Abstract
B-cell chronic lymphocytic leukemia (CLL) is the most common human leukemia. Deregulation of the T-cell leukemia/lymphoma 1 oncogene (TCL1) in mouse B cells causes a CD5+ leukemia similar to aggressive human CLL. To examine the mechanisms by which Tcl1 protein exerts its oncogenic activity in B cells, we performed proteomics experiments to identify its interacting partners. We found that Tcl1 physically interacts with de novo DNA methylthansferases Dnmt3A and Dnmt3B. We further investigated the effects of Tcl1 up-regulation on the enzymatic activity of Dnmt3A and found that Tcl1 overexpression drastically inhibits Dnmt3A function. In addition, B cells from TCL1 transgenic mice showed a significant decrease in DNA methylation compared with WT controls. Similarly, CLL samples with high Tcl1 expression showed a decrease in DNA methylation compared with CLL samples with low Tcl1 expression. Given the previous reports of inactivating mutations of DNMT3A in acute myelogenous leukemia and myelodysplastic syndrome, our results suggest that inhibition of de novo DNA methylation may be a common oncogenic mechanism in leukemogenesis.
The lymphocytes of B-cell chronic lymphocytic leukemia (CLL) are mostly resting cells with mature appearance and the B220+CD5+ phenotype (1, 2). The TCL1 oncogene has been identified as a target of chromosomal translocations and inversions at 14q31.2 in T-cell prolymphocytic leukemias (3). We previously showed that transgenic mice overexpressing TCL1 in B cells develop an aggressive form of CLL (4), and that aggressive human CLLs overexpress Tcl1 (5). These findings indicate that deregulation of TCL1 is critically important in the pathogenesis of the aggressive form of CLL. In previously work, we demonstrated that Tcl1 is a coactivator of the Akt oncoprotein (6). Akt could be robustly activated in mouse B cells by homozygous deletion of Pten (7). Surprisingly, these mice did not develop B-cell malignancies (7), suggesting that Tcl1 deregulation in B cells causes CLL by mechanisms other than Akt activation. We recently reported that Tcl1 functions as a transcriptional regulator and contributes to CLL pathogenesis by inhibiting activating protein 1 (AP-1) and activating NF-κB, and that that CLL-specific TCL1 mutants show gain of function in AP-1 inhibition (8).
Three human genes—DNMT1, DNMT3A, and DNMT3B—encode DNA methyltransferases, enzymes responsible for methylation of CpG islands (9). Whereas DNMT1 functions mainly in maintaining methylation patterns, DNMT3A and DNMT3B encode de novo DNA methyltransferases (9). Inactivating mutations of DNMT3A have been reported in acute myelogenous leukemia (AML) and myelodysplastic syndrome (MDS) (10–12). To further examine the mechanisms by which Tcl1 protein exerts oncogenic activity in B-cells, we used a proteomic approach to identify its interacting partners. This approach identified Dnmt3a and Dnmt3b as putative Tcl1 interactors.
Results
To identify putative Tcl1 interactors, we performed proteomics experiments using Omni-GST vector expressing Tcl1and Dleu7 (as a negative control) fused with GST in mammalian cells. HEK 293 cells were transfected with Omni-GST-Tcl1 or Omni-GST-Dleu7 (as a negative control), followed by GST pulldown. The resulting protein complexes were analyzed by mass spectrometry. Table S1 lists the top eight hits found in Tcl1 protein complexes but not in Dleu7 complexes. Interestingly, Pnpt1, a previously reported Tcl1 interactor (13), was most abundant in Tcl1 protein complexes (Table S1). We focused our attention on Dnmt3A and Dnmt3B, because aberrant DNA methylation has been hypothesized to contribute to cancer development, and inactivating mutations of DNMT3A were recently reported in AML and MDS (10–12).
To validate Tcl1-Dnmt3A and Tcl1-Dnmt3B interactions, we carried out a series of coimmunoprecipitation experiments in HEK 293 cells. Fig. 1 A and B shows the results of these experiments using transiently expressed proteins. Tcl1 was strongly coimmunoprecipitated with Dnmt3A in both directions, whereas no positive coimmunoprecipitates were detected between Fhit (used as a negative control) and Dnmt3A (Fig. 1A, Left vs. Middle). These results were confirmed by GST pulldown experiments, in which Dnmt3a was detected in Tcl1 complexes but not in Fhit complexes (Fig. 1A, Right). Similarly, Tcl1 was strongly coimmunoprecipitated with Dnmt3B in both directions, whereas no positive coimmunoprecipitates were detected between Fhit and Dnmt3B (Fig. 1B, Left vs. Middle).
Fig. 1.
Tcl1 interacts with Dnmt3A and Dnmt3B. (A) (Left) HEK 293 cells were cotransfected with DNMT3A-FLAG and 2xHA-TCL1 or with DNMT3A-FLAG and 2xHA-FHIT as indicated. After lysis, immunoprecipitation was carried out using anti-FLAG, IgG, or anti-HA antibodies. Western blot analysis was performed as indicated. (Right) HEK 293 cells were cotransfected DNMT3A-FLAG and Omni-GST-TCL1 or with DNMT3A-FLAG and Omni-GST-FHIT as indicated. After lysis and GST pulldown, Western blot analysis was performed as indicated. (B) Same as A, except using DNMT3B-FLAG instead DNMT3A-FLAG. (C) Daudi cells were lysed, and immunoprecipitation was carried out using anti-Tcl1, IgG, anti-Dnmt3A, or anti-Dnmt3B antibodies. Western blot analysis was performed as indicated. (D) HEK 293 cells were cotransfected with DNMT3A-FLAG and 2xHA-TCL1, 2xHA-FHIT, or 2xHA-TCL1b as indicated. After lysis, immunoprecipitation was carried out using anti-FLAG or anti-HA antibodies, and Western blot analysis was performed as indicated. (E) HEK 293 cells were cotransfected with Omni-GST-DNMT3B and 2xHA-TCL1, 2xHA-FHIT, or 2xHA-TCL1b as indicated. After lysis and GST pulldown, Western blot analysis was performed using anti-Omni (Top) or anti-HA (Middle and Bottom) antibodies. (F) Same as A, Left, but using DNMT1-FLAG instead of DNMT3A-FLAG. (G) Same as A, Left, but using DNMT3L-FLAG instead DNMT3A-FLAG.
In addition, Tcl1-Dnmt3B, but not Fhit-Dnmt3B, complexes were detected in GST pulldown experiments (Fig. 1B, Right). To prove that Tcl1-Dnmt3A and Tcl1-Dnmt3B complexes also occur at the endogenous level, we carried out coimmunoprecipitation experiments in Daudi cells expressing moderate levels of Tcl1 (8). Fig. 1C Upper shows that endogenous Tcl1 was coimmunoprecipitated with endogenous Dnmt3A in both directions. Similarly, endogenous Tcl1-Dnmt3B complexes were detected in Daudi cells (Fig. 1C, Lower). We previously identified Tcl1b, a member of the Tcl1 protein family activated in mature T-cell leukemia (6). Coimmunoprecipitation experiments in 293 cells (Fig. 1D) showed that Dnmt3A interacts with Tcl1, but not with Tcl1b. Similar GST pulldown experiments showed that Tcl1, but not Tcl1b, strongly interacts with Dnmt3B (Fig. 1E). The DNA methyltransferase protein family includes Dnmt3A, Dnmt3B, Dnmt1, and Dnmt3L (9). To investigate whether Tcl1 also interacts with Dnmt1 and Dnmt3L, we carried out similar coimmunoprecipitation experiments. Fig. 1 F and G shows that Tcl1 does not interact with Dnmt1 and Dnmt3L. Thus, our results indicate that Tcl1 strongly interacts with Dnmt3A and Dnmt3B, but not with Dnmt1 and Dnmt3L, suggesting that Tcl1 may be involved in de novo DNA methylation.
We previously showed that Tcl1 localizes in both nucleus and cytoplasm (6). In contrast, Dnmt3A and Dnmt3B are mostly nuclear proteins (9). To determine the intracellular localization of Tcl1-Dnmt3A and Tcl1-Dnmt3B complexes, we carried out immunofluorescence experiments in NIH 3T3 cells. Fig. 2 shows the intracellular locations of Tcl1, Dnmt3A, and Dnmt3B in three different fields. As expected, Dnmt3A and Dnmt3B (green) were localized in the nucleus. In contrast, Tcl1 (red) was localized in the nucleus and the cytoplasm. Fig. 2 Bottom shows Tcl1-Dnmt3A and Tcl1-Dnmt3B complexes (yellow) localized in distinct compartments within the nucleus. These findings provide additional evidence of a direct association of Tcl1 and Dnmt3A/3B.
Fig. 2.
Intracellular localization of Tcl1, Dnmt3a, and Dnmt3b. NIH 3T3 cells were cotransfected with pCMV5-TCL1, Omni-DNMT3A, and Omni-DNMT3B as indicated. Sixteen hours later, cells were fixed, permeabilized, and immunostained with mouse anti-Tcl1 and rabbit anti-Omni antibodies. Secondary antibodies goat anti-mouse Alexa Fluor 546 (red) and goat anti-rabbit Alexa Fluor 488 (green) were used to visualize intercellular locations of Tcl1 (red), Dnmt3A (green), and Dnmt3B (green). Colocalization of Tcl1 with Dnmt3A or Dnmt3B is shown in yellow.
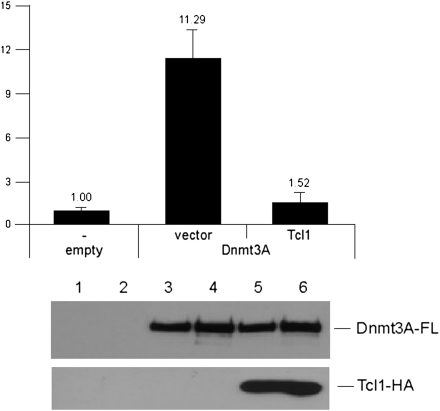
Given our finding that Tcl1 interacts with Dnmt3A and Dnmt3B (Figs. 1 and 2), and previous reports of inactivating mutations of DNMT3A in AML and MDS (10–12), it is reasonable to speculate that Tcl1 inhibits DNA methyltransferase enzymatic activity. To test this hypothesis, we performed Dnmt3A enzymatic activity assays, in which HEK 293 cells were transfected with vector and cotransfected with DNMT3A and vector or with DNMT3A and TCL1 (Fig. 3). After immunoprecipitation, Dnmt3A activity was measured using tritium-labeled AdoMet and tritium incorporation into DNA substrate. Fig. 3A shows that Tcl1 drastically inhibited Dnmt3A enzymatic activity by more than sevenfold (11.29 vs. 1.52) compared with the activity of Dnmt3A transfected with vector, whereas Dnmt3A expression was similar in both cases. This indicates that Tcl1 interacts with and strongly inhibits Dnmt3A activity.
Fig. 3.
Tcl1 inhibits Dnmt3a enzymatic activity. (Upper) HEK 293 cells were cotransfected with DNMT3A-FLAG and either 2xHA-TCL1 or vector construct. HEK 293 cells transfected only with vector were used as a negative control. Cell lysates were immunoprecipitated for 2 h with anti-FLAG. Dnmt3a enzymatic activity was measured as described in Materials and Methods. (Lower) Amounts of Dnmt3A and Tcl1 in Dnmt3a activity assay were measured by Western blot analysis using anti-HA and anti-FLAG antibodies as indicated. The experiment was carried out in duplicate. Lanes 1 and 2 correspond to the left bar, lanes 3 and 4 correspond to the middle bar, and lanes 5 and 6 correspond to the right bar.
To determine whether Tcl1 inhibits DNA methylation in vivo, we used our Eμ-TCL1 transgenic mice expressing Tcl1 exclusively in B cells (4). Because these mice develop CLL-like disease at age 8–12 mo, and given the difficulty in interpreting methylation differences in normal and malignant cells (4), we used 4- to 6-wk-old Eμ-TCL1 mice (which show no signs of disease at that age) for these experiments. To assess methylation differences in B cells of Eμ-TCL1 mice and WT controls, we sorted mouse spleen B cells from three Eμ-TCL1 and three WT mice and carried out global methylation profiling experiments using MethylCap-Seq protocol. Fig. 4 shows the average number of regions within each WT vs. Eμ-TCL1 comparison for which hypomethylation was detected by genomic feature type. Two-tailed P values across the nine comparisons (three WT and three 3 Eμ-TCL1) wre calculated using the two-sample t test. Interestingly, we observed an approximate fourfold increase in the number of hypomethylated regions in the Eμ-TCL1 samples compared with the WT samples in CpG islands (Fig. 4A, Left), as well as a twofold to fourfold increase in the number of hypomethylated regions in promoter regions (Fig. 4A, Right). These findings indicate an overall decrease in methylation within these regions. This trend held for both twofold and fourfold change cutoffs in both islands and promoters (Fig. 4A).
Fig. 4.
Tcl1 inhibits DNA methylation in vivo. (A) Tcl1 inhibits DNA methylation in Eμ-TCL1 B cells. (B) Tcl1 inhibits DNA methylation in CLL cells. (C) Tcl1 expression levels in CLL samples used in B. L, three selected samples showing low Tcl1 expression. H, three selected samples showing high Tcl1 expression. Protein load was measured using Ponceau S staining.
To examine whether our findings are relevant to human CLL, we carried out similar experiments using human CLL samples. We compared global methylation levels in three aggressive CLL samples with high Tcl1 expression and in three indolent CLL samples with low Tcl1 expression (Fig. 4B). We observed an approximate threefold to fivefold increase in the number of hypomethylated regions in samples with high Tcl1 expression vs. samples with low Tcl1 expression in CpG islands (Fig. 4B, Left), as well as a twofold to threefold increase in the number of hypomethylated regions in promoters in the same set of samples (Fig. 4B, Right). These data clearly indicate that Tcl1 expression inhibits DNA methylation in human CLL as well as in a mouse model in vivo.
Discussion
Previous studies of animal models have conclusively demonstrated that deregulation of TCL1 is a causal event in the development of mature T-cell leukemia and CLL (4, 14). Because Tcl1 protein does not contain domains or motifs pointing to its function, investigation of molecular mechanisms involved in Tcl1-induced oncogenesis faces significant challenges. We and others have previously reported that Tcl1 is a coactivator of Akt (6, 15). More recently, we found that Tcl1 functions as a transcriptional regulator and contributes to CLL pathogenesis by inhibiting AP-1 and activating NF-κB Dnmt3A, and identified Dnmt3B as Tcl1 interacting partners in proteomics experiments (8). In cancer research, Dnmts traditionally have been viewed as oncogenes, given that in many solid tumors, tumor-suppressor genes are inactivated by hypermethylation (16). This traditional view is currently changing in response to several studies reporting inactivating mutations of DNMT3A in AML and MDS (10–12).
Our data clearly indicate that Tcl1 functions as an inhibitor of de novo DNA methylation in vitro and in vivo. We previously showed that transgenic mice overexpressing TCL1 in B cells develop the aggressive form of CLL (4), and that aggressive human CLLs overexpress Tcl1 (5). Thus, our results are clearly relevant to human aggressive CLL (Fig. 5). We recently reported significantly higher miR-29a and miR-29b expression levels in indolent CLL compared with normal CD19+ B cells (17). To study the role of miR-29 in CLL, we generated Eμ-miR-29 transgenic mice overexpressing miR-29 in B cells, and found that these mice developed a disease similar to indolent CLL (17). Given that DNMT3A is a known miR-29 target (16), we analyzed its expression in Eμ-miR-29 transgenics, and found down-regulated Dnmt3A expression in Eμ-miR-29 transgenic mice (17). Thus, Dnmt3a expression is down-regulated in indolent CLL (Fig. 5).
Fig. 5.
Inhibition of Dnmt3a/3b is a common mechanism in leukemias and lymphomas.
Recent studies have reported inactivating mutations of DNMT3A in AML and MDS (10–12). Here we report that Tcl1 inhibits methylation in aggressive CLL. Taken together with our previous report of down-regulation of Dnmt3A in mouse model of indolent CLL (17) and a study showing up-regulation of TCL1 expression in Burkitt's lymphoma, follicular lymphoma, and diffuse large B-cell lymphoma (18), these data strongly indicate that inhibition of de novo DNA methylation may be a common oncogenic mechanism in leukemogenesis (Fig. 5).
Materials and Methods
DNA Constructs.
The pCMV5-TCL1 construct was described previously (8). Full-length human TCL1, TCL1B, and FHIT ORFs were cloned into pCMV-2xHA vector [based on pCMV-HA vector (BD Biosciences), with an added HA tag, creating HA tags at both 5′ and 3′ termini of all ORFs]. The resulting constructs were designated 2xHA-TCL1, 2xHA-TCL1B, and 2xHA-FHIT, respectively. Full-length human TCL1, DNMT3A, DNMT3B, and DLEU7 ORFs were cloned into a pcDNA4-HisMaxC vector (Omni-TCL1, Omni-DNMT3A, Omni-DNMT3B, and Omni-DLEU7, respectively; Invitrogen) using standard protocols. A GST-encoding DNA fragment was cloned between Omni-tag and TCL1, FHIT, DNMT3B, and DLEU7 cDNA sequences of Omni-TCL1, Omni-FHIT, Omni-DNMT3B, and Omni-DLEU7 constructs to obtain Omni-GST-TCL1, Omni-GST-FHIT, Omni-GST-DLEU7, and Omni-GST-DNMT3B constructs. Expression constructs containing Myc/FLAG-tagged ORFs of human DNMT3A, DNMT3B, DNMT3L, and DNMT1, designated DNMT3A-FLAG, DNMT3B-FLAG, DNMT3L-FLAG, and DNMT1-FLAG, respectively, were purchased from OriGene.
Cell Culture, Transfection, Western Blotting, and Immunoprecipitation.
HEK 293 and NIH 3T3 cells were grown in RPMI medium 1640 with 10% (vol/vol) FBS and 25 μg/mL of gentamicin at 37 °C in a humidified atmosphere of 5% (vol/vol) CO2 in air. FuGene 6 transfection reagent and protease inhibitor mixture tablets were obtained from Roche. Transfections, cell lysate preparations, and Western blot analysis were carried out as described previously (8). Immunoblots were developed using Pierce ECL Western Blotting Substrate or Thermo Scientific SuperSignal West Femto Maximum Sensitivity Substrate. The following antibodies were used for Western blotting and immunoprecipitation experiments: anti-Tcl1 (sc-32331), anti-Omni (sc-7270 for immunoprecipitation and Western blot analysis; sc-499 for immunofluorescence), anti-Dnmt3A (sc-373905 and sc-365769), anti-Dnmt3B (sc-130740 and sc-81252) (all from Santa Cruz Biotechnology), anti-HA (HA.11; Covance), anti-FLAG M2 and anti-FLAG M2-HRP (Sigma-Aldrich), and anti–HA-HRP (Roche).
Immunofluorescence.
NIH 3T3 cells were grown on two-well human fibronectin Cellware culture slides (BD Biosciences). Immunofluorescence experiments were carried out as described previously using a Zeiss LCM 510 confocal microscope (19). The secondary antibodies used for immunofluorescence were goat anti-mouse Alexa Fluor 546 (red) and goat anti-rabbit Alexa Fluor 488 (green) (Invitrogen).
Proteomics Experiments.
Approximately 5 × 107 of HEK 293 cells were transfected with Omni-GST-TCL1 or Omni-GST-DLEU7 constructs. After GST pulldown, protein complexes were separated in 12% (wt/vol) Criterion Tris-HCl SDS/PAGE gels (Bio-Rad) and stained with Coomassie blue (Bio-Rad). Experiments were carried out using data-dependent LC-MS3 on a LTQ Orbitrap mass spectrometer (Thermo Fisher). In gel, digested peptides were injected into the LC-MS system and eluted off the capillary HPLC column into the mass spectrometer in 2% (vol/vol) solvent B for 10 min, followed by a gradient of 2–40% (vol/vol) of solvent B over 45 min, then 40–90% of solvent B over the next 2 min at a flow rate of 2 μL/min. Solvent B was acetonitrile with 0.1% formic acid. Ions were fragmented by collision-induced dissociation. The MS3 scan was targeted at phosphorylation-neutral loss ions in the MS2 scan with mass differences of 98.0 Da, 80.0 Da, 49.0 Da, 40 Da, 32.7 Da, or 26.7 Da from the precursor mass. Protein identification was done using the Mass Matrix search engine (20). False-discovery rates for protein matches were estimated using the target-decoy search strategy (21).
Isolation of Mouse Genomic DNA from Separated B Cells.
WT and Eμ-TCL1 transgenic mice were killed at age 1 mo. White blood cells were isolated from mouse spleens as described previously (22). Mouse B cells were separated from non-B cells using the mouse B Cell Isolation Kit (Miltenyi Biotec) in accordance with the manufacturer's instructions. Genomic DNA from mouse B cells was extracted by a standard phenol/chloroform method.
CLL Samples.
CLL samples were obtained after informed consent was obtained from patients diagnosed with CLL at the CLL Research Consortium institutions (Table S2). In brief, lymphocytes were isolated from patient blood samples by Ficoll/Hypaque gradient centrifugation (Amersham) and processed for DNA extraction using a standard standard phenol/chloroform method. Protein extraction was carried out as described previously (23).
Genomic Methylation Experiments.
The methylated regions of the genome were enriched by MBD2 protein as described previously (24) using the MethylMiner Methylated DNA Enrichment Kit (Invitrogen). Illumina sequencing libraries were generated from the enriched methylated material using the TruSeq DNA Sample Prep Kit (Illumina).
Single-read 36-bp sequencing was carried out on an Illumina GA IIx sequencer. Sequence reads were analyzed using the Illumina ELAND primary analysis pipeline and subsequently aligned to the mm9 mouse genome using Bowtie software, allowing up to three mismatches. The aligned sequences were analyzed with MEDIPS software (25). The criteria for region selection were (i) ≥0.15 reads per million in at least one of the two comparisons; (ii) P ≤ 0.05 in both Wilcoxon and Student t tests; and (iii) a twofold to fourfold or greater change in reads per million. To assess methylation differences in mouse and patient methylome studies, two-tailed P values were calculated across the nine comparisons (Fig. 4A: three WT and three 3 Eμ-TCL1; Fig. 4B: low vs. high Tcl1 expression) using the two-sample t test on the average number of significantly hypomethylated CpG islands and promoters.
In Vitro Dnmt3 Activity Assay.
The DNA methyltransferase assay was as described previously (26), with modifications. In brief, HEK 293 cells were cotransfected with DNMT3A-FLAG and either 2xHA-TCL1 or vector construct. HEK 293 cells transfected only with vector served as a negative control. Cell lysates were immunoprecipitated for 2 h with anti-FLAG. Then A/G agarose beads with immunoprecipitates were washed several times in Dnmt reaction buffer as described previously (26) to remove excess Nonidet P-40 detergent. The typical Dnmt in vitro reaction was performed in 1× Dnmt reaction buffer containing A/G agarose beads, 1 mg of DNA substrate (poly-dG-dC DNA; Sigma-Aldrich), and tritium-labeled AdoMet (S-adenosyl-l-[methyl-3H] methionine; PerkinElmer). Reactions (in a total volume of 40 μL) were incubated for 2 h at 37 °C. Tritium-labeled DNA substrate from each reaction was isolated using the QIAquick PCR Purification Kit (QIAGEN). 3H radioactivity was measured using a Packard 2200CA Tri-Carb Liquid Scintillation Analyzer.
Supplementary Material
Acknowledgments
The research was supported by an American Cancer Society Research Scholar grant (to Y.P.) and National Institutes of Health Grant PO1-CA81534 (to the CLL Research Consortium; L.R., T.J.K., and C.M.C.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1200003109/-/DCSupplemental.
References
- 1.Sgambati M, Linet M, Devesa S. Chronic lymphocytic leukemia, epidemiological, familial, and genetic aspects. In: Cheson B, editor. Chronic Lymphocytic Leukemias. 2nd Ed. New York: Marcel Dekker; 2001. pp. 33–62. [Google Scholar]
- 2.Bullrich F, Croce C. Molecular biology of chronic lymphocytic leukemia. In: Cheson B, editor. Chronic Lymphocytic Leukemias. 2nd Ed. New York: Marcel Dekker; 2001. pp. 9–32. [Google Scholar]
- 3.Virgilio L, et al. Identification of the TCL1 gene involved in T-cell malignancies. Proc Natl Acad Sci USA. 1994;91:12530–12534. doi: 10.1073/pnas.91.26.12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bichi R, et al. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proc Natl Acad Sci USA. 2002;99:6955–6960. doi: 10.1073/pnas.102181599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pekarsky Y, et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66:11590–11593. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 6.Pekarsky Y, et al. Tcl1 enhances Akt kinase activity and mediates its nuclear translocation. Proc Natl Acad Sci USA. 2000;97:3028–3033. doi: 10.1073/pnas.040557697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki A, et al. Critical roles of Pten in B cell homeostasis and immunoglobulin class switch recombination. J Exp Med. 2003;197:657–667. [Google Scholar]
- 8.Pekarsky Y, et al. Tcl1 functions as a transcriptional regulator and is directly involved in the pathogenesis of CLL. Proc Natl Acad Sci USA. 2008;105:19643–19648. doi: 10.1073/pnas.0810965105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeltsch A. Molecular enzymology of mammalian DNA methyltransferases. Curr Top Microbiol Immunol. 2006;301:203–225. doi: 10.1007/3-540-31390-7_7. [DOI] [PubMed] [Google Scholar]
- 10.Ley TJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan XJ, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43:309–315. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 12.Walter MJ, et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia. 2011;25:1153–1158. doi: 10.1038/leu.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.French SW, et al. The TCL1 oncoprotein binds the RNase PH domains of the PNPase exoribonuclease without affecting its RNA-degrading activity. Cancer Lett. 2007;248:198–210. doi: 10.1016/j.canlet.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 14.Virgilio L, et al. Deregulated expression of TCL1 causes T cell leukemia in mice. Proc Natl Acad Sci USA. 1998;95:3885–3889. doi: 10.1073/pnas.95.7.3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laine J, Künstle G, Obata T, Sha M, Noguchi M. The protooncogene TCL1 is an Akt kinase coactivator. Mol Cell. 2000;6:395–407. doi: 10.1016/s1097-2765(00)00039-3. [DOI] [PubMed] [Google Scholar]
- 16.Fabbri M, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santanam U, et al. Chronic lymphocytic leukemia modeled in mouse by targeted miR-29 expression. Proc Natl Acad Sci USA. 2010;107:12210–12215. doi: 10.1073/pnas.1007186107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teitell MA. The TCL1 family of oncoproteins: Co-activators of transformation. Nat Rev Cancer. 2005;5:640–648. doi: 10.1038/nrc1672. [DOI] [PubMed] [Google Scholar]
- 19.Palamarchuk A, et al. 13q14 deletions in CLL involve cooperating tumor suppressors. Blood. 2010;115:3916–3922. doi: 10.1182/blood-2009-10-249367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu H, Freitas MA. MassMatrix: A database search program for rapid characterization of proteins and peptides from tandem mass spectrometry data. Proteomics. 2009;9:1548–1555. doi: 10.1002/pmic.200700322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 22.Costinean S, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(μ)-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palamarchuk A, et al. Akt phosphorylates Tal1 oncoprotein and inhibits its repressor activity. Cancer Res. 2005;65:4515–4519. doi: 10.1158/0008-5472.CAN-05-0751. [DOI] [PubMed] [Google Scholar]
- 24.Fraga MF, et al. The affinity of different MBD proteins for a specific methylated locus depends on their intrinsic binding properties. Nucleic Acids Res. 2003;31:1765–1774. doi: 10.1093/nar/gkg249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chavez L, et al. Computational analysis of genome-wide DNA methylation during the differentiation of human embryonic stem cells along the endodermal lineage. Genome Res. 2010;20:1441–1450. doi: 10.1101/gr.110114.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yokochi T, Robertson KD. Preferential methylation of unmethylated DNA by Mammalian de novo DNA methyltransferase Dnmt3a. J Biol Chem. 2002;277:11735–11745. doi: 10.1074/jbc.M106590200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.