Abstract
Nitric oxide (NO) is implicated in neuronal cell survival. However, excessive NO production mediates neuronal cell death, in part via mitochondrial dysfunction. Here, we report that the mitochondrial ubiquitin ligase, MITOL, protects neuronal cells from mitochondrial damage caused by accumulation of S-nitrosylated microtubule-associated protein 1B-light chain 1 (LC1). S-nitrosylation of LC1 induces a conformational change that serves both to activate LC1 and to promote its ubiquination by MITOL, indicating that microtubule stabilization by LC1 is regulated through its interaction with MITOL. Excessive NO production can inhibit MITOL, and MITOL inhibition resulted in accumulation of S-nitrosylated LC1 following stimulation of NO production by calcimycin and N-methyl-D-aspartate. LC1 accumulation under these conditions resulted in mitochondrial dysfunction and neuronal cell death. Thus, the balance between LC1 activation by S-nitrosylation and down-regulation by MITOL is critical for neuronal cell survival. Our findings may contribute significantly to an understanding of the mechanisms of neurological diseases caused by nitrosative stress-mediated mitochondrial dysfunction.
Keywords: neurodegenerative diseases, oxidative stress
Nitric oxide (NO) is a signaling molecule implicated in a variety of physiological processes (1). However, excessive NO mediates neuronal cell death by nitrosative stress, potentially contributing to the pathology of many neurodegenerative disorders (2). In the nervous system, NO is synthesized mainly by neuronal NO synthases (nNOS), which are regulated by calcium mobilization (3). One way of propagating NO signals is the S-nitrosylation of proteins, which involves the covalent attachment of a nitrogen monoxide group to the thiol side chain of a cysteine residue (4). S-nitrosylation of several proteins causes neurotoxicity, in part by triggering protein misfolding and aggregation and mitochondrial dysfunction (5, 6). Neurons are particularly vulnerable to mitochondrial dysfunction because they require high levels of energy for their survival and synaptic function (7). Accumulating evidence suggests that many mitochondrial proteins or regulatory proteins for mitochondrial function are physiological substrates for S-nitrosylation under nitrosative stress (8). S-nitrosylation of mitochondrial fission protein, Drp1, has been shown to link excessive mitochondrial fission to neuronal injury in neurodegeneration (9). Thus, nitrosative stress because of the generation of excessive NO can mediate neurotoxicity, in part via the S-nitrosylation of mitochondria-related proteins.
Microtubule-associated protein 1B (MAP1B) is a protein complex that consists of a heavy chain and light chain (LC1) (10). MAP1B plays an important role in the stability of the cytoskeleton and may have other cellular functions. In particular, MAP1S (C19ORF5), a sequence homolog of MAP1A/MAP1B, has been shown to accumulate in mitochondria and induce mitochondrial aggregation and genome destruction via a specific domain named MAGD (11). Similar to MAP1S, MAP1A and MAP1B are considered to play an important role in the regulation of mitochondrial dynamics and cell death. LC1 has recently been shown to be S-nitrosylated on cysteine 257 and translocated to microtubules via a conformational change of LC1 (12). Additionally, LC1 has been implicated in human neurological disorders, such as giant axonal neuropathy (GAN), fragile-X syndrome, spinocerebellar ataxia type 1, and Parkinson disease (10, 13, 14). Therefore, the control of S-nitrosylated LC1 levels is critical for neuronal cell survival.
We previously identified a mitochondrial ubiquitin ligase, MITOL (also known as March5), which is involved in mitochondrial dynamics and mitochondrial quality control (15–19). However, the exact roles of MITOL in mitochondria are still obscure. Here, we report that MITOL ubiquitinates LC1 in an S-nitrosylation–dependent manner and promotes LC1 degradation via the ubiquitin-proteasome pathway. The implications of mitochondrial dysfunction via S-nitrosylated LC1 in nitrosative stress-related neurodegenerative diseases are discussed.
Results
MITOL Interacts with and Ubiquitinates MAP1B-LC1 in Mitochondria.
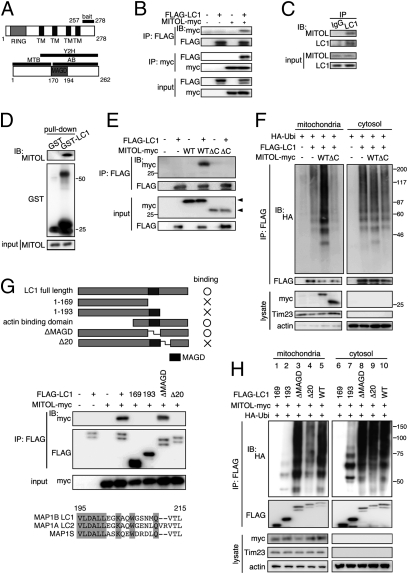
We found that MITOL knockdown induced mitochondrial aggregation and microtubule stabilization (Fig. S1). To search for microtubule-related substrate of MITOL, we performed a yeast two-hybrid screen on a mouse brain cDNA library using the C-terminal region of MITOL (amino acids 257–278) as the bait fragment (Fig. 1A) and identified MAP1B-LC1, which possesses a microtubule-binding domain (MTB), an actin-binding domain (AB), and a MAGD (Fig. 1A) (11). A subcellular fractionation and immunofluorescence analyses revealed that LC1 exists not only in the cytosol but also in the mitochondria (Fig. S2 A and B). Trypsin treatment and sodium carbonate extraction assay suggested that LC1 is not embedded in the mitochondrial outer membrane, but rather peripheral proteins on the surface of the mitochondrial outer membrane (Fig. S2 C and D). Immunoprecipitation assays indicated that MITOL-myc interacted with FLAG-LC1 in SH-SY5Y cells (Fig. 1B). Similarly, endogenous MITOL was found to be coimmunoprecipitated with anti-LC1 Ab (Fig. 1C). Moreover, a GST pull-down assay demonstrated that endogenous MITOL coprecipitated with GST-LC1 but not with GST alone (Fig. 1D). These results suggest that MITOL interacts with LC1 on the mitochondrial outer membrane.
Fig. 1.
MITOL interacts with and ubiquitinates mitochondrial LC1. (A) Schematic structures of MITOL and MAP1B-LC1 (LC1). (B) Association of MITOL with LC1 in cell expression system. Immunoprecipitation (IP) assay followed by immunoblot (IB) assay (IP-IB) was performed on SH-SY5Y cells cotransfected with indicated vectors. (C) Endogenous association of MITOL with LC1. IP-IB assay was performed on mouse brain lysates with control IgG or anti-LC1 antibody (Ab). (D) Pull-down of endogenous MITOL with GST-LC1. GST pull-down assay was performed on mouse brain lysates with GST-only or GST-LC1. (E and F) The C terminus of MITOL is essential for interaction with LC1 (E) and ubiquitination of mitochondrial LC1 (F). WT, WT MITOL-myc (1–278aa); ΔC, C terminus deleted MITOL-myc (1–256aa). The position of each MITOL-myc is indicated by an arrowhead. (G) Mapping of the binding sites of LC1 on MITOL. Structures of LC1 mutants and their binding abilities to MITOL are indicated. The sequence of 20-aa region of LC1 interacting with MITOL is compared with those of MAP1A and MAP1S. (H) MITOL fails to ubiquitinate Δ20 LC1 mutant in mitochondria.
To identify the sites of interaction between MITOL and LC1, we examined whether a C terminus- (amino acids257–278) deleted MITOL mutant (ΔC MITOL) interacts with and ubiquitinate LC1. As expected, the interaction between ΔC MITOL and LC1 was completely abolished (Fig. 1E). Moreover, ΔC MITOL failed to ubiquitinate mitochondrial LC1 (Fig. 1F). Thus, the C-terminal domain of MITOL is essential for interaction with and ubiquitination of LC1. To determine which domain of LC1 is responsible for the interaction with MITOL, various deletion mutants of LC1 were generated (Fig. 1G). Immunoprecipitation assays revealed that amino acid 1–169 and amino acid 1–193 deletion mutants of LC1 could not associate with MITOL, whereas a MAGD-deleted mutant (ΔMAGD) associated with MITOL. We identified a specific region (amino acids 195–215) located on the C-terminal side of MAGD as the potential binding site for LC1 with MITOL. As confirmation of this as the binding site, Δ20 LC1, a 20-aa deletion mutant (amino acids 195–215), could not interact with MITOL. Because this domain is well conserved among all MAP1 family proteins (Fig. 1G, Lower), it may play an important and ubiquitous role in protein–protein interactions. Finally, we examined whether MITOL can ubiquitinate Δ20 LC1. As expected, MITOL-dependent ubiquitination of mitochondrial Δ20 LC1 was severely reduced, whereas ubiquitination of cytosolic Δ20 LC1 was not affected (Fig. 1H, lanes 4 and 9). Thus, a specific interaction between the C-terminal domain of MITOL and a 20-aa domain of LC1 is critical for the recognition and ubiquitination of LC1 by MITOL.
MITOL Facilitates the Degradation of Mitochondrial LC1.
MITOL was found to ubiquitinate endogenous LC1 (Fig. S3A) and an in vitro ubiquitination assay suggested that MITOL directly ubiquitinates LC1 (Fig. S3B). MITOL ubiquitinates mitochondrial LC1 but not cytosolic LC1 in SH-SY5Y cells (Fig. S3C). A previous study suggested that ubiquitination of LC1 is, in part, required for gigaxonin, the mutations of which cause GAN (20). The ubiquitination of mitochondrial LC1 was not affected by gigaxonin-knockdown, whereas gigaxonin knockdown significantly decreased the ubiquitination of cytosolic LC1 (Fig. S3D). Therefore, gigaxonin appears to be required for the MITOL-independent ubiquitination of cytosolic LC1. The cycloheximide (CHX) chase assay revealed a rapid degradation of LC1 by MITOL overexpression compared with that of LC1 without MITOL expression, and the degradation of LC1 was inhibited by treatment with the proteasomal inhibitor, MG132 (Fig. S3E). These results demonstrate that MITOL promotes LC1 degradation via the ubiquitin-proteasome pathway. Immunofluorescence analysis indicated that expression of LC1 alone induced mitochondrial aggregation, but that coexpression with MITOL completely blocked LC1 accumulation and LC1-induced mitochondrial aggregation (Fig. S3F). Thus, MITOL may regulate LC1 function in mitochondria by controlling mitochondrial aggregation via mitochondrial LC1 levels.
We next examined the effects of MITOL knockdown on LC1 degradation and mitochondrial morphology. Specific siRNA-mediated MITOL knockdown reduced the ubiquitination of FLAG-LC1 in the mitochondrial fraction but not in the cytosolic fraction (Fig. 2A). The reduced levels of LC1 ubiquitination caused by MITOL siRNA were rescued by cotransfection of a siRNA-resistant MITOL gene, verifying the specificity of knockdown by the MITOL siRNA. Additionally, MITOL knockdown increased mitochondrial accumulation of endogenous LC1 by ∼1.5-fold, which was restored by cotransfection of a siRNA-resistant MITOL gene (Fig. 2B). In contrast, the amount of cytosolic LC1 was not altered by MITOL knockdown. Furthermore, a CHX assay indicated that MITOL knockdown caused delayed degradation of LC1, which was restored by cotransfection of a siRNA-resistant MITOL gene (Fig. 2C). These results demonstrate that MITOL knockdown causes LC1 accumulation in mitochondria by inhibiting degradation of mitochondrial LC1.
Fig. 2.
MITOL knockdown inhibits the ubiquitination and degradation of LC1. (A) MITOL knockdown decreased the ubiquitination of mitochondrial LC1. IP-IB assay was performed on mitochondrial and cytosolic fractions separated from SH-SY5Y cells cotransfected with siRNAs [sc (scramble) or si, MITOL-specific siRNA] or MITOL WT resistant to MITOL siRNA (res). (B) MITOL knockdown induced an accumulation of endogenous LC1 in mitochondria. IB assay was performed on lysates of whole cells, mitochondrial, and cytosolic fractions separated from SH-SY5Y cells cotransfected with siRNAs (sc or si, MITOL-specific siRNA) or MITOL WT resistant to MITOL siRNA (res). Statistic results are shown in the bottom. (C) MITOL knockdown caused a delayed degradation of FLAG-LC1. CHX-chase assay was performed, as described previously (18), on SH-SY5Y cells cotransfected with indicated siRNAs. Statistic data are shown on the right. (D) MITOL knockdown induced LC1 accumulation and LC1-mediated mitochondrial aggregation. SH-SY5Y cells cotransfected with DsRed-MITO and indicated siRNAs were immunostained with anti-LC1 Ab. The LC1 knockdown by shRNA are shown in the upper right panel. Statistic analysis indicates the ratio of cells with mitochondrial aggregation. (Scale bar, 10 μm.) (B–D) Error bars represent SD. *P < 0.05, n = 3, one-way ANOVA.
We previously reported that MITOL knockdown causes mitochondrial fragmentation and aggregation (18). Mitochondrial aggregation seen in MITOL-knockdown cells may be a result of LC1 accumulation. To test this possibility, immunofluorescence microscopy analysis was performed on cells in which MITOL was knocked down. Compared with the control cells, endogenous LC1 in MITOL-knockdown cells extensively accumulated in the mitochondria and showed perinuclear aggregation (Fig. 2D). Because this mitochondrial aggregation was partially blocked by LC1 knockdown, mitochondrial aggregation by MITOL knockdown is, in part, because of LC1 accumulation in mitochondria.
S-Nitrosylation Confers a Specific Signal for Ubiquitination and Degradation of LC1.
The E3 ubiquitin ligases recognize substrates, in some cases, through posttranslational modifications, such as phosphorylation, oxidation, or glycosylation (21–23). We focused on S-nitrosylation of LC1 because a previous study indicated that the S-nitrosylation of LC1 at cysteine 257 (C257) may induce a conformational change in LC1 (12). To test this possibility, we constructed nonnitrosylation mutant C257S (12) and nitrosomimetic mutant C257W (24), as illustrated in Fig. 3A, and examined the ubiquitinations of these mutants by MITOL. An in vivo ubiquitination assay revealed that MITOL-dependent ubiquitination of C257S was severely inhibited (Fig. 3B, lane 5), whereas that of C257W was drastically increased (Fig. 3B, lane 7). Thus, MITOL may ubiquitinate LC1 in an S-nitrosylation–dependent manner. A similar result was obtained with the cytosolic fraction, although the MITOL dependency was weak. We examined the effects of S-nitrosylation on the interaction between MITOL and LC1. Unexpectedly, an immunoprecipitation assay revealed that the interactions of these LC1 mutants with MITOL were not altered (Fig. S4A), suggesting that increased ubiquitination of C257W is not because of an enhanced interaction of MITOL with LC1. A CHX chase assay indicated that compared with LC1 WT, C257W rapidly degraded without MITOL cotransfection, and that its degradation was accelerated by MITOL cotransfection, whereas degradation of C257S was strongly inhibited even in the MITOL overexpressing cells (Fig. 3C). These results indicate that MITOL promotes the proteasomal degradation of S-nitrosylated LC1 by specific recognition of S-nitrosylated LC1.
Fig. 3.
S-nitrosylation–dependent ubiquitination and degradation of LC1. (A) Schematic structure of S-nitrosylation site of LC1 at cysteine 257 residue (C257) and its two mutants, C257S and C257W. (B) MITOL ubiquitinates C257W but not C257S. (C) Slow degradation of C257S and rapid degradation of C257W. CHX-chase assay was performed on SH-SY5Y cells cotransfected with indicated vectors. Error bars represent SD. (D and E) Effects of NO donor SNAP and nNOS inhibitor l-NAME (D), and calcimycin (E) on MITOL-dependent ubiquitination of LC1. SH-SY5Y cells cotransfected with indicated vectors were treated with SNAP and l-NAME. (F) Decreased S-nitrosylated LC1 by MITOL overexpression. S-nitrosylated LC1 was assessed by biotin-switch assay. Nitrosylated proteins was reduced by ascorbate and replaced with biotin. NaCl was substituted for ascorbate (asc-) as a negative control. Error bars represent SD. *P < 0.05, n = 3, Student t test. (G) Increased S-nitrosylated LC1 by MITOL knockdown. (H) Accumulation of endogenous S-nitrosylated LC1 by MITOL knockdown. Control and MITOL-knockdown cells were treated with calcimycin with or without l-NAME followed by biotin-switch assay (G and H) Error bars represent SD. *P < 0.05, n = 3, one-way ANOVA. (I) A schematic model for mechanism underlying S-nitrosylation–dependent ubiquitination of LC1 by MITOL.
We examined the effect of NO on LC1 ubiquitination by MITOL in SH-SY5Y cells. As shown in lanes 1–3 of Fig. 3D, ubiquitination levels of both mitochondrial and cytosolic LC1 were slightly increased by treatment with NO donor S-nitroso-N-acetylpenicillamine (SNAP) and decreased by treatment with N-nitro-l-arginine methyl ester( l-NAME), an arginine analog that is a potent and reversible inhibitor of NOS. Furthermore, with MITOL overexpression, this effect was greatly enhanced, particularly in the mitochondrial fraction (Fig. 3D, lanes 4–6). It is well known that intracellular calcium mobilization activates nNOS, resulting in NO generation in neuronal cells. As shown in Fig. 3E, lane 4, calcimycin stimulation enhanced the MITOL-dependent ubiquitination of LC1, whereas l-NAME inhibited calcimycin-induced ubiquitination of LC1. This result suggests that calcimycin mediates LC1 ubiquitination through NO production by nNOS activation. In addition, the association of LC1 with MITOL was not affected by either SNAP or l-NAME (Fig. S4B), consistent with our previous observation that the interaction of MITOL with LC1 is not dependent on S-nitrosylation of LC1. Taken together, these results suggest that MITOL ubiquitinates LC1 in an NO-dependent manner.
A biotin-switch assay for detecting proteins containing S-nitrosothiols (25) revealed that the amount of S-nitrosylated LC1 was specifically decreased in MITOL-overexpressing cells (Fig. 3F). Statistical analysis indicated that MITOL overexpression reduced the S-nitrosylated LC1 level by half, compared with control cells. On the other hand, MITOL knockdown increased the amount of S-nitrosylated LC1, and this increase was reversed by coexpression with an expression vector that is resistant to siMITOL (Fig. 3G). Endogenous S-nitrosylated LC1 also increased in MITOL-knockdown cells compared with control cells (Fig. 3H). Moreover, calcimycin treatment significantly enhanced the amount of S-nitrosylated LC1 in MITOL-knockdown cells, whereas l-NAME treatment decreased the amounts of S-nitrosylated LC1 in both control and MITOL-knockdown cells. Taken together, these results demonstrate that under physiological conditions, MITOL regulates the amount of S-nitrosylated LC1.
To gain a better understanding of the relationship between LC1 ubiquitination and the conformational change in LC1 caused by S-nitrosylation, LC1 was separated into two fragments, as illustrated in Fig. S5A. A previous study suggested that the fragment containing the MTB associates with the fragment containing the AB when unstimulated and dissociates from the AB fragment by S-nitrosylation at C257 (12). In the unstimulated state, an intense ubiquitination of AB by MITOL was detected to a similar degree as that of the S-nitrosylated form (Fig. S5A, lane 3). Mutation analysis suggests that three lysine sites located in AB (positions 164, 203, and 233) are major sites of MITOL-mediated ubiquitination (Fig. S5B). Thus, the AB fragment may expose these major ubiquitin sites to MITOL. Interestingly, the ubiquitination of AB was inhibited by the addition of MTB (Fig. S5A, lane 4), and this inhibition was cleared by treatment with calcimycin (Fig. S5A, lane 5). Furthermore, cotreatment with l-NAME inhibited calcimycin-induced ubiquitination of AB by MITOL. These results suggest that in the absence of stimulation of full-length LC1, ubiquitin sites in AB are masked by MTB and that S-nitrosylation of LC1 on C257 unmasks and exposes these ubiquitin sites to MITOL by a conformational change in LC1 (Fig. 3I).
MITOL Protects Neuronal Cells Against LC1-Induced Cytotoxicity.
A previous study demonstrated that overexpression of MAP1B by in utero electroporation revealed an impaired radial migration in developing cortical neurons (26). Similar to the findings of a previous study, radial migration was severely impaired by LC1 alone, but coexpression of MITOL completely rescued the LC1-mediated impaired migration (Fig. S6A). We examined the effects of MITOL knockdown on cortical migration and cell viability by a specific shRNA. MITOL knockdown induced a severe defect in cortical migration (Fig. 4A) and TUNEL-positive cell death (Fig. 4B). Statistical analyses are shown in Fig. S6 B and C, respectively. Co-knockdown of LC1 rescued the migration disorder and cell death observed in MITOL-knockdown cells, suggesting that impaired cortical migration and cell death by MITOL knockdown is, at least in part, the result of LC1 accumulation. Thus, MITOL appears to block LC1-mediated cytotoxicity in vivo.
Fig. 4.
MITOL protects neuronal cells against LC1-induced cytotoxicity. (A) Impaired cortical migration by MITOL knockdown was rescued by LC1 coknockdown. Sagittal sections of mice brains electroporlated at E14 in utero with indicated vectors plus EGFP were analyzed at postnatal day 2. CP; cortical plate, IZ; intermediate zone, VZ; ventricular zone. (Scale bar, 100 μm.) (B) Increased TUNEL-positive cortical neurons by MITOL knockdown. TUNEL assay was performed with postnatal day 2 mouse brain sections as described in A. Arrowheads indicate TUNEL-positive cells. (Scale bar, 10 μm.) (C) S-nitrosylated form of LC1 induced cell death under MITOL knockdown. SH-SY5Y cells were stained with Hoechst 33258 72 h after indicated transfection. Sc, scramble siRNA; si, siMITOL; LC1-WT, LC1 wild-type; LC1-CS, LC1 C257S mutant; LC1-CW, LC1 C257W mutant; res, siMITOL-resistant MITOL WT. Cytosolic MITOL is a mitochondrial localization-defective mutant. (D) NO-mediated cell death partially via LC1 in MITOL-knockdown cells. SH-SY5Y cells transfected with indicated vectors were treated with 100 μM SNAP for 3 h, 7 μM calcimycin for 3 h, or with 300 μM l-NAME for 1 h followed by calcimycin for 3 h. (E) NMDA stimulation induces cortical cell death partially via LC1 under MITOL knockdown. Primary cultured mouse cortical neurons were treated with 300 μM NMDA and 5 μm glycine for 10 min. After 24 h, cell death was determined by nuclear condensation. (C–E) The number of cells showing nuclear condensation in DsRed-MITO–positive cells was counted from 100 cells and each percentage was indicated. Error bars represent SD. *P < 0.05, n = 3, one-way ANOVA. (F) A schematic model depicting a role of MITOL in regulation of LC1 under nitrosative stress.
To confirm this in vitro result, the effects of MITOL knockdown on cell viability were examined in MITOL-knockdown cells. We used undifferentiated SH-SY5Y cells because differentiated SH-SY5Y cells had become more resistant to oxidative stress as previously reported (27). In MITOL-knockdown cells, LC1 induced cell death with nuclear condensation (Fig. 4C and Fig. S7A). Importantly, nitrosomimetic mutant C257W caused an intense cell death, which was completely rescued by siMITOL-resistant MITOL coexpression, whereas nonnitrosylation mutant C257S did not induce cell death (Fig. 4C). These results suggested the S-nitrosylation–dependent cytotoxicity of LC1 in MITOL-knockdown cells. Furthermore, mitochondrial localization-defective mutant of MITOL, which lacks the second loop domain (Δ159 aa–210 aa) of MITOL (Fig. S7B), failed to block the LC1-mediated cytotoxicity (Fig. 4C, lane 8). Thus, MITOL specifically blocks mitochondrial LC1-dependent toxicity. To demonstrate mitochondrial dysfunction by LC1, we measured the reactive oxygen species (ROS) production and mitochondrial membrane potential in MITOL knockdown cells. We found that LC1 WT and C257W, but not C257S, induced the ROS generation and loss of mitochondrial membrane potential before cell death (Fig. S7 C and D), suggesting that mitochondrial damage by LC1 accumulation caused cell death.
To determine whether cell vulnerability resulting from MITOL knockdown is involved in NO-mediated signaling, the effects of SNAP or calcimycin treatment on LC1-induced cell death were examined in MITOL-knockdown cells. The amount of cell death was increased by treatment with SNAP or calcimycin, and calcimycin-induced cell death was attenuated by treatment with l-NAME (Fig. 4D). This enhanced NO-induced cell toxicity in MITOL-knockdown cells was attenuated by LC1 knockdown, which was recovered by shLC1-unaffected LC1 WT coexpression but not by coexpression of C257S mutant. Similar to this observation, NMDA stimulation caused LC1-dependent cell death in MITOL-knockdown cortical neurons (Fig. 4E). Taken together, these results suggest that MITOL knockdown enhanced sensitivity to NO-induced cell toxicity, in part, via the accumulation of S-nitosylated LC1 in mitochondria.
We observed that overstimulation of calcimycin could induce cell death without MITOL knockdown (Fig. 4D), suggesting that MITOL dysfunction may occur under some physiological conditions, such as excess NO production. For example, it was reported that S-nitrosylation of parkin regulates its ubiquitin ligase activity in Parkinson disease (28, 29). We found that calcimycin treatment induced S-nitrosylation of MITOL (Fig. S8A). Furthermore, in vitro SNAP treatment drastically inactivated MITOL E3 ubiquitin ligase activity in a dose-dependent manner (Fig. S8B). Thus, S-nitrosylation of MITOL may regulate an enzymatic activity of MITOL. Because the RING domain in MITOL contains cysteine residues that are critical for its enzymatic activity, S-nitrosylation on these cysteine residues may induce MITOL inactivation. Therefore, it is thought that excess NO production in neuronal cells causes MITOL dysfunction by direct S-nitrosylation of MITOL.
In conclusion, our findings show that MITOL specifically ubiquitinates S-nitrosylated LC1 in mitochondria, thereby inhibiting mitochondrial aggregation and dysfunction induced by S-nitrosylated LC1 during NO-mediated signaling (Fig. 4F). However, excess NO production may inactivate MITOL by direct S-nitrosylation, which fails to block LC1-mediated mitochondrial dysfunction and cell death. These events may be closely linked to the pathogenesis of nitrosative stress-mediated neurodegenerative disorders.
Discussion
Mechanism of S-Nitrosylation–Dependent Ubiquitination of LC1.
In this study, we demonstrated that MITOL recognizes S-nitrosylated LC1 and facilitates the degradation of LC1 through the ubiquitin-proteasome pathway. Recently, the phosphatase PTEN was reported to be ubiquitinated by NEDD4 in an S-nitrosylation–dependent manner and degraded via the ubiqitin-proteasome pathway (30). On the other hand, S-nitrosylation of Bcl-2 was reported to inhibit Bcl-2 ubiquitination and its proteasomal degradation (31). Therefore, S-nitrosylation is not always a specific signal for protein ubiquitination and degradation. S-nitrosylation may trigger a conformational change in proteins and may affect various posttranslational modifications, such as ubiquitination and phosphorylation. It has been shown that S-nitrosylation of critical cysteine residues leads to protein misfolding, which is involved in many neurodegenerative diseases (2). However, in the case of LC1, S-nitrosylation appeared to activate LC1. A previous study suggested that S-nitrosylation converts inactive LC1, which is mainly localized to the cytosol, to active LC1, which rapidly translocates to the cytoskeleton and stabilizes microtubules (12). Thus, S-nitrosylation is involved not only in protein misfolding but also in protein activation. To down-regulate active LC1, MITOL may ubiquitinate S-nitrosylated LC1 by recognition of the active form. However, this LC1 recognition system appears not to be specific to MITOL. Even in the cytosol, an S-nitrosylation–defective LC1 mutant (C257S) revealed attenuated ubiquitination, whereas an S-nitrosylation mimetic LC1 mutant (C257W) was extensively ubiquitinated (Fig. 3B). Thus, S-nitrosylation of LC1 on C257 is the common ubiquitination signal for participating E3 ubiquitin ligases.
Mutational analysis of LC1 lysine sites identified three major ubiquitination sites (positions 164, 203, and 233) on the C-terminal region of LC1 (Fig. S5B). Indeed, one-half of the LC1 fragment (MTB) inhibited ubiquitination of the other half of the LC1 fragment (AB). This result suggests that when unstimulated, inactive LC1 forms a closed circle through an interaction between the N terminus and C terminus that masks the ubiquitination sites on the C-terminal region. Furthermore, S-nitrosylation of C257 of LC1 may block the N- and C-terminal contact and induce a conformational change in LC1, such as an open structure, which exposes the ubiquitination sites to associated ubiquitin ligases. Thus, as summarized in Fig. 4F, we propose a model for the molecular mechanisms underlying the S-nitrosylation–dependent ubiquitination of LC1 by MITOL.
Role of MITOL in LC1 Function on Microtubules.
We observed enhanced microtubule stabilization with increased acetylated tubulin in MITOL-knockdown cells (Fig. S1). This result can be explained, in part, by LC1 accumulation. MITOL may suppress excess microtubule stabilization by LC1 accumulation via LC1 ubiquitination and degradation. Neuronal cells constantly receive various extracellular stimulations, which convey intracellular signal transduction, including local calcium mobilization and subsequent NO production via nNOS activation. Therefore, it is possible that to respond to an intracellular emergency, such as NO production, S-nitrosylated LC1 may interrupt traffic on the microtubules, such as vesicle transport, by interacting with motor proteins, such as dynein, to protect neuronal cells from NO-induced toxicity. S-nitrosylated LC1 may act like a red traffic light to stop transportation by transient induction of microtubule stabilization. Thus, the primary role of S-nitrosylation of LC1 is considered to be neuroprotective. However, if microtubule stabilization occurs in excess and continues for a prolonged time, neuronal cells would not survive because all transport is severely blocked. When the intracellular emergency, such as oxidative stress, has disappeared, microtubules must be flexible to restart transportation on the microtubules. MITOL could prevent excess microtubule stabilization via LC1 degradation, thereby transmitting cell survival signals by maintenance of microtubules. Thus, MITOL appears to act like a green traffic light by removing S-nitrosylated LC1 from microtubules, although how MITOL determines the timing of ubiquitination and down-regulation of S-nitrosylated LC1 is unknown. In cells, mitochondria move vigorously along microtubules. The reason for dynamic mitochondrial movement in the cell is still uncertain; however, one attractive hypothesis is that mitochondria move on microtubules to check the condition of and to repair microtubules. Thus, mitochondria may be involved in the quality control of microtubules in cells. Previous studies indicated that local calcium mobilization transiently prevents mitochondrial movement (32) and NO prevents mitochondrial movement in primary cortical neurons with depolarized mitochondrial membrane potential (33). In addition, LC1 inhibits retrograde transport of mitochondria in the axon of neurons (34). Therefore, it is possible that calcium mobilization activates nNOS and NO-mediated S-nitrosylated LC1 suppresses mitochondrial movement via inhibition of the motor protein dynein and microtubule stabilization. Thus, MITOL may regulate not only mitochondrial morphology, such as fusion and fission, but also mitochondrial movement, at least in part, through the regulation of S-nitrosylated LC1 level.
LC1-Mediated Cell Death Pathway and Protective Pathway by MITOL.
Glutamate is the major excitatory neurotransmitter released from nerve terminals. Activation of NMDA-type glutamate receptors drives an influx of calcium, which in turn induces NO production via nNOS activation. NO can lead to protective or damaging actions in the central nervous system. For example, there is a protective feedback mechanism via down-regulation of NMDA receptor by S-nitrosylation (35, 36). Similarly, S-nitrosylation of LC1 may protect from NMDA-induced toxicity via stabilization of microtubules. Thus, under unstimulated conditions, MITOL blocks LC1-mediated mitochondrial aggregation and dysfunction, but when MITOL is inactivated, LC1-dependent mitochondrial dysfunction may occur. Indeed, in MITOL-knockdown cells, we observed that LC1 caused mitochondrial damage, including mitochondrial aggregation, loss of mitochondrial membrane potential, and ROS generation, and led to cell death with nuclear condensation. Therefore, we propose a unique NO-mediated cell death pathway that results from LC1-dependent mitochondrial dysfunction.
Under physiological conditions, what signals induce LC1-dependent mitochondrial dysfunction? In other words, when is MITOL inactivated? Previous studies have demonstrated that S-nitrosylated parkin regulates its ubiquitin ligase activity, both in Parkinson disease animal models and in patients (28, 29). In our study, we suggested that MITOL was inactivated by S-nitrosylation (Fig. S8). Indeed, NMDA stimulation caused, in part, LC1-dependent cell death in MITOL-knockdown cortical neurons (Fig. 4E). Therefore, excess NO generation may cause MITOL inactivation by S-nitrosylation and may induce neuronal cell death via LC1-dependent mitochondrial dysfunction. Thus, cytotoxicity caused by LC1 overaccumulation may be closely involved in the onset of many neurodegenerative disorders, such as Alzheimer's and Parkinson diseases. Methods of inhibiting MITOL dysfunction and LC1 accumulation may provide new strategies for disease therapy.
Materials and Methods
Biotin-Switch Assay.
Cell lysates were diluted with HEN buffer (100 mM Hepes, 1 mM EDTA, 0.1 mM neocuproine, pH 8.0) and added 25% (vol/vol) SDS and 10% (vol/vol) methylmethane thiosulfonate to final concentration of 2.5% (vol/vol) and 0.1%, respectively. Samples were incubated at 50 °C in the dark for 20 min. After incubation, samples were precipitated with cold acetone, and resuspended in HENS buffer (HEN buffer with 1% SDS). Samples were added biotin-HPDP (Pierce) and sodium ascorbate (Wako), and rotated at room temperature in the dark for 1 h. After labeling, samples were precipitated with cold acetone and pellets were resuspended in HENS/10 (HENS buffer diluted 10-fold with distilled H2O) buffer and were diluted with neutralization buffer (25 mM Hepes, 100 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, pH 7.4) followed by precipitation with indicated Abs.
Additional materials and methods are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
This study was supported in part by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology and the Japan Society for the Promotion of Science (to R.Y. and S.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. T.M.D. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1114985109/-/DCSupplemental.
References
- 1.Calabrese V, et al. Nitric oxide in the central nervous system: Neuroprotection versus neurotoxicity. Nat Rev Neurosci. 2007;8:766–775. doi: 10.1038/nrn2214. [DOI] [PubMed] [Google Scholar]
- 2.Foster MW, Hess DT, Stamler JS. Protein S-nitrosylation in health and disease: A current perspective. Trends Mol Med. 2009;15:391–404. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dawson VL, Dawson TM. Nitric oxide neurotoxicity. J Chem Neuroanat. 1996;10:179–190. doi: 10.1016/0891-0618(96)00148-2. [DOI] [PubMed] [Google Scholar]
- 4.Seth D, Stamler JS. The SNO-proteome: Causation and classifications. Curr Opin Chem Biol. 2011;15:129–136. doi: 10.1016/j.cbpa.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shahani N, Sawa A. Nitric oxide signaling and nitrosative stress in neurons: Role for S-nitrosylation. Antioxid Redox Signal. 2011;14:1493–1504. doi: 10.1089/ars.2010.3580. [DOI] [PubMed] [Google Scholar]
- 6.Nakamura T, Lipton SA. Redox modulation by S-nitrosylation contributes to protein misfolding, mitochondrial dynamics, and neuronal synaptic damage in neurodegenerative diseases. Cell Death Differ. 2011;18:1478–1486. doi: 10.1038/cdd.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benard G, et al. Mitochondrial bioenergetics and structural network organization. J Cell Sci. 2007;120:838–848. doi: 10.1242/jcs.03381. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura T, Lipton SA. Redox regulation of mitochondrial fission, protein misfolding, synaptic damage, and neuronal cell death: Potential implications for Alzheimer's and Parkinson's diseases. Apoptosis. 2010;15:1354–1363. doi: 10.1007/s10495-010-0476-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho DH, et al. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riederer BM. Microtubule-associated protein 1B, a growth-associated and phosphorylated scaffold protein. Brain Res Bull. 2007;71:541–558. doi: 10.1016/j.brainresbull.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 11.Liu L, Vo A, Liu G, McKeehan WL. Distinct structural domains within C19ORF5 support association with stabilized microtubules and mitochondrial aggregation and genome destruction. Cancer Res. 2005;65:4191–4201. doi: 10.1158/0008-5472.CAN-04-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stroissnigg H, et al. S-nitrosylation of microtubule-associated protein 1B mediates nitric-oxide-induced axon retraction. Nat Cell Biol. 2007;9:1035–1045. doi: 10.1038/ncb1625. [DOI] [PubMed] [Google Scholar]
- 13.Opal P, et al. Mapmodulin/leucine-rich acidic nuclear protein binds the light chain of microtubule-associated protein 1B and modulates neuritogenesis. J Biol Chem. 2003;278:34691–34699. doi: 10.1074/jbc.M302785200. [DOI] [PubMed] [Google Scholar]
- 14.Jensen PH, et al. Microtubule-associated protein 1B is a component of cortical Lewy bodies and binds alpha-synuclein filaments. J Biol Chem. 2000;275:21500–21507. doi: 10.1074/jbc.M000099200. [DOI] [PubMed] [Google Scholar]
- 15.Karbowski M, Neutzner A, Youle RJ. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J Cell Biol. 2007;178:71–84. doi: 10.1083/jcb.200611064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura N, Kimura Y, Tokuda M, Honda S, Hirose S. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 2006;7:1019–1022. doi: 10.1038/sj.embor.7400790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugiura A, et al. A mitochondrial ubiquitin ligase MITOL controls cell toxicity of polyglutamine-expanded protein. Mitochondrion. 2011;11:139–146. doi: 10.1016/j.mito.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Yonashiro R, et al. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 2006;25:3618–3626. doi: 10.1038/sj.emboj.7601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yonashiro R, et al. Mitochondrial ubiquitin ligase MITOL ubiquitinates mutant SOD1 and attenuates mutant SOD1-induced ROS generation. Mol Biol Cell. 2009;20:4254–4530. doi: 10.1091/mbc.E09-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allen E, et al. Gigaxonin-controlled degradation of MAP1B light chain is critical to neuronal survival. Nature. 2005;438:224–228. doi: 10.1038/nature04256. [DOI] [PubMed] [Google Scholar]
- 21.Yamanaka K, et al. Identification of the ubiquitin-protein ligase that recognizes oxidized IRP2. Nat Cell Biol. 2003;5:336–340. doi: 10.1038/ncb952. [DOI] [PubMed] [Google Scholar]
- 22.Yoshida Y, et al. E3 ubiquitin ligase that recognizes sugar chains. Nature. 2002;418:438–442. doi: 10.1038/nature00890. [DOI] [PubMed] [Google Scholar]
- 23.Orian A, et al. SCF(beta)(-TrCP) ubiquitin ligase-mediated processing of NF-kappaB p105 requires phosphorylation of its C-terminus by IkappaB kinase. EMBO J. 2000;19:2580–2591. doi: 10.1093/emboj/19.11.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palmer ZJ, et al. S-nitrosylation of syntaxin 1 at Cys(145) is a regulatory switch controlling Munc18-1 binding. Biochem J. 2008;413:479–491. doi: 10.1042/BJ20080069. [DOI] [PubMed] [Google Scholar]
- 25.Forrester MT, Foster MW, Benhar M, Stamler JS. Detection of protein S-nitrosylation with the biotin-switch technique. Free Radic Biol Med. 2009;46:119–126. doi: 10.1016/j.freeradbiomed.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawauchi T, Chihama K, Nishimura YV, Nabeshima Y, Hoshino M. MAP1B phosphorylation is differentially regulated by Cdk5/p35, Cdk5/p25, and JNK. Biochem Biophys Res Commun. 2005;331:50–55. doi: 10.1016/j.bbrc.2005.03.132. [DOI] [PubMed] [Google Scholar]
- 27.Schneider L, et al. Differentiation of SH-SY5Y cells to a neuronal phenotype changes cellular bioenergetics and the response to oxidative stress. Free Radic Biol Med. 2011;51:2007–2017. doi: 10.1016/j.freeradbiomed.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chung KK, et al. S-nitrosylation of parkin regulates ubiquitination and compromises parkin's protective function. Science. 2004;304:1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- 29.Yao D, et al. Nitrosative stress linked to sporadic Parkinson's disease: S-nitrosylation of parkin regulates its E3 ubiquitin ligase activity. Proc Natl Acad Sci USA. 2004;101:10810–10814. doi: 10.1073/pnas.0404161101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwak YD, et al. NO signaling and S-nitrosylation regulate PTEN inhibition in neurodegeneration. Mol Neurodegener. 2010;5:49. doi: 10.1186/1750-1326-5-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azad N, et al. S-nitrosylation of Bcl-2 inhibits its ubiquitin-proteasomal degradation. A novel antiapoptotic mechanism that suppresses apoptosis. J Biol Chem. 2006;281:34124–34134. doi: 10.1074/jbc.M602551200. [DOI] [PubMed] [Google Scholar]
- 32.Jeyaraju DV, Cisbani G, Pellegrini L. Calcium regulation of mitochondria motility and morphology. Biochim Biophys Acta. 2009;1787:1363–1373. doi: 10.1016/j.bbabio.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 33.Rintoul GL, Bennett VJ, Papaconstandinou NA, Reynolds IJ. Nitric oxide inhibits mitochondrial movement in forebrain neurons associated with disruption of mitochondrial membrane potential. J Neurochem. 2006;97:800–806. doi: 10.1111/j.1471-4159.2006.03788.x. [DOI] [PubMed] [Google Scholar]
- 34.Jiménez-Mateos EM, González-Billault C, Dawson HN, Vitek MP, Avila J. Role of MAP1B in axonal retrograde transport of mitochondria. Biochem J. 2006;397:53–59. doi: 10.1042/BJ20060205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakamura T, Lipton SA. S-Nitrosylation and uncompetitive/fast off-rate (UFO) drug therapy in neurodegenerative disorders of protein misfolding. Cell Death Differ. 2007;14:1305–1314. doi: 10.1038/sj.cdd.4402138. [DOI] [PubMed] [Google Scholar]
- 36.Brown GC. Nitric oxide and neuronal death. Nitric Oxide. 2010;23:153–165. doi: 10.1016/j.niox.2010.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.