted in cleavage of all glycosidic bonds [117]. Moreover, IRMPD of alkali metal-adducted oligosaccharides generates different fragmentation patterns compared to those from SORI-CAD, e.g., IRMPD readily yields monosaccharide residues, whereas multiple CAD MS/MS stages are required to achieve similar results [24, 59]. Therefore, IRMPD is not only a fast and efficient MS/MS method but it can also potentially serve as a complementary technique to CAD for structural characterization of glycans.
c) Electron Capture Dissociation (ECD)
ECD is performed by irradiating at least doubly positively charged precursor ions with low energy (<1 eV) electrons, generating charge-reduced radical species from electron capture and product ions from radical-driven fragmentation [81, 119–121]. Since its first introduction in 1998 [81], the application of ECD for biomolecular structural analysis has been rapidly expanding due to its complementary nature compared to CAD and IRMPD [122]. For example, ECD shows unique analytical utility in PTM characterization because labile PTMs are retained and can thus be localized within a molecule. ECD has been successfully utilized in this manner for characterizing, e.g., protein N-glycosylation [123–124], O-glycosylation [125], and phosphorylation [126–127].
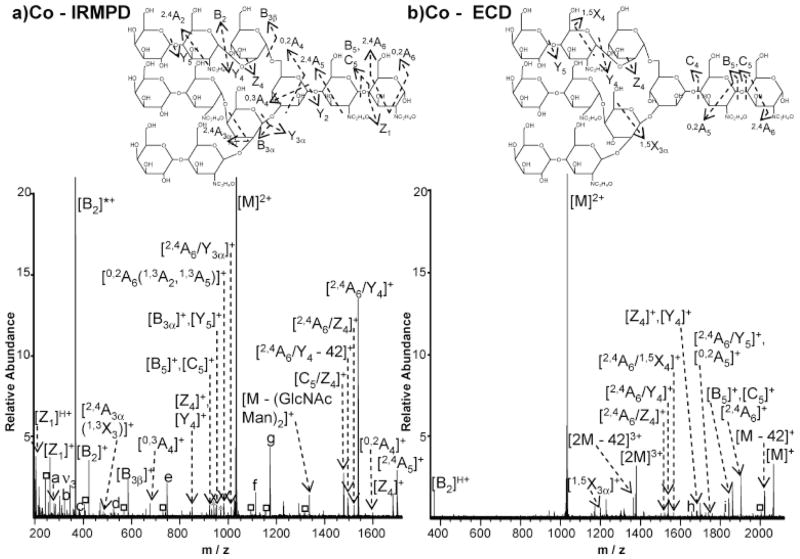
While ECD has been widely applied to peptides and proteins, its application towards carbohydrates has just begun to emerge. Due to the lack of basic groups in carbohydrates and their frequent relatively small size, it is often difficult to obtain multiply protonated precursor ions (multiple positive charges are required for ECD). The first application of ECD towards carbohydrate analysis circumvented this issue by involving aminoglycans, which can easily be multiply protonated. However, this work reported only glycosidic cleavages from ECD [128]. Our group utilized alkali, alkaline earth, and transition metals to generate multiply positively charged ions for model oligosaccharides and observed complementary fragmentation patterns from IRMPD and ECD of these species, as shown in Fig. (3) [60]. Here, IRMPD (Fig. (3a)) is shown to yield extensive glycosidic cleavages, as well as several less abundant cross-ring fragments. Following ECD (Fig. (3b)), fewer product ions are observed compared to IRMPD, however, cross-ring cleavage is the dominant fragmentation pathway, providing valuable information regarding linkage type. We also examined ECD and IRMPD of metal-adducted N-linked glycans released from a glycoprotein and found that ECD generates unique fragments, complementary to those from IRMPD [129]. ECD of different metal-adducted species showed different fragmentation patterns, but sulfate groups were retained in all ECD spectra, providing valuable sulfate location information. O’Connor and co-workers explored the application of CAD and hot ECD (i.e., ECD with ~10 eV electrons) towards permethylated oligosaccharides [130]. For linear oligosaccharides, CAD and hot ECD resulted in similar product ions. In contrast, CAD generated A-, B-, and Y-type ions for branched N-glycans, while hot ECD generated C-, Z- and complementary A- and X-type cross-ring fragment pairs. All these studies show that ECD has great potential to serve as a glycan structural analysis technique.
Fig. (3).
IRMPD (a) and ECD (b) spectra of Co2+-adducted N-linked glycan in FT-ICR MS. Complementary product ions are observed in IRMPD and ECD spectra [60]. Reprinted with permission from [60]. Copyright (2007) American Chemical Society.
d) Electron Detachment Dissociation (EDD)
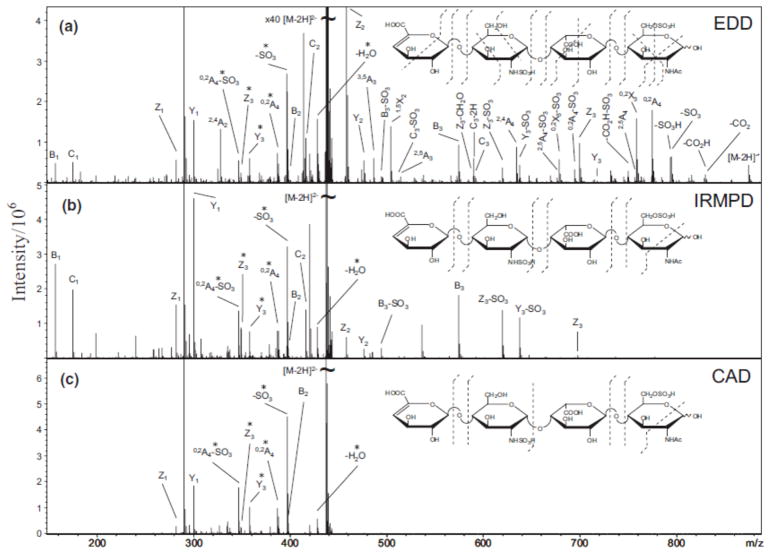
EDD was first introduced for the characterization of peptide anions in 2001 [82]. In EDD, polyanions are irradiated with >10 eV electrons to yield electron detachment, forming radical anions which undergo further radical-driven fragmentation. EDD has been applied to peptides [82, 131–132], oligonucleotides [133–134], gangliosides [135], model oligosaccharides [136], and GAG-derived oligosaccharides [137–143]. Our work demonstrated that EDD of neutral and sialylated oligosaccharides results in fragments complementary to those from IRMPD and CAD, including extensive cross-ring product ions such as 1,5A, 3,5A, 1,5X and 3,5X ions [136]. When applying EDD to sialylated N-glycans released from glycoproteins, EDD preferentially yielded X-, Y- and Z-type ions while IRMPD resulted in mostly A-, B- and C-type ions. Cross-ring cleavages generated by EDD were completely complementary to IRMPD in each case studied [144]. The extensive work by Amster and co-workers on EDD of GAGs has shown that extensive glycosidic and cross-ring cleavages are generated, as exemplified in Fig. (4). In contrast, IRMPD and CAD of GAGs mainly generate glycosidic cleavages. EDD was also shown to be able to differentiate the isomers glucuronic acid and iduronic acid [138]. The complementarity of EDD and IRMPD/CAD makes them highly promising approaches for structural characterization of carbohydrates, particularly acidic carbohydrates in negative-ion mode.
Fig. (4).
Tandem mass spectra of [M - 2H]2− precursor ions of a tetrasaccharide, obtained by (a) EDD, (b) IRMPD, and (c) CAD [137]. EDD of GAGs generates extensive glycosidic and cross-ring cleavages, including some unique cross-ring fragments compared to IRMPD and CAD. Reprinted with permission from [137]. Copyright (2007) Springer.
e) Electron Induced Dissociation (EID)
In electron induced dissociation (EID), precursor ions are irradiated with electrons of energy exceeding the ionization energy of the precursor neutral molecules, but below their second ionization energy. EID generally results in extensive fragmentation [128, 143] and has been utilized to characterize aminoglycans [128], GAG tetrasaccharides [143], phosphorylated metabolites [145], and fluorescently labeled oligosaccharides [146]. EID of GAG tetrasaccharides yielded EDD-like fragmentation, including cleavages of all glycosidic bonds, and abundant cross-ring product ions, highly different from the product ions resulting from IRMPD and CAD [143]. EID of singly protonated oligosaccharides provided complementary structural information compared to SORI-CAD, including B-, C-, Y-, Z-, and 1,5X-type ions [146]. These results show great potential of EID as an alternative strategy to characterize carbohydrates. Particularly, EID is compatible with singly charged analytes.
CONCLUSIONS
In summary, tandem mass spectrometry is a highly valuable tool for structural determination of carbohydrates. The high resolution and high mass accuracy of Fourier transform instruments enable detailed carbohydrate structural analysis even from low abundance species. High mass accuracy allows unique identification of carbohydrate elemental composition and high resolution/high resolving power is essential for achieving accurate interpretation of MS/MS spectra. Permethylation, reductive amination, and metal adduction of carbohydrates serve to increase ionization responses and often aid in providing rich structural information following MS/MS fragmentation. Alternative high resolution MS/MS methods, such as IRMPD, ECD, and EDD, generate highly informative fragments, complementary to those from CAD. Thus, high resolution tandem mass spectrometry is continuing to evolve for improved carbohydrate structural analysis.
Table 1.
Tandem Mass Spectrometric Techniques for Glycans
| MS/MS Techniques | Vibrational Activation | Ion-Electron Reactions | |||||
|---|---|---|---|---|---|---|---|
| CAD | IRMPD | SORI-CAD | HCD | ECD | EDD | EID | |
| Activation energy source | Slow heating by inelastic collisions between reactant ions and neutral gas molecules | Absorption of multiple infra-red photons | Collisions between neutral gas and ions in cyclotron motion undergoing alternating radial acceleration and deceleration as a result of an off-resonance rf pulse | Same as CAD | Polycations capture low energy electrons (<1 eV) | Polyanions irradiated with >10 eV electrons to yield electron detachment | Cat- or anions are irradiated with > 10 eV electrons to cause excitation but not ionization |
| Precursor ion charge states | Any charge state | Any charge state | Any charge state | Any charge state | ≥ 2+ | ≥ 2− | Any charge state |
| Glycan fragmentation features | Tend to generate glycosidic cleavages but 0,2A and 2,4A-type ions are also commonly observed. | Generate complementary product ions compared to vibrational activation techniques and often yield more cross-ring cleavages | |||||
| Available mass analyzers | Most types | FT-ICR, ion trap | FT-ICR | Orbitrap | FT-ICR, ion trap | FT-ICR, ion trap | FT-ICR |
Acknowledgments
This work was supported by Award Number R21CA138331 from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. W.Z. was partially supported by a George Ashworth Analytical Chemistry Fellowship from the University of Michigan.
ABBREVIATIONS
- 2-AA
2-aminobenzoic acid
- 2-AB
2-aminobenzamide
- 2-AP
2-aminopyridine
- AGC
Automatic gain control
- CAD
Collision activated dissociation
- DMSO
Dimethyl sulfoxide
- ECD
Electron capture dissociation
- EDD
Electron detachment dissociation
- EID
Electron induced dissociation
- Endo-H
Endoglycosidase-H
- ESI
Electrospray ionization
- ETD
Electron transfer dissociation
- FTICR-MS
Fourier transform ion cyclotron resonance mass spectrometry
- GAG
Glycosaminoglycan
- GC
Gas chromatography
- GlcNAc
N-acetyl glucosamine
- HCD
High-energy C-trap dissociation
- HPLC
High-performance liquid chromatography
- MALDI
Matrix-assisted laser desorption/ionization
- MS
Mass spectrometry
- MS/MS or MSn
Tandem mass spectrometry
- Nano ESI
Nano electrospray ionization
- NETD
Negative electron transfer dissociation
- NMR
Nuclear magnetic resonance
- PNGase A
Peptide-N-glycosidase A
- PNGase F
Peptide-N-glycosidase F
- PTM
Posttranslational modification
- SORI-CAD
Sustained off-resonance irradiation collision activated dissociation
- TOF
Time of flight
- UVPD
Ultraviolet photodissociation
References
- 1.Varki A. Biological roles of oligosaccharides - All of the theories are correct. Glycobiol. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rudd PM, Elliott T, Cresswell P, Wilson IA, Dwek RA. Glycosylation and the immune system. Science. 2001;291:2370–2376. doi: 10.1126/science.291.5512.2370. [DOI] [PubMed] [Google Scholar]
- 3.Bertozzi CR, Kiessling LL. Chemical glycobiology. Science. 2001;291:2357–2364. doi: 10.1126/science.1059820. [DOI] [PubMed] [Google Scholar]
- 4.Krishnamoorthy L, Mahal LK. Glycomic analysis: an array of technologies. ACS Chem Biol. 2009;4:715–732. doi: 10.1021/cb900103n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dwek RA. Glycobiology: Toward understanding the function of sugars. Chem Rev. 1996;96:683–720. doi: 10.1021/cr940283b. [DOI] [PubMed] [Google Scholar]
- 6.Raman R, Raguram S, Venkataraman G, Paulson JC, Sasisekharan R. Glycomics: an integrated systems approach to structure-function relationships of glycans. Nat Methods. 2005;2:817–824. doi: 10.1038/nmeth807. [DOI] [PubMed] [Google Scholar]
- 7.Mechref Y, Novotny MV. Structural investigations of glycoconjugates at high sensitivity. Chem Rev. 2002;102:321–369. doi: 10.1021/cr0103017. [DOI] [PubMed] [Google Scholar]
- 8.Zaia J. Mass spectrometry and the emerging field of glycomics. Chem Biol. 2008;15:881–892. doi: 10.1016/j.chembiol.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harvey DJ. Proteomic analysis of glycosylation: structural determination of N- and O-linked glycans by mass spectrometry. Expert Rev Proteomics. 2005;2:87–101. doi: 10.1586/14789450.2.1.87. [DOI] [PubMed] [Google Scholar]
- 10.Domon B, Costello CE. A systematic nomenclature for carbohydrate fragmentations in FAB-MS MS spectra of glycoconjugates. Glycoconj J. 1988;5:397–409. [Google Scholar]
- 11.Ashline D, Singh S, Hanneman A, Reinhold V. Congruent strategies for carbohydrate sequencing. 1. Mining structural details by MSn. Anal Chem. 2005;77:6250–6262. doi: 10.1021/ac050724z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashline DJ, Lapadula AJ, Liu YH, Lin M, Grace M, Pramanik B, Reinhold VN. Carbohydrate structural isomers analyzed by sequential mass spectrometry. Anal Chem. 2007;79:3830–3842. doi: 10.1021/ac062383a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prien JM, Ashline DJ, Lapadula AJ, Zhang H, Reinhold VN. The high mannose glycans from bovine ribonuclease B isomer characterization by ion trap MS. J Am Soc Mass Spectrom. 2009;20:539–556. doi: 10.1016/j.jasms.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huzarska M, Ugalde I, Kaplan DA, Hartmer R, Easterling ML, Polfer NC. Negative electron transfer dissociation of deprotonated phosphopeptide anions: choice of radical cation reagent and competition between electron and proton transfer. Anal Chem. 2010;82:2873–2878. doi: 10.1021/ac9028592. [DOI] [PubMed] [Google Scholar]
- 15.Wolff JJ, Leach FE, Laremore TN, Kaplan DA, Easterling ML, Linhardt RJ, Amster IJ. Negative electron transfer dissociation of glycosaminoglycans. Anal Chem. 2010;82:3460–3466. doi: 10.1021/ac100554a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devakumar A, Mechref Y, Kang P, Novotny MV, Reilly JP. Laser-induced photofragmentation of neutral and acidic glycans inside an ion-trap mass spectrometer. Rapid Commun Mass Spectrom. 2007;21:1452–1460. doi: 10.1002/rcm.2981. [DOI] [PubMed] [Google Scholar]
- 17.Devakumar A, Mechref Y, Kang P, Novotny MV, Reilly JP. Identification of isomeric N-glycan structures by mass spectrometry with 157 nm laser-induced photofragmentation. J Am Soc Mass Spectrom. 2008;19:1027–1040. doi: 10.1016/j.jasms.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson JJ, Brodbelt JS. Ultraviolet photodissociation at 355 nm of fluorescently labeled oligosaccharides. Anal Chem. 2008;80:5186–5196. doi: 10.1021/ac800315k. [DOI] [PubMed] [Google Scholar]
- 19.Harvey DJ. Matrix-assisted laser desorption/ionization mass spectrometry of carbohydrates. Mass Spectrom Rev. 1999;18:349–450. doi: 10.1002/(SICI)1098-2787(1999)18:6<349::AID-MAS1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 20.Harvey DJ. Identification of protein-bound carbohydrates by mass spectrometry. Proteomics. 2001;1:311–328. doi: 10.1002/1615-9861(200102)1:2<311::AID-PROT311>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 21.Zaia J. Mass spectrometry of oligosaccharides. Mass Spectrom Rev. 2004;23:161–227. doi: 10.1002/mas.10073. [DOI] [PubMed] [Google Scholar]
- 22.Park YM, Lebrilla CB. Application of Fourier transform ion cyclotron resonance mass spectrometry to oligosaccharides. Mass Spectrom Rev. 2005;24:232–264. doi: 10.1002/mas.20010. [DOI] [PubMed] [Google Scholar]
- 23.Huberty MC, Vath JE, Yu W, Martin SA. Site-specific carbohydrate identification in recombinant proteins using MALDI-TOF MS. Anal Chem. 1993;65:2791–2800. doi: 10.1021/ac00068a015. [DOI] [PubMed] [Google Scholar]
- 24.Lancaster KS, An HJ, Li BS, Lebrilla CB. Interrogation of N-linked oligosaccharides using infrared multiphoton dissociation in FT-ICR mass spectrometry. Anal Chem. 2006;78:4990–4997. doi: 10.1021/ac0600656. [DOI] [PubMed] [Google Scholar]
- 25.Karas M, Bahr U, Dulcks T. Nano-electrospray ionization mass spectrometry: addressing analytical problems beyond routine. Fresenius J Anal Chem. 2000;366:669–676. doi: 10.1007/s002160051561. [DOI] [PubMed] [Google Scholar]
- 26.Kebarle P, Verkerk UH. Electrospray: from ions in solution to ions in the gas phase, what we know now. Mass Spectrom Rev. 2009;28:898–917. doi: 10.1002/mas.20247. [DOI] [PubMed] [Google Scholar]
- 27.Pabst M, Kolarich D, Poltl G, Dalik T, Lubec G, Hofinger A, Altmann F. Comparison of fluorescent labels for oligosaccharides and introduction of a new postlabeling purification method. Anal Biochem. 2009;384:263–273. doi: 10.1016/j.ab.2008.09.041. [DOI] [PubMed] [Google Scholar]
- 28.Yoshino K, Takao T, Murata H, Shimonishi Y. Use of the derivatizing agent 4-aminobenzoic acid 2-(diethylamino)ethyl ester for high-sensitivity detection of oligosaccharides by electrospray-ionization mass-spectrometry. Anal Chem. 1995;67:4028–4031. doi: 10.1021/ac00117a034. [DOI] [PubMed] [Google Scholar]
- 29.Bahr U, Pfenninger A, Karas M, Stahl B. High sensitivity analysis of neutral underivatized oligosaccharides by nanoelectrospray mass spectrometry. Anal Chem. 1997;69:4530–4535. doi: 10.1021/ac970624w. [DOI] [PubMed] [Google Scholar]
- 30.Bereman MS, Williams TI, Muddiman DC. Development of a nanoLC LTQ orbitrap mass spectrometric method for profiling glycans derived from plasma from healthy, benign tumor control, and epithelial ovarian cancer patients. Anal Chem. 2009;81:1130–1136. doi: 10.1021/ac802262w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo QZ, Rejtar T, Wu SL, Karger BL. Hydrophilic interaction 10 mu m ID porous layer open tubular columns for ultratrace glycan analysis by liquid chromatography-mass spectrometry. J Chromatogr A. 2009;1216:1223–1231. doi: 10.1016/j.chroma.2008.09.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomsson KA, Backstrom M, Larsson JMH, Hansson GC, Karlsson H. Enhanced detection of sialylated and sulfated glycans with negative ion mode nanoliquid chromatography/mass spectrometry at high pH. Anal Chem. 2010;82:1470–1477. doi: 10.1021/ac902602e. [DOI] [PubMed] [Google Scholar]
- 33.Wohlgemuth J, Karas M, Jiang W, Hendriks R, Andrecht S. Enhanced glyco-profiling by specific glycopeptide enrichment and complementary monolithic nano-LC (ZIC-HILIC/RP18e)/ESI-MS analysis. J Sep Sci. 2010;33:880–890. doi: 10.1002/jssc.200900771. [DOI] [PubMed] [Google Scholar]
- 34.Tretter V, Altmann F, Marz L. Peptide-N4-(N-acetyl-beta-glycosaminyl)asparagine amidase-F cannot release glycans with fucose attached alpha-1-3 to the asparagine-linked N-acetylglucosamine residue. Eur J Biochem. 1991;199:647–652. doi: 10.1111/j.1432-1033.1991.tb16166.x. [DOI] [PubMed] [Google Scholar]
- 35.Tarentin Al, Plummer TH, Maley F. Release of intact oligosaccharides from specfic glycoproteins by endo-beta-N-acetylglucosaminidase H. J Biol Chem. 1974;249:818–824. [PubMed] [Google Scholar]
- 36.Adolf GR, Kalsner I, Ahorn H, Maurerfogy I, Cantell K. Natural human interferon-alpha-2 is O-glycosylated. Biochem J. 1991;276:511–518. doi: 10.1042/bj2760511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel T, Bruce J, Merry A, Bigge C, Wormald M, Jaques A, Parekh R. Use of hydrazine to release in intact and unreduced form both N-linked and O-linked oligosaccharides from glycoproteins. Biochemistry (Mosc) 1993;32:679–693. doi: 10.1021/bi00053a037. [DOI] [PubMed] [Google Scholar]
- 38.Harvey DJ. Electrospray mass spectrometry and collision-induced fragmentation of 2-aminobenzamide-labelled neutral N-linked glycans. Analyst. 2000;125:609–617. [Google Scholar]
- 39.Wheeler SF, Harvey DJ. Extension of the in-gel release method for structural analysis of neutral and sialylated N-linked glycans to the analysis of sulfated glycans: Application to the glycans from bovine thyroid-stimulating hormone. Anal Biochem. 2001;296:92–100. doi: 10.1006/abio.2001.5199. [DOI] [PubMed] [Google Scholar]
- 40.Maniatis S, Zhou H, Reinhold V. Rapid de-O-glycosylation concomitant with peptide labeling using microwave radiation and an alkyl amine base. Anal Chem. 2010;82:2421–2425. doi: 10.1021/ac902734w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anumula KR. Advances in fluorescence derivatization methods for high-performance liquid chromatographic analysis of glycoprotein carbohydrates. Anal Biochem. 2006;350:1–23. doi: 10.1016/j.ab.2005.09.037. [DOI] [PubMed] [Google Scholar]
- 42.Goetz JA, Novotny MV, Mechref Y. Enzymatic/chemical release of O-glycans allowing MS analysis at high sensitivity. Anal Chem. 2009;81:9546–9552. doi: 10.1021/ac901363h. [DOI] [PubMed] [Google Scholar]
- 43.Ruhaak LR, Zauner G, Huhn C, Bruggink C, Deelder AM, Wuhrer M. Glycan labeling strategies and their use in identification and quantification. Anal Bioanal Chem. 2010;397:3457–3481. doi: 10.1007/s00216-010-3532-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mechref Y, Kang P, Novotny MV. Differentiating structural isomers of sialylated glycans by matrix-assisted laser desorption/ionization time-of-flight/time-of-flight tandem mass spectrometry. Rapid Commun Mass Spectrom. 2006;20:1381–1389. doi: 10.1002/rcm.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheeley DM, Reinhold VN. Structural characterization of carbohydrate sequence, linkage, and branching in a quadrupole ion trap mass spectrometer: Neutral oligosaccharides and N-linked glycans. Anal Chem. 1998;70:3053–3059. doi: 10.1021/ac9713058. [DOI] [PubMed] [Google Scholar]
- 46.Ciucanu I, Kerek F. A simple and rapid method for the permethylation of carbohydrates. Carbohydr Res. 1984;131:209–217. [Google Scholar]
- 47.Kang P, Mechref Y, Klouckova I, Novotny MV. Solid-phase permethylation of glycans for mass spectrometric analysis. Rapid Commun Mass Spectrom. 2005;19:3421–3428. doi: 10.1002/rcm.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kang P, Mechref Y, Novotny MV. High-throughput solid-phase permethylation of glycans prior to mass spectrometry. Rapid Commun Mass Spectrom. 2008;22:721–734. doi: 10.1002/rcm.3395. [DOI] [PubMed] [Google Scholar]
- 49.Lei M, Mechref Y, Novotny MV. Structural analysis of sulfated glycans by sequential double-permethylation using methyl iodide and deuteromethyl iodide. J Am Soc Mass Spectrom. 2009;20:1660–1671. doi: 10.1016/j.jasms.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 50.Wuhrer M, Deelder AM. Negative-mode MALDI-TOF/TOF-MS of oligosaccharides labeled with 2-aminobenzamide. Anal Chem. 2005;77:6954–6959. doi: 10.1021/ac051117e. [DOI] [PubMed] [Google Scholar]
- 51.Harvey DJ. Collision-induced fragmentation of negative ions from N-linked glycans derivatized with 2-aminobenzoic acid. J Mass Spectrom. 2005;40:642–653. doi: 10.1002/jms.836. [DOI] [PubMed] [Google Scholar]
- 52.Wuhrer M, Koeleman CAM, Deelder AM. Hexose rearrangements upon fragmentation of N-glycopeptides and reductively aminated N-glycans. Anal Chem. 2009;81:4422–4432. doi: 10.1021/ac900278q. [DOI] [PubMed] [Google Scholar]
- 53.Lamari FN, Kuhn R, Karamanos NK. Derivatization of carbohydrates for chromatographic, electrophoretic and mass spectrometric structure analysis. J Chromatogr B. 2003;793:15–36. doi: 10.1016/s1570-0232(03)00362-3. [DOI] [PubMed] [Google Scholar]
- 54.Lattova E, Snovida S, Perreault H, Krokhin O. Influence of the labeling group on ionization and fragmentation of carbohydrates in mass spectrometry. J Am Soc Mass Spectrom. 2005;16:683–696. doi: 10.1016/j.jasms.2005.01.021. [DOI] [PubMed] [Google Scholar]
- 55.Hofmeister GE, Zhou Z, Leary JA. Linkage position determination in lithium-cationized disaccharides - tandem mass-sepctrometry and semiempirical calculations. J Am Chem Soc. 1991;113:5964–5970. [Google Scholar]
- 56.Sible EM, Brimmer SP, Leary JA. Interaction of first row transition metals with alpha 1-3, alpha 1-6 mannotriose and conserved trimannosyl core oligosaccharides: A comparative electrospray ionization study of doubly and singly charged complexes. J Am Soc Mass Spectrom. 1997;8:32–42. [Google Scholar]
- 57.Konig S, Leary JA. Evidence for linkage position determination in cobalt coordinated pentasaccharides using ion trap mass spectrometry. J Am Soc Mass Spectrom. 1998;9:1125–1134. doi: 10.1016/S1044-0305(98)00096-8. [DOI] [PubMed] [Google Scholar]
- 58.Cancilla MT, Penn SG, Carroll JA, Lebrilla CB. Coordination of alkali metals to oligosaccharides dictates fragmentation behavior in matrix assisted laser desorption ionization Fourier transform mass spectrometry. J Am Chem Soc. 1996;118:6736–6745. [Google Scholar]
- 59.Xie YM, Lebrilla CB. Infrared multiphoton dissociation of alkali metal-coordinated oligosaccharides. Anal Chem. 2003;75:1590–1598. doi: 10.1021/ac026009w. [DOI] [PubMed] [Google Scholar]
- 60.Adamson JT, Hakansson K. Electron capture dissociation of oligosaccharides ionized with alkali, alkaline earth, and transition metals. Anal Chem. 2007;79:2901–2910. doi: 10.1021/ac0621423. [DOI] [PubMed] [Google Scholar]
- 61.Harvey DJ. Ionization and collision-induced fragmentation of N-linked and related carbohydrates using divalent canons. J Am Soc Mass Spectrom. 2001;12:926–937. doi: 10.1016/S1044-0305(01)00268-9. [DOI] [PubMed] [Google Scholar]
- 62.Liu HC, Hakansson K. Divalent metal ion-peptide interactions probed by electron capture dissociation of trications. J Am Soc Mass Spectrom. 2006;17:1731–1741. doi: 10.1016/j.jasms.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 63.Liu H, Hakansson K. Analysis of sulfated oligosaccharides with combination of divalent metal complexation and electron capture dissociation. Int J Mass Spectrom. 2011 In press. [Google Scholar]
- 64.Harvey DJ, Mattu TS, Wormald MR, Royle L, Dwek RA, Rudd PM. “Internal residue loss”: Rearrangements occurring during the fragmentation of carbohydrates derivatized at the reducing terminus. Anal Chem. 2002;74:734–740. doi: 10.1021/ac0109321. [DOI] [PubMed] [Google Scholar]
- 65.Franz AH, Lebrilla CB. Evidence for long-range glycosyl transfer reactions in the gas phase. J Am Soc Mass Spectrom. 2002;13:325–337. doi: 10.1016/S1044-0305(02)00343-4. [DOI] [PubMed] [Google Scholar]
- 66.Ernst B, Muller DR, Richter WJ. False sugar sequence ions in electrospray tandem mass spectrometry of underivatized sialyl-Lewis-type oligosaccharides. Int J Mass Spectrom Ion Processes. 1997;160:283–290. [Google Scholar]
- 67.Wuhrer M, Koeleman CAM, Hokke CH, Deelder AM. Mass spectrometry of proton adducts of fucosylated N-glycans: fucose transfer between antennae gives rise to misleading fragments. Rapid Commun Mass Spectrom. 2006;20:1747–1754. doi: 10.1002/rcm.2509. [DOI] [PubMed] [Google Scholar]
- 68.Harvey DJ. Fragmentation of negative ions from carbohydrates: Part 1. Use of nitrate and other anionic adducts for the production of negative ion electrospray spectra from N-linked carbohydrates. J Am Soc Mass Spectrom. 2005;16:622–630. doi: 10.1016/j.jasms.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 69.Zhu JH, Cole RB. Ranking of gas-phase acidities and chloride affinities of monosaccharides and linkage specificity in collision-induced decompositions of negative ion electrospray-generated chloride adducts of oligosaccharides. J Am Soc Mass Spectrom. 2001;12:1193–1204. doi: 10.1016/S1044-0305(01)00305-1. [DOI] [PubMed] [Google Scholar]
- 70.Cai Y, Jiang YJ, Cole RB. Anionic adducts of oligosaccharides by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Chem. 2003;75:1638–1644. doi: 10.1021/ac0205513. [DOI] [PubMed] [Google Scholar]
- 71.Jiang YJ, Cole RB. Oligosaccharide analysis using anion attachment in negative mode electrospray mass spectrometry. J Am Soc Mass Spectrom. 2005;16:60–70. doi: 10.1016/j.jasms.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 72.Marshall AG, Hendrickson CL, Jackson GS. Fourier transform ion cyclotron resonance mass spectrometry: A primer. Mass Spectrom Rev. 1998;17:1–35. doi: 10.1002/(SICI)1098-2787(1998)17:1<1::AID-MAS1>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 73.Perry RH, Cooks RG, Noll RJ. Orbitrap mass spectrometry: instrumentation, ion motion and applications. Mass Spectrom Rev. 2008;27:661–699. doi: 10.1002/mas.20186. [DOI] [PubMed] [Google Scholar]
- 74.Hu QZ, Noll RJ, Li HY, Makarov A, Hardman M, Cooks RG. The Orbitrap: a new mass spectrometer. J Mass Spectrom. 2005;40:430–443. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]
- 75.Yates JR, Ruse CI, Nakorchevsky A. Proteomics by mass spectrometry: approaches, advances, and applications. Annu Rev Biomed Eng. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- 76.Marshall AG, Hendrickson CL. High-resolution mass spectrometers. Annu Rev Anal Chem. 2008;1:579–599. doi: 10.1146/annurev.anchem.1.031207.112945. [DOI] [PubMed] [Google Scholar]
- 77.Scigelova M, Makarov A. Orbitrap mass analyzer - Overview and applications in proteomics. Proteomics. 2006:16–21. doi: 10.1002/pmic.200600528. [DOI] [PubMed] [Google Scholar]
- 78.Makarov A, Denisov E, Lange O, Horning S. Dynamic range of mass accuracy in LTQ Orbitrap hybrid mass spectrometer. J Am Soc Mass Spectrom. 2006;17:977–982. doi: 10.1016/j.jasms.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 79.Packer NH, Karlsson NG. There are no facts, only interpretations. J Proteome Res. 2006;5:1291–1292. doi: 10.1021/pr0627286. [DOI] [PubMed] [Google Scholar]
- 80.Sleno L, Volmer DA, Marshall AG. Assigning product ions from complex MS/MS spectra: The importance of mass uncertainty and resolving power. J Am Soc Mass Spectrom. 2005;16:183–198. doi: 10.1016/j.jasms.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 81.Zubarev RA, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein cations. A nonergodic process. J Am Chem Soc. 1998;120:3265–3266. [Google Scholar]
- 82.Budnik BA, Haselmann KF, Zubarev RA. Electron detachment dissociation of peptide di-anions: an electron-hole recombination phenomenon. Chem Phys Lett. 2001;342:299–302. [Google Scholar]
- 83.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bushey JM, Baba T, Glish GL. Simultaneous collision induced dissociation of the charge reduced parent ion during electron capture dissociation. Anal Chem. 2009;81:6156–6164. doi: 10.1021/ac900627n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Silivra OA, Kjeldsen F, Ivonin IA, Zubarev RA. Electron capture dissociation of polypeptides in a three-dimensional quadrupole ion trap: Implementation and first results. J Am Soc Mass Spectrom. 2005;16:22–27. doi: 10.1016/j.jasms.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 86.Baba T, Hashimoto Y, Hasegawa H, Hirabayashi A, Waki I. Electron capture dissociation in a radio frequency ion trap. Anal Chem. 2004;76:4263–4266. doi: 10.1021/ac049309h. [DOI] [PubMed] [Google Scholar]
- 87.Ding L, Brancia FL. Electron capture dissociation in a digital ion trap mass spectrometer. Anal Chem. 2006;78:1995–2000. doi: 10.1021/ac0519007. [DOI] [PubMed] [Google Scholar]
- 88.McCullough BJ, Entwistle A, Konishi I, Buffey S, Hasnain SS, Brancia FL, Grossmann JG, Gaskell SJ. Digital ion trap mass spectrometer for probing the structure of biological macromolecules by gas phase X-ray scattering. Anal Chem. 2009;81:3392–3397. doi: 10.1021/ac802662d. [DOI] [PubMed] [Google Scholar]
- 89.Wiesner J, Premsler T, Sickmann A. Application of electron transfer dissociation (ETD) for the analysis of posttranslational modifications. Proteomics. 2008;8:4466–4483. doi: 10.1002/pmic.200800329. [DOI] [PubMed] [Google Scholar]
- 90.McAlister GC, Berggren WT, Griep-Raming J, Horning S, Makarov A, Phanstiel D, Stafford G, Swaney DL, Syka JEP, Zabrouskov V, Coon JJ. A proteomics grade electron transfer dissociation-enabled hybrid linear ion trap-orbitrap mass spectrometer. J Proteome Res. 2008;7:3127–3136. doi: 10.1021/pr800264t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McAlister GC, Phanstiel D, Good DM, Berggren WT, Coon JJ. Implementation of electron-transfer dissociation on a hybrid linear ion trap-orbitrap mass spectrometer. Anal Chem. 2007;79:3525–3534. doi: 10.1021/ac070020k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Carroll JA, Ngoka L, Beggs CG, Lebrilla CB. Liquid secondary-ion mass-spectrometry Fourier-transform mass-spectrometry of oligosaccharide anions. Anal Chem. 1993;65:1582–1587. doi: 10.1021/ac00059a017. [DOI] [PubMed] [Google Scholar]
- 93.Harvey DJ. Fragmentation of negative ions from carbohydrates: Part 2. Fragmentation of high-mannose N-linked glycans. J Am Soc Mass Spectrom. 2005;16:631–646. doi: 10.1016/j.jasms.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 94.Harvey DJ. Fragmentation of negative ions from carbohydrates: Part 3. Fragmentation of hybrid and complex N-linked glycans. J Am Soc Mass Spectrom. 2005;16:647–659. doi: 10.1016/j.jasms.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 95.Seymour JL, Costello CE, Zaia J. The influence of sialylation on glycan negative ion dissociation and energetics. J Am Soc Mass Spectrom. 2006;17:844–854. doi: 10.1016/j.jasms.2006.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Harvey DJ, Bateman RH, Green MR. High-energy collision-induced fragmentation of complex oligosaccharides ionized by matrix-assisted laser desorption/ionization mass spectrometry. J Mass Spectrom. 1997;32:167–187. doi: 10.1002/(SICI)1096-9888(199702)32:2<167::AID-JMS472>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 97.Mechref Y, Novotny MV, Krishnan C. Structural characterization of oligosaccharides using MALDI-TOF/TOF tandem mass spectrometry. Anal Chem. 2003;75:4895–4903. doi: 10.1021/ac0341968. [DOI] [PubMed] [Google Scholar]
- 98.Yu SY, Wu SW, Khoo KH. Distinctive characteristics of MALDI-Q/TOF and TOF/TOF tandem mass spectrometry for sequencing of permethylated complex type N-glycans. Glycoconj J. 2006;23:355–369. doi: 10.1007/s10719-006-8492-3. [DOI] [PubMed] [Google Scholar]
- 99.Maslen SL, Goubet F, Adam A, Dupree P, Stephens E. Structure elucidation of arabinoxylan isomers by normal phase HPLC-MALDI-TOF/TOF-MS/MS. Carbohydr Res. 2007;342:724–735. doi: 10.1016/j.carres.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 100.Hakansson K, Cooper HJ, Hudgins RR, Nilsson CL. High resolution tandem mass spectrometry for structural biochemistry. Curr Org Chem. 2003;7:1503–1525. [Google Scholar]
- 101.Gauthier JW, Trautman TR, Jacobson DB. Sustained off-resonance irradiation for collision-activated dissociation involving Fourier-transform mass spectrometry - collision-activated dissociation technique that emulates infrared multiphoton dissociation. Anal Chim Acta. 1991;246:211–225. [Google Scholar]
- 102.Solouki T, Reinhold BB, Costello CE, O’Malley M, Guan SH, Marshall AG. Electrospray ionization and matrix-assisted laser desorption/ionization Fourier transform ion cyclotron resonance mass spectrometry of permethylated oligosaccharides. Anal Chem. 1998;70:857–864. doi: 10.1021/ac970562+. [DOI] [PubMed] [Google Scholar]
- 103.Aguilan JT, Dayrit FM, Zhang JH, Ninonuevo MR, Lebrilla CB. Structural analysis of alpha-carrageenan sulfated oligosaccharides by positive mode nano-ESI-FTICR-MS and MS/MS by SORI-CID. J Am Soc Mass Spectrom. 2006;17:96–103. doi: 10.1016/j.jasms.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 104.Leavell MD, Kruppa GH, Leary JA. Determination of phosphate position in hexose monosaccharides using an FTICR mass spectrometer: ion/molecule reactions, labeling studies, and dissociation mechanisms. Int J Mass Spectrom. 2003;222:135–153. [Google Scholar]
- 105.Cancilla MT, Wang AW, Voss LR, Lebrilla CB. Fragmentation reactions in the mass spectrometry analysis of neutral oligosaccharides. Anal Chem. 1999;71:3206–3218. doi: 10.1021/ac9813484. [DOI] [PubMed] [Google Scholar]
- 106.Pitteri SJ, McLuckey SA. Recent developments in the ion/ion chemistry of high-mass multiply charged ions. Mass Spectrom Rev. 2005;24:931–958. doi: 10.1002/mas.20048. [DOI] [PubMed] [Google Scholar]
- 107.Huang TY, McLuckey SA. Annu Rev Anal Chem. 2010;3:365–385. doi: 10.1146/annurev.anchem.111808.073725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Olsen JV, Macek B, Lange O, Makarov A, Horning S, Mann M. Higher-energy C-trap dissociation for peptide modification analysis. Nat Methods. 2007;4:709–712. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- 109.Zhang Y, Ficarro SB, Li SJ, Marto JA. Optimized Orbitrap HCD for quantitative analysis of phosphopeptides. J Am Soc Mass Spectrom. 2009;20:1425–1434. doi: 10.1016/j.jasms.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 110.Wu JA, Warren P, Shakey Q, Sousa E, Hill A, Ryan TE, He T. Integrating titania enrichment, iTRAQ labeling, and Orbitrap CID-HCD for global identification and quantitative analysis of phosphopeptides. Proteomics. 2010;10:2224–2234. doi: 10.1002/pmic.200900788. [DOI] [PubMed] [Google Scholar]
- 111.Brodbelt JS, Wilson JJ. Infrared multiphoton dissociation in quadrupole ion traps. Mass Spectrom Rev. 2009;28:390–424. doi: 10.1002/mas.20216. [DOI] [PubMed] [Google Scholar]
- 112.Vala M, Szczepanski J, Oomens J, Steill JD. H-2 Ejection from Polycyclic Aromatic Hydrocarbons: Infrared Multiphoton Dissociation Study of Protonated 1,2-Dihydronaphthalene. J Am Chem Soc. 2009;131:5784–5791. doi: 10.1021/ja808965x. [DOI] [PubMed] [Google Scholar]
- 113.Stefan SE, Eyler JR. Differentiation of methyl-glucopyranoside anomers by infrared multiple photon dissociation with a tunable CO2 laser. Anal Chem. 2009;81:1224–1227. doi: 10.1021/ac802067d. [DOI] [PubMed] [Google Scholar]
- 114.Vala M, Szczepanski J, Dunbar R, Oomens J, Steill JD. Infrared multiphoton dissociation spectrum of isolated protonated 1-azapyrene. Chem Phys Lett. 2009;473:43–48. [Google Scholar]
- 115.Polfer NC, Valle JJ, Moore DT, Oomens J, Eyler JR, Bendiak B. Differentiation of isomers by wavelength-tunable infrared multiple-photon dissociation-mass spectrometry: Application to glucose-containing disaccharides. Anal Chem. 2006;78:670–679. doi: 10.1021/ac0519458. [DOI] [PubMed] [Google Scholar]
- 116.Hakansson K, Klassen JS. In: Electrospray and MALDI Mass Spectrometry: Fundamentals, Instrumentation, Practicalities, and Biological Applications. 2. Cole RB, editor. John Wiley & Sons, Inc; Hoboken, New Jersey: 2010. [Google Scholar]
- 117.Zhang JH, Schubothe K, Li BS, Russell S, Lebrilla CB. Infrared multiphoton dissociation of O-linked mucin-type oligosaccharides. Anal Chem. 2005;77:208–214. doi: 10.1021/ac0489824. [DOI] [PubMed] [Google Scholar]
- 118.An HJ, Miyamoto S, Lancaster KS, Kirmiz C, Li BS, Lam KS, Leiserowitz GS, Lebrilla CB. Profiling of glycans in serum for the discovery of potential biomarkers for ovarian cancer. J Proteome Res. 2006;5:1626–1635. doi: 10.1021/pr060010k. [DOI] [PubMed] [Google Scholar]
- 119.Zubarev RA, Horn DM, Fridriksson EK, Kelleher NL, Kruger NA, Lewis MA, Carpenter BK, McLafferty FW. Electron capture dissociation for structural characterization of multiply charged protein cations. Anal Chem. 2000;72:563–573. doi: 10.1021/ac990811p. [DOI] [PubMed] [Google Scholar]
- 120.McLafferty FW, Horn DM, Breuker K, Ge Y, Lewis MA, Cerda B, Zubarev RA, Carpenter BK. Electron capture dissociation of gaseous multiply charged ions by Fourier-transform ion cyclotron resonance. J Am Soc Mass Spectrom. 2001;12:245–249. doi: 10.1016/S1044-0305(00)00223-3. [DOI] [PubMed] [Google Scholar]
- 121.Zubarev RA. Reactions of polypeptide ions with electrons in the gas phase. Mass Spectrom Rev. 2003;22:57–77. doi: 10.1002/mas.10042. [DOI] [PubMed] [Google Scholar]
- 122.Cooper HJ, Hakansson K, Marshall AG. The role of electron capture dissociation in biomolecular analysis. Mass Spectrom Rev. 2005;24:201–222. doi: 10.1002/mas.20014. [DOI] [PubMed] [Google Scholar]
- 123.Hakansson K, Cooper HJ, Emmett MR, Costello CE, Marshall AG, Nilsson CL. Electron capture dissociation and infrared multiphoton dissociation MS/MS of an N-glycosylated tryptic peptide to yield complementary sequence information. Anal Chem. 2001;73:4530–4536. doi: 10.1021/ac0103470. [DOI] [PubMed] [Google Scholar]
- 124.Hakansson K, Chalmers MJ, Quinn JP, McFarland MA, Hendrickson CL, Marshall AG. Combined electron capture and infrared multiphoton dissociation for multistage MS/MS in a Fourier transform ion cyclotron resonance mass spectrometer. Anal Chem. 2003;75:3256–3262. doi: 10.1021/ac030015q. [DOI] [PubMed] [Google Scholar]
- 125.Mirgorodskaya E, Roepstorff P, Zubarev RA. Localization of O-glycosylation sites in peptides by electron capture dissociation in a fourier transform mass spectrometer. Anal Chem. 1999;71:4431–4436. doi: 10.1021/ac990578v. [DOI] [PubMed] [Google Scholar]
- 126.Stensballe A, Jensen ON, Olsen JV, Haselmann KF, Zubarev RA. Electron capture dissociation of singly and multiply phosphorylated peptides. Rapid Commun Mass Spectrom. 2000;14:1793–1800. doi: 10.1002/1097-0231(20001015)14:19<1793::AID-RCM95>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 127.Shi SDH, Hemling ME, Carr SA, Horn DM, Lindh I, McLafferty FW. Phosphopeptide/phosphoprotein mapping by electron capture dissociation mass spectrometry. Anal Chem. 2001;73:19–22. doi: 10.1021/ac000703z. [DOI] [PubMed] [Google Scholar]
- 128.Budnik BA, Haselmann KF, Elkin YN, Gorbach VI, Zubarev RA. Applications of electron-ion dissociation reactions for analysis of polycationic chitooligosaccharides in Fourier transform mass spectrometry. Anal Chem. 2003;75:5994–6001. doi: 10.1021/ac034477f. [DOI] [PubMed] [Google Scholar]
- 129.Zhou W, Hakansson K. Electron capture dissociation of metal-adducted N-linked oligosaccharides released from bovine thyroid stimulating hormone. Proc. 56th ASMS Conference on Mass Spectrometry and Allied Topics; Denver, CO. 2008. [Google Scholar]
- 130.Zhao C, Xie B, Chan SY, Costello CE, O’Connor PB. Collisionally activated dissociation and electron capture dissociation provide complementary structural information for branched permethylated oligosaccharides. J Am Soc Mass Spectrom. 2008;19:138–150. doi: 10.1016/j.jasms.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 131.Kalli A, Hakansson K. Preferential cleavage of S-S and C-S bonds in electron detachment dissociation and infrared multiphoton dissociation of disulfide-linked peptide anions. Int J Mass Spectrom. 2007;263:71–81. [Google Scholar]
- 132.Haselmann KF, Budnik BA, Kjeldsen F, Nielsen ML, Olsen JV, Zubarev RA. Electronic excitation gives informative fragmentation of polypeptide cations and anions. Eur J Mass Spectrom. 2002;8:117–121. [Google Scholar]
- 133.Yang J, Mo JJ, Adamson JT, Hakansson K. Characterization of oligodeoxynucleotides by electron detachment dissociation Fourier transform ion cyclotron resonance mass spectrometry. Anal Chem. 2005;77:1876–1882. doi: 10.1021/ac048415g. [DOI] [PubMed] [Google Scholar]
- 134.Yang J, Hakansson K. Fragmentation of oligoribonucleotides from gas-phase ion-electron reactions. J Am Soc Mass Spectrom. 2006;17:1369–1375. doi: 10.1016/j.jasms.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 135.McFarland MA, Marshall AG, Hendrickson CL, Nilsson CL, Fredman P, Mansson JE. Structural characterization of the GM1 ganglioside by infrared multiphoton dissociation/electron capture dissociation, and electron detachment dissociation electrospray ionization FT-ICR MS/MS. J Am Soc Mass Spectrom. 2005;16:752–762. doi: 10.1016/j.jasms.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 136.Adamson JT, Hakansson K. Electron detachment dissociation of neutral and sialylated oligosaccharides. J Am Soc Mass Spectrom. 2007;18:2162–2172. doi: 10.1016/j.jasms.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 137.Wolff JJ, Amster IJ, Chi LL, Linhardt RJ. Electron detachment dissociation of glycosaminoglycan tetrasaccharides. J Am Soc Mass Spectrom. 2007;18:234–244. doi: 10.1016/j.jasms.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wolff JJ, Chi LL, Linhardt RJ, Amster IJ. Distinguishing glucuronic from iduronic acid in glycosaminoglycan tetrasaccharides by using electron detachment dissociation. Anal Chem. 2007;79:2015–2022. doi: 10.1021/ac061636x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wolff JJ, Laremore TN, Busch AM, Linhardt RJ, Amster IJ. Electron detachment dissociation of dermatan sulfate oligosaccharides. J Am Soc Mass Spectrom. 2008;19:294–304. doi: 10.1016/j.jasms.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wolff JJ, Laremore TN, Busch AM, Linhardt RJ, Amster IJ. Influence of charge state and sodium cationization on the electron detachment dissociation and infrared multiphoton dissociation of glycosaminoglycan oligosaccharides. J Am Soc Mass Spectrom. 2008;19:790–798. doi: 10.1016/j.jasms.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wolff JJ, Laremore TN, Leach FE, Linhardt RJ, Amster IJ. Electron capture dissociation, electron detachment dissociation and infrared multiphoton dissociation of sucrose octasulfate. Eur J Mass Spectrom. 2009;15:275–281. doi: 10.1255/ejms.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chi LL, Wolff JJ, Laremore TN, Restaino OF, Xie J, Schiraldi C, Toida T, Amster IJ, Linhardt RJ. Structural analysis of bikunin glycosaminoglycan. J Am Chem Soc. 2008;130:2617–2625. doi: 10.1021/ja0778500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wolff JJ, Laremore TN, Aslam H, Linhardt RJ, Amster IJ. Electron-induced dissociation of glycosaminoglycan tetrasaccharides. J Am Soc Mass Spectrom. 2008;19:1449–1458. doi: 10.1016/j.jasms.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhou W, Håkansson K. Ion-electron reactions of sialylated N-linked glycans released from glycoproteins. Proc. 57th ASMS Conference on Mass Spectrometry and Allied Topics; Philadelphia, PA. 2009. [Google Scholar]
- 145.Yoo HJ, Liu HC, Hakansson K. Infrared multiphoton dissociation and electron-induced dissociation as alternative MS/MS strategies for metabolite identification. Anal Chem. 2007;79:7858–7866. doi: 10.1021/ac071139w. [DOI] [PubMed] [Google Scholar]
- 146.Gao D, Zhou W, Hakansson K. Electron induced dissociation (EID) of singly protonated glycans. Proc. 58th ASMS Conference on Mass Spectrometry and Allied Topics; Salt Lake City, UT. 2010. [Google Scholar]