Abstract
Mature B-cell exit from germinal centers is controlled by a transcriptional regulatory module that integrates antigen and T-cell signals and, ultimately, leads to terminal differentiation into memory B cells or plasma cells. Despite a compact structure, the module dynamics are highly complex because of the presence of several feedback loops and self-regulatory interactions, and understanding its dysregulation, frequently associated with lymphomagenesis, requires robust dynamical modeling techniques. We present a quantitative kinetic model of three key gene regulators, BCL6, IRF4, and BLIMP, and use gene expression profile data from mature human B cells to determine appropriate model parameters. The model predicts the existence of two different hysteresis cycles that direct B cells through an irreversible transition toward a differentiated cellular state. By synthetically perturbing the interactions in this network, we can elucidate known mechanisms of lymphomagenesis and suggest candidate tumorigenic alterations, indicating that the model is a valuable quantitative tool to simulate B-cell exit from the germinal center under a variety of physiological and pathological conditions.
Keywords: B cell differentiation, gene regulatory networks, immunity, master regulators
Immune response to pathogens requires rapid maturation of naïve B cells into memory B cells (MCs) and antibody-producing plasma cells (PCs) in highly specialized environments of the lymphoid organs, the germinal centers (GCs). The GC comprises two different compartments, the dark zone and the light zone. In the dark zone, B cells (called centroblasts) undergo class switch recombination (CSR) and somatic hypermutation (SHM) of genes encoding the transmembrane Ig receptors, the B-cell receptors (BCRs). After a few days, B cells migrate to the light zone, where they are exposed to antigen. B cells (centrocytes at this stage) are committed to apoptosis and compete for survival and differentiation signals, which can be provided in vitro by stimulation through the BCRs and the cell-surface receptors CD40 (1, 2). In vivo, these signals are delivered by high-affinity antigen cross-linking and T cells respectively.
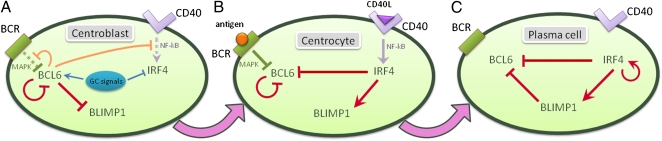
Exit from GCs is tightly regulated by a small transcriptional module that integrates physiologic signals from pathways sensing antibody affinity and interactions with T cells (Fig. 1). An essential regulator of the GC reaction is B-cell lymphoma 6 (BCL6), a potent transcriptional repressor required for the establishment and maintenance of GCs (3). Maturation of GC B cells toward MCs and PCs requires the down-regulation of BCL6, which is achieved in the light zone by several mechanisms. Antigen BCR activation leads to BCL6 rapid proteasomal degradation (4). In addition, T-cell–mediated stimulation through the CD40 pathway leads to NF-κB–mediated induction of IFN regulatory factor 4 (IRF4), an essential regulator of plasma-cell development (5). IRF4 leads to the transcriptional repression of BCL6 (6) and to the transactivation of B-lymphocyte–induced maturation protein 1 (BLIMP1) (7), which drives the regulatory program associated with plasmacytic differentiation and Ig secretion (8).
Fig. 1.
Time-dependent regulatory network of GC B cells. (A) Regulatory network at the centroblast stage. Upstream signals promote the expression of BCL6, a potent transcriptional repressor that controls the regulatory program of the GC. BCL6 directly represses BLIMP1, a key regulator necessary for plasma cell establishment. (B) At the centrocyte stage, the B cells compete for survival signals delivered by the BCRs and T cells, which lead to degradation of BCL6 protein and up-regulation of IRF4. (C) In the plasma cell stage, BLIMP1 and IRF4 are expressed and contribute to the transcriptional silencing of BCL6. The cell is locked in this terminally differentiated stage by a self-positive regulatory loop on IRF4.
BCL6, IRF4, and BLIMP1 interact with each other in a tightly regulated module, responsible for both normal and aberrant dynamics of the GC physiologic development. BCL6 binds its own promoter and inhibits its own transcription, thus implementing an autoregulatory loop (9). Furthermore, BCL6 represses several genes encoding molecules involved in the signal transduction of the BCR and CD40 pathways (10), some of which (MAPK, NF-κB) have been shown to regulate BCL6, implying the existence of additional regulatory loops. BCL6 also prevents GC B cells from differentiating into PCs by transcriptionally repressing prdm1, the gene encoding BLIMP1 (11, 12). Acetylation of BCL6 disrupts its transcriptional repression activity (13), allowing the induction of BLIMP1 expression by a multifactorial mechanism that includes IRF4. In turn, BLIMP1 contributes to the silencing of BCL6 by binding to the bcl6 promoter (14). Regarding IRF4, its expression is repressed in GCs, presumably by the MITF transcription factor (15). In PCs, IRF4 binds to its own promoter, supporting a positive feedback mechanism by which PCs can maintain high IRF4 expression (16).
The correct functioning of the complex circuitry underlying the B-cell maturation process is crucial for an efficient immune response, yet the molecular mechanisms governing the transition from a naive B-cell to a terminally differentiated MC and PC are poorly characterized from a quantitative perspective. To address the GC exit pathway dynamical behavior, we have developed a small, yet surprisingly complex quantitative model that elucidates the subtle mechanistic processes that make the normal B-cell development both robust and irreversible and underlie the dysregulation and block of maturation in GC-derived lymphomas.
Results
GC-Exit Pathway Model.
To understand the effect of BCR and CD40 signals on the network dynamics, we decompose the GC B-cell regulatory network into two submodules, each one associated with one signaling pathway. The corresponding kinetic models, which are discussed in SI Materials and Methods, provide a valuable framework to study GC B-cell dynamics under independent stimulation of BCR and CD40. We then consider the combined effect in physiological GC B cells.
BCR Signaling Module.
We first consider a GC B-cell stimulated only by BCR signaling because of high affinity antigen binding. Steady-state exploration of the BCL6 and BLIMP1 expression levels at different levels of protein synthesis, degradation and BCR stimulation, shows that the system has a bistable regime characterized by the existence of three stationary points, two stable and one unstable. The unstable critical point cannot be observed biologically because even an infinitesimal amount of noise would move the cell away toward the stable states. Thus, there are two homeostatic states: one characterized by high levels of BCL6 and low levels of BLIMP1, and another one by the opposite pattern, i.e., low levels of BCL6 and high levels of BLIMP1. Physiologically, these states represent steady-state GC B cells and PCs, respectively.
Fig. S1 shows the nullclines of the BCR module, i.e., the curves obtained by setting both the BCL6 and the BLIMP1 rates of change to zero, and the critical points, located at the intersection of the two nullclines. In the bistable regime, a hysteresis loop is established when a GC B-cell is activated through BCR signaling (Fig. S1, Inset). Specifically, a GC B-cell stimulated via BCR starts moving down along the upper GC branch (characterized by high levels of BCL6). When the branch becomes unstable (pink points), the cell jumps to the lower PC branch (characterized by lower levels of BCL6). As BCR signaling decreases, the cell retraces its dynamics through the lower branch and jumps back to the upper branch, eventually returning to the same initial state. In the absence of additional interactions, this reversible process can be repeated many times by increasing and decreasing the BCR stimulation. Thus, in isolation, it does not explain the irreversible nature of GC B-cell maturation.
CD40 Signaling Module.
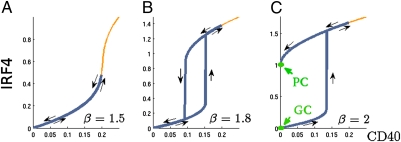
To investigate the dynamics of this submodule, we assume that BCL6 is already down-regulated by independent signals and, thus, repression on the CD40 pathway has been abrogated (see corresponding kinetic model in SI Materials and Methods). Quantitative steady-state analysis shows another bistable regime that notably, depends only on a dimensionless parameter β that comprises all IRF4 kinetic parameters (Eqs. S9–S11). β has a straightforward interpretation as a measurement of the ratio of all IRF4 synthesis contributions (basal, induced, and CD40-stimulated transcription) to degradation contributions (degradation rate and dissociation constant to the IRF4 self-promoting binding site). Fig. 2 shows the stationary expression levels of IRF4 as a function of CD40 stimulation and for different values of β. In the bistable regime, a hysteresis curve comprising two branches is observed: the lower branch corresponds to GC-like steady states, whereas the upper branch accounts for PCs. At low levels of β, CD40-stimulated B cells move along the lower branch toward high IRF4 levels. Upon cessation of CD40 signals, the cells return to their initial state through the upper branch, therefore experiencing a reversible transition (Fig. 2 A and B). However, as the IRF4 transcription increases (corresponding to higher values of β), the higher branch will intersect the y axis, at which point the lower branch is no longer dynamically accessible and cells are permanently trapped in the PC stage even after CD40 signaling is abrogated (Fig. 2C). Positive feedbacks have been shown to play major roles in developmental processes characterized by a point of no return (17). In B-cell physiology, an IRF4 positive feedback loop makes the terminal differentiation into PCs an irreversible event.
Fig. 2.
Hysteretic behavior of IRF4 for different values of β. The blue line shows the evolutionary steady states reached by a cell stimulated through the CD40 pathway. (A and B) After the signaling process is over, the cell reverts back to the initial GC steady state. (C) Above a critical ratio of production and degradation of IRF4, the GC state is no longer accessible and the GC B cells differentiate into a plasma cell. Therefore, the differentiation process has become a terminal, irreversible event.
The appearance of the dimensionless parameter β that governs the global dynamical behavior of the system is indicative of the critical role of IRF4 homeostatic level in the developmental pathway. Thus, independent or small coordinated changes affecting IRF4 homeostasis may disrupt normal B-cell GC exit dynamics in a similar way and induce an aberrant immune response and tumorigenesis.
Cosignaling and Irreversibility in B-Cell Differentiation.
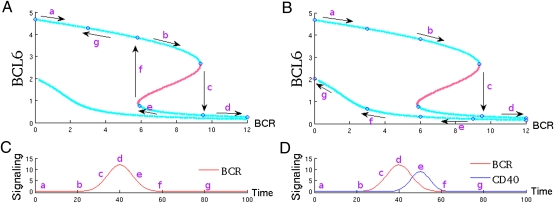
We consider now the combined effect of both signaling pathways acting together. Fig. 3 A and B show respectively BCL6 expression after BCR signaling alone and following coordinated BCR and CD40 signaling. Figs. 3C and 3D show the time-dependent signal intensity of each pathway, modeled as two partially overlapping bell curves. In our model, CD40 becomes activated some time after the initiation of BCR signaling to account for the physiological delay necessary for T cells to recognize antigen-bound B cells and to stimulate B cells. The model parameters have been fitted by using microarray gene expression data sets from normal, transformed and experimentally manipulated GC B cells and PCs (Table S1).
Fig. 3.
Irreversibility due to cosignaling of BCR and CD40. A and B show the stationary points of a B-cell at different levels of BCR stimulation. Blue and red dotted lines indicate stable and unstable stationary points respectively. C and D show the signaling intensity through the BCR (C) and the BCR and CD40 (D) pathways as a function of time. (A) After BCR signaling, BCL6 protein is degraded (arrows a–d) but after the cessation of the signal, returns to levels prior to stimulation. (B) Coordinated BCR and CD40 stimulation promotes a jump to a different branch of stationary points leading to an irreversible plasma cell phenotype.
In Fig. 3A, BCL6 is degraded after BCR stimulation (arrows a–c). Degradation proceeds until a sharp reduction of BCL6 protein is observed, indicating transition to the lower branch (PC). If the CD40 pathway is inactive, however, the transition is reversible and, upon BCR signals cessation, the B-cell returns to its previous GC state, thus failing to differentiate into a PC (arrows e–g). In Fig. 3B, BCL6 is similarly degraded after initial BCR stimulation (arrows a–d); however, when the cell jumps to the PC branch, the transition is made irreversible because of the activation of the CD40 pathway. As both signals decay, the cell moves along the lower branch (arrows e–g) that leads to a PC state. Thus, cooperativity between BCR and the CD40 signaling has produced an irreversible differentiation into a PC.
A bistability-related parameter sensitivity study shows that the system exhibits the correct bistable behavior for a wide range of the parameters that regulate BCL6 and BLIMP1 dynamics. However, the parameters that regulate IRF4 expression are constrained to a small range of variation in order for the system to be bistable, according to the analytical conditions of bistability (Table S2 and Eqs. S9–S11). Fig. S2 shows a bifurcation study for BCL6, i.e., a study of the changes of BCL6 homeostatic levels as we change the values of each model parameter. The figure shows that the kinetic parameters associated to IRF4 dynamics control the bistable switch in the system. Taken together, these findings support the notion that IRF4 dynamics play a critical role in the bistable switching behavior that allows terminal differentiation of GC B cells into PCs.
Memory B-Cell Differentiation.
The regulatory mechanisms that trigger GC B cells transition into the memory compartment are poorly understood. Several models have emerged over the years: Sustained exposure to CD40L has been suggested to direct GC B cells toward the memory B-cell compartment (18); survival in the GC has been postulated to be sufficient to allow differentiation into a memory B cell, whereas PCs would require a more-stringent affinity-based signal (19). Recent data suggest a more dynamic model where PCs and memory B cells emerge at all stages of the GC reaction (20). Consistent with this finding, the idea that memory B cells are stochastically selected from the pool of available GC B cells has gained support (21).
We interrogated our GC exit model about the kinetic changes that could allow the development of a stable population of MCs, defined by low levels of IRF4 and BCL6 and lack of BLIMP1 expression. Our model suggests that abrogation of IRF4 transcriptional program after centrocyte activation may be necessary for maturation into memory B cells. Specifically, removal of the IRF4-mediated BLIMP1 activation in the model leads to a steady state after BCR and CD40 signaling that recapitulates the memory B-cell phenotype (Fig. S3). This observation is in agreement with experimental findings suggesting that IRF4 may not be required for memory B-cell generation (22).
Abrogation of the IRF4 regulatory program may be achieved by different mechanisms: lowered expression levels of IRF4 binding cofactors, changes of IRF4 concentration as a result of asymmetric cell divisions or stochastic fluctuations, epigenetic silencing of IRF4 responsive elements in key target promoters, or additional unknown signals delivered at the GC. Regardless, our model suggests that small changes in the regulatory program of IRF4 are sufficient to induce a memory B-cell phenotype, further stressing the key role of IRF4 kinetics in GC B-cell development.
Comparison with Other Models in the Literature.
The gene regulatory network summarized in Fig. 1 represents a minimal model that captures the complex developmental process of B-cell maturation. However, other models have been proposed in the literature, several of them aiming to address the earlier developmental decision of undergoing CSR and SHM versus progressing to further differentiation. PAX5, a transcription factor required for both the establishment of the B-cell lineage identity and the process of CSR and SHM (23), and BLIMP1 have been shown to mutually repress each other's expression in a double-negative feedback loop (24). This binary switch has been proposed to produce a delay in BLIMP1 induction necessary to undergo CSR (25). Additionally, because IRF4-deficient B cells lack CSR (26), it has been proposed that a graded expression of IRF4 controlled by the BCR signaling intensity underlies two mutually antagonistic programs, where low IRF4 levels promote CSR and SHM, and higher concentrations induce BLIMP1 transcription and PC differentiation (7, 27). It has also been postulated that a double-positive feedback loop between BLIMP1 and IRF4 would be responsible for keeping the PC status (7).
We have investigated whether these additional regulatory interactions could potentially modify the qualitative behavior of the minimal model defined in Fig. 1. To do so, we have defined three extended models that include the additional regulatory interactions defined above, and compare them with the minimal model (Fig. S4 and SI Materials and Methods). The simulations run in all models show that for a broad range of biologically relevant parameters, the additional interactions of the extended models do not modify the qualitative behavior of the minimal model, as they only reinforce dynamical patterns that are already taken into account.
GC-Derived Malignancies and Associated Lymphomagenic Mechanisms.
The mechanisms that mediate the remodeling of antigen receptors in the GCs involve potentially mutagenic DNA double-strand breaks and suppression of the apoptotic machinery by BCL6 (28, 29). Failure to reactivate apoptosis upon exit from the GC has been established as a key mechanism of lymphomagenesis, and it has been specifically linked to diffuse large B cell lymphoma (DLBCL), an aggressive GC-derived malignancy that accounts for ≈35% of all non-Hodgkin lymphoma (NHL) cases. DLBCL is a heterogeneous disease with two major subtypes: GC B cell-like (GCB) subtype, characterized by an expression signature more similar to normal GC B cells, and a poor prognosis activated B-cell like (ABC) subtype, expressing genes typically induced after in vitro BCR stimulation (30).
DLBCL has been linked to an aberrant block of the GC exit program (2), among other mechanisms, via translocations of BCL6 (31, 32) and inactivation of BLIMP1 (33–35). In addition, constitutive activation of the NF-κB pathway, which activates IRF4 in the ABC-DLBCL tumor cells (36), is required for survival of the ABC cells (37). Although the functional role of IRF4 in ABC-DLBCL cells remains to be fully elucidated, the IRF4-interacting factor SPIB has emerged as an oncogene in the ABC subtype, but not in the GCB (38). The major oncogenic roles played by the key regulators of the GC exit pathway in DLBCL suggest that our model can be used for a systematic elucidation of previously identified lymphomagenic mechanisms and the determination of new candidate alterations. We define and study nine aberrant versions of the model that recapitulate the most common genetic alterations involved in DLBCL tumorigenesis. For each model, we simulate its kinetics by using parameters inferred from normal B-cell assays (SI Materials and Methods). Starting from a resting GC stage, we simulate stimulation by BCR and CD40 signals and evaluate the stability of the transition to PCs. For simplicity and because the model is not validated, we do not study exit into the memory B-cell compartment. Results are shown in Fig. 4. Our model shows significant agreement with experimental data for samples characterized by the corresponding alteration, suggesting that the proposed regulatory model could be valuable in elucidating novel mechanisms of GC-related lymphomagenesis.
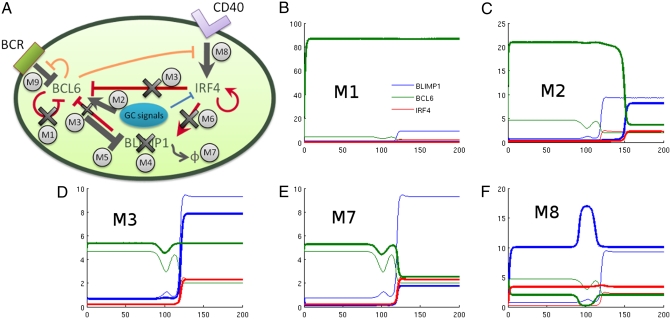
Fig. 4.
(A) Schematic representation of the most common genetic alterations in DLBCL. (B–F) Simulations of the different cancer models mimicking the genetic alterations in DLBCL. M1, loss of BCL6 auto-regulation; M2, constitutive high expression of BCL6; M3, synergistic loss of IRF4 and BLIMP1-mediated BCL6 silencing; M7, reduced BLIMP1 protein stability; M8, NF-κB constitutive signaling. Thick lines show protein levels in the cancer models, whereas thin lines show protein levels in the normal GC exit pathway model for comparison.
Models 1 and 2: BCL6 Dysregulation.
The most common genetic alterations in DLBCL affect the BCL6 promoter region and comprise mutations in the 5′ noncoding region (73% of the cases) and chromosomal translocations (45% of cases). Translocations are more frequent in the ABC-DLBCL (2), where the rearrangement breakpoints cluster within the promoter region and result in constitutive BCL6 expression (31, 32). Mutations in the BCL6 autoregulatory elements are almost exclusively seen in the GCB subtype (39) and may be responsible for the inactivation of the BCL6 negative regulatory circuit (9, 40). Somatic mutations in the regulatory region may also prevent IRF4-mediated down-regulation (10). This possibility will be explored in model 3.
The inactivation of BCL6 autoregulation is seen in many DLBCL cases but not in other GC-derived NHL (9) which suggests a DLBCL-specific regulatory mechanism. To better understand the mechanistic role of BCL6 self-regulation in lymphomagenesis, we define model 1 (Fig. 4A), where negative autoregulation of BCL6 is abrogated. We complement it with model 2, where BCL6 is constitutively expressed. Models 1 and 2 recapitulate the BCL6 alterations of the GCB and ABC subtype respectively. Specifically, in model 1 we eliminate the Hill function associated with BCL6 self-repression (Eq. S2), whereas in model 2 we increase BCL6 basal transcription by 10-fold. All other parameters are unchanged.
Fig. 4 B and C show numerical simulations run in both models. As expected, both models show up-regulation in the levels of BCL6 expression; however, the increase is much more pronounced in model 1 where it reaches levels 20-fold higher than normal cells, compared with a more modest fourfold increase in model 2. The key difference between these models is the robustness of BCL6 overexpression in model 1, where the levels of BCL6 are virtually unaffected by BCR and CD40 signals. In this model, high expression of BCL6 prevents transcriptional activation of IRF4 and BLIMP1 after physiologic GC signals and, thus, locks the cell in a GC stage where it may accumulate additional mutations over time. Conversely, model 2 shows a phenotype intermediate between GC and PC, more consistent with the ABC-DLBCL subtype that often shows coexpression of BCL6, IRF4, and BLIMP1. In this pathogenic scenario, some cells may be stochastically selected to leave the GC, whereas the cells left in undifferentiated states may contribute to lymphomagenesis because of additional mutations acquired at the GC.
Model 3: Loss of IRF4- and BLIMP1-Mediated Transcriptional Silencing of BCL6.
The BCL6 promoter is rich in IRF4-binding sites, which make this interaction prone to dysregulation in DLBCL. Chromosomal translocations and mutations can disrupt the IRF4-responsive region in the BCL6 promoter and block its down-regulation after CD40 signaling (6). Complete or partial inactivation of CD40–mediated IRF4 regulation of BCL6 expression affects predominantly the ABC subtype.
This pathogenic scenario can be modeled by eliminating the Hill function associated with IRF4-mediated repression of BCL6 (Eq. S2). Simulations do not show a significant change in expression levels in any of the three proteins before or after stimulation. Similar results are obtained when BLIMP1-mediated BCL6 repression is dysregulated. We then analyze the simultaneous loss of IRF4- and BLIMP1-mediated BCL6 repression (model 3), leading to an unexpected synergistic interaction (Fig. 4D) that abrogates BCL6 repression and prevents BLIMP1 up-regulation. The redundancy in gene regulation makes the system robust against losses of individual BCL6 repression by either BLIMP1 or IRF4 but not both. This robustness seems necessary to counter the high incidence of mutations targeting the regulatory elements in the BCL6 promoter during the physiological process of SHM.
Models 4–7: Mutations Targeting BLIMP1.
A vast majority of ABC-DLBCL malignancies fail to express a competent BLIMP1 protein despite normal IRF4 expression. BLIMP1 function is dysregulated by multiple mechanisms in ≈53% of ABC-DLBCL, including inactivating mutations (33, 34), transcriptional repression by constitutively active BCL6, and mutations affecting the protein stability and its transrepression activity (35). It has also been suggested that BCL6 may prevent high-affinity DNA binding by IRF4 and/or maintain IRF4 in an autoinhibitory state (41), preventing IRF4-mediated BLIMP1 activation. Furthermore, an additional fraction of ABC-DLBCL cases fails to express BLIMP1 at the protein level, despite normal prdm1, irf4, and bcl6 gene function, suggesting additional posttranscriptional and posttranslational mechanisms (35).
Models 4–7 explore different mechanisms of BLIMP1 inactivation (Fig. 4A), including loss-of-function mutations (model 4), transcriptional silencing by BCL6 (model 5), impairment of IRF4-mediated BLIMP1 activation (model 6), and mutations leading to an increased turnover of BLIMP1 protein (model 7). The latter also recapitulate posttranscriptional inactivation by microRNA. Analysis of these models after BCR and CD40 signaling consistently shows that BLIMP1 protein expression levels fail to reach the normal levels necessary for PC differentiation. For instance, Fig. 4E shows model 7 dynamics, where a fivefold increase of BLIMP1 turnover results in BLIMP1 protein levels comparable to those of normal GC B cells before BCR and CD40 activation.
Models 8 and 9: Mutations Targeting the Signaling Pathways.
A hallmark of ABC-DLBCL is constitutive activation of the NF-κB pathway (37). ABC cells often harbor point mutations targeting genes involved in the regulation of NF-κB, leading to its constitutive activation (42, 43). In some ABC cell lines, BCR is also found to be chronically activated by somatic mutations, leading to NF-κB activation through a different pathway (44).
Model 8 explores constitutively activation of the NF-κB pathway. NF-κB activation induces enhanced transactivation of IRF4 in ABC-DLBCL cell lines (6, 36). We simulate this effect by a 10-fold increase in IRF4 transcription rates. In our model, this change accelerates GC B-cell differentiation toward the PC compartment (Fig. 4F) by rapidly achieving post-GC levels of BLIMP1 and IRF4. Although higher levels of IRF4 may lead to premature GC exit before completing affinity maturation, our model suggests that this mechanism is not sufficient for tumorigenesis and would require additional mutations to be pathogenic. Whereas NF-κB may promote lymphomagenesis by providing antiapoptotic and proproliferative signals, our model suggests that its role is mostly associated with tumor subtype progression rather than with initiation, as also suggested by the fact that NF-κB activation is frequently associated with BLIMP1 mutations in ABC-DLBCL (33, 35).
Simultaneous dysregulation of both CD40 and BCR pathways is observed in some ABC-DLBCL cases. Model 9 explores this situation. Similar to the previous case, the cell acquires an early terminally differentiated phenotype. However, this time the cell is characterized by the near-total absence of BCL6 expression, because of the combined effect of BCR-promoted degradation and CD40-mediated transcriptional silencing by IRF4.
Discussion
We have developed a kinetic model to quantitatively characterize B-cell exit from the GC phase and terminal differentiation into plasma and memory B cells.
In mature B cells, our model predicts the existence of two different hysteresis cycles associated to the BCR and the CD40 signaling pathways. Cooperativity between these two signaling events ensures the correct maturation into a terminally differentiated B-cell. This finding suggests a variety of experimentally testable hypotheses aimed at determining the minimum duration of BCR signals necessary to induce a preplasma-cell state, and persistence of this state in the absence or presence of additional CD40 signals. Although the model is not complete because it does not include all regulators and signals contributing to B-cell maturation, its power lies in its minimality: Comprising only three key gene regulators and two signaling pathways, the model can recapitulate the normal and pathological GC B-cell exit and, furthermore, it can elucidate a variety of mechanisms, leading to partially or fully incompetent immune response after antigen presentation.
At the molecular level, the stimulus response of IRF4 plays a central role in the differentiation pathway. The kinetics of the differentiation module is controlled by a dimensionless parameter β that accounts for the homeostatic level of IRF4. Low levels of β keep the cell in the GC stage, whereas an increase prompts the cell toward the PC fate, where it is prevented from dedifferentiating by a self-positive regulatory loop on IRF4. Interestingly, our model suggests that the correct differentiation process behavior is very sensitive to small changes in the parameters incorporated in β, i.e., the parameters that regulate IRF4 dynamics. During the normal development of a GC B cell, β is increased only through the CD40-mediated stimulation of IRF4, but a variety of tumorigenic alterations may directly or indirectly target IRF4 expression or protein stability. For instance, after an aberrant change in the IRF4 self-induced transcription rate, GC B cells lose the ability to transit through the normal developmental pathway and become prematurely locked into the GC or PC state (Fig. S5). Supporting this idea, constitutive high expression of IRF4 due to an aberrant regulatory network is observed in multiple myeloma, a malignancy of PCs (16). Conversely, we hypothesize that mutations in the IRF4 promoter site that abrogate the IRF4 self-binding site could prevent GC B cells from fully differentiating into PCs. IRF4 could also play a role in directing cells toward the memory B-cell compartment. Abrogation of IRF4-mediated BLIMP1 activation in our model leads to an irreversible state that closely recapitulates the memory B-cell expression signature. This experimentally testable hypothesis suggests that IRF4 could be a master regulator whose expression levels and regulatory interactions direct GC B cells maturation into either memory B or PCs, possibly as a result of stochastic fluctuations of the levels of these proteins in individual cells.
We have also explored the impact of some of the most common genetic aberrations in DLBCL, a GC-derived malignancy where BCL6 and BLIMP1 alterations play a key role. Our model shows that abrogation of the self-regulatory loop of BCL6 has a very strong impact in the dynamical evolution of the cell, resulting not only in an increased expression of BCL6 but also in the cell's inability to respond to differentiation signals, which could explain the high incidence of DLBCL cases that bypass the autoregulatory circuit. Additionally, the BCL6 promoter region is particularly prone to mutations and chromosomal rearrangements that can result in the loss of transcriptional interactions aimed at silencing BCL6. Our model shows that B cells can still undergo normal differentiation after losing either IRF4- or BLIMP1-mediated repression of BCL6, but not both of them.
Our model also shows that mutations or alteration targeting the expression of BLIMP1 have a drastic impact in the production of competent BLIMP1 protein and seriously compromises the cell's ability to exit the GC stage. A cell blocked in the GC is potentially exposed to additional genetic stress (e.g., DNA double-strand breaks through SHM), whereas DNA-break sensing genes, most notably p53, are transcriptionally silenced by BCL6 (28), therefore greatly increasing the risk of acquiring unchecked additional tumorigenic lesions (2). We have also explored the importance of the malfunction of additional signaling pathways recently reported in the literature. Alterations of the NF-κB or of the BCR signaling pathway aberrantly accelerate transition toward a terminally differentiated stage, but do not appear to block differentiation. When these are combined with additional mutations targeting the GC exit pathway, such as BLIMP1 inactivation, those alterations can increase tumorigenesis by further repressing apoptosis and increasing cell proliferation.
Materials and Methods
The kinetics of the GC exit pathway gene regulatory module was modeled by using ordinary differential equations. The time course simulations and the bifurcation analysis were performed by using Matlab and the Matcont continuation package for Matlab, respectively. Model parameters we estimated by using microarray gene expression datasets from normal, transformed, and experimentally manipulated human B cells related to the GC reaction (GEO accession no. GSE12195), and gene expression profiling of B lymphocytes and PCs (GEO GSE6691). Additional information can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Riccardo Dalla Favera, Laura Pasqualucci, Katia Basso, Yishai Shimoni, and Gabrielle Rieckhof for insightful discussions on the B-cell physiology and many useful comments on the manuscript. We also thank the generous support of National Cancer Institute Grant 1R01CA109755-01A1, National Institute of Allergy and Infectious Diseases Grant 1R01AI066116-01, and National Institutes of Health Roadmap Grants 1U54CA121852-01A1 and 2U54CA121852-06.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113019109/-/DCSupplemental.
References
- 1.Liu Y-J, et al. Mechanism of antigen-driven selection in germinal centres. Nature. 1989;342:929–931. doi: 10.1038/342929a0. [DOI] [PubMed] [Google Scholar]
- 2.Klein U, Dalla-Favera R. 2008 Germinal centres: Role in B-cell physiology and malignancy. Nat Rev Immunol 8:22–33. [Google Scholar]
- 3.Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of Inflammation, Cytokine Expression, and Germinal Center Formation by BCL-6 Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 4.Niu H, Ye BH, Dalla-Favera R. Antigen receptor signaling induces MAP kinase-mediated phosphorylation and degradation of the BCL-6 transcription factor. Genes Dev. 1998;12:1953–1961. doi: 10.1101/gad.12.13.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mittrücker H-W, et al. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 1997;275:540–543. [PubMed] [Google Scholar]
- 6.Saito M, et al. A signaling pathway mediating downregulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer Cell. 2007;12:280–292. doi: 10.1016/j.ccr.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 7.Sciammas R, et al. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 2006;25:225–236. doi: 10.1016/j.immuni.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Turner CA, Jr, Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77:297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- 9.Pasqualucci L, et al. Mutations of the BCL6 proto-oncogene disrupt its negative autoregulation in diffuse large B-cell lymphoma. Blood. 2003;101:2914–2923. doi: 10.1182/blood-2002-11-3387. [DOI] [PubMed] [Google Scholar]
- 10.Basso K, et al. 2009. Integrated biochemical and computational approach identifies BCL6 direct target genes controlling multiple pathways in normal germinal-center B cells. Blood, 2009-06-227017.
- 11.Shaffer AL, et al. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13:199–212. doi: 10.1016/s1074-7613(00)00020-0. [DOI] [PubMed] [Google Scholar]
- 12.Pasqualucci L, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphomas. Nature. 2008;471:189–195. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bereshchenko OR, Gu W, Dalla-Favera R. Acetylation inactivates the transcriptional repressor BCL6. Natl Genet. 2002;32:606–613. doi: 10.1038/ng1018. [DOI] [PubMed] [Google Scholar]
- 14.Shaffer AL, et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 15.Lin L, Gerth AJ, Peng SL. Active inhibition of plasma cell development in resting B cells by microphthalmia-associated transcription factor. J Exp Med. 2004;200:115–122. doi: 10.1084/jem.20040612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaffer AL, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tyson JJ, Chen KC, Novak B. Sniffers, buzzers, toggles and blinkers: Dynamics of regulatory and signaling pathways in the cell. Curr Opin Cell Biol. 2003;15:221–231. doi: 10.1016/s0955-0674(03)00017-6. [DOI] [PubMed] [Google Scholar]
- 18.Arpin C, et al. Generation of memory B cells and plasma cells in vitro. Science. 1995;268:720–722. doi: 10.1126/science.7537388. [DOI] [PubMed] [Google Scholar]
- 19.Tarlinton DM, Smith KGC. Dissecting affinity maturation: A model explaining selection of antibody-forming cells and memory B cells in the germinal centre. Immunol Today. 2000;21:436–441. doi: 10.1016/s0167-5699(00)01687-x. [DOI] [PubMed] [Google Scholar]
- 20.Blink EJ, et al. Early appearance of germinal center-derived memory B cells and plasma cells in blood after primary immunization. J Exp Med. 2005;201:545–554. doi: 10.1084/jem.20042060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarlinton D. B-cell memory: Are subsets necessary? Nat Rev Immunol. 2006;6:785–790. doi: 10.1038/nri1938. [DOI] [PubMed] [Google Scholar]
- 22.Klein U, et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- 23.Schebesta A, et al. Transcription factor Pax5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration, and immune function. Immunity. 2007;27:49–63. doi: 10.1016/j.immuni.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 24.Mora-López F, Reales E, Brieva JA, Campos-Caro A. Human BSAP and BLIMP1 conform an autoregulatory feedback loop. Blood. 2007;110:3150–3157. doi: 10.1182/blood-2007-05-092262. [DOI] [PubMed] [Google Scholar]
- 25.Muto A, et al. Bach2 represses plasma cell gene regulatory network in B cells to promote antibody class switch. EMBO J. 2010;29:4048–4061. doi: 10.1038/emboj.2010.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein U, et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- 27.Sciammas R, et al. An incoherent regulatory network architecture that orchestrates B cell diversification in response to antigen signaling. Mol Syst Biol. 2011;7:495. doi: 10.1038/msb.2011.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- 29.Ranuncolo SM, et al. Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat Immunol. 2007;8:705–714. doi: 10.1038/ni1478. [DOI] [PubMed] [Google Scholar]
- 30.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 31.Ye BH, et al. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science. 1993;262:747–750. doi: 10.1126/science.8235596. [DOI] [PubMed] [Google Scholar]
- 32.Lo Coco F, et al. Rearrangements of the BCL6 gene in diffuse large cell non-Hodgkin's lymphoma. Blood. 1994;83:1757–1759. [PubMed] [Google Scholar]
- 33.Pasqualucci L, et al. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma. J Exp Med. 2006;203:311–317. doi: 10.1084/jem.20052204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tam W, et al. Mutational analysis of PRDM1 indicates a tumor-suppressor role in diffuse large B-cell lymphomas. Blood. 2006;107:4090–4100. doi: 10.1182/blood-2005-09-3778. [DOI] [PubMed] [Google Scholar]
- 35.Mandelbaum J, et al. BLIMP1 is a tumor suppressor gene frequently disrupted in activated B cell-like diffuse large B cell lymphoma. Cancer Cell. 2010;18:568–579. doi: 10.1016/j.ccr.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lam LT, et al. Small molecule inhibitors of IkappaB kinase are selectively toxic for subgroups of diffuse large B-cell lymphoma defined by gene expression profiling. Clin Cancer Res. 2005;11:28–40. [PubMed] [Google Scholar]
- 37.Davis RE, et al. Constitutive Nuclear Factor κB activity is required for survival of activated B cell–like diffuse large B cell lymphoma cells. J Exp Med. 2001;194:1861–1874. doi: 10.1084/jem.194.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lenz G, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci USA. 2008;105:13520–13525. doi: 10.1073/pnas.0804295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iqbal J, et al. Leukemia/Lymphoma Molecular Profiling Project Distinctive patterns of BCL6 molecular alterations and their functional consequences in different subgroups of diffuse large B-cell lymphoma. Leukemia. 2007;21:2332–2343. doi: 10.1038/sj.leu.2404856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, Li Z, Naganuma A, Ye BH. Negative autoregulation of BCL-6 is bypassed by genetic alterations in diffuse large B cell lymphomas. Proc Natl Acad Sci USA. 2002;99:15018–15023. doi: 10.1073/pnas.232581199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gupta S, et al. Lineage-specific modulation of interleukin 4 signaling by interferon regulatory factor 4. J Exp Med. 1999;190:1837–1848. doi: 10.1084/jem.190.12.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Compagno M, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459:717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lenz G, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319:1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 44.Davis RE, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463:88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.