Abstract
Processing of microRNA (miRNA) precursors results in the release of a double-stranded miRNA/miRNA* duplex. The miRNA or guide strand, is loaded onto the Argonaute (AGO) effector, and the miRNA* or passenger strand is typically degraded. The loaded AGO-containing RNA-induced silencing complex specifically recognizes a target mRNA, leading to its degradation or translational inhibition. In plants, miRNA-mediated cleavage of a target triggers in some cases the production of secondary small interfering RNAs (siRNAs), which in turn can silence other genes in trans. This alternative pathway depends on the length of the miRNA and the specific AGO in the effector complex. However, 22-nt miRNAs are sufficient, but not essential for this pathway. Using a combination of computational and experimental approaches, we show that transitivity can be triggered when the small RNA that is not retained in AGO is 22-nt long. Moreover, we demonstrate that asymmetrically positioned bulged bases in the miRNA:miRNA* duplex, regardless of miRNA or miRNA* length, are sufficient for the initiation of transitivity. We propose that the RNA-induced silencing complex reprogramming occurs during the early steps of miRNA loading, before the miRNA duplex is disassembled and the guide strand is selected.
Keywords: Arabidopsis thaliana, RNAi, gene silencing, tasiRNA, phasiRNA
MicroRNAs (miRNAs), which are typically 21 nt in length, posttranscriptionally regulate gene expression. In plants, Dicer-like 1 (DCL1), together with accessory proteins Hyponastic Leaves 1 (HYL1) and Serrate (SE), processes longer primary transcripts into miRNA/miRNA* duplexes with a 2-nt 3′ overhang (1). The duplexes are 2′-O-methylated at their 3′ ends by HUA Enhancer 1, which prevents their degradation, and exported out of the nucleus (2, 3). The mature miRNA duplexes are then sorted and loaded into one of the 10 Argonaute (AGO) proteins. In contrast to other small RNAs, miRNAs are preferentially loaded into AGO1, mainly based on recognition of a 5′ uracil (4, 5). During maturation of the AGO-containing RNA-induced silencing complex (RISC), the miRNA strand with the less thermodynamically stable 5′ end is retained, and the other is removed or cleaved by AGO. This process leads to different steady-state levels of the single-stranded components of the original duplex. Strand sorting depends on the AGO complex onto which it is loaded (6), an example being the HYL1-assisted mechanism of AGO1 loading (7). The AGO-retained strand, also called the guide strand, is often more abundant and is typically the miRNA; the less abundant passenger strand is generally the miRNA*. Nevertheless, even the passenger strand can be retained in the AGO1 complex or sorted into an alternative AGO complex (8, 9).
Once loaded, RISC is guided by sequence complementarity to a target mRNA, leading to target cleavage and degradation or to repression of its translation (1). In some instances, miRNA-mediated cleavage of an RNA can trigger the production of secondary siRNAs, a phenomenon called transitivity (10, 11). miRNA-dependent transacting siRNAs (tasiRNAs), also known as phased siRNAs, are generated from noncoding TAS as well as protein-coding transcripts, and the secondary siRNAs can silence additional genes (12–16). This amplification mechanism allows miRNAs to regulate large gene families, such as PPR genes in Arabidopsis thaliana and NB-LRR genes in other dicots, through a limited number of primary targets (12, 17).
After miRNA-guided target cleavage, the next step during transitivity is the conversion of the cleaved target transcript into double-stranded RNA (dsRNA) by the specialized RNA-dependent RNA polymerase 6 (RDR6) and cofactors, such as Suppressor of Gene Silencing 3 (SGS3). The newly synthesized dsRNA molecules are then further processed by DCL4 into tasiRNAs. What is not yet completely understood is why only some miRNAs trigger transitivity. MiR390 is loaded onto AGO7/ZIPPY (ZIP), and interaction with two separate miRNA recognition sites on the TAS3 transcript is required for tasiRNA formation (5, 18). In contrast, miR173 and miR828, which are loaded onto AGO1, can trigger tasiRNA formation from a single target site (5, 19). Both miRNAs differ from the majority of other miRNAs by being 22 nt instead of 21-nt long (20, 21). It has been proposed that the larger size of these miRNAs promotes the reprogramming of AGO1-containing RISC, such that RDR6 and SGS3 are recruited, which convert the targets into dsRNA. However, there are also cases where AGO1-loaded 21-nt miRNAs trigger secondary siRNA formation.
Here, we have used a combination of computational and experimental approaches to identify siRNA-producing mRNAs targeted by miRNAs. We demonstrate the length of the miRNA* strand, which is not part of active RISC, can influence miRNA-triggered transitivity. Thus, either a miRNA or mRNA* of 22 nt in length is sufficient for the initiation of transitivity. However, our results suggest that it is not the length per se, but asymmetry in the duplex structure that triggers transitivity. Together, our observations suggest that secondary siRNA production is a consequence of early reprogramming of the immature AGO1-RISC. This modification of the RISC, which results in the recruitment of RDR6/SGS3, occurs apparently before the miRNA duplex is disassembled and the guide strand is selected, and it is maintained even after the passenger strand has been removed.
Results and Discussion
Initiation of Transitivity and Asymmetric miRNA Duplexes.
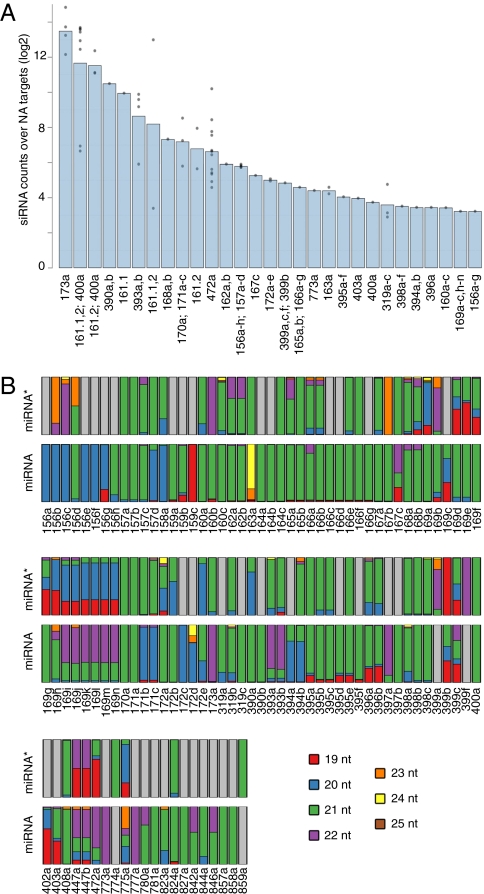
To survey the extent of miRNA-induced transitivity, we mapped 21-nt siRNAs from wild-type A. thaliana plants to known miRNA targets. Close to one-third (67 of 208) miRNA targets produced such siRNAs, which we suspected to be secondary, or tasiRNAs. The results were consistent across replicates (Fig. S1), and those with more than five reads in each replicate were analyzed further. TasiRNAs are processed in a phased fashion (17, 22), but the often low number of siRNAs precluded the reliable determination of phasing scores. The low abundance of some siRNAs could be a consequence of the relative expression level of a miRNA or of its target, of the cleavage efficiency of the miRNA, of limited coexpression of miRNA and target, or of degradation of nonfunctional secondary siRNAs. Location and sequence complementarity of the miRNA target sites also can affect transitivity (23).
We confirmed that mRNAs targeted by AGO7-loaded miR390 and by four 22-nt long miRNAs—miR168, miR173, miR393, and miR472—each generated large numbers of secondary siRNAs (20, 21). SiRNAs were also produced from several mRNAs that were neither targeted by AGO7-associated miRNAs nor by 22-nt miRNAs (Fig. 1, Table 1, and Dataset S1). Most of these siRNA-spawning mRNAs were targeted by miRNA families that contain at least one member with a predominantly 22-nt miRNA* (Table 1 and Dataset S1). This observation suggested that length differences between the two strands of the miRNA:miRNA* duplex may be sufficient for the initiation of secondary siRNA production, independently of which strand is 22 nt in length.
Fig. 1.
Twenty-one–nucleotide small RNAs produced in wild-type A. thaliana from miRNA targets. (A) Bars indicate average number of Illumina reads from two biological replicates mapping to miRNA targets. Gray dots indicate average for individual targets among one miRNA family. (B) Size distribution of reads mapping to miRNA or miRNA*. Gray indicates miRNAs or miRNA*s that were not detected. For miRNA or miRNA* not detected in our data, other datasets were consulted (45).
Table 1.
Secondary siRNA-triggering miRNAs
| MiRNA | Distinguishing feature | See also |
| miR167a,b,c | 22-nt miRNA | Dataset S1 |
| miR168a,b | 22-nt miRNA | Dataset S1 |
| miR173a | 22-nt miRNA | Dataset S1 |
| miR393a,b | 22-nt miRNA | Dataset S1 |
| miR396a,b | 22-nt miRNA | Ref. 45 |
| miR472a | 22-nt miRNA | Dataset S1 |
| miR773a | 22-nt miRNA | Dataset S1 |
| miR156a-h, miR157a-d | 22-nt miRNA* | Dataset S1 |
| miR160a,b,c | 22-nt miRNA* | Dataset S1 |
| miR162a,b | 22-nt miRNA* | Dataset S1 |
| miR165a,b: miR166a-g | 22-nt miRNA* | Dataset S1 |
| miR169a-n | 22-nt miRNA* | Dataset S1 |
| miR398a,b,c | 22-nt miRNA* | Dataset S1 |
| miR399a,b,c | 22-nt miRNA* | Dataset S1 |
| miR403a | 22-nt miRNA* | Ref. 45 |
| miR172a-e | AGO2-associated miRNA | Ref. 8 |
| miR395a-f | AGO2-associated miRNA | Ref. 8 |
| miR161.1a, miR161.2a, miR400 | AGO7-associated miRNA | Ref. 5 |
| miR390a,b | AGO7-associated miRNA | Ref. 5 |
| miR170a, miR171a,b,c | Partially AGO7-associated | Ref. 5 |
| miR163a | Structural bulge | Fig. 4B |
| miR319a,b,c | Structural bulge | Fig. 4B |
| miR394a,b | Structural bulge | Fig. 4B |
Twenty-Two–Nucleotide miRNA*s Trigger Secondary siRNA Production from Guide-Strand Targets.
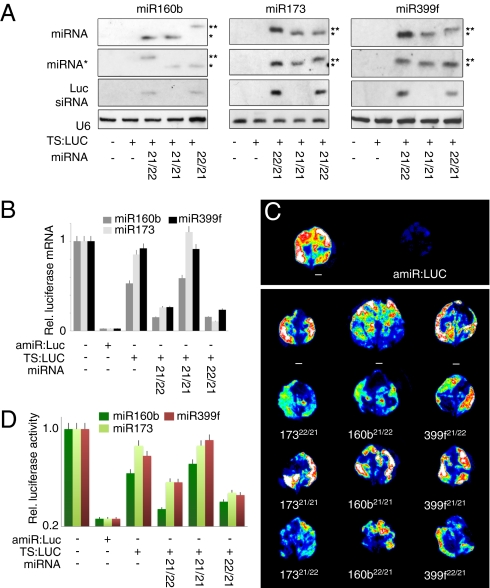
To test the hypothesis that length asymmetries are the primary cause of secondary siRNA formation, we manipulated the asymmetry of three miRNA duplexes: miR160b:miR160b* (21/22 nt), miR173a:miR173a* (22/21 nt), and miR399f:miR399f* (21/22 nt). In each of these cases, there is a bulge of one unpaired base on one strand that produces the asymmetries. We altered the lengths of each miRNA and miRNA* to produce 22-nt miRNA, 22-nt miRNA*, or symmetric 21/21-nt duplexes by closing or opening bulges in the duplex sequence (Fig. S2). To evaluate the efficiency of these constructs as secondary siRNA triggers, we transiently cotransformed Nicotiana benthamiana leaves with a fragment of the firefly luciferase cDNA linked to miRNA target sites (TS:Luc), the mutated miRNA precursor, and full-length luciferase cDNA (Luc) as a reporter of silencing in trans (Fig. S3A). Blot analysis of total or AGO1-coimmunoprecipitaton (co-IP) RNA confirmed that miRNAs of the predicted size were correctly produced and loaded onto AGO1 (Fig. 2A and Fig. S3B). Endogenous miR160 was detectable in transiently transformed N. benthamiana leaves (Fig. 2A and Fig. S3B), and miR160b transitivity was inferred from increased activity relative to background.
Fig. 2.
Secondary siRNAs triggered by 22-nt miRNA*s. (A) Small RNA blots to detect unmodified and modified miRNAs and miRNA*s expressed in transiently transformed N. benthamiana leaves. Secondary siRNAs produced from the TS:Luc reporter were also monitored. U6 was used as loading control. One asterisk (*) represents 21-nt miRNAs; two asterisks (**) represent 22-nt miRNAs. (B) Relative luciferase full-length mRNA levels, indicating effectiveness of trans silencing activity of secondary siRNAs. RNA levels were determined by quantitative RT-PCR using primers that did not amplify the TS:Luc fragment containing target sites for the miRNAs shown on top. Maximal silencing was defined using an artificial miRNA against luciferase (amiR:Luc); error bars are for SD. (C) Images of spatial distribution of luciferase intensity in N. benthamiana leaves. White, red, green, and blue indicate highest to lowest luciferase activity. Lower panel; leaves co-transformed with TS:LUC and miRNA constructs. (D) Quantification of luciferase activity in the same leaves.
All modified miRNAs could guide cleavage of the TS:Luc target, demonstrating their functionality (Fig. S3C). Transformation of constructs with either a 22-nt miRNA or miRNA* strand was sufficient to induce accumulation of LUC-derived siRNAs (Fig. 2A) with a concomitant reduction of both mRNA and protein levels of the full-length Luc transitivity sensor (Fig. 2 B–D and Fig. S4). These results support our hypothesis that miRNA duplex asymmetry, where either strand is 22-nt long, is sufficient to promote transitive siRNA production.
Twenty-One–Nucleotide miR173* Can Cause Transitivity.
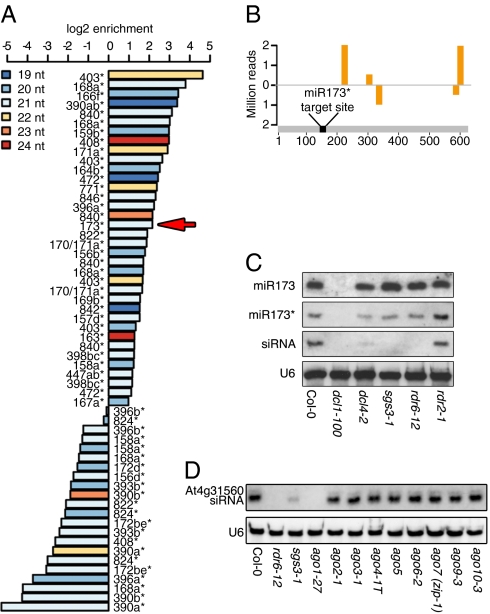
Studies in both plants and animals have shown that, in addition to the miRNA strand, the passenger strand can be loaded onto the RISC complex and trigger mRNA silencing (8, 9). Therefore, if our working hypothesis was correct, we could expect an AGO1-loaded 21-nt miRNA* to promote secondary siRNA production on its own target, as long as the miRNA is 22-nt long. To identify miRNA/miRNA* duplex suitable for testing this hypothesis, we re-examined published data, searching for miRNA*s enriched by AGO1 co-IP (20).
Among 264 unique miRNA or miRNA* species enriched in the AGO1-IP fraction, we found 59 distinct miRNA* sequences. Of these sequences, 36 were enriched at least twofold in AGO1–co-IP material relative to the total small RNA population (Fig. 3A). Several miRNAs*, such as miR390*, miR393b*, and miR396b*, have been shown to be associated with other AGOs (4, 6, 8), and they are accordingly underrepresented in the AGO1–co-IP fraction. Among the miRNAs for which we could identify a passenger strand loaded onto AGO1, we decided to use the miRNA173/173* duplex as a test case, because miR173 can trigger transitivity (22), and has a 21-nt miRNA* abundantly loaded onto AGO1 (Fig. 3A).
Fig. 3.
Secondary siRNAs produced from targets of miRNAs* with a 22-nt miRNA. (A) Enrichment of miRNAs* in AGO1–co-IP fractions. Only miRNA*s that are enriched or depleted at least twofold in AGO1–co-IP fractions compared with the total small RNA population are shown. MiR173* is indicated with a red arrow. (B) Small RNAs mapping to At4g31560 cDNA. (C) RNA blots to detect miR173, miR173* and At4g31560-derived small RNAs (“siRNA”) in transitivity defective mutants. (D) RNA blot to detect At4g31560-derived small RNAs in different ago mutants.
Using established criteria (17, 24), we predicted At4g31560 as a potential target for miR173*. By 5′ RACE-PCR, we detected At4g31560 products indicative of miR173*-triggered cleavage, along with a spectrum of additional, shorter products (Fig. S5). The shorter products could be the consequence of further processing of the target into siRNAs. Supporting such a scenario, small RNAs derived from At4g31560 mRNA are found in our deep-sequencing data (Fig. 3B) and could be detected on RNA blots (Fig. 3C). One feature of AGO1-generated tasiRNAs is their phasing (22). Unfortunately, a clear phasing of At4g31560-derived siRNAs could not be detected because of the low number of reads derived from this locus. However, the observed siRNA production required the activity of proteins known to be essential for secondary siRNA formation, RDR6, SGS3, and DCL1 and DCL4 (10–12, 25) (Fig. 3C), confirming its nature.
The two strands in a miRNA:miRNA* duplex can have different fates and be loaded into alternative AGO proteins (6, 8). We could therefore not exclude that miR173*, despite its enrichment in AGO1–co-IP material, triggers transitivity because it is loaded into AGO proteins other than AGO1. We therefore monitored siRNA production from At4g31560 in plants with mutations in all of the A. thaliana AGO genes, except AGO8, which is consider to be a pseudogene. Only plants compromised in AGO1 activity lacked At4g31560 derived siRNAs (Fig. 3D).
These observations support the hypothesis that either the guide or passenger strand being 22-nt long is sufficient for secondary siRNA production. This finding might seem surprising, given that only one of the miRNA (guide strand) is retained in RISC. However, the entire miRNA duplex associates with the AGO1-containing complex before the miRNA* (passenger strand) is removed and the complex matures (7, 26, 27). The fact that the passenger strand is present only during the early steps of RISC maturation suggests that RISC is programmed for secondary siRNA production before the guide strand is selected and the miRNA duplex disassembled.
RISC Reprogramming Is Caused by Structural Asymmetry of the miRNA Duplex.
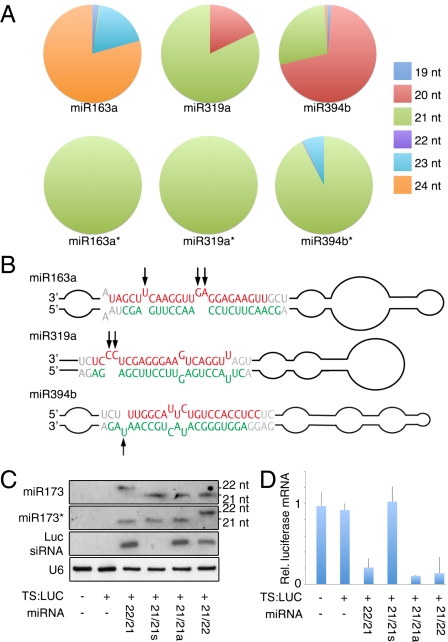
An alternative to the hypothesis of transitivity being determined by the miRNA:miRNA* duplex size, is that the structural asymmetry produced by the bulge of an unpaired base is the primary determinant of RISC reprogramming. The two scenarios are difficult to distinguish because a single bulge normally leads to a 21/22-nt duplex. To find an exception to the 21/22-nt rule, we sought to identify siRNA-producing loci without evidence for either the miRNA trigger or the corresponding miRNA* being 22 nt. From analysis of our deep-sequencing small-RNA data, we could find such loci (Table 1). These loci include targets of miR161 and miR390, which are preferentially loaded onto AGO7 (5), and miR170/171, which also appears to be partially enriched in AGO7–co-IP material, according to published data (5). In addition, miR172 and miR395, which lack a uracil at their 5′ end, are preferentially loaded onto AGO2 (8), and thus may also initiate transitivity through an AGO1-independent pathway. The remaining group comprises miR163, miR319, and miR394. Twenty-two–nucleotide miRNAs or miRNA*s for these families were not detected in our data (Fig. 4A and Dataset S1). To evaluate if asymmetry in the structure of these miRNA duplexes could explain their ability to trigger secondary siRNA production, we compared the predicted secondary structure of the different family members. In all three cases, there was at least one family member with asymmetric bulges in the miRNA/miRNA* region of the precursor (Fig. 4B and Fig. S6). MiR163a contains two bulges of 1 and 2 nt that result in the formation of an unusual 24/21-nt duplex. MiR319a has a 2-nt bulge at the 3′ end of the miRNA that leads to a 21/21-nt duplex. MiR394b contains one bulge at the end of the miRNA* that results in the formation of a 20/21-nt duplex.
Fig. 4.
Effects of structural asymmetry on transitivity. (A) Size distribution of mature miRNA and miRNA* in wild-type A. thaliana plants. (B) Predicted secondary structure of miR163a, miR319a, and miR394b precursors. Red indicates miRNA guide strand and green miRNA* passenger strand. Black arrows mark asymmetric bulges. (C) RNA blot with material from transiently transformed N. benthamiana leaves. Symmetric 21-nt–producing miR173 is noted as miR17321/21s, and asymmetric is miR17321/21a. (D) Relative luciferase mRNA levels in infiltrated leaves.
In addition, we scrutinized the secondary structure of the remaining siRNA-generating miRNAs for asymmetric bulges (Fig. S6), focusing on all miRNA and miRNA* sequences present in our deep-sequencing data. As expected for miRNA:miRNA* duplexes containing one 22-nt strand, we observed asymmetric bulges in most of them. Two miRNAs, miR394a/b and miR472, have asymmetric bulges only in the context of the unprocessed precursor, as the corresponding nucleotides are at the unpaired 3′ end, leading to an unusual 3-nt overhang (Fig. S6). This finding may have several implications. The transitivity-triggering step could occur already during dicing, or the unusually long 3′ overhang could reprogram RISC. It is also possible that minor, offset variants of these miRNAs include the bulge in a double-stranded region of the duplex.
These observations are compatible with the asymmetric structure of an miRNA duplex causing RISC reprogramming and triggering siRNA formation, independently of miRNA or miRNA* length. To challenge this hypothesis, we engineered a version of miR173 that includes an additional bulge in the miRNA* (Fig. S7A). Such an asymmetric duplex, naturally found in MIR159C (Fig. S7 B and C), should only generate 21-nt miRNA or miRNA*. In N. benthamiana, the engineered precursor produced miRNA and miRNA* of the expected size (Fig. 4C), and we detected the expected cleavage product of the reporter target (Fig. S7D). The asymmetric 21/21-nt miR173/miR173* duplex was as efficient in triggering transitivity as the wild-type 22/21-nt duplex (Fig. 4 C and D), but no transitivity was observed with the symmetric 21/21-nt duplex.
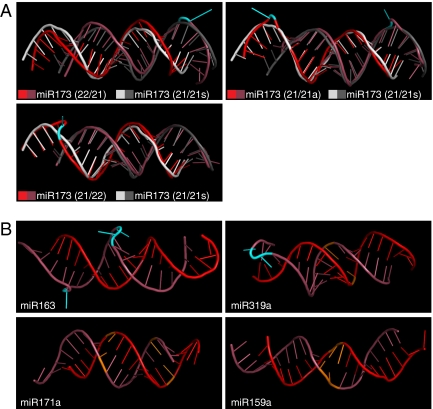
Secondary-structure predictions (Fig. S6) suggested that the asymmetric bulges may affect the shape of the duplexes. We therefore modeled the 3D structure (28) of the endogenous and engineered miR173 duplexes along with several symmetric and asymmetric endogenous duplexes (Fig. 5). The asymmetric duplexes not only had a different torsion (Fig. 5A), but also differed in the unpaired nucleotides having protruding base moieties, which we speculate could affect conformation of RISC (Fig. 5).
Fig. 5.
Predicted 3D structures of symmetric and asymmetric duplexes. (A) Comparison of endogenous and engineered miR173 duplexes. (B) Endogenous symmetric and asymmetric miRNA duplexes. Darker colors indicate miRNA strands. Nucleotides at asymmetric bulges are cyan and nucleotides at symmetric bulges orange.
We propose that asymmetric bulges in the miRNA:miRNA* duplex can specify the initiation of transitivity, although other features, such as position and arrangement of symmetric bulges, may also affect RISC programming. That miR162, which has a miRNA* of 22 nt, triggers transitivity even without obvious predicted structural asymmetry, points to alternative mechanisms for transitivitiy, perhaps because this miRNA is loaded not only onto AGO1.
Conclusions
We conclude that the AGO1-RISC engages the machinery responsible for siRNA production but both strands of the small RNA duplex are still associated with AGO1. The most parsimonious explanation for our results is that an asymmetric miRNA duplex alters the conformation of AGO in a manner that supports recruitment of RDR6/SGS3 and the ensuing transitivity. Such reprogramming seems to be a consequence of structural asymmetry, rather than a strand-size–dependent phenomenon, although we cannot exclude that both features promote transitivity independently. Our results imply that RISC reprogramming for transitivity occurs early during maturation and that this property is maintained through the later events of target recognition and cleavage. One possibility is that an asymmetric duplex triggers structural changes that allow AGO1 to recruit the transitivity proteins RDR6 and SGS3. It is also possible that such RISC cofactors are recruited during duplex processing, and that these are transferred onto RISC upon loading of the duplexes.
Materials and Methods
Bioinformatic Analyses.
After adapter trimming, reads from Illumina small RNA libraries were mapped without mismatches to miRNA precursors and the The Arabidopsis Information Resource (TAIR) 10 representative gene models of predicted miRNA targets (http://asrp.cgrb.oregonstate.edu/) using the Bowtie aligner (29). A minimum of at least five 21-nt reads in both biological replicates was required to classify a miRNA target as siRNA-producing. miRNA size distributions were determined from reads overlapping the predicted miRNA or miRNA* sites (http://mpss.udel.edu/common/web/starExamples.php?SITE=at_pare) by at least 9 nt and present in both replicates. For miRNA or miRNA* sequences present in multiple locations in the genome, the total number of reads was normalized to the number of potential source loci. AGO1 enriched miRNA*s were identified from published data using analogous methods (20). Statistical analysis and data visualization was accomplished in the R statistical programming environment. Data processing was accomplished using custom Perl scripts. Mir173* targets were predicted using WMD3 (http://wmd3.weigelworld.org).
Plant Material.
A. thaliana plants, accession Col-0, and N. benthamiana were grown on soil at 23 °C in long days (16-h light/8-h dark) in growth rooms with 65% humidity under a 2:1 mixture of cool white and warm fluorescent light. Alternatively, plants were grown on MS medium plates with 0.4% agar in growth chambers (Percival Scientific). Mutants and plants expressing HA-AGO1 in ago1-25 have been previously described (25, 30–39). The ago5 mutant SALK_118422 was ordered from the European Arabidopsis Stock Centre (http://arabidopsis.info/).
Expression Analysis.
Total RNA was extracted from 12-d-old seedlings and 30-d-old inflorescences using TRIzol Reagent (Invitrogen). CDNA was produced with the RevertAid First Strand cDNA Synthesis Kit (Fermentas), using 1 μg of total RNA pretreated with DNase I (Fermentas). Real time RT-PCR and small RNAs blots were done with total RNA extracted from 15-d-old A. thaliana seedlings or N. benthamiana leaves 3 d after Agrobacterium infiltration. Biological triplicates and technical duplicates were used. In the case of N. benthamiana experiments, samples with similar luciferase activity were selected for RNA analyses. Technical and biological replicates were treated as independent samples. Averages and SD were calculated after obtaining the 2(-ΔΔCt) value. A P value <0.05 in a t test with Bonferroni's correction was considered significant. Expression levels were normalized against β-TUBULIN2 (At5g62690). MiRNA-guided 3′ cleavage products were detected using RNA ligase-mediated 5′ RACE (40). For blot analyses, 1–5 μg of total RNA was resolved on 17% (vol/vol) polyacrylamide gels under denaturing conditions with 7 M urea. RNA was transferred to HyBond-N+ membranes (Amersham) by semidry electroblotting, and membranes were hybridized to DNA oligonucleotides probes labeled with digoxigenin using the second generation DIG oligonucleotide 3′-end labeling kit (Roche). Small RNA blots were repeated at least once with independent biological samples. See Table S1 for oligonucleotide primers and small RNA probes.
Transgenes.
Mutated versions of the MIR160B, MIR173, and MIR399F precursors were obtained by PCR-mediated mutagenesis, cloning into pCR8GW-TOPO and recombination behind the CaMV 35S promoter into a modified pGREEN vector (pFK210). See Table S2 for a detailed list of constructs, names, and descriptions, and Table S1 for a detailed list of PCR primers. The artificial miRNA against luciferase and the luciferase-expressing constructs were a gift of Pierre Hilson (VIB Department of Plant Systems Biology, Ghent University, Gent, Belgium). A. thaliana plants were transformed using the flower-dip method (41). Transgenic seedlings were selected with 50 mg/mL kanamycin on plates or 0.1% ammonium glufosinate (BASTA) on soil. At least 15 T1 seedlings were analyzed for each construct. Transient infiltration of N. benthamiana leaves was performed as previously described (42).
Luciferase Activity Measurement.
Ten-day-old A. thaliana seedlings or N. benthamiana leaves infiltrated with luciferase constructs were imaged using an Orca 2-BT cooled CCD camera (Hamamatsu). Ten hours before measuring luciferase activity, plants were sprayed or infiltrated with 1 mM d-luciferin potassium salt (PJK), and this was repeated 20 min before measurements. Luciferase activity was detected using integration times of 1 min for A. thaliana seedlings and 1–10 s for N. benthamiana leaves. Signal from chlorophyll autofluorescence was removed using a luciferase-specific filter. Light signals were quantified within a region of interest corresponding to whole seedlings (for A. thaliana) or the entire leaf (for N. benthamiana), and background was subtracted and normalized by area unit to obtain net light emission. An average of 50 seedlings or 20 leaves were analyzed for each combination of constructs.
Immunoprecipitation.
Leaves of transgenic 20-d-old ago1-25 mutants or Agrobacterium-infiltrated N. benthamiana plants expressing HA-AGO1 (20) were collected and 1.5 g of material were ground in liquid nitrogen. The following steps were performed at 4 °C. The frozen powder was homogenized by vortexing in 4 mL of lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% (vol/vol) glycerol, 5 mM MgCl2, 0.1% Nonidet P-40) containing 1 tablet/10 mL of protease inhibitor mixture (Roche). The homogenate was incubated under rotation for 1 h, and cell debris was removed by centrifugation at 12,000 × g for 30 min. Twenty microliters of Protein G Plus-Agarose (Santa Cruz Biotechnology) was added and samples were incubated with rotation for 30 min. Protein G Plus-agarose was removed by centrifugation at 6,000 × g for 1 min; this step reduced recovery of unspecifically antibody-bound material. The supernatant was incubated with 40 μL of Monoclonal Anti−HA-Agarose (Sigma) overnight under rotation. Immunoprecipitates were washed four times with lysis buffer before extracting RNA and protein with TRIzol. Protein was precipitated with ice-cold acetone from the organic phase.
Small RNA Libraries and Sequencing.
Illumina sequencing and analysis of duplicate small RNA libraries from 21-d-old A. thaliana rosette leaves was performed as previously described (43).
Secondary and Tertiary miRNA Structures.
Predictions of miRNA precursor secondary structures were obtained from RNAfold on the Vienna RNA WebServer (http://www.tbi.univie.ac.at/). Predictions of miRNA duplex secondary structures were obtained from the RNAhybrid Web service (44). The MC-Fold/MC-Sym pipeline was used to confirm secondary structures and predict tertiary structures (28). A 2-nt linker was added to the miRNA strand to produce a linear molecule. After generating the MC-Sym scripts the linkers were removed and tertiary structures built. The obtained structures were refined and scored according to internal energy. Three-dimensional images of the structures with the best energy scores were obtained using PyMol (http://www.pymol.org/).
Supplementary Material
Acknowledgments
We thank James Carrington and Olivier Voinnet for seeds of HA-Argonaute plants and rdr6-12 mutants, and Pierre Hilson for the luciferase constructs. This work was supported by Human Frontier Science Program Long-Term Fellowships (to P.A.M. and D.K.); by FP7 Collaborative Project Acquired environmental epigenetics advances: From Arabidopsis to maize (AENEAS) Contract KBBE-2009-226477; and the Max Planck Society (D.W.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1200169109/-/DCSupplemental.
References
- 1.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 2.Yu B, et al. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mi SJ, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montgomery TA, et al. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 6.Takeda A, Iwasaki S, Watanabe T, Utsumi M, Watanabe Y. The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol. 2008;49:493–500. doi: 10.1093/pcp/pcn043. [DOI] [PubMed] [Google Scholar]
- 7.Eamens AL, Smith NA, Curtin SJ, Wang MB, Waterhouse PM. The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. RNA. 2009;15:2219–2235. doi: 10.1261/rna.1646909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, et al. Arabidopsis Argonaute 2 regulates innate immunity via miRNA393(*)-mediated silencing of a Golgi-localized SNARE gene, MEMB12. Mol Cell. 2011;42:356–366. doi: 10.1016/j.molcel.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devers EA, Branscheid A, May P, Krajinski F. Stars and symbiosis: microRNA- and microRNA*-mediated transcript cleavage involved in arbuscular mycorrhizal symbiosis. Plant Physiol. 2011;156:1990–2010. doi: 10.1104/pp.111.172627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howell MD, et al. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell. 2007;19:926–942. doi: 10.1105/tpc.107.050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen HM, Li YH, Wu SH. Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:3318–3323. doi: 10.1073/pnas.0611119104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fahlgren N, et al. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol. 2006;16:939–944. doi: 10.1016/j.cub.2006.03.065. [DOI] [PubMed] [Google Scholar]
- 15.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Si-Ammour A, et al. miR393 and secondary siRNAs regulate expression of the TIR1/AFB2 auxin receptor clade and auxin-related development of Arabidopsis leaves. Plant Physiol. 2011;157:683–691. doi: 10.1104/pp.111.180083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhai J, et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011;25:2540–2553. doi: 10.1101/gad.177527.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 19.Felippes FF, Weigel D. Triggering the formation of tasiRNAs in Arabidopsis thaliana: The role of microRNA miR173. EMBO Rep. 2009;10:264–270. doi: 10.1038/embor.2008.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cuperus JT, et al. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat Struct Mol Biol. 2010;17:997–1003. doi: 10.1038/nsmb.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen HM, et al. 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc Natl Acad Sci USA. 2010;107:15269–15274. doi: 10.1073/pnas.1001738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montgomery TA, et al. AGO1-miR173 complex initiates phased siRNA formation in plants. Proc Natl Acad Sci USA. 2008;105:20055–20062. doi: 10.1073/pnas.0810241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang C, Ng DW, Lu J, Chen ZJ. Roles of target site location and sequence complementarity in trans-acting siRNA formation in Arabidopsis. Plant J. 2012;69:217–226. doi: 10.1111/j.1365-313X.2011.04783.x. [DOI] [PubMed] [Google Scholar]
- 24.Ossowski S, Schwab R, Weigel D. Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 2008;53:674–690. doi: 10.1111/j.1365-313X.2007.03328.x. [DOI] [PubMed] [Google Scholar]
- 25.Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawamata T, Yoda M, Tomari Y. Multilayer checkpoints for microRNA authenticity during RISC assembly. EMBO Rep. 2011;12:944–949. doi: 10.1038/embor.2011.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okamura K, Liu N, Lai EC. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol Cell. 2009;36:431–444. doi: 10.1016/j.molcel.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parisien M, Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 29.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harvey JJ, et al. An antiviral defense role of AGO2 in plants. PLoS ONE. 2011;6:e14639. doi: 10.1371/journal.pone.0014639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hunter C, Sun H, Poethig RS. The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr Biol. 2003;13:1734–1739. doi: 10.1016/j.cub.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 32.Laubinger S, et al. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105:8795–8800. doi: 10.1073/pnas.0802493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morel JB, et al. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002;14:629–639. doi: 10.1105/tpc.010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mourrain P, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 35.Olmedo-Monfil V, et al. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature. 2010;464:628–632. doi: 10.1038/nature08828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie Z, Allen E, Wilken A, Carrington JC. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2005;102:12984–12989. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie Z, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng X, Zhu J, Kapoor A, Zhu JK. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 2007;26:1691–1701. doi: 10.1038/sj.emboj.7601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu H, et al. Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell. 2011;145:242–256. doi: 10.1016/j.cell.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 41.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 42.de Felippes FF, Weigel D. Transient assays for the analysis of miRNA processing and function. Methods Mol Biol. 2010;592:255–264. doi: 10.1007/978-1-60327-005-2_17. [DOI] [PubMed] [Google Scholar]
- 43.de Felippes FF, Ott F, Weigel D. Comparative analysis of non-autonomous effects of tasiRNAs and miRNAs in Arabidopsis thaliana. Nucleic Acids Res. 2011;39:2880–2889. doi: 10.1093/nar/gkq1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krüger J, Rehmsmeier M. RNAhybrid: MicroRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34(Web Server issue):W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gustafson AM, et al. ASRP: The Arabidopsis Small RNA Project Database. Nucleic Acids Res. 2005;33(Database issue):D637–D640. doi: 10.1093/nar/gki127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.