Abstract
Classical nonhomologous DNA end-joining (C-NHEJ), which is a major DNA double-strand break (DSB) repair pathway in mammalian cells, plays a dominant role in joining DSBs during Ig heavy chain (IgH) class switch recombination (CSR) in activated B lymphocytes. However, in B cells deficient for one or more requisite C-NHEJ factors, such as DNA ligase 4 (Lig4) or XRCC4, end-joining during CSR occurs by a distinct alternative end-joining (A-EJ) pathway. A-EJ also has been implicated in joining DSBs found in oncogenic chromosomal translocations. DNA ligase 3 (Lig3) and its cofactor XRCC1 are widely considered to be requisite A-EJ factors, based on biochemical studies or extrachromosomal substrate end-joining studies. However, potential roles for these factors in A-EJ of endogenous chromosomal DSBs have not been tested. Here, we report that Xrcc1 inactivation via conditional gene-targeted deletion in WT or XRCC4-deficient primary B cells does not have an impact on either CSR or IgH/c-myc translocations in activated B lymphocytes. Indeed, homozygous deletion of Xrcc1 does not impair A-EJ of I-SceI–induced DSBs in XRCC4-deficient pro–B-cell lines. Correspondingly, substantial depletion of Lig3 in Lig4-deficient primary B cells or B-cell lines does not impair A-EJ of CSR-mediated DSBs or formation of IgH/c-myc translocations. Our findings firmly demonstrate that XRCC1 is not a requisite factor for A-EJ of chromosomal DSBs and raise the possibility that DNA ligase 1 (Lig1) may contribute more to A-EJ than previously considered.
Double-strand breaks (DSBs) can be caused by environmental factors (e.g., ionizing radiation, UV exposure), metabolic byproducts (free radicals), replication stress, and programmed gene rearrangements in developing lymphocytes (1, 2). In vertebrates, there are two major DSB repair pathways, namely, homologous recombination (HR) and classical nonhomologous DNA end-joining (C-NHEJ) (1). HR requires a long, intact DNA template to initiate repair (2). In contrast, C-NHEJ directly joins DNA ends without overlapping nucleotides as well as ends with very short stretches of complementary nucleotides, referred to as microhomologies (MHs) (1–3). During DSB repair by C-NHEJ, DSB recognition is provided by the Ku70/Ku80 complex and joining is mediated by the XRCC4/ligase 4 (Lig4) ligation complex (1). These four factors are evolutionarily conserved in their roles in C-NHEJ and considered to be core C-NHEJ factors. In the absence of core C-NHEJ factors, DSBs still can be repaired at reduced efficiencies by an alternative end-joining (A-EJ) process (2, 4). A-EJ was initially identified based on experiments that showed linear extrachromosomal plasmid substrates could recircularize in C-NHEJ–deficient cells (5, 6). Subsequently, A-EJ was implicated in generating recurrent oncogenic chromosomal translocations found in progenitor B-cell tumors from mice doubly deficient for XRCC4 or Lig4 and the p53 tumor suppressor (7, 8).
Ig heavy chain (IgH) class switch recombination (CSR) in activated B cells replaces the Cμ constant region exons with one of a series of sets of downstream exons (“CH genes”) that encode different IgH constant regions to carry out switching from IgM to a different IgH isotype, such as IgG1 or IgA. During CSR, DSBs in the donor switch region upstream of Cμ (Sμ) are joined to DSBs in a downstream acceptor S region, followed by the end-joining of the two broken S regions (9). In WT activated B cells, CSR DSBs are joined largely by C-NHEJ via either direct or short MH-mediated joins (10). However, in cells deficient in one or more of the core C-NHEJ factors, CSR breaks are joined at reduced levels, but still robustly, by an A-EJ process that is heavily biased toward MH-mediated joins (10, 11). A-EJ also repairs yeast endonuclease I-SceI–generated DSBs within chromosomally integrated substrates in Ku80- or XRCC4-deficient cell lines (12, 13). Although joins formed by A-EJ tend to be more biased toward MHs than those of C-NHEJ, both in frequency and in length of MHs (14), MH use is not an absolute criterion for A-EJ (2). For example, direct joins can comprise up to 20–50% of the total joins of CSR-associated or I-SceI–mediated chromosomal DSBs in Ku-deficient B cells or CHO cells, respectively (11–13).
The precise nature of A-EJ has been enigmatic. There may be more than one A-EJ pathway, and A-EJ pathways may potentially vary in the absence of particular C-NHEJ factors, with some even representing variant C-NHEJ pathways (1, 11). However, A-EJ operates even in the combined absence of C-NHEJ recognition (Ku70) and joining (Lig4) components, clearly demonstrating independence from C-NHEJ (11, 15). Thus, a working definition of A-EJ has been suggested to be any form of end-joining occurring in the absence of a core C-NHEJ factor (2). Factors that have been reported to function in chromosomal A-EJ in the context of CSR, V(D)J recombination, I-SceI substrates, and/or chromosomal translocations include Nbs1 (16), Mre11 (17–19), and CtIP (20, 21). These factors are thought to influence the choice between C-NHEJ and A-EJ pathways by mediating DNA end resection to uncover MHs and promote A-EJ. In addition, recent studies have indicated that Lig3 and, to lesser extent, Lig1 can mediate A-EJ associated with particular chromosomal translocations (22). Lig4 and its requisite cofactor XRCC4 are absolutely required for C-NHEJ during V(D)J recombination (2, 23). Therefore, A-EJ in the absence of Lig4 or XRCC4 might be expected to use one of the other cellular ligases, namely, Lig1 or Lig3 (2).
Lig1 is the replicative ligase and is involved in joining Okazaki fragments during lagging strand synthesis (24). Lig1 has also been implicated in long-patch base excision repair (BER) and nucleotide excision repair (25). The XRCC1/Lig3 complex operates in general short-patch BER and in single-strand break repair (26). Lig3 has both nuclear and mitochondrial isoforms (27), with the mitochondrial isoform, but not the nuclear isoform, operating independent of XRCC1 and being required for cell viability via maintenance of mtDNA integrity (28, 29). Both Lig3 and Lig1 have been implicated in A-EJ based on extrachromosomal plasmid reporter assays, biochemical studies (30–33), and assays using chromosomal endonuclease sites (22); all these studies concluded that Lig3 is a key ligase for A-EJ. The XRCC1 BER factor serves as a scaffold to recruit other BER factors, including nuclear Lig3, which it also stabilizes (25, 34). XRCC1 was described as a mammalian A-EJ factor, based on biochemical experiments (30). Whereas XRCC1 might be hypothesized to function in A-EJ via stabilizing Lig3, the XRCC1 plant ortholog has been proposed to be involved in A-EJ, although plants lack Lig3 (35). In this regard, XRCC1 also has nonoverlapping roles with Lig3 in nuclear DNA repair, because XRCC1, but not Lig3, depletion leads to sensitivity to DNA-alkylating agents (28, 29). Recently, studies of XRCC1 haploinsufficient B cells led to the conclusion that XRCC1 functions in A-EJ during CSR and formation of IgH/c-myc translocations (36).
To define potential roles of XRCC1 and Lig3 in A-EJ further, we have now assayed A-EJ in the context of CSR, joining of I-SceI DSBs, or chromosomal translocations in primary B cells and B-cell lines in which the functional Xrcc1 gene was eliminated by gene-targeted mutation or in which Lig3 protein was reduced to nearly undetectable levels by Lig3-specific shRNA.
Results
Conditional Inactivation of Xrcc1 in Activated B Cells Causes Genotoxic Stress Sensitivity.
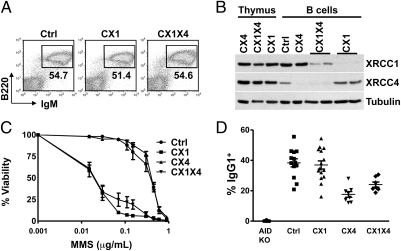
To investigate a potential role of XRCC1 in CSR in mice, we conditionally inactivated Xrcc1 alone or together with Xrcc4 specifically in mature B cells. Mice that conditionally delete Xrcc1 in B cells (“CX1 mice”) were generated by crossing mice carrying either two copies of a loxP-flanked (“floxed”) Xrcc1 allele (37) or one copy of a floxed Xrcc1 allele and one copy of an Xrcc1 null allele with CD21-Cre transgenic mice (38). For combined deletion of both Xrcc1 and Xrcc4 (“CX1X4”) in B cells, CX1 mice were crossed with conditional Xrcc4 KO mice (“CX4”; mice either carrying 2 floxed Xrcc4 alleles or 1 floxed Xrcc4 allele and 1 copy of an Xrcc4 null allele) (10). In comparison to controls (Ctrls; either WT mice or mice with floxed alleles without CD21-Cre, because they give indistinguishable results), CX1 and CX1X4 mice had similar fractions of splenic B cells (Fig. 1A). XRCC1 protein levels were generally below detection levels in mature CX1 B cells and reduced to low levels in CX1X4 B cells (Fig. 1B and Fig. S1 A and B). XRCC4 levels were dramatically reduced in both CX4 and CX1X4 activated B cells. Consistent with B cell-specific deletion, protein levels of these factors were normal in mutant and double-mutant thymocytes (Fig. 1B and Fig. S1A). XRCC1 is required for the stability of nuclear Lig3 (37). Conditional deletion of Xrcc1 caused a substantial reduction in Lig3 protein levels in CX1 and CX1X4 B cells; however, consistent with less complete XRCC1 inactivation, residual Lig3 levels were higher in the CX1X4 B-cell population (Fig. S1A). Lig1 protein levels were normal in both CX1 and CX1X4 B cells (Fig. S1B).
Fig. 1.
XRCC1 is dispensable for normal and A-EJ–mediated CSR to IgG1. (A) Splenic B cells from Ctrl, CX1, or CX1X4 mice were stained with IgM and B220 antibodies and analyzed by flow cytometry. (B) Deletion levels in CX1 and CX1X4 splenic B cells. Extracts from purified activated B cells or thymocytes from CX1 (n = 2), CX1X4 (n = 2), or Ctrl mice were immunoblotted with the indicated antibodies. (C) Elevated genotoxic stress sensitivity of mature CX1 and CX1X4 B cells activated with αCD40/IL-4 for 3 d. B cells from at least three mice per genotype were used for each MMS concentration. (D) Surface IgG1 expression of B cells stimulated with αCD40/IL-4 for 4 d. A total of 13 Ctrl, 15 CX1, 8 CX4, and 7 CX1X4 mice were analyzed in independent experiments. AID−/− (AID KO) mice were included as negative Ctrls. The results of IgG1 time course experiments are presented in Fig. S1D. Data are means ± SEM.
Cellular sensitivity to the alkylating agent methane methyl sulfonate (MMS) is a hallmark of XRCC1 deficiency (34, 37). To test whether CX1 or CX1X4 B cells are functionally deficient for XRCC1, we examined MMS sensitivity. Isolated mature B cells from either CX1 or CX1X4 spleens were stimulated with αCD40 plus IL-4 (αCD40/IL-4) for 1, 2, or 3 days, and MMS was added at the indicated concentrations for 6 h (Fig. 1C and Fig. S1C). Xrcc1 deletion rendered CX1 B cells highly sensitive to MMS (Fig. 1C and Fig. S1C), indicating that these cells are functionally XRCC1-deficient. In contrast, Xrcc4 deletion (CX4) did not render cells sensitive to MMS (Fig. 1C and Fig. S1C). In comparison to both Ctrl and CX4 B cells, combined Xrcc1 and Xrcc4 deletion (CX1X4) also rendered a large portion of the B cells sensitive to MMS (Fig. 1C and Fig. S1C). However, especially at early time points, CX1X4 B cells displayed a biphasic MMS sensitivity curve (Fig. 1C and Fig. S1C), consistent with the presence of B cells with either complete or incomplete Xrcc1 deletion. Analyses of the MMS curves suggested that 50–80% of the CX1X4 cells were fully MMS-sensitive at day 1, with the number increasing to 80% and 90%, respectively, at days 2 and 3 (Fig. 1C and Fig. S1C). We conclude that CD21-Cre–mediated Xrcc1 deletion is more efficient in B cells carrying only floxed Xrcc1 alleles than in B cells carrying both floxed Xrcc1 and Xrcc4 alleles. However, most CX1X4 B cells are functionally XRCC1-deficient by day 2 or day 3 of in vitro stimulation when CSR occurs.
Conditional Xrcc1 Inactivation in B Cells Does Not Impair C-NHEJ– or A-EJ–Mediated CSR or IgH/c-myc Translocation Formation.
To test the effect of Xrcc1 inactivation on end-joining in the context of CSR, we assayed CX1 or CX1X4 splenic B cells for surface IgG1 expression after in vitro stimulation with αCD40/IL-4. B cells from CX1, CX1X4, CX4, and Ctrl mice were analyzed in independent time course experiments at days 2, 3, and 4 of stimulation (Fig. 1D and Fig. S1D). Despite XRCC1 deficiency (Fig. 1B) and functional inactivation (Fig. 1C), CX1 B cells did not display significant IgG1 CSR defects at early or late time points (Fig. 1D and Fig. S1D). In contrast, CX4 B cells showed reduced IgG1 CSR levels (Fig. 1D and Fig. S1D), as previously demonstrated (10). CX1X4 B cells underwent CSR to IgG1 at levels similar to CX4 cells, at both early and late time points (Fig. 1D and Fig. S1D), indicating that Xrcc1 inactivation in a large fraction of the B-cell population did not affect A-EJ–mediated CSR. We also stimulated CX1 and CX1X4 B cells with bacterial LPS plus αIgD-dextran to induce CSR to IgG3 and obtained similar results as for switching to IgG1 (Fig. S1E).
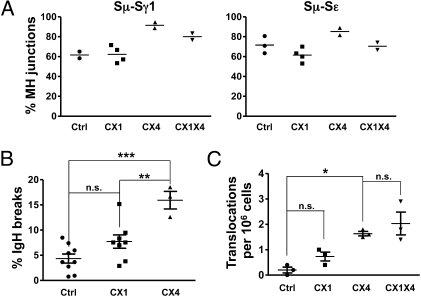
To address a potential role of XRCC1 in CSR and A-EJ further, we analyzed CSR junction sequences. In WT B cells, up to half of the S region joins are direct and the rest are mediated by short, ≤4-nt MHs (10). In CX4 B cells, nearly 90% of junctions were MH-mediated as described (10). On the other hand, Sμ-Sγ1 and Sμ-Sε junctions from αCD40/IL-4–activated CX1 B cells showed similar levels of direct and MH-mediated junctions as Ctrl cells (Fig. 2A and Table S1). CX1X4 Sμ-Sγ1 and Sμ-Sε junctions also had increased MHs compared with Ctrl or CX1 junctions but not to the extent of CX4 junctions (Fig. 2A and Table S1). The latter finding is consistent with the presence of a small population of B cells in CX1X4 mice that did not undergo Cre-mediated deletion of Xrcc4.
Fig. 2.
XRCC1 deficiency does not affect switch region junctions, IgH genomic instability, or IgH/c-myc translocations. (A) Levels of MH-mediated Sμ-Sγ1 joins (Left) and Sμ-Sε joins (Right) in Ctrl, CX1, CX4, and CX1X4 activated B cells. S region junction data are shown in detail in Table S1. (B) CX1 B cells do not display altered levels of IgH locus breaks. The percentage of unrepaired IgH locus breaks in activated Ctrl, CX1, and CX4 B cells is shown. Data are means ± SEM. A summary of all FISH data is included in Table S2. (C) XRCC1 is dispensable for IgH/c-myc translocations in CX1 and CX1X4 B cells. IgH/c-myc translocations were amplified from B cells cultured with αCD40/IL-4 for 4 d. Data are means ± SEM. One-way ANOVA analysis with a Tukey posttest was used for calculation of statistical significance. n.s., not significant; *P < 0.05; **P < 0.001; ***P < 0.0001.
Absence of DNA repair factors, such as Artemis, causes activation-induced cytidine deaminase (AID)-dependent IgH locus breaks in B cells activated for CSR without markedly diminishing overall CSR levels (39). Thus, we tested whether B cell-specific Xrcc1 deletion leads to unrepaired AID-mediated IgH breaks or translocations. For this purpose, we used two sensitive DSB repair defect assays, namely, metaphase FISH (39) and PCR-mediated amplification of IgH/c-myc translocations (40). Compared with Ctrl, CX1 B cells did not show significant differences in frequency of IgH breaks (Fig. 2B and Table S2), in general translocations involving the IgH locus (Table S2), or in IgH/c-myc translocations (Fig. 2C). In contrast, CX4 B cells, as expected (10, 41), had elevated IgH breaks, IgH translocations, and IgH/c-myc translocations (Fig. 2 B and C and Table S2). Although FISH data were not available for CX1X4 B cells, these cells showed a similar frequency of IgH/c-myc translocations in comparison to CX4 cells (Fig. 2C). Because the majority of CX1X4 B cells are functionally inactivated for XRCC1 by day 2 of activation (Fig. S1C), the finding that CX1X4 B cells have similar IgH/c-myc translocation levels as CX4 B cells indicates that XRCC1 is not required for A-EJ in this context.
Xrcc1 Deletion Does Not Have an Impact on Joining of I-SceI Chromosomal DSB Substrates.
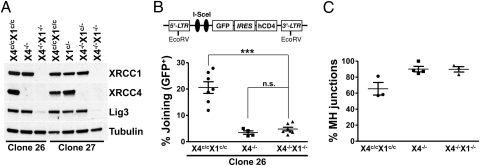
Our studies of CX1 and CX1X4 B cells indicate that XRCC1-independent A-EJ mechanisms can mediate CSR and chromosomal translocations. However, because residual XRCC1 protein remains in a subset of CX1X4 B cells (Fig. 1B and Fig. S1 A and B), we sought to confirm this observation by a different approach. For this purpose, we generated Abelson murine leukemia virus (v-Abl)–transformed pro–B-cell lines (hereafter referred to as v-Abl–transformed pro-B lines) from progenitor B cells derived from bone marrow of mice carrying conditional Xrcc4 and Xrcc1 KO alleles (X4c/cX1c/c) and introduced a single retroviral end-joining reporter construct containing two I-SceI target sites. Joining of the two I-SceI sites eliminates an out-of-frame translation start site and results in expression of GFP (19). Following retroviral transduction, clones with unique genomic integration sites of the reporter substrate were identified by Southern blotting and three different X4c/cX1c/c reporter lines (clones 26, 27, and 150), each with a unique single-copy substrate integration, were treated with recombinant Tat-Cre protein to generate subclones that were either null for XRCC4 (X4−/−) or double-deficient for both XRCC1 and XRCC4 (X4−/−X1−/−). Absence of XRCC4 protein or of both XRCC1 and XRCC4 proteins in the respective lines was confirmed by Western blot analysis (Fig. 3A and Fig. S2A). Ablation of XRCC1 protein also resulted in a >80% reduction in total cellular Lig3 protein in X4−/−X1−/− cells (Fig. 3A and Fig. S2B), consistent with prior findings showing that XRCC1 stabilizes Lig3 (25, 34).
Fig. 3.
Xrcc4−/−Xrcc1−/−-transformed pro-B lines do not display XRCC1-dependent end-joining defects. (A) Western blot analysis revealed complete absence of XRCC1 and XRCC4 proteins in subclones from two independent v-Abl–transformed pro-B lines. Lig3 levels are dramatically reduced in XRCC1 null cells. (B) (Upper) Illustration of I-SceI chromosomal reporter. (Lower) Quantification of I-SceI chromosomal joining for X4c/cX1c/c (n = 7), X4−/− (n = 4), and X4−/−X1−/− (n = 7) v-Abl clonal lines with single-copy I-SceI chromosomal substrates integrated in the same location (clone 26). Data are means ± SEM. One-way ANOVA analysis with a Tukey posttest was used for calculation of statistical significance. n.s., not significant; ***P < 0.0001. Tables S3 and S4 list details on I-SceI joining experiments. (C) Xrcc1 deletion does not affect the nature of I-SceI junctions in XRCC4 null cells. I-SceI junctions were amplified from v-Abl cells of the indicated genotypes. Data are means ± SEM.
To test DSB repair in X4−/−X1−/− v-Abl–transformed pro-B lines, we transduced them with retroviral vectors encoding an I-SceI–glucocorticoid receptor (GR) fusion protein and subsequently induced nuclear translocation of the I-SceI–GR fusion protein by treatment of cells with triamcinolone acetonide (42). Average joining levels, based on the GFP reporter, were 20.6 ± 2.2% for the X4c/cX1c/c clone 26 line, whereas in X4−/− and X4−/−X1−/− clone 26 sublines, joining levels decreased to 3.5 ± 0.7% and 4.8 ± 0.7%, respectively, which were not significantly different (Fig. 3B, Fig. S3, and Table S3). We obtained similar results for two additional lines with independent integration sites (Table S4). Therefore, abrogation of XRCC1 in XRCC4-deficient lines did not have a measurable impact on A-EJ activity.
To test whether XRCC1 loss affects the use of MH-mediated vs. direct joins in XRCC4 null cells, we PCR-amplified I-SceI junctions from X4−/−X1−/−, X4−/−, and X4c/cX1c/c v-Abl transformants. We used primers flanking the I-SceI sites (located 650 bp apart) to allow for amplification of joins with relatively large resections. Because the two I-SceI sites in the reporter construct are in opposite orientations, joining involves DNA ends with noncomplementary 4-nt overhangs. We analyzed 237 independent X4c/cX1c/c junctions, excluding junctions with short (<10 bp) insertions, which could be mediated by terminal deoxynucleotidyl transferase (43), and found ∼34% direct junctions vs. 66% MH-mediated junctions (Fig. 3C). For X4−/− cells, we observed that the frequency of MH-mediated joins increased to ∼90%, with only about 10% of the junctions being direct (Fig. 3C). We analyzed 119 junctions from X4−/−X1−/− cells and found levels of MH-mediated vs. direct joins (∼90% vs. 10%, respectively) similar to those of X4−/− cells (Fig. 3C). These findings indicate that XRCC1 is not required to achieve the MH-mediated A-EJ repair of DSBs observed in XRCC4-deficient B cells.
Lig3 Inactivation in WT or Lig4-Deficient B-Lineage Cells Does Not Have a Measurable Impact on CSR or Translocations.
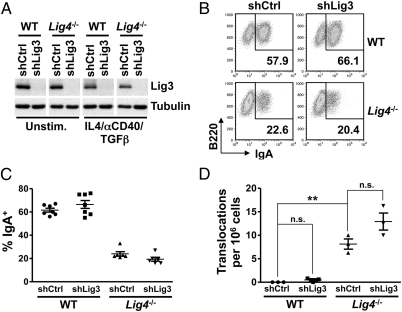
XRCC1 and Lig3 are often considered to function together to mediate A-EJ (23, 31, 44–47). Conditional Lig3 inactivation in primary B cells gave results very similar to those we described above for conditional Xrcc1 inactivation (Fig. S4 and Table S1), but we had no functional assay for Lig3 ablation. We also tried to inactivate Lig3 conditionally alone or in combination with Lig4 in v-Abl–transformed pro-B lines carrying conditional Lig3 KO alleles (Lig3c/c) or both conditional Lig3 and Lig4 KO alleles (Lig3c/cLig4c/c) by Tat-Cre treatment. Screening of 412 Tat-Cre–treated Lig3c/c subclones and ∼600 Lig3c/cLig4c/c subclones did not yield any Lig3 null subclones, indicating that total Lig3 inactivation is cell-lethal. Thus, to address a potential role of Lig3 in chromosomal A-EJ further, we assayed IgA switching in WT and Lig4−/− CH12F3 cells transduced with lentiviruses expressing either Lig3-specific or scrambled Ctrl shRNAs. After 3 d of stimulation with αCD40/IL-4/TGF-β, ∼60% of WT (transduced with a scrambled Ctrl shRNA) and Lig3-depleted WT CH12F3 cells had switched to IgA as evidenced by surface-staining flow cytometry (Fig. 4 B and C), despite depletion of cellular Lig3 in both by more than 90% (Fig. 4A and Fig. S5A). Like Lig4- or XRCC4-deficient primary B cells, 3-d stimulated Lig4−/− CH12F3 cells infected with a Ctrl shRNA undergo CSR via A-EJ at about 30–50% of WT levels (10, 48) (Fig. 4 B and C). Notably, Lig4−/− CH12F3 cells expressing Lig3 shRNA also still underwent CSR at similar levels to the Ctrl shRNA-expressing Lig4−/− CH12F3 cells based on accumulation of IgA+ cells (Fig. 4 B and C), despite depletion of Lig3 levels by more than 90% (Fig. 4A and Fig. S5A). We did note, however, ∼40% lower median fluorescence intensity of the IgA+ fraction of Lig4−/− shLig3 cells in comparison to that of Lig4−/− shCtrl cells (Fig. 4B). In this regard, we found similar relative levels of IgA-switched Ctrl and Lig3-depleted Lig4−/− CH12F3 cells at days 2 and 3 of stimulation, indicating the absence of a kinetic delay in switching in the latter (Fig. 4C and Fig. S5B). Thus, the decreased surface IgA expression most likely reflects general adverse effects of Lig3 depletion in Lig4−/− CH12F3 cells, as also reflected by slightly lower cell viability of Lig3-depleted vs. Ctrl Lig4−/− CH12F3 cells [mean ± SEM (%): 34.2 ± 3.8 vs. 45.8 ± 2.3; P = 0.025, unpaired two-tailed t test].
Fig. 4.
shRNA-mediated depletion of Lig3 in B-cell lines does not affect CSR or frequency of IgH/c-myc translocations. (A) Lig3 protein levels in CH12F3 cells of the indicated genotypes expressing either Ctrl or Lig3 shRNA. Blots were stripped and probed for tubulin as a loading Ctrl. Unstim., unstimulated. (B) Representative example of flow cytometry results for IgA expression of WT or Lig4−/− CH12F3 cells stably expressing either Ctrl or Lig3 shRNA on day 3 of stimulation with IL-4/αCD40/TGF-β. Cultures propagated without cytokines and analyzed in parallel contained <1% IgA+ cells. (C) Summary of CSR experiments in CH12F3 B-cell lines. Means ± SEM from seven (WT shCtrl and WT shLig3) or six (Lig4−/− shCtrl and Lig4−/− shLig3) independent experiments are shown. (D) Lig3 depletion does not affect the frequency of IgH/c-myc translocations. Translocations were analyzed in CH12F3 cells stimulated for 3 d with IL-4/αCD40/TGF-β. Results from three independent knockdown experiments using WT shCtrl, WT shLig3, Lig4−/− shCtrl, or Lig4−/− shLig3 CH12F3 cells are shown. One-way ANOVA analysis with a Tukey posttest was used to assess statistical significance. Data are means ± SEM. n.s., not significant; **P < 0.01.
Lig4−/− CH12F3 cells transduced with either Ctrl or Lig3 shRNA showed similar levels of direct and MH-mediated Sμ-Sα junctions (Table S5). Together, these findings suggest that substantial Lig3 depletion does not measurably influence the level or quality of A-EJ–mediated CSR in CH12F3 cells. To assess a potential role of Lig3 in chromosomal end-joining further, we analyzed levels of IgH/c-myc translocations in stimulated WT or Lig4−/− CH12F3 cells stably expressing either scrambled Ctrl or Lig3 shRNA (Fig. 4D). Deletion of Lig4 significantly increased the frequency of IgH/c-myc translocations in CH12F3 cells (Fig. 4D), consistent with findings in primary B cells (11, 15). In contrast, Lig3 knockdown did not detectably alter IgH/c-myc translocation frequency in WT CH12F3 cells (Fig. 4D). Moreover, Lig3 depletion did not reduce the frequency of IgH/c-myc translocations in the absence of Lig4 (Fig. 4D).
Discussion
In mammalian cells, A-EJ has been shown to mediate both intrachromosomal joining for CSR (10) and interchromosomal translocation junctions (8, 22). XRCC1, a Lig3-stabilizing cofactor, and Lig3 itself have been very widely assumed to provide major end-ligation functions in A-EJ (22, 23, 31, 36, 44–47). We have now unequivocally established that XRCC1 is not required for A-EJ repair of I-SceI DSBs in XRCC4-deficient pro-B lines. We also found that conditional depletion of XRCC1 in XRCC4-deficient primary B cells, of which the vast majority were functionally XRCC1-deficient based on MMS sensitivity, led to no measurable impact on the formation of CSR junctions by A-EJ and did not alter the abundant use of MHs for their formation. Conditional depletion of XRCC1 in XRCC4-deficient primary B cells also had no apparent effect on the frequency of IgH/c-Myc translocations. We conclude that XRCC1 is not a requisite factor for major pathways of chromosomal A-EJ.
XRCC1 was previously implicated in A-EJ based on plasmid assays and biochemical experiments (30, 31, 33). Our current finding of no requisite role for XRCC1 in A-EJ might be rationalized with these earlier results if joining of DSBs in transient DNA substrates has different requirements than joining of chromosomal DSBs. In this context, cells lacking the C-NHEJ factor XLF have a V(D)J defect in extrachromosomal V(D)J recombination substrate assays but not for chromosomally integrated V(D)J recombination substrates or for V(D)J recombination of endogenous Ig and T-cell receptor loci (49, 50). A recent study found normal CSR in LPS-stimulated Xrcc1+/− splenic B cells but noted a reduction in MH length at Sμ-Sγ3 switch junctions and also found reduced levels of IgH/c-myc translocations in Xrcc1+/− B cells, leading to the conclusion that XRCC1 plays a role in A-EJ in both processes (36). However, although we did not analyze Sμ-Sγ3 junctions, we did not find altered Sμ-Sγ1 or Sμ-Sε junctions in XRCC1-deficient B cells, where XRCC1 protein levels were reduced to almost undetectable levels. Likewise, we found no reduction in translocations upon conditional Xrcc1 inactivation in XRCC4-proficient or -deficient B cells. Another recent study reported that nuclear Lig3 is the primary ligase for translocation formation in the context of zinc finger endonuclease-mediated chromosomal DSBs in mouse ES cells (22). In our experiments, depletion of Lig3 to extremely low levels did not affect CSR, CSR junctions, or the frequency of IgH/c-myc translocations. However, because complete Lig3 ablation appears cell-lethal, we cannot unequivocally rule out the possibility that very low residual Lig3 levels contribute to the A-EJ we observe in Lig4-deficient cells.
Overall, our findings demonstrate that XRCC1 is not required for major known forms of A-EJ, and thus cannot be considered a chromosomal A-EJ factor at this time. Our results also rule out a requisite role for the XRCC1/Lig3 complex in A-EJ and raise the further possibility that there may be significant A-EJ mechanisms that function independent of both Lig3 and Lig4. Because eukaryotic cells contain only three known enzymes that ligate DNA ends (Lig1, Lig3, and Lig4) (25), the major candidate for this role would be Lig1, which has already been suggested to play a role in A-EJ, albeit a more modest role than Lig3 (22, 51). Future experiments will need to address a direct role for Lig1 in A-EJ in Lig3/4-deficient B lymphocytes, as well as the relative contributions of Lig1 and Lig3 to A-EJ in XRCC4/Lig4-deficient and WT B cells.
Materials and Methods
Mice.
All experiments involving mice were performed according to protocols approved by the Institutional Animal Care Facility of Children's Hospital Boston. Xrcc1c/c, Lig3c/c, Xrcc4c/c, and CD21CreLig4c/c (CL4) mice were reported (15, 28, 37).
B-Cell Purification and Culture.
Splenic B cells were isolated and stimulated as described elsewhere (52).
Generation of v-Abl–Transformed Pro-B Lines.
The v-Abl kinase-transformed pro-B cells were generated (53) and used as described in SI Materials and Methods.
CH12F3 Cell Culture and shRNA-Mediated Knockdown.
Details on WT and Lig4−/− (48) CH12F3 cell culture and shRNA experiments are provided in SI Materials and Methods.
Switch Region Junction Analysis.
Sμ-Sγ1 and Sμ-Sε junctions were analyzed as described previously (10). Conditions for Sμ-Sα junction analysis in CH12F3 cells are described in SI Materials and Methods.
Two-Color IgH FISH and IgH/c-myc Translocations.
Metaphases were prepared and processed as described elsewhere (39). All FISH samples were analyzed in a blinded manner. IgH/c-myc translocation analysis in primary B cells was performed as described elsewhere (40). Details on IgH/c-myc translocation analysis in CH12F3 cells are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Maria-Vivienne Boboila for stimulating discussions and critical reading of the manuscript; Erica Hansen, Grace Yuen, and Duane Wesemann for help with experiments and suggestions; Ralph Scully for providing the I-SceI vectors; and Kefei Yu for providing the Lig4 null CH12F3 cells. This work was supported by National Institutes of Health Grants AI031541 and CA092625 (to F.W.A.), NS-37956 and CA-21765 (to P.J.M.), and AI037526 (to M.C.N.). B.S. was supported by National Institutes of Health Training Grant 5T32CA009382. J.H.W. was supported by a Leukemia and Lymphoma Society of America Special Fellowship and by National Institutes of Health Training Grant 5T32CA009382-26. C.B. was supported by a Cancer Research Institute training grant. Y.Z. was supported by a Cancer Research Institute postdoctoral fellowship. M.G. is a V Foundation Scholar. F.W.A. and M.C.N. are investigators of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1121470109/-/DCSupplemental.
References
- 1.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y, et al. The role of mechanistic factors in promoting chromosomal translocations found in lymphoid and other cancers. Adv Immunol. 2010;106:93–133. doi: 10.1016/S0065-2776(10)06004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haber JE. Alternative endings. Proc Natl Acad Sci USA. 2008;105:405–406. doi: 10.1073/pnas.0711334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zha S, Boboila C, Alt FW. Mre11: Roles in DNA repair beyond homologous recombination. Nat Struct Mol Biol. 2009;16:798–800. doi: 10.1038/nsmb0809-798. [DOI] [PubMed] [Google Scholar]
- 5.Boulton SJ, Jackson SP. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- 6.Kabotyanski EB, Gomelsky L, Han JO, Stamato TD, Roth DB. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res. 1998;26:5333–5342. doi: 10.1093/nar/26.23.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roth DB. Amplifying mechanisms of lymphomagenesis. Mol Cell. 2002;10:1–2. doi: 10.1016/s1097-2765(02)00573-7. [DOI] [PubMed] [Google Scholar]
- 8.Zhu C, et al. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
- 9.Chaudhuri J, et al. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv Immunol. 2007;94:157–214. doi: 10.1016/S0065-2776(06)94006-1. [DOI] [PubMed] [Google Scholar]
- 10.Yan CT, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 11.Boboila C, et al. Alternative end-joining catalyzes class switch recombination in the absence of both Ku70 and DNA ligase 4. J Exp Med. 2010;207:417–427. doi: 10.1084/jem.20092449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guirouilh-Barbat J, et al. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol Cell. 2004;14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 13.Guirouilh-Barbat J, Rass E, Plo I, Bertrand P, Lopez BS. Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends. Proc Natl Acad Sci USA. 2007;104:20902–20907. doi: 10.1073/pnas.0708541104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McVey M, Lee SE. MMEJ repair of double-strand breaks (director's cut): Deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boboila C, et al. Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proc Natl Acad Sci USA. 2010;107:3034–3039. doi: 10.1073/pnas.0915067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deriano L, Stracker TH, Baker A, Petrini JHJ, Roth DB. Roles for NBS1 in alternative nonhomologous end-joining of V(D)J recombination intermediates. Mol Cell. 2009;34:13–25. doi: 10.1016/j.molcel.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinkelmann M, et al. Multiple functions of MRN in end-joining pathways during isotype class switching. Nat Struct Mol Biol. 2009;16:808–813. doi: 10.1038/nsmb.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rass E, et al. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol. 2009;16:819–824. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 19.Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol. 2009;16:814–818. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee-Theilen M, Matthews AJ, Kelly D, Zheng S, Chaudhuri J. CtIP promotes microhomology-mediated alternative end joining during class-switch recombination. Nat Struct Mol Biol. 2011;18:75–79. doi: 10.1038/nsmb.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Jasin M. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat Struct Mol Biol. 2011;18:80–84. doi: 10.1038/nsmb.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simsek D, et al. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 2011;7:e1002080. doi: 10.1371/journal.pgen.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gostissa M, Alt FW, Chiarle R. Mechanisms that promote and suppress chromosomal translocations in lymphocytes. Annu Rev Immunol. 2011;29:319–350. doi: 10.1146/annurev-immunol-031210-101329. [DOI] [PubMed] [Google Scholar]
- 24.Tomkinson AE, Vijayakumar S, Pascal JM, Ellenberger T. DNA ligases: Structure, reaction mechanism, and function. Chem Rev. 2006;106:687–699. doi: 10.1021/cr040498d. [DOI] [PubMed] [Google Scholar]
- 25.Ellenberger T, Tomkinson AE. Eukaryotic DNA ligases: Structural and functional insights. Annu Rev Biochem. 2008;77:313–338. doi: 10.1146/annurev.biochem.77.061306.123941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair (Amst) 2003;2:955–969. doi: 10.1016/s1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- 27.Lakshmipathy U, Campbell C. The human DNA ligase III gene encodes nuclear and mitochondrial proteins. Mol Cell Biol. 1999;19:3869–3876. doi: 10.1128/mcb.19.5.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao Y, et al. DNA ligase III is critical for mtDNA integrity but not Xrcc1-mediated nuclear DNA repair. Nature. 2011;471:240–244. doi: 10.1038/nature09773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simsek D, et al. Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature. 2011;471:245–248. doi: 10.1038/nature09794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 31.Della-Maria J, et al. Human Mre11/human Rad50/Nbs1 and DNA ligase IIIalpha/XRCC1 protein complexes act together in an alternative nonhomologous end joining pathway. J Biol Chem. 2011;286:33845–33853. doi: 10.1074/jbc.M111.274159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang L, et al. Human DNA ligases I and III, but not ligase IV, are required for microhomology-mediated end joining of DNA double-strand breaks. Nucleic Acids Res. 2008;36:3297–3310. doi: 10.1093/nar/gkn184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H, et al. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 2005;65:4020–4030. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- 34.Tebbs RS, et al. Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Dev Biol. 1999;208:513–529. doi: 10.1006/dbio.1999.9232. [DOI] [PubMed] [Google Scholar]
- 35.Charbonnel C, Gallego ME, White CI. Xrcc1-dependent and Ku-dependent DNA double-strand break repair kinetics in Arabidopsis plants. Plant J. 2010;64:280–290. doi: 10.1111/j.1365-313X.2010.04331.x. [DOI] [PubMed] [Google Scholar]
- 36.Saribasak H, et al. XRCC1 suppresses somatic hypermutation and promotes alternative nonhomologous end joining in Igh genes. J Exp Med. 2011;208:2209–2216. doi: 10.1084/jem.20111135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee Y, et al. The genesis of cerebellar interneurons and the prevention of neural DNA damage require XRCC1. Nat Neurosci. 2009;12:973–980. doi: 10.1038/nn.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kraus M, Alimzhanov MB, Rajewsky N, Rajewsky K. Survival of resting mature B lymphocytes depends on BCR signaling via the Igalpha/beta heterodimer. Cell. 2004;117:787–800. doi: 10.1016/j.cell.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 39.Franco S, et al. DNA-PKcs and Artemis function in the end-joining phase of immunoglobulin heavy chain class switch recombination. J Exp Med. 2008;205:557–564. doi: 10.1084/jem.20080044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramiro AR, et al. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 41.Wang JH, et al. Mechanisms promoting translocations in editing and switching peripheral B cells. Nature. 2009;460:231–236. doi: 10.1038/nature08159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiarle R, et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147:107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Desiderio SV, et al. Insertion of N regions into heavy-chain genes is correlated with expression of terminal deoxytransferase in B cells. Nature. 1984;311:752–755. doi: 10.1038/311752a0. [DOI] [PubMed] [Google Scholar]
- 44.Ciccia A, Elledge SJ. The DNA damage response: Making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mladenov E, Iliakis G. Induction and repair of DNA double strand breaks: The increasing spectrum of non-homologous end joining pathways. Mutat Res. 2011;711(1-2):61–72. doi: 10.1016/j.mrfmmm.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 46.Neal JA, Meek K. Choosing the right path: Does DNA-PK help make the decision? Mutat Res. 2011;711(1-2):73–86. doi: 10.1016/j.mrfmmm.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stavnezer J, Björkman A, Du L, Cagigi A, Pan-Hammarström Q. Mapping of switch recombination junctions, a tool for studying DNA repair pathways during immunoglobulin class switching. Adv Immunol. 2010;108:45–109. doi: 10.1016/B978-0-12-380995-7.00003-3. [DOI] [PubMed] [Google Scholar]
- 48.Han L, Yu K. Altered kinetics of nonhomologous end joining and class switch recombination in ligase IV-deficient B cells. J Exp Med. 2008;205:2745–2753. doi: 10.1084/jem.20081623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li G, et al. Lymphocyte-specific compensation for XLF/cernunnos end-joining functions in V(D)J recombination. Mol Cell. 2008;31:631–640. doi: 10.1016/j.molcel.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zha S, Alt FW, Cheng HL, Brush JW, Li G. Defective DNA repair and increased genomic instability in Cernunnos-XLF-deficient murine ES cells. Proc Natl Acad Sci USA. 2007;104:4518–4523. doi: 10.1073/pnas.0611734104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simsek D, Jasin M. DNA ligase III: A spotty presence in eukaryotes, but an essential function where tested. Cell Cycle. 2011;10:3636–3644. doi: 10.4161/cc.10.21.18094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng HL, et al. Integrity of the AID serine-38 phosphorylation site is critical for class switch recombination and somatic hypermutation in mice. Proc Natl Acad Sci USA. 2009;106:2717–2722. doi: 10.1073/pnas.0812304106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bredemeyer AL, et al. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 2006;442:466–470. doi: 10.1038/nature04866. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.