Abstract
Pnkp is the end-healing and end-sealing component of an RNA repair system present in diverse bacteria from ten different phyla. To gain insight to the mechanism and evolution of this repair system, we determined the crystal structures of the ligase domain of Clostridium thermocellum Pnkp in three functional states along the reaction pathway: apoenzyme, ligase•ATP substrate complex, and covalent ligase-AMP intermediate. The tertiary structure is composed of a classical ligase nucleotidyltransferase module that is embellished by a unique α-helical insert module and a unique C-terminal α-helical module. Structure-guided mutational analysis identified active site residues essential for ligase adenylylation. Pnkp defines a new RNA ligase family with signature structural and functional properties.
Keywords: covalent catalysis, enzyme evolution, RNA repair
RNA breakage by site-specific “ribotoxins” is an ancient mechanism by which microbes respond to cellular stress and distinguish self from nonself (1–3). Ribotoxins are transesterifying endonucleases that generate 5′-OH and 2′,3′ cyclic phosphate termini. Repair of this type of RNA damage is feasible via sequential enzymatic end-healing and end-sealing steps (4, 5). In the healing phase, the 2′,3′ cyclic phosphate end is hydrolyzed to a 3′-OH and the 5′-OH end is phosphorylated. The healed 3′-OH and 5′-PO4 termini are then suitable substrates for sealing by an RNA ligase that restores the 3′,5′ phosphodiester backbone. However, in the event that a ribotoxin is constitutively activated, the RNA repair system faces an uphill battle against relentless RNA cleavage. An ingenious way to evade the vicious cycle is to install a 2′-OCH3 “mark” at the repair junction (as depicted in Fig. 1B), which then protects the marked RNAs from recurrent damage (6).
Fig. 1.
The Pnkp-Hen1 RNA repair pathway confers protective immunity to recurrent RNA damage. (A) The adjacent cooriented ORFs encoding the CthHen1 (465-aa) and CthPnkp (870-aa) polypeptides comprise a bacterial RNA repair cassette conserved in > 60 taxa from 10 different bacterial phyla. Pnkp is a trifunctional RNA repair enzyme composed of 5′ OH polynucleotide kinase, 2′,3′ phosphoesterase, and ligase-like adenylyltransferase domains. Hen1 is composed of a distinctive N-terminal domain and a C-terminal methyltransferase domain (MTase). (B) RNA cleavage by site-specific transesterifying endonucleases results in 2′,3′ cyclic phosphate and 5′-OH ends that are substrates for healing and sealing by Pnkp and Hen1. The 5′ end is phosphorylated by the Pnkp kinase module and the 2′,3′ cyclic phosphate is removed by the Pnkp phosphoesterase module. The Hen1 methyltransferase domain installs a 2′-OCH3 mark at the terminal ribonucleoside prior to ligation of the ends in a reaction that requires both Pnkp (via its LIG domain) and Hen1. The repair junction with the methyl mark is then resistant to scission by transesterification.
The capacity for protective immunity during RNA repair is illustrated by the many bacterial species that encode the repair enzymes Pnkp (polynucleotide 5′-kinase/3′-phosphatase) and Hen1 in an operon-like gene cassette (6, 7) (Fig. 1A). Clostridium thermocellum Pnkp (CthPnkp) is an 870-aa polypeptide composed of three catalytic domains: N-terminal kinase, central phosphoesterase, and C-terminal adenylyltransferase (8–11). The kinase module catalyzes phosphoryl transfer from ATP to the 5′-OH RNA end. The phosphoesterase domain releases Pi from 2′-PO4, 3′-PO4 or 2′,3′ cyclic phosphate ribonucleotides. The adenylyltransferase domain reacts with ATP to form a covalent enzyme-AMP adduct, just as RNA ligases do during strand joining, but it is unable per se to seal RNA strands (8). The sealing function of the Pnkp ligase-like domain is activated by the Hen1 protein (6) encoded by a flanking genomic ORF. C. thermocellum Hen1 is a a 465-aa polypeptide. The C-terminal half of CthHen1 is an autonomous manganese-dependent 3′-terminal ribose 2′O-methyltransferase (7, 12, 13). The N-terminal half of CthHen1 has no known enzymatic activity.
Raven Huang’s lab has studied Anabaena variabilis Hen1 methyltransferase and its connection to Anabaena Pnkp; they showed that an AvaPnkp-AvaHen1 complex has end-healing and sealing activities on a broken tRNA substrate and that ribose 2′O-methylation of the 3′ end prior to sealing can protect the repair junction from further damage by transesterifying ribotoxins (6) (Fig. 1B). The native size of the Pnkp-Hen1 complex, gauged by gel filtration, was consistent with a (Pnkp)2(Hen1)2 tetramer. A model for the repair complex was suggested, whereby the Hen1 N-domain interacts physically with the ligase-like domain of Pnkp (6).
The classical RNA ligase pathway entails three sequential nucleotidyl transfer steps, similar to those of DNA ligases, as follows: (i) RNA ligase reacts with ATP to form a covalent ligase-(lysyl-N)-AMP intermediate plus pyrophosphate; (ii) AMP is transferred from ligase-adenylate to the 5′-PO4 RNA end to form an RNA-adenylate intermediate (AppRNA); and (iii) ligase catalyzes attack by an RNA 3′-OH on the RNA-adenylate to seal the two ends via a phosphodiester bond and release AMP. The step 1 autoadenylylation reaction of RNA ligases is performed by a nucleotidyltransferase (NTase) domain that is conserved among RNA ligases and is thought to be the progenitor of the homologous NTase modules found in present-day DNA ligases and mRNA capping enzymes (14). It is proposed that modern RNA ligases evolved from an ancestral standalone NTase domain similar to that of T4 RNA ligase 2 (Rnl2) (shown in Fig. 2 with AMP bound). The AMP-binding pocket is formed by a cage of β-strands and interstrand loops that includes the defining peptide motifs of the covalent NTase superfamily (ligases and capping enzymes) (Fig. S1). Biochemical specificity and dedication to a particular biological pathway of RNA sealing appear to have evolved by fusions of the NTase domain to structurally diverse C-terminal domain modules, which are depicted in Fig. 2 for each of the three structurally characterized RNA ligase clades, exemplified by: T4 RNA ligase 1 (Rnl1) (15); T4 Rnl2 (16); and Pyrococccus abyssi RNA ligase (17), the prototype of a newly characterized ligase family (hereby christened the “Rnl3 family”) found in many archaea (18).
Fig. 2.
Three distinct RNA ligase families. Three structurally characterized clades of RNA ligases evolved separately by fusion of unique C-terminal domain modules to a common conserved nucleotidyltransferase domain. The fold of an ancestral NTase is suggested to resemble that of T4 Rnl2 as shown (from PDB ID 1S68). The structurally diverse C-terminal domains are shown for T4 Rnl1 (PDB ID 2C5U), T4 Rnl2 (PDB ID 2HVQ), and Pyrococccus abyssi RNA ligase (PDB ID 2VUG), the prototypes of the Rnl1, Rnl2, and Rnl3 families.
The physical and functional properties of the CthPnkp ligase are distinctive vis à vis all other ATP-dependent RNA ligases that have been characterized. First, there is no instance in which RNA strand sealing by a covalently adenylylated NTase module requires a separate protein catalyst. Rather, the known RNA ligases rely on their distinctive structural domains linked in cis to the nucleotidyltransferase to impart RNA substrate recognition and catalysis of adenylate transfer to the RNA 5′-phosphate (19, 20). Second, whereas inspection of the primary structure of the CthPnkp adenylyltransferase domain suggested retention of certain motifs that define the covalent NTase superfamily, the spacing between some of the motifs was quite deviant from the norm (8). In particular, CthPnkp has an unusually large interval separating motifs III and IV (160-aa; compared to 66-aa in T4 Rnl1) that has no apparent sequence similarity to known ligases. Also, the segment of the CthPnkp adenylyltransferase domain distal to motif V has no recognizable primary structure similarity to the C-terminal domains of Rnl1, Rnl2, or Rnl3 family members, which raised the prospect that CthPnkp might exemplify a novel ligase family.
To explore this issue, we crystallized the CthPnkp adenylyltransferase domain and determined structures of the protein in three functional states along the ligase reaction pathway: apoenzyme, ligase•ATP substrate complex, and covalent ligase-AMP intermediate. The tertiary structure is indeed unique, being composed of a “classical” ligase NTase module that is embellished by a distinctive α-helical insert and C-terminal α-helical domains. Whereas the insert domain is conformationally flexible in different crystal forms, the position of the C-terminal segment is fixed. The adenylyltransferase active site comprises the five NTase motifs plus constituents specific to bacterial Pnkp ligases. We conclude that Pnkp defines a ligase family with signature structural and functional properties.
Results and discussion
Crystallization of the LIG domain of CthPnkp.
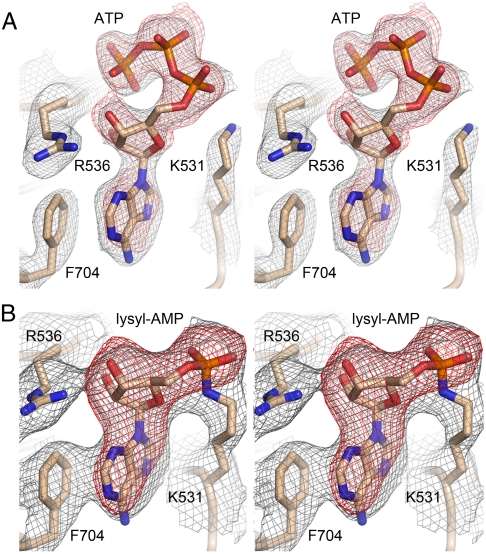
It had been shown previously that two versions of the recombinant His-tagged C-terminal domain of CthPnkp, spanning residues 433–870 and 462–870, respectively, were active as covalent adenylyltransferases in vitro (8). Here we conducted parallel crystallization trials with His10-CthPnkp-(433–870) and His10-CthPnkp-(462–870), and with a shorter variant— His10-CthPnkp-(479–870)—that also evinced adenylyltransferase activity in vitro. Only the shortest version yielded diffracting crystals in our hands; henceforth, we will refer to the CthPnkp-(479–870) protein as the LIG domain. We initially obtained crystals of the His10-tagged LIG protein that contained a single fortuitous mutation (E529G); the recombinant protein was incubated with 10 mM ATP prior to crystal growth by vapor diffusion against a precipitant solution containing PEG-3350 and tartrate. These crystals diffracted to 2.5 Å and belonged to space group P21. SIRAS phases from crystals of a mercury derivative of E529G were exploited to derive an initial model, which was then refined using the diffraction data from native crystals. The refined model of LIG•ATP at 2.5 Å resolution (Table S1) contained two protomers in the asymmetric unit. Protomer A had ATP in the active site (Fig. 3A); protomer B was modeled with ADP bound as there was no interpretable density for the γ phosphate. We also crystallized the wild-type His10-tagged LIG protein by vapor diffusion against PEG-3350 and tartrate, albeit without prior exposure to ATP. These P21 crystals were sufficiently similar to those of E529G to permit straightforward refinement of the LIG apoenzyme structure (apoLIG) at 2.6 Å resolution (Table S1).
Fig. 3.
Adenylate electron density in LIG•ATP and LIG-AMP complexes. (A) Stereo view of the ATP-bound LIG-(E529G) protomer A showing maps of both composite omit electron density (in gray mesh contoured at 1.5 σ) and Fo-Fc difference density calculated prior to the inclusion of ATP in the model (in red mesh contoured at 3.0 σ). Both maps were calculated using X-ray diffraction data to a limiting resolution of 2.5 Å sampled on a 0.3 Å grid. (B) Stereo view of the wild-type LIG-AMP protomer A showing 4-fold NCS-averaged maps of composite omit electron density (gray mesh contoured at 1.5 σ) and Fo-Fc difference density calculated before covalent AMP was modeled (red mesh contoured at 8 σ). Both maps were calculated using diffraction data to a limiting resolution of 3.12 Å sampled on a 0.5 Å grid.
A tag-free version of wild-type LIG that had been reacted with ATP and magnesium prior to gel filtration to promote enzyme adenylylation was crystallized in 20% hexylene glycol. The tag-free LIG crystals diffracted to 3.12 Å and belonged to space group P212121. All four protomers in the asymmetric unit had a lysyl-(Nζ)-adenylate adduct in the active site (Fig. 3B). Thus, the structural ensemble captures the enzyme before and after the reaction with ATP to form a covalent LIG-AMP intermediate.
Overview of the LIG Structure.
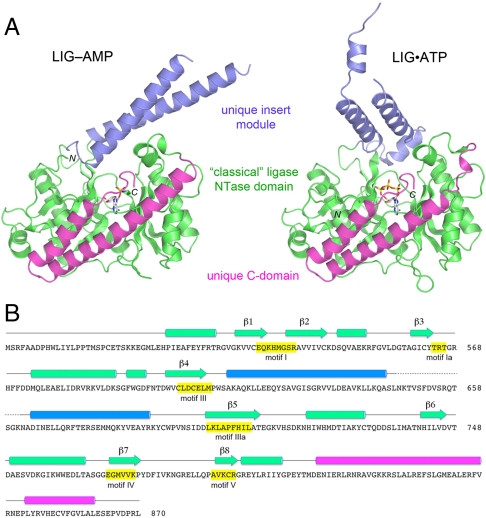
The tertiary structure of CthLIG is composed of three domains (Fig. 4A). The largest module, colored green and embracing residues 479–612 and 693–808, is homologous to the “classical” NTase domain of DNA and RNA ligases. The NTase domain consists of two central 4-stranded antiparallel β-sheets, (with topologies β3↑β2↓β4↑β5↓ and β6↓β1↑β7↓β8↑, respectively) that form the adenylate-binding pocket. Seven α helices decorate the lateral surfaces of the NTase domain (Fig. 4A). A unique feature of CthLIG is the en bloc insertion of an α-helical domain (aa 613–693; colored blue) between the β4 and β5 stands of the NTase fold. The helical insert module is located on the surface of the NTase domain and projects outward like a tower, making little contact with the rest of the LIG protein. Moreover, as can be appreciated from the side-by-side comparisons of the structures of the LIG-AMP and LIG•ATP complexes (Fig. 4A), the insert module is positionally and conformationally flexible. The C-terminal domain (aa 809–870; colored magenta) is also unique and consists of an antiparallel two-helix bundle with an extended tail peptide. Unlike the insert module, the C-domain packs tightly against an extended peptide segment (aa 489–501) near the N-terminus of the NTase domain and its position is invariant in the several structures of CthLIG captured in different functional states (Fig. 5). The secondary structure elements are aligned to the CthLIG amino acid sequence in Fig. 4B.
Fig. 4.
Tertiary structure and component domains of CthLIG. (A) CthLIG is composed of three domains: a classical ligase nucleotidyl transferase domain (green), a unique insert domain (blue) within the NTase, and a unique C-terminal domain (magenta). The LIG-AMP and LIG•ATP (protomer A) structures were superimposed and then offset horizontally. N and C specify the amino and carboxyl termini of the LIG polypeptides. The nucleotide in the active site is shown as a stick model. (B) The secondary structure elements of LIG-AMP [colored by domain as in (A)] are aligned above the amino acid sequence of CthLIG, with β strands rendered as arrows and α helices as cylinders. The nucleotidyltransferase motifs are highlighted in yellow boxes.
Fig. 5.
Conformational variability of the helical insert module. The structures of the A and B protomers of LIG•ATP, the A and B protomers of apoLIG, and the LIG-AMP A protomer were superimposed and then offset horizontally as shown. The NTase domain is colored gray and the C-terminal domain is magenta. The helical insert domain in its various conformations is colored blue. The numbers indicate the modeled amino acids at the margins of disordered segments of the insert domain.
A DALI search (21) with the complete LIG•ATP structure identified ATP-dependent DNA ligases, ATP-dependent RNA ligases, and NAD+-dependent DNA ligases as top scoring hits, trailed by GTP-dependent RNA capping enzymes (Table S2). The homologies were confined in all cases to the NTase domain; separate DALI searches with the insert domain and C-terminal domain yielded no convincing homologs. Thus, the helical insert and C-terminal domains of CthLIG are distinctive modules acquired by fusion to the NTase catalytic core. We discuss below the unique structural features of CthLIG and then exploit the structures to guide a mutational analysis of the active site.
Conformational Flexibility of the Insert Module.
The fold and orientation of the helical insert domain differs significantly in the A and B protomers of the LIG•ATP crystal structure (Fig. 5), reflecting inherent conformational flexibility. In the A protomer, the distal α-helix—which is continuous from aa 662–688 in the LIG-AMP structure (Fig. 4A)—is broken into two shorter α-helices (aa 664–673 and 674–690) arranged orthogonally to one another (Fig. 4A and 5). Similarly, the proximal α-helix of the LIG•ATP A protomer (which continuous from aa 613–646 in LIG-AMP) is broken into two shorter α-helices (aa 619–624 and 631–645), also orthogonal to one another. The peptide segment from aa 648–660 is disordered in the A protomer (Fig. 5). In the B promoter of the LIG•ATP crystal, the insert domain undergoes a large reorientation with respect to the NTase domain (compared to the A protomer) and the disordered segments within the insert domain (aa 618–620 and 652–658) are different (Fig. 5). The positional shift of the insert module in the A and B promoters of LIG•ATP is on the order of 41 Å at the Asn662 Cα atom and 23 Å at the Asn647 Cα atom. The conformational differences between the helical insert domains in the A and B protomers of the apoLIG crystal (Fig. 5) generally reflect those seen in the LIG•ATP A and B protomers, with some subtle variations in the positions of the helices and the sizes and locations of the disordered segments.
By contrast, the structure and orientation of the helical insert domain in the LIG-AMP crystal is virtually identical in all four protomers of the asymmetric unit (as exemplified by A promoter shown in Fig. 4A and 5). In LIG-AMP, the two long helices form a tight antiparallel bundle that projects in a completely different orientation relative to the NTase domain (e.g., into the plane of the page in Fig. 5) compared to their positions in the A and B protomers of LIG•ATP and apoLIG. It is worth noting that the disordered segment between the long helices in the LIG-AMP structure is actually missing (i.e., deleted) from many other bacterial Pnkp proteins, including A. variabilis Pnkp for which RNA ligase activity has been demonstrated (6). The surface location and the positional and conformational heterogeneity of the helical insert domain raise the prospect that this module mediates the interaction of the Pnkp LIG component with the Hen1 protein that enables the RNA sealing activity of the Pnkp-Hen1 complex. Alternatively (or in addition) the insert domain might interact with the broken RNA substrate.
The LIG Active Site.
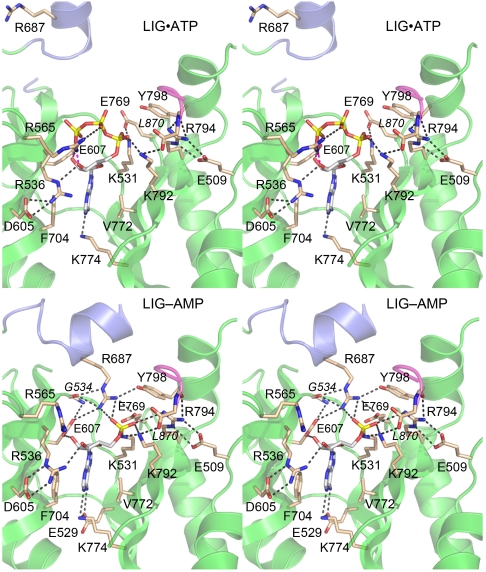
The architecture and atomic contacts to ATP and AMP in the active sites of the LIG•ATP and LIG-AMP structures are shown in Fig. 6. The adenylate-binding pocket is composed of a cage of β strands and interstrand loops that includes NTase motifs I, Ia, III, IIIa, IV, and V, which are highlighted by yellow shading in the LIG primary structure in Fig. 4B. The adenosine nucleosides of LIG•ATP and LIG-AMP are in the anti conformation (Fig. 3). The adenine base is sandwiched between the aromatic ring of Phe704 in motif IIIa and the hydrophobic side chain of Val772 in motif III (Fig. 6). Adenine specificity is apparently conferred by purine contacts to the Glu529 (motif I) and Lys774 (motif IV) side chains, which are located on adjacent β strands and comprise an ion pair (Fig. 6, Bottom). Adenine-N6 donates a hydrogen bond to Glu529 Oϵ1 and adenine-N1 accepts a hydrogen bond from Lys774 Nζ. The ribose 2′-OH of ATP and AMP is coordinated by Glu607 in motif III. The ribose 3′-OH of ATP is engaged by the side chain of Arg536 (in motif I), which is itself held in place by a bifurcated salt-bridge from NH1 to Asp605 (in motif III) (Fig. 6, Top). In the LIG-AMP structure, the ribose 3′-OH is still coordinated by Arg536, albeit via the NH1 atom instead of Nϵ, and the Arg536-Asp605 interaction shifts to a bidentate salt-bridge to Nϵ and NH2 (Fig. 6, Bottom).
Fig. 6.
Atomic contacts in the active sites of LIG•ATP and LIG-AMP. Stereo views of the active site of the LIG•ATP complex (Top) and the LIG-AMP complex (Bottom). The main-chain cartoon traces are colored by domain as in Fig. 4. Amino acids and ATP are shown as stick models with beige and gray carbons, respectively. Atomic contacts are indicated by dashed lines.
The ATP phosphates are engaged by three basic side chains in the active site. Arg565 (motif Ia) makes bidentate ionic interactions with nonbridging β and γ phosphate oxygens while Lys792 (motif V) and Lys531 (the motif I nucleophile) engage the two nonbridging oxygens of the α phosphate (Fig. 6, Top). Lys531 forms an ion pair with Glu769 (motif IV), a residue implicated in metal-binding; this ion pair is conserved in many structures of covalent NTase superfamily enzymes (14). A distinctive feature of the LIG•ATP active site is the engagement of motif V Lys792 and Arg794 in a bonding network with the main-chain atoms of the C-terminal dipeptide of LIG (colored magenta in Fig. 6). Lys792 forms an ion pair with the main-chain carboxylate of the C-terminal residue Leu870, while Arg794 makes a bifurcated hydrogen bond to the main-chain carbonyl of the penultimate residue. Arg794 also forms a bidentate salt bridge to Glu509, a conserved Pnkp side chain located in a loop immediately preceding the first α-helix of the NTase domain (Fig. 6, Top).
The transition from LIG•ATP to LIG-AMP entails changes in the active site. The electron density maps validate the covalent linkage of Lys531 Nζ to the AMP phosphorus (Fig. 3B). Upon covalent adenylylation, multiple new atomic contacts are established to the AMP phosphate. For example, the lysyl-AMP phosphate moves closer to motif V, so that Lys792 makes a bifurcated contact to bridging and nonbridging phosphate oxygens, while Arg794 gains a contact to a nonbridging phosphate oxygen; the interactions of Lys792 and Arg794 with the LIG C-terminus are maintaining during this transition. A more dramatic conformational switch is seen for the Arg687 side chain in the distal helix of the insert domain, which, in conjunction with the domain movements discussed above, undergoes a 17 Å shift in the position of its terminal guanidinium nitrogens, resulting in the incorporation of Arg687 into the LIG active site (Fig. 6, Bottom). In its new location, Arg687 coordinates a nonbridging AMP phosphate oxygen and anchors an extended bonding network: to the Glu607, Glu769, and Tyr798 side chains and the Gly534 carbonyl.
Structure-Guided Mutational Analysis of the LIG Active Site.
The initial characterization of CthPnkp demonstrated that motif I residues Lys531 and Arg536 are essential for covalent adenylyltransferase activity (8). Here we used the LIG•ATP and LIG-AMP structures to guide a new mutational analysis of the ligase active site. We tested the effects of nine single alanine mutations on adenylyltransferase activity. The residues targeted for alanine scanning were Arg565 (motif Ia), Asp605 and Glu607 (motif III), Glu769 and Lys774 (motif IV), Lys792 and Arg794 (motif V), Glu509 (the loop residue that engages Arg794), and Arg687 (the mobile insert domain residue that joins the active site in LIG-AMP). We also queried the role of the motif V contacts to the C-terminal peptide, by deleting the Leu870 residue singly and deleting the C-terminal  tripeptide en bloc. The wild-type and mutated proteins were produced in E. coli as His10 fusions and purified from soluble lysates by Ni-affinity chromatography (Fig. 7A).
tripeptide en bloc. The wild-type and mutated proteins were produced in E. coli as His10 fusions and purified from soluble lysates by Ni-affinity chromatography (Fig. 7A).
Fig. 7.
Structure-guided mutagenesis. (A) Aliquots (8 μg) of the Ni-agarose preparations of the indicated His10CthPnkp-(462–870) proteins (purified as described in Supplemental Information) were analyzed by SDS-PAGE. The polypeptides were visualized by staining with Coomassie blue dye. The positions and sizes (kDa) of marker polypeptides are indicated. (B) Adenylyltransferase activity. Reaction mixtures (20 μL) containing 50 mM Tris-acetate (pH 7.0), 5 mM DTT, 5 mM MgCl2, 20 μM [α32P]ATP, and 2 μg of wild-type or mutated His10CthPnkp were incubated at 37 °C for 15 min. The reactions were quenched with SDS and the products were analyzed by SDS-PAGE. The enzyme-[32P]AMP adduct was quantified by scanning the gel with a Fujix imager. The extents of label transfer from [α32P]ATP to the respective CthPnkp-(462–870) polypeptides are shown. Each datum in the bar graphs is the average of three separate experiments ± SEM.
The proteins were reacted with [α32P]ATP and magnesium and the yields of enzyme-[32P]AMP adduct were gauged by SDS-PAGE analysis of the reaction products (Fig. 7B). In the case of the wild-type protein, 30–35% of the input Pnkp molecules were labeled with [32P]AMP. Adenylyltransferase activity was virtually ablated by mutations R565A, D605A, E607A, E769A, and K774A; their normalized yields of covalent AMP adduct were 1.2%, ≤ 0.1%, 1.1%, 0.7%, and 0.8%, of the wild-type yield, respectively. These results highlight the importance of Lys774 as a critical determinant of adenine recognition, of Glu607 in ribose recognition (and of Asp605 in positioning the other ribose-binding residue, Arg536), and the essential role of motif Ia Arg565 in binding and orienting the PPi leaving group of the ATP substrate. The essentiality of the Glu769 is consistent with the proposed roleof the conserved motif IV carboxylate side chain in binding the divalent cation cofactor (14).
Instructive mutational results were obtained for the network of interactors with the AMP phosphate (Fig. 7B). The motif V mutations K792A and R794A reduced adenylyltransferase activity to 0.8% and 5.6% of the wild-type levels, respectively. The E509A change that erased the salt bridge to motif V from the adjacent loop also reduced activity to 2.4% of the wild-type level. By contrast, the R687A mutant retained adenylyltransferase activity (75% of the wild-type level), signifying that the extensive contacts made by Arg687 in the LIG-AMP active site are not necessary during formation of the covalent ligase-adenylate intermediate. The C-terminal peptide of LIG and its contacts to motif V also appear to be inessential for ligase adenylylation, insofar as the ΔL870 and  proteins were 47–55% as active as the full-length enzyme (Fig. 7B). Our working criterion for an essential RNA ligase residue is a ≥10-fold decrement in activity when substituted by alanine (22). Our results do not rule out a role for Arg687 or the C-terminal segment in downstream steps of the ligation reaction.
proteins were 47–55% as active as the full-length enzyme (Fig. 7B). Our working criterion for an essential RNA ligase residue is a ≥10-fold decrement in activity when substituted by alanine (22). Our results do not rule out a role for Arg687 or the C-terminal segment in downstream steps of the ligation reaction.
Concluding Remarks.
The present study provides new insights to the structure and evolution of the ligase domain of bacterial Pnkp and the essential active site moieties that bind the ATP substrate and are required for generation of the covalent ligase-adenylate intermediate. The distinctive domain composition of Pnkp LIG fortifies the argument that multiple different clades of RNA ligases emerged by fusions of diverse accessory modules to a shared NTase domain. Extending the scheme depicted in Fig. 2, we consider Pnkp LIG to be the founding member of an “Rnl4” family of RNA ligases. Each of the four RNA ligase families has a defining C-terminal module with no structural homology to the other RNA ligase C-domains or to the OB domains that are linked to the NTase of DNA ligases and RNA capping enzymes. Pnkp LIG is further distinguished by the acquisition of a second accessory module inserted into a surface loop of the NTase domain. Pyrococcus RNA ligase (the Rnl3 prototype) also has two accessory domains added to its NTase (one at the N-terminus and one at the C-terminus), but they have different folds and different spatial relationships to the core NTase domain compared to what is seen in CthLIG (Fig. S2). Based on the structural diversity of the relatively small number of RNA ligases for which structures are available, we predict that additional RNA ligase families are out there awaiting discovery and/or structural characterization (23, 24).
Methods
Crystallization of CthLIG.
Proteins for crystallization were purified as described in Supplemental Information. A His10CthPnkp-(479–870)-E529G protein solution (13 mg/mL) was adjusted to 10 mM ATP, 5 mM MgCl2 and 20 mM DTT prior to mixture with a equal volume (2 μL) of precipitant solution containing 0.2 M di-ammonium tartrate and 20% PEG3350. Crystals grown at 22 °C by the hanging drop vapor diffusion method were transferred to a solution of 0.2 M di-ammonium tartrate, 20% PEG3350, 10 mM ATP, 5 mM MgCl2, 10% ethylene glycol prior to rapid freezing in liquid nitrogen. Alternatively, the His10CthPnkp-(479–870)-E529G crystals were reacted with Hg by soaking them in a solution of 0.25 M di-ammonium tartrate, 25% PEG3350, 10 mM ATP, 5 mM MgCl2, 10% ethylene glycol and 1% saturated thimerosal for 7 min prior to rapid freezing in liquid nitrogen. A wild-type His10CthPnkp-(479–870) protein solution (15 mg/mL; 2 μL) was mixed with an equal volume of reservoir buffer containing 0.24 M di-ammonium tartrate, 20% PEG3350. Crystals grown at 22 °C by hanging drop vapor diffusion were transferred to a solution of 0.24 M di-ammonium tartrate, 20% PEG3350, 10% ethylene glycol prior to freezing in liquid nitrogen. The tag-free wild-type CthPnkp-(479–870) protein (22 mg/mL; 2 μL) was mixed with an equal volume of reservoir buffer containing 100 mM Hepes (pH 7.1), 20% hexylene glycol. Crystals grown at 22 °C by hanging drop vapor diffusion were harvested and frozen directly in liquid nitrogen.
Diffraction Data Collection and Structure Determination.
Diffraction data were collected continuously from single crystals mounted in a nitrogen cooling stream at National Synchrotron Light Source beamline X25 equipped with an ADSC-Q315 CCD detector. Crystals of His10CthPnkp-(479–870) and His10CthPnkp-(479–870)-E529G had similar monoclinic P21 lattices and diffracted to 2.5 Å and 2.6 Å, respectively. Diffraction data at 3.0 Å resolution collected from a thiomerosal-soaked His10CthPnkp-(479–870)-E529G crystal were used for phase determination. Crystals of the tag-free CthPnkp-(479–870) belonged to spacegroup P212121 and diffracted to 3.12 Å. The data statistics are provided in Table S1. The methods employed for phasing, model building, and refinement are described in detail in Supplemental Information. Refinement statistics and models contents are compiled in Table S1.
Supplementary Material
Acknowledgments.
This research was supported by National Institutes of Health Grant GM42498. S.S. is an American Cancer Society Research Professor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates for the refined models of LIG•ATP, apoLIG, and LIG-AMP have been deposited in the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank, www.rcsb.org (PDB ID codes 3TY5, 3TY8, and 3TY9).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1116827109/-/DCSupplemental.
References
- 1.Ogawa T, et al. A cytotoxic ribonuclease targeting specific tRNA anticodons. Science. 1999;283:2097–2100. doi: 10.1126/science.283.5410.2097. [DOI] [PubMed] [Google Scholar]
- 2.Lu J, Huang B, Esberg A, Johanson M, Byström AS. The Kluyveromyces lactis gamma-toxin targets tRNA anticodons. RNA. 2005;11:1648–1654. doi: 10.1261/rna.2172105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winther KS, Gerdes K. Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc Natl Acad Sci USA. 2011;108:7403–7407. doi: 10.1073/pnas.1019587108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amitsur M, Levitz R, Kaufman G. Bacteriophage T4 anticodon nuclease, polynucleotide kinase, and RNA ligase reprocess the host lysine tRNA. EMBO J. 1987;6:2499–2503. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nandakumar J, Schwer B, Schaffrath R, Shuman S. RNA repair: An antidote to cytotoxic eukaryal RNA damage. Molecular Cell. 2008;31:278–286. doi: 10.1016/j.molcel.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan CM, Zhou C, Huang R. Reconstituting bacterial RNA repair and modification in vitro. Science. 2009;326:247. doi: 10.1126/science.1179480. [DOI] [PubMed] [Google Scholar]
- 7.Jain R, Shuman S. Bacterial Hen1 is a 3′ terminal RNA ribose 2′O-methyltransferase component of a bacterial RNA repair cassette. RNA. 2010;16:316–323. doi: 10.1261/rna.1926510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martins A, Shuman S. An end-healing enzyme from Clostridium thermocellum with 5′ kinase, 2′,3′ phosphatase, and adenylyltransferase activities. RNA. 2005;11:1271–1280. doi: 10.1261/rna.2690505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keppetipola N, Shuman S. Mechanism of the phosphatase component of Clostridium thermocellum polynucleotide kinase-phosphatase. RNA. 2006;12:73–82. doi: 10.1261/rna.2196406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keppetipola N, Shuman S. Distinct enzymic functional groups are required for the phosphomonoesterase and phosphodiesterase activities of Clostridium thermocellum polynucleotide kinase-phosphatase. J Biol Chem. 2006;281:19251–19259. doi: 10.1074/jbc.M602549200. [DOI] [PubMed] [Google Scholar]
- 11.Keppetipola N, Shuman S. Characterization of the 2′,3′ cyclic phosphodiesterase activities of Clostridium thermocellum polynucleotide kinase-phosphatase and bacteriophage lambda phosphatase. Nucleic Acids Res. 2007;35:7721–7732. doi: 10.1093/nar/gkm868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan CM, Zhou C, Brunzelle JS, Huang RH. Structural and biochemical insights into 2′-O-methylation at the 3′-terminal nucleotide of RNA by Hen1. Proc Natl Acad Sci USA. 2009;106:17699–17704. doi: 10.1073/pnas.0907540106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jain R, Shuman S. Active site mapping and substrate specificity of bacterial Hen1, a manganese-dependent 3′ terminal RNA ribose 2′O-methyltransferase. RNA. 2011;17:429–438. doi: 10.1261/rna.2500711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shuman S, Lima CD. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr Opin Struct Biol. 2004;14:757–764. doi: 10.1016/j.sbi.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 15.El Omari K, et al. Molecular architecture and ligand recognition determinants for T4 RNA ligase. J Biol Chem. 2005;281:1573–1579. doi: 10.1074/jbc.M509658200. [DOI] [PubMed] [Google Scholar]
- 16.Nandakumar J, Shuman S, Lima C.D. RNA ligase structures reveal the basis for RNA specificity and conformational changes that drive ligation forward. Cell. 2006;127:71–84. doi: 10.1016/j.cell.2006.08.038. [DOI] [PubMed] [Google Scholar]
- 17.Brooks MA, et al. The structure of an archaeal homodimeric ligase which has RNA circularization activity. Protein Sci. 2008;17:1336–1345. doi: 10.1110/ps.035493.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torchia C, Takagi Y, Ho CK. Archaeal RNA ligase is a homodimeric protein that catalyzes intramolecular ligation of single-stranded RNA and DNA. Nucleic Acids Res. 2008;36:6218–6227. doi: 10.1093/nar/gkn602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho CK, Wang LK, Lima CD, Shuman S. Structure and mechanism of RNA ligase. Structure. 2004;12:327–339. doi: 10.1016/j.str.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 20.Wang LK, Nandakumar J, Schwer B, Shuman S. The C-terminal domain of T4 RNA ligase 1 confers specificity for tRNA repair. RNA. 2007;13:1235–1244. doi: 10.1261/rna.591807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:1780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang LK, Ho CK, Pei Y, Shuman S. Mutational analysis of bacteriophage T4 RNA ligase 1: Different functional groups are required for the nucleotidyl transfer and phosphodiester bond formation steps of the ligation reaction. J Biol Chem. 2003;278:29454–29462. doi: 10.1074/jbc.M304320200. [DOI] [PubMed] [Google Scholar]
- 23.Martins A, Shuman S. An RNA ligase from Deinococcus radiodurans. J Biol Chem. 2004;279:50654–50661. doi: 10.1074/jbc.M407657200. [DOI] [PubMed] [Google Scholar]
- 24.Raymond A, Shuman S. Deinococcus radiodurans RNA ligase exemplifies a novel ligase clade with a distinctive N-terminal module that is important for 5′-PO4 nick sealing and ligase adenylylation but dispensable for phosphodiester formation at an adenylylated nick. Nucleic Acids Res. 2007;35:839–849. doi: 10.1093/nar/gkl1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.