Abstract
Photosynthesis has recently gained considerable attention for its potential role in the development of renewable energy sources. Optimizing photosynthetic organisms for biomass or biofuel production will therefore require a systems understanding of photosynthetic processes. We reconstructed a high-quality genome-scale metabolic network for Synechocystis sp. PCC6803 that describes key photosynthetic processes in mechanistic detail. We performed an exhaustive in silico analysis of the reconstructed photosynthetic process under different light and inorganic carbon (Ci) conditions as well as under genetic perturbations. Our key results include the following. (i) We identified two main states of the photosynthetic apparatus: a Ci-limited state and a light-limited state. (ii) We discovered nine alternative electron flow pathways that assist the photosynthetic linear electron flow in optimizing the photosynthesis performance. (iii) A high degree of cooperativity between alternative pathways was found to be critical for optimal autotrophic metabolism. Although pathways with high photosynthetic yield exist for optimizing growth under suboptimal light conditions, pathways with low photosynthetic yield guarantee optimal growth under excessive light or Ci limitation. (iv) Photorespiration was found to be essential for the optimal photosynthetic process, clarifying its role in high-light acclimation. Finally, (v) an extremely high photosynthetic robustness drives the optimal autotrophic metabolism at the expense of metabolic versatility and robustness. The results and modeling approach presented here may promote a better understanding of the photosynthetic process. They can also guide bioengineering projects toward optimal biofuel production in photosynthetic organisms.
Keywords: constraint-based modeling, photobioenergetic, metabolic engineering, biosustainability, metabolic robustness
The recent emphasis on biosustainability has brought attention to photosynthetic microorganisms. Microphototrophs, including cyanobacteria, represent efficient biological systems for producing biomass from inorganic carbon (Ci) and high-value products such as carotenoids, and they are also viewed as a potential source of biofuel (1, 2). On the other hand, photosynthesis is an inherently inefficient process, and photoautotrophic growth is limited by environmental perturbations (3). The success of future light-driven bioengineering approaches requires a systems understanding of the photosynthetic processes, including their bioenergetics and robustness.
Optimal photosynthetic performance requires fine-tuning the light/energy conversion by the photosystems and metabolic reactions, with the ATP/NADPH ratio being a particularly important parameter (4, 5). Carbon dioxide (CO2) fixation through the Calvin cycle is the main ATP and NADPH sink in the autotrophic metabolism, requiring an ATP/NADPH ratio of 1.5. Because the photosynthetic linear electron flow (LEF) pathway generates an ATP/NADPH ratio of 1.28, additional ATP is needed (4, 5). Phototrophs have developed a large arsenal of alternate electron flow (AEF) pathways, which assist in modulating the ATP/NADPH ratio as a function of the metabolic demand. Although the in vivo physiological roles of the AEF pathways are not completely known because of their cyclical nature, they are believed to work cooperatively with LEF (4, 6). The development of genome-scale metabolic models is therefore desirable for systematic understanding of the photosynthetic process (2, 4). To date, analysis of photosynthetic processes at the genome-scale has received limited attention (7).
The cyanobacterium Synechocystis sp. PCC6803, henceforth called Synechocystis, is a model photosynthetic prokaryotic organism capable of growing phototrophically because of oxygenic photosynthesis and heterotrophically at the expense of reduced carbon sources in the dark. These features make Synechocystis an interesting microorganism for biotechnology applications. Although several central-metabolic and genome-scale reconstructions are available for Synechocystis (8–12), they do not describe photosynthetic and heterotrophic metabolism in detail. Moreover, pathways relevant for biotechnological endeavors, such as the biosynthesis of lipids and photosynthetic pigments, are currently poorly represented. To fill these gaps, we reconstructed a high-quality genome-scale metabolic network of Synechocystis named iJN678. Here we describe iJN678 and the properties of the metabolic network it represents.
Results
Reconstruction Content, Enhancements, and Validation.
We assembled a genome-scale metabolic reconstruction of Synechocystis based on available genomic and biochemical data by using an established reconstruction protocol (13). The final reconstruction, iJN678, contains 678 genes, 863 metabolic and transport reactions, and 795 nonunique metabolites, which are distributed over 54 subsystems and four different cellular compartments: extracellular, periplasm, cytoplasm, and thylakoid (SI Appendix, Table S1 and Dataset S1).
To enable systems analysis of the photosynthetic processes, an extensive effort was made to construct a complete and compartmentalized model of photosynthesis that exceeds previously published efforts in detail and coverage (8–11). The LEF reconstruction includes individual reactions for photosystem II (PSII), cytochrome b6f (CytBF), photosystem I (PSI), and ferredoxin NADP+ oxidoreductase (FNR), which were connected through plastoquinone (PQ), plastocyanin/cytochrome C (cytC), and ferredoxin, respectively (5). The AEF modeling includes the ferredoxin PQ reductase (FQR) and the NAD(P)H dehydrogenase complexes (14, 15), the aa3-type terminal oxidase (CYO), the PQ oxidase (CydBD), the MEHLER (16), and the hydrogenase (H2ase) (17) reactions. We additionally included the three photorespiration (PHOTOR) branches (18) and succinate dehydrogenase (SDH). The CYO, CydBD, and SDH reactions were also placed in the thylakoid membrane-enabling interaction between respiration and photosynthesis (19). The modeled photosynthetic apparatus is depicted in SI Appendix, Fig. S1. The complete biosynthesis pathways for fatty acids, including ω-3 polyunsaturated fatty acids, and for photosynthetic-specific lipids were captured as well as the biosynthetic pathways for photosynthetic pigments, including chlorophyll a, phylloquinone, lycopene, β-carotenoids, zeaxanthin, echinenone, and α-, β-, γ- and δ-tocopherols. This level of detail allowed us to define an accurate biomass objective function (BOF) (Dataset S2). It also opens up the possibility for future biotechnology ventures to focus on the overproduction of these high-value compounds. Finally, the modeling of the heterotrophic metabolism was enhanced by including all transport reactions with genetic or physiological evidence (SI Appendix, Fig. S2). A detailed description of the major areas of enhancement of iJN678 was compared with published reconstructions (SI Appendix, Table S2).
Genome-scale reconstructions need to be validated by assessing their ability to compute physiological states (13). We validated iJN678 by comparing growth predictions with experimental values. The prediction error was less than 5% under mixotrophic and autotrophic conditions (SI Appendix, Table S3). Finally, the model predicted flux distributions, which correlated well with experimentally determined fluxes under the heterotrophic, mixotrophic, and autotrophic conditions (τ = 0.89, 0.92, and 0.96, respectively) (refs. 20 and 21; SI Appendix, Figs. S5–S7 and Dataset S3). A validated model can be used to characterize the metabolic states that underlie observed phenotypic functions.
Optimal Autotrophic Metabolism Requires the Contribution of Alternative Photosynthetic Electron Flow Pathways.
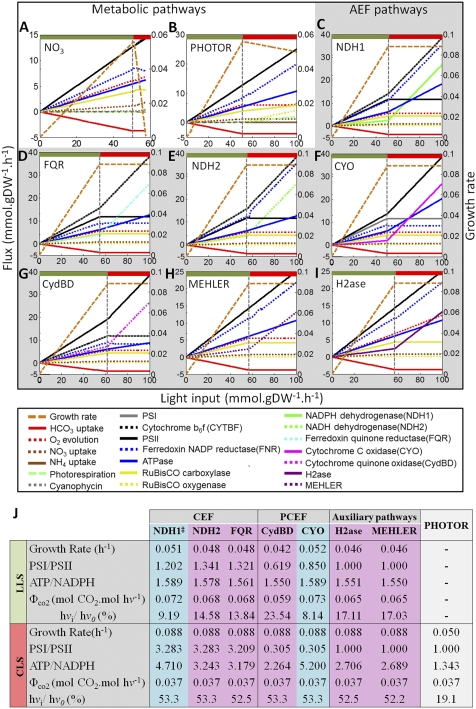
We studied the photosynthetic process as a function of Ci and light availability. Maximal possible growth rates were computed under varying light conditions while maintaining a fixed Ci uptake rate (Fig. 1). We identified two unique states: (i) an initial state in which light, not Ci, was the growth-limiting factor [light-limited state (LLS)] and (ii) a final state in which Ci was growth-limiting instead of light [Ci-limited state (CLS)]. Under LLS, the fluxes through the LEF pathway, ATPase, and the HCO3 uptake increased with light availability. The attainment of maximum Ci uptake [3.7 mmol⋅g dry wt (gDW)−1⋅h−1] marked the beginning of CLS, which was characterized by light excess and Ci limitation. The increasing light availability led to more photosynthetic activity and, thus, to higher levels of reducing equivalents (PSII, PSI, or FNR) and ATP (ATPase) (Fig. 1).
Fig. 1.
Impact of Ci (i.e., HCO3−) and light availability on the autotrophic metabolism of Synechocystis. Shown are the functional states found in the absence of AEF pathways and PHOTOR (A), the absence of AEF pathways allowing PHOTOR (B), and the presence of one AEF pathway (indicated by the caption in each diagram) (C–I). The growth rate (orange, right y axis) was computed under autotrophic conditions by using HCO3− as Ci source (uptake rate = 3.7 mmol⋅gDW−1⋅h−1) and NO3 as nitrogen source (unconstrained uptake rate) as a function of light availability by varying the photon uptake rate from 0 to 100 mmol⋅gDW−1⋅h−1 (x axis). The computed flux rates for each relevant reaction are shown (left y axis). The boundary between LLS (green) and CLS (red) states is shown by a vertical gray line. (J) Main photosynthetic and bioenergetics parameters yielded by the AEF pathways and PHOTOR. Light uptake rates of 30 and 100 mmol⋅gDW−1⋅h−1 were taken as representative of the LLS (green) and CLS (red), respectively. The parameters from high and low photosynthetic-yield AEF pathways are shown in blue and pink, respectively. Because NDH-13, NDH-14, and NDH-1 exhibited identical parameters in both LLS and CLS, only NDH-1 is shown.
By constraining the AEF and PHOTOR pathways, we found that the balancing of the ATP/NADPH ratio and the prevention of internal overreduction were achieved by the reduction of an extra NO3 (Fig. 1A). The excess of NH4 produced was fixed via synthesis of cyanophycin, a cyanobacteria-specific nitrogen and carbon storage polymer, and led to a suboptimal growth rate. Under CLS, the high level of reducing equivalents produced exceeded the energy-dissipation capabilities of the NO3 reduction pathway, and the model could not grow. The inclusion of PHOTOR in the simulation resulted in a suboptimal growth rate under CLS (Fig. 1B). This result suggests that PHOTOR could act as an energy-dissipation pathway under these conditions, which is in good agreement with the participation of PHOTOR in high-light acclimation (22).
When the growth rate was computed allowing one AEF at the time, we found that, in all cases, optimal growth rate was achieved. Under LLS, AEF pathways guaranteed optimal growth by consuming reducing equivalents and increasing the ATP levels (Fig. 1 C–I). The computed ATP/NADPH ratios were greater than 1.55 in all cases (Fig. 1J). They were higher than the ratio provided by LEF alone (i.e., 1.28) and agreed with the reported optimal requirement for CO2 fixation (i.e., 1.5) (5). Under CLS, AEF pathways primarily acted as photoenergy-dissipation pathways by consuming reducing equivalents and dissipating more than 50% of available photons (hvi/hvo) (Fig. 1J).
Our analysis showed that the LEF pathway alone did not enable maximal growth under suboptimal light conditions but that the AEF pathways were also required, in agreement with previous observations (5, 6, 23). Furthermore, the results highlight that growth under environmental perturbations, such as high light or Ci depletion, also highly depended on the AEF pathways and PHOTOR.
Quantification and Classification of Alternative Photosynthetic Pathways.
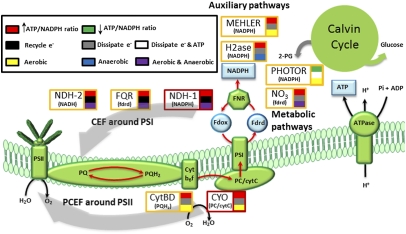
Cyclic electron flow (CEF) around PSI has been suggested to be the main AEF pathway in phototrophs and to be primarily responsible for supplying the additional ATP required for CO2 fixation (5, 6). We identified five CEF pathways assisting the LEF in ATP/NADPH balancing: Three pathways were involved in returning electrons to the PQ pool from NADPH (NDH-1, NDH-13, and NDH-14), and the NDH-2 and FQR pathways used NADH and reduced ferredoxin as electron donors, respectively (Figs. 1 C–E and 2). The flux across CEF pathways resulted in PSI/PSII ratios above 1.20, which agrees with theoretical estimates (Fig. 1J) (5).
Fig. 2.
Functional characterization of the alternative photosynthetic pathways involved in ATP/NADPH balancing and photoenergy dissipation. The LEF pathway (PSII, PQ pool, CytBF, PSI, and FNR), the ATPase, and the Calvin cycle are shown in green. The final products from LEF and photophosphorylation [reduced ferrodoxin (Fdrd), NADPH, and ATP] are shown in blue. The high and low light-yield pathways and their electron donors are shown with red and yellow boxes, respectively. Furthermore, the alternative pathways were color-coded based as shown based on their contribution to the ATP/NADPH ratio, the mechanism of dissipation, and by dependence on O2 availability.
The computed quantum yield (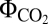 , i.e., moles of photons absorbed per mole of CO2 fixed) varied as a function of the used CEF pathways. The NDH-1–related pathways showed higher quantum yield (
, i.e., moles of photons absorbed per mole of CO2 fixed) varied as a function of the used CEF pathways. The NDH-1–related pathways showed higher quantum yield (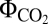 = 0.072) than NDH-2 and FQR did (
= 0.072) than NDH-2 and FQR did (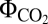 = 0.068) (Fig. 1J). The
= 0.068) (Fig. 1J). The 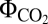 , measured under light-limited conditions, is a standard measure of photosynthetic efficiency, with mean
, measured under light-limited conditions, is a standard measure of photosynthetic efficiency, with mean 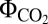 values of 0.052 for C3 plants and 0.057 for the more efficient C4 plants (24). Based on our computed
values of 0.052 for C3 plants and 0.057 for the more efficient C4 plants (24). Based on our computed 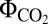 , we suggest that CEF pathways are not equivalent. Using the arbitrary
, we suggest that CEF pathways are not equivalent. Using the arbitrary 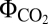 value of 0.070 as cutoff, we suggest that NDH-2 and FQR pathways are low photosynthetic-yield pathways, whereas NDH-1–related CEF pathways can be classified as high photosynthetic-yield pathways (Fig. 1J). Under CLS, a large increase in flux through the CEF pathways was predicted, which indirectly consumed more than 52% of available photons (Fig. 1J). Consequently, the PSI/PSII and ATP/NADPH ratios increased more than three- and fourfold, respectively (Fig. 1). These results are consistent with reports that light excess and low CO2 levels induce CEF around PSI (6, 14).
value of 0.070 as cutoff, we suggest that NDH-2 and FQR pathways are low photosynthetic-yield pathways, whereas NDH-1–related CEF pathways can be classified as high photosynthetic-yield pathways (Fig. 1J). Under CLS, a large increase in flux through the CEF pathways was predicted, which indirectly consumed more than 52% of available photons (Fig. 1J). Consequently, the PSI/PSII and ATP/NADPH ratios increased more than three- and fourfold, respectively (Fig. 1). These results are consistent with reports that light excess and low CO2 levels induce CEF around PSI (6, 14).
The PQ oxidase CydBD and the terminal cytC oxidase CYO were identified as AEF pathways involved in a pseudocyclic electron flow (PCEF) around PSII (Fig. 2). Under LLS, the flow of electrons around PSII resulted in a PSI/PSII ratio of less than 1 but an optimal ATP/NADPH ratio (Fig. 1 F, G, and J). The CYO and CydBD pathways could be classified as high and low photosynthetic-yield pathways, respectively, based on the calculated 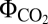 values similar to CEF pathways (Fig. 1J). Under CLS, PCEF also enabled optimal growth by acting as an electron sink, indirectly consuming more than 53% of the available photons and preventing internal overreduction (Fig. 1 F, G, and J). In agreement with our in silico data, genetic evidence showed that, in the absence of PSI or under high-light conditions, terminal oxidases in Synechocystis used PSII-generated electrons and provided extra ATP levels (15, 25).
values similar to CEF pathways (Fig. 1J). Under CLS, PCEF also enabled optimal growth by acting as an electron sink, indirectly consuming more than 53% of the available photons and preventing internal overreduction (Fig. 1 F, G, and J). In agreement with our in silico data, genetic evidence showed that, in the absence of PSI or under high-light conditions, terminal oxidases in Synechocystis used PSII-generated electrons and provided extra ATP levels (15, 25).
In addition to CEF and PCEF, our in silico analysis identified the NADPH-dependent reactions MEHLER (reduction of O2) and H2ase as reactions assisting the LEF. These pathways yielded a PSI/PSII ratio of 1, low 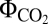 values, and reduced growth rates under LLS (Fig. 1 H–J). The difference between the H2ase and the MEHLER pathways was the O2 evolution and larger flux through the H2ase pathway. Our analysis thus suggests that the H2ase and MEHLER reactions may be complementary low photosynthetic-yield systems whose activity depends on O2 availability. Indeed, experimental evidence indicates that H2ase only operates in anaerobic or microaerobic conditions, whereas MEHLER pathways require aerobic conditions (17, 26).
values, and reduced growth rates under LLS (Fig. 1 H–J). The difference between the H2ase and the MEHLER pathways was the O2 evolution and larger flux through the H2ase pathway. Our analysis thus suggests that the H2ase and MEHLER reactions may be complementary low photosynthetic-yield systems whose activity depends on O2 availability. Indeed, experimental evidence indicates that H2ase only operates in anaerobic or microaerobic conditions, whereas MEHLER pathways require aerobic conditions (17, 26).
In summary, our analysis suggests that Synechocystis has a large number of AEF pathways involved to preserve optimal photosynthetic processes under environmental perturbation, such as Ci or light availability (photosynthetic robustness). The quantitative analysis of photosynthetic parameters exhibited by iJN678 allowed us to classify these AEF pathways based on (i) photosynthetic yield, (ii) net contribution to the ATP/NADPH ratio, (iii) mechanisms used for ATP/NADPH balancing and/or energy dissipation, (iv) dependency on O2 availability, and (v) LEF target components on which they acted (Fig. 2). This classification represents a valuable tool for better understanding how phototrophs might select among their large array of accessory photosynthetic pathways those that are best suited for efficient response to environmental perturbations.
PHOTOR Plays an Active Role as an ATP-Consuming Pathway Under CLS.
PHOTOR recycles 2-phosphoglycolate, which is produced by the oxygenic reaction of ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO). This process leads to a net loss of carbons and reduces the efficiency of photosynthesis because one glyceraldehyde 3-phosphate molecule and one CO2 molecule are produced from two 2-phosphoglycolate molecules. We found that PHOTOR was not active under LLS but that it enabled growth under CLS in absence of AEF (Fig. 1B). PHOTOR acted as an alternative energy-dissipation pathway under CLS, yielding a unitary PSI/PSII ratio and suboptimal ATP/NADPH ratio (Fig. 1J). The latter observation is consistent with the high energetic cost of PHOTOR (two ATP and one NADPH are necessary for the production of one glyceraldehyde 3-phosphate molecule from two 2-phosphoglycolate molecules) (23) and strongly suggests that PHOTOR uses a photoenergy-dissipation mechanism without extra ATP production. This suggestion is particularly interesting because excessive ATP levels is a major problem in phototrophs, leading to metabolic congestion (4). The environmental conditions in which PHOTOR is activated (18, 22), together with our in silico results, suggest a primarily role for PHOTOR as an ATP-consuming pathway under CLS. Nonetheless, an involvement in ATP-consuming futile cycles (SI Appendix, Fig. S10) and/or other regulatory mechanisms cannot be discounted (4).
Integrative Analysis of Photosynthesis Performance.
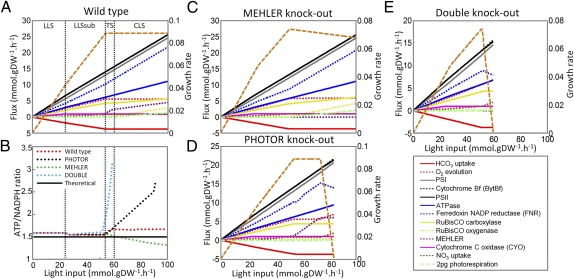
To better understand how AEF and metabolic pathways collaborate in response to environmental perturbations, we evaluated results from our in silico analysis against published experimental observations. In particular, we investigated the reported close collaboration between the MEHLER and PHOTOR pathways in response to high-light conditions (22). In silico growth rates were first computed while having an unconstrained flux through the PHOTOR pathways and bounding the maximal possible flux through MEHLER and CYO pathways based on experimental values (Fig. 3) (27). Subsequently, we deleted either the PHOTOR or MEHLER pathways and observed significantly decreased maximal possible growth rates under CLS. The double knockout of these two pathways reduced the in silico growth rate even further (Fig. 3), which is in good agreement with experimental reports (22). These results illustrate that both pathways were required for optimal growth rates under CLS.
Fig. 3.
Cooperativity between the MEHLER and PHOTOR pathways under high-light conditions. (A) The wild-type strain was simulated by constraining all fluxes in the AEF pathways to 0 mmol⋅gDW−1⋅h−1, except the CYO and MEHLER reactions, which were constrained to an upper bound of 1 and 6 mmol⋅gDW−1⋅h−1, respectively, corresponding to 6% and 40% of the electrons from PSII at the end of CLS (27). PHOTOR remained unconstrained. (B) ATP/NADPH ratio as a function of the light input (SI Appendix and Dataset S6). The theoretical optimum ratio for CO2 fixation is indicated by a solid black line. (C) The MEHLER knockout was simulated by constraining the flux to 0 mmol⋅gDW−1⋅h−1. (D) PHOTOR knockout was simulated by constraining the photorespiratory metabolism (SI Appendix). (E) In the double knockout, the MEHLER and PHOTOR mutant constraints were applied. The growth rate (orange, right y axis) was computed under autotrophic conditions as a function of the light availability by varying the photon uptake rate from 0 to 100 mmol⋅gDW−1⋅h−1 (x axis). The computed reaction fluxes are shown (left y axis). The functional states are denoted by LLS, LLSsub (suboptimal LLS), TS (transition state), and CLS. The boundaries between the different functional states are indicated by dotted, vertical black lines.
We then studied the collaboration between MEHLER and PHOTOR at a mechanistic level. By using the above constraints, the fluxes across the main photosynthetic reactions and the ATP/NADPH ratio were computed as a function of light availability (Fig. 3). As expected, the high photosynthetic-yield pathway CYO was the AEF pathway used by the model under LLS for ATP/NADPH balancing. The attainment of maximum flux through CYO triggered the activation of the MEHLER pathway as a new LEF-assisting pathway, leading to new suboptimal LLS. The growth rate and ATP/NADPH ratio were lower in this state because of the low photosynthetic yield of the MEHLER pathway (Fig. 3A). These observations indicate that low-yield pathways cannot replace the role of high-yield pathways under LLS. Before CLS, we identified a transition state in which the flux across the MEHLER reaction increased significantly. Under CLS, the flux across the MEHLER pathway increased in response to light availability, and PHOTOR was activated as an additional assistant pathway. We found that PHOTOR dissipated excess ATP to maintain ATP/NADPH balance in the wild type because no ATP dissipation was observed in PHOTOR-deficient and double-mutant strains (Fig. 3).
Our in silico analysis demonstrated the needed balancing act between MEHLER (primarily responsible for NADPH dissipation) and PHOTOR (involved in reestablishing optimal ATP/NADPH ratio via ATP dissipation) to preserve photosynthetic optimality under Ci depletion and/or high-light conditions.
Insights into the Heterotrophic Metabolism of Synechocystis.
We evaluated the growth capabilities of iJN678 under mixotrophic and heterotrophic conditions with all of the carbon and nitrogen sources for which transport reactions were present. The model predicted growth on 10 organic carbon sources under heterotrophic conditions (SI Appendix, Table S4). The amino acids alanine, glutamine, and serine sustained growth in silico only under mixotrophic conditions. We characterized metabolic precursors present in iJN678 and identified 31 additional candidate carbon sources that could support growth in silico when specific transporters were included (in silico metabolic engineering) (SI Appendix, Table S5). Therefore, iJN678 exhibited low metabolic versatility compared with other heterotrophic bacteria, such as Escherichia coli (28). This analysis also suggests the presence of potential latent heterotrophic pathways encoded in the genome that could be exploited in future biotechnology applications.
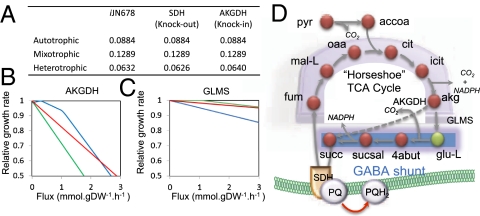
iJN678 was used to elucidate unknown aspects of the heterotrophic metabolism in Synechocystis, such as the missing pyruvate oxidation pathway caused by the lack of a 2-oxoglutarate dehydrogenase (AKGDH). Computed fluxes under heterotrophic conditions indicate that glucose was metabolized to pyruvate through the oxidative branch of the pentose phosphate pathway, which agreed with experimental observations (20) (SI Appendix, Fig. S5). We found that pyruvate was subsequently oxidized by the combined action of the GABA shunt and the SDH complex (Fig. 4 and SI Appendix, Fig. S5). The contribution of SDH under heterotrophic conditions was highlighted by performing an in silico deletion of SDH that led to a reduced growth rate in the dark, but no effect on the growth rate was predicted under either mixotrophic or autotrophic conditions (Fig. 4). In the line with our results, SDH has been suggested to be the main reaction respiratory pathway providing electrons to the PQ pool in darkness but not under light conditions, where PSII plays this role (Fig. 2) (19).
Fig. 4.
Analysis of the heterotrophic metabolism in Synechocystis. (A) Effect on the growth rate (h−1) of SDH knockout and AKGDH knockin. (B and C). Relative growth rate as a function of the flux through AKGDH (B) and GLMS (C) reactions on autotrophic (green), mixotrophic (red), and heterotrophic (blue) metabolisms. Optimal light conditions (photon uptake of 54.5 mmol⋅gDW−1⋅h−1) were used for both the mixotrophic and autotrophic metabolism. HCO3− (3.7 mmol⋅gDW−1⋅h−1) and glucose (0.85 mmol⋅gDW−1⋅h−1) were used as carbon sources to simulate autotrophic and heterotrophic conditions, respectively. Glucose (0.38 mmol⋅gDW−1⋅h−1) and HCO3− (3.7 mmol⋅gDW−1⋅h−1) were used as carbon sources under mixotrophic conditions. (D) Proposed complete oxidation pathway of pyruvate in Synechocystis under heterotrophic conditions, which involves an incomplete TCA cycle, a GABA shunt, and SDH complex. Glutamate as a common metabolite of the nitrogen assimilation pathway and the GABA shunt is indicated in green. The nonnative AKGDH reaction is indicated by a dashed arrow. See Dataset S1 for abbreviations.
We then investigated the role of the GABA shunt on heterotrophic metabolism and whether its presence is advantageous over having a complete tricarboxylic acid (TCA) cycle, which is incomplete in Synechocystis. We found that the inclusion of the AKGDH complex in the model, which completes the TCA cycle, increased the growth rate under heterotrophic conditions, whereas it negatively influenced the mixotrophic and autotrophic metabolism, with a stronger effect on the latter (Fig. 4 A and B). The funneling of 2-oxoglutarate (akg) to the GABA shunt through glutamate synthase (GLMS) showed the opposite effect (Fig. 4C). Thus, having a GABA shunt instead of a complete TCA cycle via AKGDH inclusion may represent an evolutionary advantage for Synechocystis in autotrophic conditions at the expense of reduced growth performance in heterotrophic conditions. Genome analysis of other cyanobacteria strengthens this hypothesis because the lack of AKGDH was preserved in this phylogenetic group, whereas the GABA shunt was found in several species (Dataset S4).
Synechocystis Metabolism Exhibits Low Metabolic Robustness Under Genetic Perturbations.
No experimental studies addressing essential genes and metabolic robustness from phototrophs are available. We carried out a gene essentiality and synthetic lethality analysis (Dataset S5). The highest number of essential metabolic genes was predicted under autotrophic conditions (350) followed by heterotrophic and mixotrophic conditions with 261 and 259 essential genes, respectively (SI Appendix, Fig. S9). Thus, a core set of 259 genes, i.e., 38% of the included genes, was found to be essential under all tested conditions, which is consistent with the 34% of essential genes reported in a previous in silico study (8). We compared our results with the mutants available in CyanoMutants (http://genome.kazusa.or.jp/cyanobase/mutants). Despite the fact that only a low number of genes could be compared, our analysis (P < 10−3, Fisher's exact test), showed an accuracy of 79%, although a significant false positive rate (21%) was found (SI Appendix, Fig. S9 and Dataset S5). Many of these false positives participate in the unique pathways modeled in iJN678, such as photosynthetic pigments and lipid biosynthetic pathways. For instance, the genes slr0089 and slr1508 which are involved in the biosynthesis of tocopherols and digalactolipids, respectively, were predicted to be essential based on the presence of these metabolites in the biomass objective functions. Interestingly, in vivo data indicate that, although dispensable for growth, these metabolites are essential for acclimation to changing light conditions (29, 30). These results strongly suggest that Synechocystis devote many resources to preserve an optimal photosynthetic process under changing environmental perturbations, thus increasing their photosynthetic robustness.
Synthetic gene lethality is often used as a measure of metabolic robustness because synthetic essential gene products are interchangeable (isoenzymes) or operate in two separate pathways with redundant and/or complementary essential functions. This synthetic lethality analysis predicted the highest number of lethal gene deletions under mixotrophic conditions with 274, which was expected because heterotrophic and autotrophic conditions concur in mixotrophic metabolism. Conversely, the heterotrophic and autotrophic metabolic network exhibited only 234 and 158 synthetic lethal genes, respectively.
Overall, our gene essentiality and synthetic lethality analyses indicate that autotrophic metabolism requires higher numbers of genes than heterotrophic or mixotrophic metabolisms do. Therefore, the high number of metabolic essential genes in all computed growth conditions (38%) compared with other bacteria, such as E. coli (14.5%) (28), strongly suggests a reduced metabolic robustness in Synechocystis, especially in autotrophic conditions. This reduced metabolic robustness under genetic perturbation contrasts with the high photosynthetic robustness predicted (including multiple lipids, photosynthetic pigments, AEF, and metabolic pathways involved in optimal photosynthetic process). Interestingly, it has been suggested that highly evolved and specialized systems acquire high robustness against regular perturbations that but they are extremely fragile against rare perturbations (31). For Synechocystis, a highly specialized autotroph organism, Ci and/or light availability could be considered as a regular perturbation. In contrast, genetic perturbations resulting in lethal phenotypes would be rare perturbations, which only affect individual cells. Thus, the tradeoff between high photosynthetic robustness and the overall low metabolic robustness suggested by our analysis could be a consequence of the highly specialized autotrophic metabolism.
Discussion
The bottom-up metabolic network reconstruction, iJN678(Dataset S7), presented here summarizes the current knowledge about the metabolism of Synechocystis. We investigated the optimality of photoautotrophic metabolism under both environmental and genetic perturbations and gained insight on Synechocystis’ metabolism, including (i) the identification of two main states of the photosynthetic apparatus (CLS and LLS); (ii) the categorizing of nine AEF pathways present in Synechocystis; (iii) the high degree of cooperativity between AEF pathways required for optimizing the autotrophic metabolism; (iv) the critical role of PHOTOR in high-light acclimation; and (v) the inherently reduced metabolic versatility and robustness together with the high photosynthetic robustness derived from the photoautotrophic metabolism-specialization process.
By studying the impact of Ci and light availability, we have proved that multiple AEF pathways assist LEF in optimal photosynthesis performance. While under LLS, the AEF pathways provided extra ATP levels; under CLS, they dissipated the photoenergy excess (Figs. 1 and 2). The analysis of the photosynthetic and bioenergetics parameters found in iJN678 allowed us to classify these AEF pathways into different functional categories and may thus contribute to a better understanding of the photosynthetic process (Fig. 2). For instance, the CYO pathway had the highest 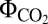 , suggesting that this pathway is the best candidate for ATP/NADPH optimization under suboptimal light availability. Interestingly, recent studies support this nonintuitive finding, and high respiration rates together with low PSI/PSII ratios have been reported in response to low-light conditions in the deep-sea green algae Ostreococcus RCC809 (32). Furthermore, we showed that additional high photosynthetic-yield CEF pathways were necessary to ensure maximum growth under LLS (Fig. 3). This result is consistent with the PSI/PSII ratios greater than 1 that have been reported in phototrophs as well as with the proposed role of the NDH-1 pathway as being the main CEF pathway under suboptimal light availability (5, 14). We propose that a strong cooperativity between high photosynthetic-yield pathways assisting the LEF around PSI and PSII is required to sustain maximum growth under suboptimal light conditions. Under light excess and/or Ci-limited conditions, light optimization was no longer a priority but instead became a problem. These conditions led to an overreduced state, preventing growth in the absence of AEF (Fig. 1). We found that all AEF pathways identified in this study dissipated excess energy by increasing their fluxes, resulting in high ATP levels (Fig. 1). A high ATP/ADP ratio leads to acid photodamage of PSII, and several mechanisms have been proposed for ATP dissipation, including down-regulation of ATPase, controlled proton leak, metabolic futile cycles, and ATP hydrolyzing enzymes (4). Our results strongly suggest an advantage in energy dissipation of low, rather than high, photosynthetic-yield pathways for minimization of the increased ATP levels (Fig. 1J). Consistently, CytBD (15), FQR (14), MEHLER (16, 22), and H2ase pathways (17) are only involved in energy dissipation under high-light or Ci-depleted conditions (CLS). Our analysis also identified that an optimal response to CLS required the additional participation of PHOTOR as a counterpart to low photosynthetic-yield AEF pathways, leading to energy dissipation by consuming the high ATP levels generated by these pathways (Fig. 3).
, suggesting that this pathway is the best candidate for ATP/NADPH optimization under suboptimal light availability. Interestingly, recent studies support this nonintuitive finding, and high respiration rates together with low PSI/PSII ratios have been reported in response to low-light conditions in the deep-sea green algae Ostreococcus RCC809 (32). Furthermore, we showed that additional high photosynthetic-yield CEF pathways were necessary to ensure maximum growth under LLS (Fig. 3). This result is consistent with the PSI/PSII ratios greater than 1 that have been reported in phototrophs as well as with the proposed role of the NDH-1 pathway as being the main CEF pathway under suboptimal light availability (5, 14). We propose that a strong cooperativity between high photosynthetic-yield pathways assisting the LEF around PSI and PSII is required to sustain maximum growth under suboptimal light conditions. Under light excess and/or Ci-limited conditions, light optimization was no longer a priority but instead became a problem. These conditions led to an overreduced state, preventing growth in the absence of AEF (Fig. 1). We found that all AEF pathways identified in this study dissipated excess energy by increasing their fluxes, resulting in high ATP levels (Fig. 1). A high ATP/ADP ratio leads to acid photodamage of PSII, and several mechanisms have been proposed for ATP dissipation, including down-regulation of ATPase, controlled proton leak, metabolic futile cycles, and ATP hydrolyzing enzymes (4). Our results strongly suggest an advantage in energy dissipation of low, rather than high, photosynthetic-yield pathways for minimization of the increased ATP levels (Fig. 1J). Consistently, CytBD (15), FQR (14), MEHLER (16, 22), and H2ase pathways (17) are only involved in energy dissipation under high-light or Ci-depleted conditions (CLS). Our analysis also identified that an optimal response to CLS required the additional participation of PHOTOR as a counterpart to low photosynthetic-yield AEF pathways, leading to energy dissipation by consuming the high ATP levels generated by these pathways (Fig. 3).
Although some microphototrophs, including Synechocystis, can grow heterotrophically, most of them are obligate photoautotrophs. Lesions in the central metabolism, such as an incomplete TCA cycle or the lack of organic carbon uptake systems, have been proposed as the primary causes of this obligate autotrophy (33). Thus, interesting questions arise as to why and how microphototrophs have maintained an incomplete TCA cycle and a reduced metabolic versatility even when being obligate autotrophs. We computationally showed here that the presence of the AKGDH, which may present an evolutionary advantage under heterotrophic conditions, hampered the photoautotrophic metabolism (Fig. 4). In addition, we showed that the heterotrophic metabolism of Synechocystis could be enhanced by the inclusion of nonnative transporters (SI Appendix, Table S5), suggesting missing evolutionary pressure to acquire or maintain organic carbon uptake systems. Interestingly, both the presence of the AKGDH and the inclusion of nonnative transporters have been suggested as negatively impacting the autotrophic metabolism (33–35). Our results suggest that Synechocystis is a highly specialized phototroph that has preserved and enhanced the photoautotrophic metabolism mainly by retaining or acquiring genes involved in controlling the light and Ci availability. In contrast, genes involved in the metabolism of organic carbon sources and/or potentially deleterious for autotrophic metabolism either have not been acquired or have been lost, contributing to low metabolic versatility and robustness. Thus, phototroph organisms are likely to require not only a high photosynthetic robustness but also an overall reduced metabolic robustness, avoiding reactions and pathways potentially deleterious for optimal autotrophic metabolism. The tradeoff between high photosynthetic robustness and low metabolic robustness suggested here must therefore be taken into account when designing efficient light-driven metabolic engineering processes.
Materials and Methods
A detailed description of methods, model setup, and in silico constraints used in this study can be found in SI Appendix. Briefly, the Synechocystis reconstruction was created according to published procedures (13, 36). Flux balance analysis (FBA) (37, 38) was used for model analysis. Essential and synthetic lethal essentiality analyses were performed according to the procedures described elsewhere (39). All computational simulations were performed with the COBRA toolbox (40) in the MATLAB environment (The MathWorks Inc.). The GNU Linear Programming Kit (GLPK) (http://www.gnu.org/software/glpk) was used to solve linear optimization problems.
Supplementary Material
Acknowledgments
We thank Ronan M. T. Fleming for valuable discussions, Marc Abrams for critical reading of the paper, and Almut Heinken for help in compiling the SBML file. This work was supported by the US Department of Energy, Offices of Advanced Scientific Computing Research and Biological and Environmental Research as part of the Scientific Discovery Through Advanced Computing program Grant DE-SC0002009. J.N. was funded, in part, by the Spanish Ministry of Education through the National Plan for Scientific Research, Development, and Technological Innovation 2008–2011.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117907109/-/DCSupplemental.
References
- 1.Liu X, Sheng J, Curtiss R., 3rd Fatty acid production in genetically modified cyanobacteria. Proc Natl Acad Sci USA. 2011;108:6899–6904. doi: 10.1073/pnas.1103014108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmidt BJ, Lin-Schmidt X, Chamberlin A, Salehi-Ashtiani K, Papin JA. Metabolic systems analysis to advance algal biotechnology. Biotechnol J. 2010;5:660–670. doi: 10.1002/biot.201000129. [DOI] [PubMed] [Google Scholar]
- 3.Blankenship RE, et al. Comparing photosynthetic and photovoltaic efficiencies and recognizing the potential for improvement. Science. 2011;332:805–809. doi: 10.1126/science.1200165. [DOI] [PubMed] [Google Scholar]
- 4.Kramer DM, Evans JR. The importance of energy balance in improving photosynthetic productivity. Plant Physiol. 2011;155:70–78. doi: 10.1104/pp.110.166652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen JF. Cyclic, pseudocyclic and noncyclic photophosphorylation: New links in the chain. Trends Plant Sci. 2003;8:15–19. doi: 10.1016/s1360-1385(02)00006-7. [DOI] [PubMed] [Google Scholar]
- 6.Peltier G, Tolleter D, Billon E, Cournac L. Auxiliary electron transport pathways in chloroplasts of microalgae. Photosynth Res. 2010;106:19–31. doi: 10.1007/s11120-010-9575-3. [DOI] [PubMed] [Google Scholar]
- 7.Oberhardt MA, Palsson BO, Papin JA. Applications of genome-scale metabolic reconstructions. Mol Syst Biol. 2009;5:320. doi: 10.1038/msb.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montagud A, Navarro E, Fernández de Córdoba P, Urchueguía JF, Patil KR. Reconstruction and analysis of genome-scale metabolic model of a photosynthetic bacterium. BMC Syst Biol. 2010;4:156. doi: 10.1186/1752-0509-4-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knoop H, Zilliges Y, Lockau W, Steuer R. The metabolic network of Synechocystis sp. PCC 6803: Systemic properties of autotrophic growth. Plant Physiol. 2010;154:410–422. doi: 10.1104/pp.110.157198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montagud A, et al. Flux coupling and transcriptional regulation within the metabolic network of the photosynthetic bacterium Synechocystis sp. PCC6803. Biotechnol J. 2011;6:330–342. doi: 10.1002/biot.201000109. [DOI] [PubMed] [Google Scholar]
- 11.Fu P. Genome-scale modeling of Synechocystis sp. PCC6803 and prediction of pathway insertion. J Chem Technol Biotechnol. 2009;84:473–483. [Google Scholar]
- 12.Shastri AA, Morgan JA. Flux balance analysis of photoautotrophic metabolism. Biotechnol Prog. 2005;21:1617–1626. doi: 10.1021/bp050246d. [DOI] [PubMed] [Google Scholar]
- 13.Thiele I, Palsson BO. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protoc. 2010;5:93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ooyabu J, Ohtsuka M, Kashino Y, Koike H, Satoh K. The expression pattern of NAD(P)H oxidases and the cyclic electron transport pathway around photosystem I of Synechocystis sp. PCC6803 depend on growth conditions. Biosci Biotechnol Biochem. 2008;72:3180–3188. doi: 10.1271/bbb.80370. [DOI] [PubMed] [Google Scholar]
- 15.Berry S, Schneider D, Vermaas WFJ, Rögner M. Electron transport routes in whole cells of Synechocystis sp. strain PCC 6803: The role of the cytochrome bd-type oxidase. Biochemistry. 2002;41:3422–3429. doi: 10.1021/bi011683d. [DOI] [PubMed] [Google Scholar]
- 16.Helman Y, et al. Genes encoding A-type flavoproteins are essential for photoreduction of O2 in cyanobacteria. Curr Biol. 2003;13:230–235. doi: 10.1016/s0960-9822(03)00046-0. [DOI] [PubMed] [Google Scholar]
- 17.Appel J, Phunpruch S, Steinmüller K, Schulz R. The bidirectional hydrogenase of Synechocystis sp. PCC 6803 works as an electron valve during photosynthesis. Arch Microbiol. 2000;173:333–338. doi: 10.1007/s002030000139. [DOI] [PubMed] [Google Scholar]
- 18.Eisenhut M, et al. The photorespiratory glycolate metabolism is essential for cyanobacteria and might have been conveyed endosymbiontically to plants. Proc Natl Acad Sci USA. 2008;105:17199–17204. doi: 10.1073/pnas.0807043105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooley JW, Vermaas WF. Succinate dehydrogenase and other respiratory pathways in thylakoid membranes of Synechocystis sp. strain PCC 6803: Capacity comparisons and physiological function. J Bacteriol. 2001;183:4251–4258. doi: 10.1128/JB.183.14.4251-4258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang C, Hua Q, Shimizu K. Metabolic flux analysis in Synechocystis using isotope distribution from 13C-labeled glucose. Metab Eng. 2002;4:202–216. doi: 10.1006/mben.2002.0226. [DOI] [PubMed] [Google Scholar]
- 21.Young JD, Shastri AA, Stephanopoulos G, Morgan JA. Mapping photoautotrophic metabolism with isotopically nonstationary 13C flux analysis. Metab Eng. 2011;13:656–665. doi: 10.1016/j.ymben.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hackenberg C, et al. Photorespiratory 2-phosphoglycolate metabolism and photoreduction of O2 cooperate in high-light acclimation of Synechocystis sp. strain PCC 6803. Planta. 2009;230:625–637. doi: 10.1007/s00425-009-0972-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noctor G, Foyer CH. A re-evaluation of the ATP:NADPH budget during C3 photosynthesis: A contribution from nitrate assimilation and its associated respiratory activity? J Exp Bot. 1998;49:1895–1908. [Google Scholar]
- 24.Skillman JB. Quantum yield variation across the three pathways of photosynthesis: Not yet out of the dark. J Exp Bot. 2008;59:1647–1661. doi: 10.1093/jxb/ern029. [DOI] [PubMed] [Google Scholar]
- 25.Vermaas WF, Shen G, Styring S. Electrons generated by photosystem II are utilized by an oxidase in the absence of photosystem I in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 1994;337:103–108. doi: 10.1016/0014-5793(94)80638-1. [DOI] [PubMed] [Google Scholar]
- 26.Gutthann F, Egert M, Marques A, Appel J. Inhibition of respiration and nitrate assimilation enhances photohydrogen evolution under low oxygen concentrations in Synechocystis sp. PCC 6803. Biochim Biophys Acta. 2007;1767:161–169. doi: 10.1016/j.bbabio.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 27.Helman Y, Barkan E, Eisenstadt D, Luz B, Kaplan A. Fractionation of the three stable oxygen isotopes by oxygen-producing and oxygen-consuming reactions in photosynthetic organisms. Plant Physiol. 2005;138:2292–2298. doi: 10.1104/pp.105.063768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feist AM, et al. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol Syst Biol. 2007;3:121. doi: 10.1038/msb4100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizusawa N, Sakurai I, Sato N, Wada H. Lack of digalactosyldiacylglycerol increases the sensitivity of Synechocystis sp. PCC 6803 to high light stress. FEBS Lett. 2009;583:718–722. doi: 10.1016/j.febslet.2009.01.021. [DOI] [PubMed] [Google Scholar]
- 30.Schäfer L, Vioque A, Sandmann G. Functional in situ evaluation of photosynthesis-protecting carotenoids in mutants of the cyanobacterium Synechocystis PCC6803. J Photochem Photobiol B. 2005;78:195–201. doi: 10.1016/j.jphotobiol.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 31.Kitano H. Biological robustness. Nat Rev Genet. 2004;5:826–837. doi: 10.1038/nrg1471. [DOI] [PubMed] [Google Scholar]
- 32.Cardol P, et al. An original adaptation of photosynthesis in the marine green alga Ostreococcus. Proc Natl Acad Sci USA. 2008;105:7881–7886. doi: 10.1073/pnas.0802762105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wood AP, Aurikko JP, Kelly DP. A challenge for 21st century molecular biology and biochemistry: What are the causes of obligate autotrophy and methanotrophy? FEMS Microbiol Rev. 2004;28:335–352. doi: 10.1016/j.femsre.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 34.Zhang C-C, Jeanjean R, Joset F. Obligate phototrophy in cyanobacteria: More than a lack of sugar transport. FEMS Microbiol Lett. 1998;161:285–292. doi: 10.1111/j.1574-6968.1998.tb12959.x. [DOI] [PubMed] [Google Scholar]
- 35.Vázquez-Bermúdez MF, Herrero A, Flores E. Uptake of 2-oxoglutarate in Synechococcus strains transformed with the Escherichia coli kgtP gene. J Bacteriol. 2000;182:211–215. doi: 10.1128/jb.182.1.211-215.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorleifsson SG, Thiele I. rBioNet: A COBRA toolbox extension for reconstructing high-quality biochemical networks. Bioinformatics. 2011;27:2009–2010. doi: 10.1093/bioinformatics/btr308. [DOI] [PubMed] [Google Scholar]
- 37.Varma A, Palsson BO. Metabolic flux balancing: Basic concepts, scientific and practical use. Nat Biotechnol. 1994;12:994–998. [Google Scholar]
- 38.Orth JD, Thiele I, Palsson BO. What is flux balance analysis? Nat Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pál C, et al. Chance and necessity in the evolution of minimal metabolic networks. Nature. 2006;440:667–670. doi: 10.1038/nature04568. [DOI] [PubMed] [Google Scholar]
- 40.Schellenberger J, et al. Quantitative prediction of cellular metabolism with constraint-based models: The COBRA Toolbox v2.0. Nat Protoc. 2011;6:1290–1307. doi: 10.1038/nprot.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.