Abstract
Archaeological and genetic evidence concerning the time and mode of wild horse (Equus ferus) domestication is still debated. High levels of genetic diversity in horse mtDNA have been detected when analyzing the control region; recurrent mutations, however, tend to blur the structure of the phylogenetic tree. Here, we brought the horse mtDNA phylogeny to the highest level of molecular resolution by analyzing 83 mitochondrial genomes from modern horses across Asia, Europe, the Middle East, and the Americas. Our data reveal 18 major haplogroups (A–R) with radiation times that are mostly confined to the Neolithic and later periods and place the root of the phylogeny corresponding to the Ancestral Mare Mitogenome at ∼130–160 thousand years ago. All haplogroups were detected in modern horses from Asia, but F was only found in E. przewalskii—the only remaining wild horse. Therefore, a wide range of matrilineal lineages from the extinct E. ferus underwent domestication in the Eurasian steppes during the Eneolithic period and were transmitted to modern E. caballus breeds. Importantly, now that the major horse haplogroups have been defined, each with diagnostic mutational motifs (in both the coding and control regions), these haplotypes could be easily used to (i) classify well-preserved ancient remains, (ii) (re)assess the haplogroup variation of modern breeds, including Thoroughbreds, and (iii) evaluate the possible role of mtDNA backgrounds in racehorse performance.
Keywords: horse mitochondrial genome, mtDNA haplogroups, origin of Equus caballus, Przewalski's horse, animal domestication
Farming and animal domestication were fundamental steps in human development, contributing to the rise of larger settlements and more stratified societies and eventually, great civilizations. One of the key events in this process was the domestication of the wild horse (Equus ferus), which was profoundly connected to numerous human activities in both prehistorical and historical times. The horse served as a means to provide food, facilitate transportation, and (since the Bronze age) enhance warfare capabilities. A question that has not yet been completely addressed concerns the timing and mode in which humans began to benefit from the employment of these animals (1–6).
From a genetic point of view, animal domestication can be reconstructed by performing phylogeographic analyses of both nuclear and mitochondrial genomic data (2, 7). A draft sequence for the horse nuclear genome was recently published (8), whereas the first complete mtDNA sequence for this species has been available since 1994 (9). Despite this information, nearly all of the molecular and evolutionary studies of horse mtDNA have generally focused on ∼350–650 bp from two HyperVariable Segments (HVS-I: nucleotide position 15,469–15,834; HVS-II: np 16,351–16,660) within the control region (d-loop: np 15,469–16,660) (7, 10–13). In many other mammalian species, the variation seen in this short segment of the mitochondrial genome is often accompanied by high levels of recurrent mutations, thus blurring the structure of the tree and rendering the distinction between some important ancient branches within the tree virtually impossible (14–16). To address this issue and improve mtDNA phylogeny, we analyzed a total of 83 horse mitochondrial genomes (GenBank accession nos. JN398377–JN398457).
Results and Discussion
Phylogeny of Horse Mitochondrial Genomes.
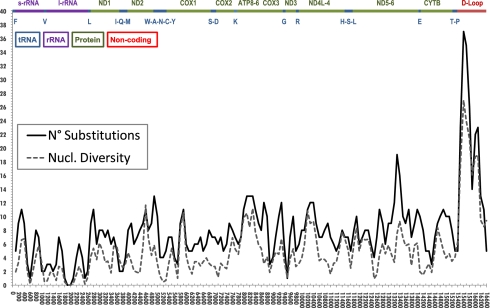
For complete sequencing, mtDNAs were collected from a rather wide range of horse breeds across Asia, Europe, the Middle East, and the Americas (SI Appendix, Table S1). Excluding ambiguous sites and the 16129–16360 short tandem repeat (STR), we identified a total number of 667 varied sites (Table 1): 549 in the coding region (15,476 nt) and 118 in the noncoding regions (1,032 nt), mostly represented by the d-loop (113 of 960 nucleotides). Overall, we observed an average number of 82.0 ± 30.7 nucleotide differences between two randomly chosen sequences. Fig. 1 illustrates the distribution of the total number of mutations along the genome (continuous line). A similar plot was obtained when considering the variation of nucleotide diversity (π) (17) along the entire mtDNA by assessing windows of 200 bp (step size = 100 bp) centered at the midpoint (Fig. 1, dotted line). As expected, the highest diversity was observed around the HVS-I segment (from np 15,500 to np 15,800), with a peak of 0.02699. The latter value is in agreement with the values previously reported, which range from 0.03 to 0.05 (12). Within the coding region, the highest proportion of varied sites was found in protein-coding genes (Table 1) where, consistent with previous reports on the human mitochondrial genome (18–20), there was an excess of synonymous mutations.
Table 1.
Distribution and recurrence of mutations in the 83 horse mtDNA sequences
| Noncoding |
Coding |
||||||
| d-loop (excluding STR) | Other | rRNA | tRNA | mRNA | Nonsynonymous | Synonymous | |
| Length (bp) | 960 | 72 | 2,556 | 1,522 | 11,400 | 8,522 | 2,845 |
| Unvaried sites | 847 | 67 | 2,496 | 1,496 | 10,937 | 8,434 | 2,469 |
| Number of varied sites | 113 | 5 | 60 | 26 | 463 | 88* | 376* |
| Proportion of varied sites | 0.133 | 0.075 | 0.024 | 0.017 | 0.042 | 0.010 | 0.152 |
| Sites with a single hit | 63 | 3 | 59 | 25 | 412 | 77 | 335 |
| Sites with two hits | 18 | 2 | 1 | 1 | 42 | 7 | 36 |
| Sites with three or more hits | 32 | 0 | 0 | 0 | 9 | 4 | 5 |
| Indels | 6 | 1 | 5 | 1 | 0 | 0 | 0 |
| Transitions | 104 | 3 | 50 | 25 | 447 | 85 | 363 |
| Transversions | 3 | 1 | 5 | 0 | 16 | 3 | 13 |
| Transition/transversion ratio | 34.7 | 3.0 | 10.0 | n.a. | 27.9 | 28.3 | 27.9 |
*A unique transition at np 8,007 was counted two times: 8007(ATP8) and 8007(V15M ATP6).
Fig. 1.
Sequence variation within the horse mitochondrial genome. Nucleotide diversity (π) and total number of substitutions by considering windows of 200 bp (step size = 100 bp) centered at the midpoint. A schematic (linearized) genetic map of the mitochondrial genome is presented.
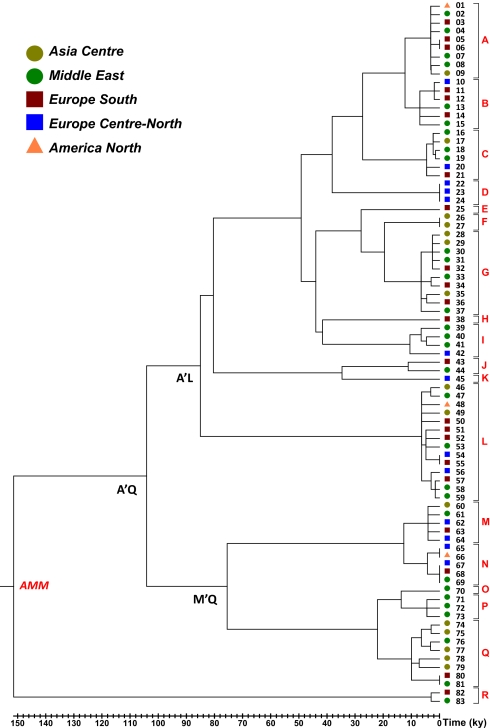
The evolutionary history of the 83 complete mtDNA sequences (81 different haplotypes) was inferred by a parsimony approach (SI Appendix, Fig. S1). To translate the molecular divergence of the major tree nodes into time, the 83 complete mtDNA sequences were compared with the single available complete mtDNA sequence (donkey reference sequence) from a donkey (E. asinus). A maximum likelihood (ML) tree was then estimated based on synonymous mutations alone (Fig. 2). Despite the number of potential factors causing time-dependent rates (21), the synonymous molecular clock is most likely to remain linear over time, although saturation can also be an issue with long time frames. The deepest node, corresponding to the Ancestral Mare Mitogenome (AMM) from which all modern horse mtDNAs derive, was dated at ∼153 ± 30 thousand years ago (kya). Two branches depart from this node; one encompasses only two mtDNAs (82 and 83), and the other comprises all other sequences. This latter branch is further split at two nodes (nodes A'L and M'Q) characterized by very similar average sequence divergences (Fig. 2). Considering the molecular separation of the derived subbranches, a series of different, relatively young nodes of ages lower than 11 ky were observed (Fig. 2 and SI Appendix, Tables S2 and S3). This value was then considered as an arbitrary threshold to classify and (re)name E. caballus haplogroups on the basis of entire mtDNA sequences.
Fig. 2.
Schematic phylogeny of complete mtDNAs from modern horses. This tree includes 81 sequences and was rooted by using a published donkey (E. asinus) mitochondrial genome (not displayed). AMM indicates the reconstructed AMM, whereas horse reference sequence (HRS) is the newly proposed horse reference sequence (GenBank JN398377). The topology was inferred by a maximum parsimony (MP) approach, whereas an ML time divergence scale (based on synonymous substitutions) is shown on the bottom. Additional details concerning samples, mutational motifs, and ages are given in SI Appendix, Fig. S1 and Tables S1 and S2.
Horse mtDNA Haplogroups.
A total of 18 major haplogroups were labeled in alphabetical order (from A to R), each defined by a specific motif encompassing both the coding and control regions. The direct comparison of the new and old nomenclatures shows that all deep nodes in the horse phylogeny and most haplogroups were not resolved by control region data alone (SI Appendix, Table S4). After these new haplogroups were defined, we were able to compare the above-mentioned mutational patterns within and outside the established haplogroups (Table 2). A sign of purifying selection was evident when comparing the nonsynonymous/synonymous ratio, which was significantly lower (P value < 0.001; Fisher exact test) in the deep portion of the tree.
Table 2.
Distribution pattern of mutations along the horse mtDNA phylogeny
| d-loop (excluding STR) | Other noncoding | rRNA | tRNA | mRNA | MN* | MS* | MN/MS | |
| Within haplogroups | 133 | 4 | 23 | 11 | 176 | 55 | 121 | 0.455 |
| Outside haplogroups | 105 | 3 | 36 | 14 | 299 | 36 | 263 | 0.137 |
| Singletons† | 19 | 0 | 2 | 2 | 49 | 16 | 33 | 0.485 |
| Total | 257 | 7 | 61 | 27 | 524 | 107 | 417 | 0.257 |
*MN and MS are measures of nonsynonymous and synonymous changes inferred from the phylogenetic tree (SI Appendix, Fig. S1).
†These are mutations in haplogroups/branches represented by a single haplotype, which are shown in Fig. 2 and SI Appendix, Fig. S1.
As for the Przewalski's horse (E. przewalskii), with a karyotype of 2N = 66 vs. 2N = 64 for E. caballus (22), our two identical mtDNAs (26 and 27) belonged to a haplogroup (F) that is distinct from the haplogroups of domestic horses (Fig. 2). This finding is in agreement with the observation that virtually all Przewalski's horse control region haplotypes belong to haplogroup F, with one notable exception (AF055879) (23). Recently, four complete Przewalski mtDNAs were published (24). Although three mtDNAs belonged to haplogroup F, the fourth mtDNA shared the control region motif with the above-mentioned sample AF055879, radiating before the J'K node in our phylogeny (SI Appendix, Fig. S1 and SI Appendix, Table S5).
Horse MtDNA Molecular Clock.
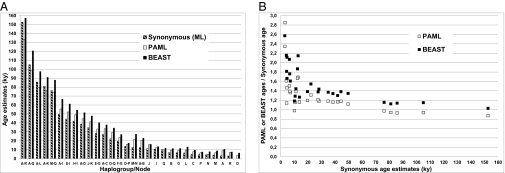
To convert mutational distances into time over the entire mitochondrial genome, we used diverse ML and Bayesian analyses as described in Materials and Methods. Age estimates based on the complete genome with five partitions (first, second, and third codon positions, RNA genes, and noncoding regions) performed with PAML (Phylogenetic Analysis by Maximum Likelihood) and BEAST (Bayesian Evolutionary Analysis Sampling Trees) provided values that were not drastically different for the AMM (133 and 157 ky, respectively) (SI Appendix, Table S2). However, for the younger clades, the age values were higher than in the synonymous estimates as shown in Fig. 3A. Haplogroups A–R date from 4.9 to 15.5 ky with PAML and from 7.5 to 16.2 ky with BEAST. This finding is expected taking into account that all of the negatively selectable characters are included in the analysis and the effect of purifying selection is incomplete for the younger haplogroups (20). In any case, these discrepancies do not affect our conclusions based on the synonymous clock, and they are only relevant to the exact dating of the rather recent time when the horse mtDNA pool rapidly expanded.
Fig. 3.
Age estimates (A) calculated on the protein coding genes (by considering only synonymous mutations) and the entire genome partitioned into five datasets (first, second, and third positions of the codons, RNAs, and noncoding region). B shows a comparison of BEAST and PAML ages relative to the synonymous estimates. Additional details are given in SI Appendix, Table S2.
Fig. 3B shows that BEAST ages for younger haplogroups are substantially higher than those ages obtained with PAML when weighted on the synonymous estimates. For branch lengths lower than 16 ky, which include all of the established haplogroups, BEAST estimates have an average ratio relative to synonymous ages of 1.92 ± 0.52, whereas PAML estimates present an average ratio of 1.57 ± 0.42. When considering all ages, the average ratio is 1.60 ± 0.51 for BEAST and 1.34 ± 0.43 for PAML.
Comparing the Ages of the Five Partitions.
Independent analyses of each partition (first, second, and third codon positions, RNA genes, and noncoding regions) (SI Appendix, Table S3) relative to synonymous ages could provide answers for the observed patterns in both analyses. The PAML results were intuitive: the ratio of the third codon position was almost linear, because most mutations are synonymous; partitions based on the first and second codon positions showed much higher ratio values for young haplogroups considering that most mutations are nonsynonymous (all nonsynonymous in the case of the second position). The same trend was obtained for the RNA partition, also explained by selection, and the noncoding partition, probably mainly caused by saturation.
In the BEAST analysis, the patterns were less plausible. The ratio to synonymous ages, when considering only the third codon position, revealed an average of 1.8 and a higher ratio in young clades, which is surprising in a partition mainly containing synonymous mutations. Apart from the noncoding partition, this class is the class that presents the highest overall average ratio relative to synonymous ages. The noncoding partition in BEAST also displays higher values than the same partition in PAML, suggesting that BEAST deals with saturation differently than PAML. Third position partition and noncoding mutations should correspond to about 80% of the mutations, and they also present the higher substitution rates per nucleotide. This finding means that they are the most prone to saturation.
In human mtDNA studies, BEAST age estimates (25) correlated well with estimates based on a clock estimated with PAML from the human/chimpanzee split (20), but the calibration points used in BEAST were internal; therefore, saturation was not an issue, and the time dependence of evolution rates was mitigated. Our analyses of horse mitochondrial genomes suggest that BEAST is best used when internal calibration points are available, which were impossible to infer in the horse case since no reliable archeological data with a connection to age for any given haplogroup are available.
Geographic Distributions of Horse MtDNA Haplogroups.
An analysis of the geographic distribution of these haplogroups in Eurasia (SI Appendix, Fig. S3) based on the information currently available in the literature or deposited in GenBank revealed that both European and Middle Eastern samples lacked representatives of lineages F and (pre)JK, which were only detected in the Przewalski's horse (SI Appendix, Fig. S1 and Tables S5 and S6). This finding also supports evidence from other studies that the Przewalski's horse is not the mtDNA source for the domestic horse (8, 11). The Asian continent displays the highest haplogroup variation, and it is the only geographic area to include representatives of all haplogroups defined on the basis of HVS-I motifs (SI Appendix, Table S7). Some haplogroups (such as G, Q, and A) showed frequency peaks in Asia (16.35%, 13.80%, and 11.93%, respectively) and a decline to the Middle East (8.33%, 10.42%, and 7.81%, respectively) and Europe (8.73%, 3.85%, and 4.49%, respectively). Intriguingly, an opposite distribution pattern was observed for haplogroup L, which seems to be most frequent in Europe (38.06%), with a progressive reduction to the east (22.4% in the Middle East and 13.46% in Asia). The modern geographical distribution of L is supported by control region data from ancient DNA samples. Indeed, analysis of ancient DNA indicates that haplogroup frequency distributions are overlapping in modern and ancient samples, with most haplogroups already present in ancient times at least in Europe and Asia. In particular, haplogroup L mtDNAs were present in Iberia since the Neolithic Bronze Age (SI Appendix, Fig. S3 and Tables S8 and S9).
Domestication of Wild Mares.
DNA analyses for other domesticated species (e.g., cattle, sheep, and goat) revealed that modern livestock has derived from a limited number of animals that were domesticated in just a few places 8–10 kya (2, 15, 26, 27). This finding is also substantiated by today's reduced genetic variation in these animals compared with their ancient forebears. For instance, mtDNA of modern taurine cattle falls into a few distinct haplogroups, suggesting that (almost) only the offspring of original livestock were used to establish herds elsewhere (14, 28). However, horse mtDNA tells a different story. Modern horse mitochondrial genomes, when analyzed at the highest level of molecular resolution, show a high diversity in terms of haplogroups. Moreover, most of the 17 haplogroups identified in domestic breeds are spread over different geographic areas. This finding should, however, be compared with the genetic traces of the male lines of modern horses: according to a recent study (29), there is virtually no sequence diversity in the male-inherited Y chromosome. This finding implies that the domestication of wild horses was a process that involved only closely related male horses but allowed for more variation in the female lineages.
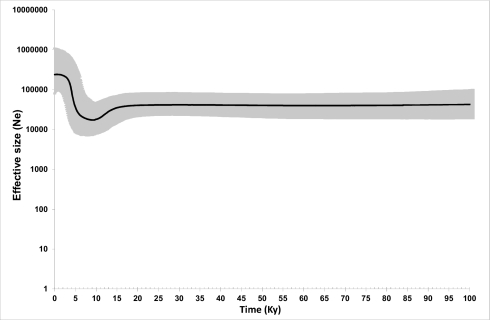
Archaeological evidence dates horse domestication no earlier than the Eneolithic period (6–7 kya) (4). Therefore, our data indicate that multiple female horse lines were domesticated. Most likely, the minimum number was 17, which corresponds to one founding haplotype for each of the identified haplogroups (A–E and G–R), but it is likely that this figure is much higher. Indeed, more than one founding mtDNA for each haplogroup and other (uncommon) haplogroups might also have been involved in the process. The horse tree shows a steep increase in the effective population size of the Bayesian skyline plot (BSP) (Fig. 4) after 5–8 ky, suggesting that most of the coalescence points in the tree date to the Neolithic period. However, these results should be interpreted carefully; the horse data do not represent random sequences from a population, and the clock in BEAST is probably overestimated.
Fig. 4.
Bayesian skyline plot (BSP) showing the horse population size trend with a generation time of 8 y (52). The y axis indicates the effective number of females. The thick solid line is the median estimate, and the gray shading shows the 95% highest posterior density limits. The time axis is limited to 100 ky; beyond that time, the curve remains linear.
Paleontological and archeological data indicate that wild horses were widely distributed throughout Eurasia during the Upper Paleolithic (35–10 kya), but it is likely that many lineages did not survive the drastic climatic changes during the Last Glacial Maximum (∼25–19.5 kya) and the Younger Dryas glacial relapse (∼12.7–11.5 kya) (30, 31). Consequently, the abundance of horse remains in Neolithic sites of Southern Ukraine and Turkestan may be the result of expansions that occurred from an eastern refuge area, perhaps in the Ukraine, where lineages of wild horses were preserved, thus providing the necessary substrate of variability (most of the 18 mtDNA haplogroups) for later horse husbandry in a broad region of the Eurasian steppe. At the same time, horses persisted continuously in the Iberian Peninsula, even during the Mesolithic era when the horse became extinct north of the Pyrenees (32–34). This mountain range effectively separated the Iberian Peninsula from the rest of Eurasia, turning it into a glacial refuge for many species, including humans (16, 35). Haplogroup L is a direct derivative of the deep A–L node in the phylogeny (Fig. 2), and it harbors the highest frequency in Europe in both modern and ancient samples (SI Appendix, Fig. S3). Moreover, its nucleotide (0.0070 ± 0.0003) and haplotype (0.809 ± 0.011) diversities in Europe are virtually identical to those diversities observed for the same haplogroup in Asia (0.0071 ± 0.0004 and 0.848 ± 0.016, respectively). These observations raise the possibility that at least one horse domestication event occurred in Western Europe, possibly in the Iberian Peninsula, which was also suggested by autosomal microsatellite variation in European horse breeds (36).
In conclusion, phylogenetic analyses of mitochondrial genomes reveal that (i) the most recent common female ancestor of all modern horses lived around the Late Saalian glacial maximum ∼140 kya (37, 38) and (ii) many haplogroups underwent domestication since the Eneolithic period in multiple Eurasian locations. Our data also show that the traditional restriction of horse mtDNA studies to a minor fragment of the control region, which is sometimes still practiced (12), and the proposed specificity of certain mtDNA clusters to pony breeds (six distinct haplogroups in our dataset) (SI Appendix, Table S1) (11) are no longer justifiable. Most importantly, now that a large number of horse haplogroups have been defined, each with diagnostic mutational motifs (in both coding and control regions), these haplotypes could be easily used to (i) classify well-preserved ancient remains, also taking into account the recent results obtained by second (and third) generation sequencing approaches (39), (ii) (re)assess the haplogroup variation of modern breeds, including Thoroughbreds (40), and (iii) evaluate the possible role of mtDNA backgrounds (41) in racehorse performance (42).
Materials and Methods
Sequencing of Horse Mitochondrial Genomes.
Before the sequencing of entire mtDNAs, a preliminary sequence analysis of the control regions was performed. This analysis allowed for the selection of the mtDNAs, which were then completely sequenced. Selected mtDNAs encompassed the widest range of mutational motifs, which was previously described for humans and cattle (43, 44). For complete sequencing, we used the Sanger sequencing approach with a sequencing protocol (SI Appendix), which is similar to the approaches previously and successfully used for human and cattle mitochondrial genomes (43–45). All experimental procedures were reviewed and approved by the Animal Research Ethics Committee of the University of Pavia (Prot. 2/2007; April 17th, 2007). Several mtDNA sequence variation parameters were estimated by using DnaSP 5.1. For an estimation of the synonymous/nonsynonymous sites, we created an alignment containing only the protein-coding genes, with the ND6 gene adjusted to present the same reading direction as the other genes. The stop codons AGA and AGG were excluded from the analysis, because it has been recently shown that mitochondria in humans use only UAA and UAG as stop codons (46).
Phylogeny Construction and Molecular Divergences.
The phylogeny construction was performed as described elsewhere (43, 47) and confirmed using an adapted version of mtPhyl 3.0 (default settings) for a maximum parsimony (MP) analysis, which was initially designed to analyze human mtDNA sequences (48). The modified .txt files are available on request. We built an MP tree, including 83 horse mtDNAs and the donkey reference sequence, and the topology was also double-checked by a Bayesian approach (Fig. 2 and SI Appendix, Fig. S1).
Molecular divergence was calculated using both the PAML 4.4 (49) and BEAST software (50). The noncoding region between np 16,129 and np 16,360 was excluded from the analysis because of the short tandem repeats present in the mitochondrial genome of both horse (29 repetitions of GTGCACCT) and donkey (10.5 repetitions of CACACCCACACACCCATGCGCGCA). The ML analysis was performed three ways using the predefined tree obtained by the MP and Bayesian approach: one tree considering only the protein coding genes (and synonymous mutations), one tree considering five partitions in the molecule (first, second, and third positions of the codons, RNAs, and noncoding regions), and one tree considering each partition independently. For the first analysis (protein coding genes), the alignment was modified with the ND6 gene adjusted to present the same reading direction as the other genes, and under the mitochondrial genetic code, the nonsynonymous substitutions were excluded from the alignment and replaced with the ancestral base pair. This alignment was analyzed with CODEML, which calculates a synonymous mutation rate by taking into account the mitochondrial genetic code. Analyses were performed assuming an HKY85 (Hasegawa-Kashino-Yano 1985) mutation model, a molecular clock, and γ-distributed rates (approximated by a discrete distribution with 32 categories). To check if the HKY85 model and the clock hypothesis were viable, we also ran the five-partition analysis with the GTR (Generalized Time Reversible) model and a second model not considering a molecular clock. A likelihood ratio test comparing the HKY85 with the GTR analyses did not yield significance (P = 0.9799), indicating that a model more complex than HKY85 was not necessary to explain the evolution of the data. A test comparing the clock and the nonclock was also insignificant (P = 0.2786), and therefore, the clock hypothesis could not be rejected.
BEAST analyses were performed in a similar manner as the last two PAML analyses (for the overall molecule containing five partitions and considering each partition separately). We used a relaxed molecular clock (lognormal in distribution across branches and uncorrelated between them) and a HKY85-type model (two parameters in the model of DNA evolution) with γ-distributed rates. We ran 50,000,000 iterations, with samples drawn every 5,000 steps.
We also obtained a BSP from the horse phylogeny (51) by using a generation time of 8 y (52). BSPs estimate effective population size through time from random sequences of a population. The horse data are especially distant from representing a population under panmixia. Still, the BSP provided good visualization of the increase in diversity in the tree of the sampled horses (mainly domestic). Additionally, we checked whether a Bayesian skyline model would actually perform better than a constant population size model by calculating a Bayes factor comparing the two analyses using Tracer 1.5.0 (http://beast.bio.ed.ac.uk/Tracer). The Bayesian skyline analysis performed positively better than the constant size model when interpreting the Bayes factor using Table 1 in ref. 53.
Calibrating the Horse MtDNA Molecular Clock.
For the calibration point in the ML and Bayesian analyses, we assumed an estimated bifurcation time between donkey and horse of 2 My (assuming a 95% interval of 1.9–2.1 My in the BEAST analysis), which is the approximate date for the first fossil evidence of caballoid horses (10, 54, 55). Considering the time dependency of molecular rate estimates (21), the use of a paleontological calibration point means that we are prone to possible biases mainly generated by nonsynonymous substitutions and mutations affecting tRNA and rRNA genes (purifying selection) and the control region (tendency to saturation because of the high rate of evolution). The available data are, however, not enough to allow the development of a mutation curve as performed already in humans (20). Internal calibration points in BEAST could be used to calculate a rate more directed at recent clades (25). However, we could not discern any viable calibration point, and therefore, our estimates relied only on the fossil record concerning the first appearance of caballoids. This long-term calibration point probably represents an underestimate of the true donkey–horse divergence; thus, it could have a considerable impact on the date estimates.
Synonymous mutations are virtually neutral and the effect of purifying selection is usually not an issue. Using CODEML and the mtDNA genetic code, we estimated 661.45 synonymous mutations from the donkey/horse ancestral split leading to a synonymous rate of 3.31 × 10−4 substitutions/y (at 3,789 codons) or one substitution every 3,024 y. The complete molecule considering five partitions yielded a rate of 4.48 × 10−8 substitutions per nucleotide/y using PAML and 6.13 × 10−8 substitutions per nucleotide/y using BEAST or one mutation every 1,358 and 992 y, respectively. The value obtained for BEAST was possibly caused by an artifact, and it did not fit the observed age estimates, which were higher than in PAML. We ran BEAST without partitions, and we obtained a substitution rate of 3.25 × 10−8 substitutions per nucleotide/y or one mutation every 1,871 y for very similar age estimates as the one with partitions (less than 5% differences).
Supplementary Material
Acknowledgments
We thank Nikolay Eltsov for his valuable help with the mtPhyl program; Ernest Bailey, Laura Morelli, Katia Cappelli, Mohammad Reza Farshid Pour, Seyed Hamid Aghajanzadeh, Rana Al-Ajouri, and Mahmoud Golchin for their help in the collection of samples; Laurene M. Kelly for proofreading the manuscript; and three anonymous reviewers for their helpful comments and suggestions. This research received support from the Italian Ministry of Education, University and Research: Fondo per gli Investimenti della Ricerca di Base (FIRB) Futuro in Ricerca 2008 (to A.A. and A.O.), Progetti Ricerca Interesse Nazionale 2008 (to E.G.) and 2009 (to A.A., O.S., and A.T.); L'ORÉAL Italia per le Donne e la Scienza (to A.O.), Fundação para a Ciência e Tecnologia (FCT), Grant SFRH/BPD/64233/2009 (to P.S.); Consorzio Interuniversitario di Biotecnologie (to M. Santagostino); Fondazione Alma Mater Ticinensis (O.S. and A.T.); and Progetto Ricerca e INNOVAzione nelle attività di miglioramento GENetico animale (INNOVAGEN), Italian Ministry of Agriculture (M. Silvestrelli). Instituto de Patologia e Imunologia Molecular da Universidade do Porto (IPATIMUP) is an Associate Laboratory of the Portuguese Ministry of Science, Technology, and Higher Education and is partially supported by FCT.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JN398377–JN398457).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111637109/-/DCSupplemental.
References
- 1.Diamond J. Evolution, consequences and future of plant and animal domestication. Nature. 2002;418:700–707. doi: 10.1038/nature01019. [DOI] [PubMed] [Google Scholar]
- 2.Groeneveld LF, et al. Genetic diversity in farm animals—a review. Anim Genet. 2010;41(Suppl 1):6–31. doi: 10.1111/j.1365-2052.2010.02038.x. [DOI] [PubMed] [Google Scholar]
- 3.Outram AK, et al. The earliest horse harnessing and milking. Science. 2009;323:1332–1335. doi: 10.1126/science.1168594. [DOI] [PubMed] [Google Scholar]
- 4.Anthony DW. The Horse, the Wheel, and Language: How Bronze-Age Riders from the Eurasian Steppes Shaped the Modern World. Princeton: Princeton University Press; 2007. [Google Scholar]
- 5.Ludwig A, et al. Coat color variation at the beginning of horse domestication. Science. 2009;324:485. doi: 10.1126/science.1172750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levine MA. In: The Domestic Horse: The Origins, Development and Management of its Behaviour. Mills DS, McDonnell SM, editors. Cambridge, UK: Cambridge University Press; 2005. pp. 5–22. [Google Scholar]
- 7.Kavar T, Dovč P. Domestication of the horse: Genetic relationships between domestic and wild horses. Livest Sci. 2008;116:1–14. [Google Scholar]
- 8.Wade CM, et al. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science. 2009;326:865–867. doi: 10.1126/science.1178158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu X, Arnason U. The complete mitochondrial DNA sequence of the horse, Equus caballus: Extensive heteroplasmy of the control region. Gene. 1994;148:357–362. doi: 10.1016/0378-1119(94)90713-7. [DOI] [PubMed] [Google Scholar]
- 10.Vilà C, et al. Widespread origins of domestic horse lineages. Science. 2001;291:474–477. doi: 10.1126/science.291.5503.474. [DOI] [PubMed] [Google Scholar]
- 11.Jansen T, et al. Mitochondrial DNA and the origins of the domestic horse. Proc Natl Acad Sci USA. 2002;99:10905–10910. doi: 10.1073/pnas.152330099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cieslak M, et al. Origin and history of mitochondrial DNA lineages in domestic horses. PLoS One. 2010;5:e15311. doi: 10.1371/journal.pone.0015311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gurney SM, et al. Developing equine mtDNA profiling for forensic application. Int J Legal Med. 2010;124:617–622. doi: 10.1007/s00414-010-0506-9. [DOI] [PubMed] [Google Scholar]
- 14.Achilli A, et al. The multifaceted origin of taurine cattle reflected by the mitochondrial genome. PLoS One. 2009;4:e5753. doi: 10.1371/journal.pone.0005753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taberlet P, et al. Are cattle, sheep, and goats endangered species? Mol Ecol. 2008;17:275–284. doi: 10.1111/j.1365-294X.2007.03475.x. [DOI] [PubMed] [Google Scholar]
- 16.Torroni A, Achilli A, Macaulay V, Richards M, Bandelt H-J. Harvesting the fruit of the human mtDNA tree. Trends Genet. 2006;22:339–345. doi: 10.1016/j.tig.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Nei M. Molecular Evolutionary Genetics. New York: Columbia University Press; 1987. [Google Scholar]
- 18.Mishmar D, et al. Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci USA. 2003;100:171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kivisild T, et al. The role of selection in the evolution of human mitochondrial genomes. Genetics. 2006;172:373–387. doi: 10.1534/genetics.105.043901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soares P, et al. Correcting for purifying selection: An improved human mitochondrial molecular clock. Am J Hum Genet. 2009;84:740–759. doi: 10.1016/j.ajhg.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho SY, et al. Time-dependent rates of molecular evolution. Mol Ecol. 2011;20:3087–3101. doi: 10.1111/j.1365-294X.2011.05178.x. [DOI] [PubMed] [Google Scholar]
- 22.Benirschke K, Malouf N, Low RJ, Heck H. Chromosome complement: Differences between Equus caballus and Equus przewalskii, Poliakoff. Science. 1965;148:382–383. doi: 10.1126/science.148.3668.382. [DOI] [PubMed] [Google Scholar]
- 23.Oakenfull EA, Ryder OA. Mitochondrial control region and 12S rRNA variation in Przewalski's horse (Equus przewalskii) Anim Genet. 1998;29:456–459. doi: 10.1046/j.1365-2052.1998.296380.x. [DOI] [PubMed] [Google Scholar]
- 24.Goto H, et al. A massively parallel sequencing approach uncovers ancient origins and high genetic variability of endangered Przewalski's horses. Genome Biol Evol. 2011;3:1096–1106. doi: 10.1093/gbe/evr067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pereira L, et al. Population expansion in the North African late Pleistocene signalled by mitochondrial DNA haplogroup U6. BMC Evol Biol. 2010;10:390. doi: 10.1186/1471-2148-10-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Driscoll CA, Macdonald DW, O'Brien SJ. From wild animals to domestic pets, an evolutionary view of domestication. Proc Natl Acad Sci USA. 2009;106(Suppl 1):9971–9978. doi: 10.1073/pnas.0901586106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeder MA. Domestication and early agriculture in the Mediterranean Basin: Origins, diffusion, and impact. Proc Natl Acad Sci USA. 2008;105:11597–11604. doi: 10.1073/pnas.0801317105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonfiglio S, et al. The enigmatic origin of bovine mtDNA haplogroup R: Sporadic interbreeding or an independent event of Bos primigenius domestication in Italy? PLoS One. 2010;5:e15760. doi: 10.1371/journal.pone.0015760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lippold S, et al. Discovery of lost diversity of paternal horse lineages using ancient DNA. Nat Commun. 2011;2:450. doi: 10.1038/ncomms1447. [DOI] [PubMed] [Google Scholar]
- 30.Clutton-Brock J. A Natural History of Domesticated Mammals. Cambridge, UK: Cambridge University Press; 1999. [Google Scholar]
- 31.MacFadden BJ. Fossil Horses: Systematics, Paleobiology, and Evolution of the Family Equidae. Cambridge, UK: Cambridge University Press; 1992. [Google Scholar]
- 32.Gonzaga PG, editor. A History of the Horse: The Iberian Horse from Ice Age to Antiquity. London, UK: JA Allen & Co. Ltd; 2004. [Google Scholar]
- 33.Lira J, et al. Ancient DNA reveals traces of Iberian Neolithic and Bronze Age lineages in modern Iberian horses. Mol Ecol. 2010;19:64–78. doi: 10.1111/j.1365-294X.2009.04430.x. [DOI] [PubMed] [Google Scholar]
- 34.Lopes MS, et al. The Lusitano horse maternal lineage based on mitochondrial D-loop sequence variation. Anim Genet. 2005;36:196–202. doi: 10.1111/j.1365-2052.2005.01279.x. [DOI] [PubMed] [Google Scholar]
- 35.Soares P, et al. The archaeogenetics of Europe. Curr Biol. 2010;20:R174–R183. doi: 10.1016/j.cub.2009.11.054. [DOI] [PubMed] [Google Scholar]
- 36.Warmuth V, et al. European domestic horses originated in two holocene refugia. PLoS One. 2011;6:e18194. doi: 10.1371/journal.pone.0018194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richter J. In: Neanderthal Lifeways, Subsistence and Technology: One Hundred Fifty Years of Neanderthal Study. Vertebrate Paleobiology and Paleoanthropology. Conard NJ, Richter J, editors. Berlin: Springer; 2011. [Google Scholar]
- 38.Colleoni F. PhD thesis. Stockholm: Stockholm University; 2009. On the Late Saalian glaciation (160-140 ka)-a climate modeling study. [Google Scholar]
- 39.Orlando L, et al. True single-molecule DNA sequencing of a pleistocene horse bone. Genome Res. 2011;21:1705–1719. doi: 10.1101/gr.122747.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bower MA, et al. The cosmopolitan maternal heritage of the Thoroughbred racehorse breed shows a significant contribution from British and Irish native mares. Biol Lett. 2011;7:316–320. doi: 10.1098/rsbl.2010.0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carelli V, et al. Haplogroup effects and recombination of mitochondrial DNA: Novel clues from the analysis of Leber hereditary optic neuropathy pedigrees. Am J Hum Genet. 2006;78:564–574. doi: 10.1086/501236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harrison SP, Turrion-Gomez JL. Mitochondrial DNA: An important female contribution to thoroughbred racehorse performance. Mitochondrion. 2006;6:53–63. doi: 10.1016/j.mito.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 43.Achilli A, et al. Mitochondrial genomes of extinct aurochs survive in domestic cattle. Curr Biol. 2008;18:R157–R158. doi: 10.1016/j.cub.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 44.Olivieri A, et al. The mtDNA legacy of the Levantine early Upper Palaeolithic in Africa. Science. 2006;314:1767–1770. doi: 10.1126/science.1135566. [DOI] [PubMed] [Google Scholar]
- 45.Torroni A, et al. Do the four clades of the mtDNA haplogroup L2 evolve at different rates? Am J Hum Genet. 2001;69:1348–1356. doi: 10.1086/324511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Temperley R, Richter R, Dennerlein S, Lightowlers RN, Chrzanowska-Lightowlers ZM. Hungry codons promote frameshifting in human mitochondrial ribosomes. Science. 2010;327:301. doi: 10.1126/science.1180674. [DOI] [PubMed] [Google Scholar]
- 47.Achilli A, et al. The molecular dissection of mtDNA haplogroup H confirms that the Franco-Cantabrian glacial refuge was a major source for the European gene pool. Am J Hum Genet. 2004;75:910–918. doi: 10.1086/425590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eltsov NP, Volodko NV. MtPhyl: Software tool for human mtDNA analysis and phylogeny reconstruction. 2011. Available at http://eltsov.org. Accessed March 2, 2011.
- 49.Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 50.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol. 2005;22:1185–1192. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- 52.Thirstrup JP, Bach LA, Loeschcke V, Pertoldi C. Population viability analysis on domestic horse breeds (Equus caballus) J Anim Sci. 2009;87:3525–3535. doi: 10.2527/jas.2008-1760. [DOI] [PubMed] [Google Scholar]
- 53.Nylander JA, Ronquist F, Huelsenbeck JP, Nieves-Aldrey JL. Bayesian phylogenetic analysis of combined data. Syst Biol. 2004;53:47–67. doi: 10.1080/10635150490264699. [DOI] [PubMed] [Google Scholar]
- 54.Oakenfull EA, Lim H, Ryder O. A survey of equid mitochondrial DNA: Implications for the evolution, genetic diversity and conservation of Equus. Conserv Genet. 2000;1:341–355. [Google Scholar]
- 55.Forstén A. Mitochondrial-DNA time-table and the evolution of Equus: Comparison of molecular and paleontological evidence. Ann Zool Fenn. 1992;28:301–309. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.