Abstract
ThnT is a pantetheine hydrolase from the DmpA/OAT superfamily involved in the biosynthesis of the β-lactam antibiotic thienamycin. We performed a structural and mechanistic investigation into the cis-autoproteolytic activation of ThnT, a process that has not previously been subject to analysis within this superfamily of enzymes. Removal of the γ-methyl of the threonine nucleophile resulted in a rate deceleration that we attribute to a reduction in the population of the reactive rotamer. This phenomenon is broadly applicable and constitutes a rationale for the evolutionary selection of threonine nucleophiles in autoproteolytic systems. Conservative substitution of the nucleophile (T282C) allowed determination of a 1.6-Å proenzyme ThnT crystal structure, which revealed a level of structural flexibility not previously observed within an autoprocessing active site. We assigned the major conformer as a nonreactive state that is unable to populate a reactive rotamer. Our analysis shows the system is activated by a structural rearrangement that places the scissile amide into an oxyanion hole and forces the nucleophilic residue into a forbidden region of Ramachandran space. We propose that conformational strain may drive autoprocessing through the destabilization of nonproductive states. Comparison of our data with previous reports uncovered evidence that many inactivated structures display nonreactive conformations. For penicillin and cephalosporin acylases, this discrepancy between structure and function may be resolved by invoking the presence of a hidden conformational state, similar to that reported here for ThnT.
Keywords: crystallography, mechanism, reactive rotamer effect, self-cleavage
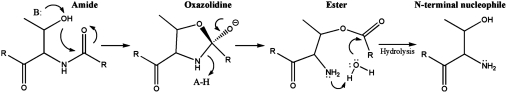
In stark contrast to the evolutionary selection for chemically stable polypeptides, there exist several classes of intrinsically reactive proteins that cleave an internal peptide bond as part of their cellular function (1). Many cis-autoprocessing events proceed through an N-O(S) acyl shift, where a threonine (Thr), serine (Ser), or cysteine (Cys) attacks the peptide bond of the preceding residue (Fig. 1). Collapse of the resultant (thia)oxazolidine exchanges the amide for a reactive (thio)ester that undergoes different chemical reactions, depending on biological context. This chemistry has been implicated in such diverse phenomena as protein folding, nucleoporin biogenesis, Hedgehog and G-protein coupled receptor signaling, protein splicing, and apoptosis (2–6). Additionally, cis-autoprocessing is necessary for maturation of many enzymes including the proteasome β-subunit, pyruvoyl-dependent decarboxylases, and the industrially used penicillin and cephalosporin acylases (PA and CA, respectively) (1, 7, 8). Enzymes use the N-terminal nucleophile (Ntn) liberated by autoproteolysis as a noncanonical catalytic diad (9). Central to these systems is the challenge of hydrolyzing an internal peptide bond, whose half-life at 25 °C and neutral pH is approximately 600 y, with no assistance from an external catalyst (10).
Fig. 1.
General mechanism of autoproteolysis. The nucleophilic residue attacks into its N-terminal amide bond. The resultant 5-membered oxazolidine ring collapses with concomitant protonation of the nitrogen leaving group. The nascent amine then acts as a general base to activate a water molecule, which hydrolyzes the ester intermediate. The new C-terminal residue dissociates, exposing the catalytic N-terminal nucleophile. “A-H” and “B:” represent a general acid and general base, respectively. A comparable mechanism also occurs for autoproteolytic systems that utilize Cys and Ser nucleophiles.
Although the chemical reactions underlying self-cleavage have been appreciated for the past decade, understanding how these reactions are facilitated is hampered by the inapplicability of many enzymological techniques. Because the active site is not returned to its original state, autoprocessing is not formally catalytic; this limits the utility of traditional kinetic assays. The large size and intrinsic reactivity of most systems precludes labeling schemes and makes nuclear magnetic resonance experiments particularly difficult. These challenges were recently overcome for the 15-kDa Muc1 SEA domain, where it was shown that protein folding introduces torsional strain at the scissile bond, lowering the barrier to cleavage by 7 kcal/mol (11, 12). The Ntn hydrolases were the first structurally characterized cis-autoproteolytic enzymes (13), but their size and complexity hinder similar progress. Enzymes from this superfamily are known to utilize all three possible nucleophiles (Thr, Ser, or Cys); however, the mechanistic constraints imparted by a given residue are entirely unknown. Once described as belonging to the Ntn superfamily, the autoactivation of the less numerous DmpA/OAT (D/O) superfamily has not been studied (14). Understanding the autoproteolytic protein scaffold is of significant practical interest because enzymes from the D/O or Ntn superfamiles are implicated in the biosynthesis of all classes of the naturally occurring bicyclic β-lactam antibiotics (15–19).
Here, we describe our investigation into the autoproteolytic activation of a D/O enzyme, ThnT, a pantetheine hydrolase from Streptomyces cattleya involved in the biosynthesis of the β-lactam antibiotic thienamycin (19). We identify specific structural constraints imparted by the γ-methyl of a Thr nucleophile, providing a common mechanistic link between the Ntn and D/O enzymes and a rationale for the evolutionary selection of this residue. The X-ray crystal structure of an uncleaved ThnT precursor showed unusual structural flexibility in the active site, allowing interconversion between inactive and active states. Evaluation of previously reported uncleaved Ntn structures found that they often contain comparable active site geometries to the inactive state of ThnT, and they are unlikely to promote the requisite chemistry. Application of the insights gleaned from analysis of ThnT may be used to address these deficiencies, providing a strengthened basis for continued research into autoprocessing.
Results and Discussion
The Reactive Rotamer Effect Accelerates the N-O Acyl Shift.
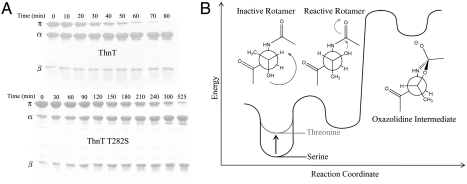
ThnT matures with a half-life (t1/2) = 43 min at 37 °C, as measured by a kinetic SDS-PAGE assay. Conservative mutation of the nucleophile, T282S, resulted in a 4.3-fold reduction in the self-cleavage rate (Fig. 2A). An extensive review of the literature revealed that comparable Thr → Ser mutations always impede autoproteolysis, suggesting a common cause underlies this phenomenon (20–27). Temperature dependent analysis of the autoproteolysis of the Ntn-hydrolase glycosylasparaginase (GA) showed the Thr → Ser rate deceleration is caused by an increase in the activation entropy; however, the molecular interaction giving rise to this effect remained elusive (22).
Fig. 2.
The reactive rotamer effect in autoproteolysis. (A) Time course of the autoproteolysis reactions of ThnT and T282S shown on 15% SDS-PAGE. As the reaction progresses, there is a decrease in the intensity of the uncleaved (π) band and a corresponding increase in the intensity of the two cleavage products (α and β). Quantification with first-order kinetics shows a 4.3-fold rate deceleration caused by removal of the γ-methyl. (B) Hypothetical reaction coordinate diagram showing how threonine accelerates nucleophilic attack into the N-terminal amide bond by minimizing the number of gauche interactions for the reactive rotamer. This effect is specific to the 3R stereochemistry of the β-carbon and is also influenced by the geometry of the local protein backbone.
The N-O acyl shift proceeds through a sterically disfavored gauche-rotamer of the nucleophile (7, 28). The reactive g- rotamer of Thr offsets this instability by placing the γ-methyl in its anti-position (Fig. 2B). The rotamer distribution of the nucleophile is also affected by interactions with the local backbone. Such interactions specifically accelerate autoproteolysis with Thr when the γ-methyl forms unfavorable steric interactions in nonproductive rotamers. This reactive rotamer effect (RRE) is disrupted by the Thr → Ser mutation, which shifts the ground state rotamer distribution away from a reactive state, accounting for the entropically derived rate deceleration (29). The RRE may contribute to the previously unexplainable observation that Thr nucleophiles are rare outside of cis-autoproteolytically activated systems, but common among the Ntn and D/O hydrolases (14, 30, 31). We suggest the evolutionary selection for different nucleophiles between Ser and Thr hydrolases reflects, in part, the propensity of a γ-methyl to assist in efficient maturation through the RRE.
The X-Ray Crystal Structure of ThnT T282C.
Although a Thr → Ser mutation has a relatively small effect on the rate of autoproteolysis, previous studies have established that mutational exchange of a hydroxyl nucleophile for a thiol has a strong adverse effect (23, 26). Chemical rescue of cleavage is traditionally accomplished by the addition of the strong nucleophile hydroxylamine (32). We were unable to detect autoproteolysis of ThnT T282C, but addition of NH2OH induced a small amount of hydroxyaminolysis after 24 h (SI Appendix, Fig. S1). This result demonstrated that T282C is a chemically stable variant that is capable of undergoing a slow N-S acyl shift and was, therefore, an ideal candidate to investigate the structure of a minimally altered autoprocessing site.
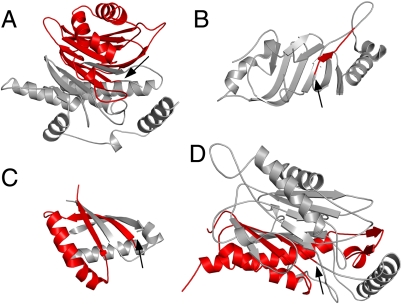
We identified crystallization conditions in which crystals of T282C grew overnight, before measurable autoproteolysis could occur. These crystals diffracted to high resolution and we report here the 1.6-Å X-ray crystal structure of this mutant, which allowed for the analysis of a D/O enzyme in its uncleaved form. ThnT is dimeric both by native PAGE (19) and within the asymmetric unit of the crystal. Like other D/O and Ntn proteins, ThnT has an αββα architecture (Fig. 3). The connectivity of the secondary structural elements differs between these superfamilies, indicating they are related through convergent evolution (14). We compared the architecture of ThnT with representative members of three other topologically unique cis-autoproteolytic systems and identified a simple feature common among them: The scissile bond is located at the end of a β-strand (12, 24, 33). Standard α-helix geometry prevents the γ-atom of a residue from approaching its N-terminal peptide bond, accounting for the general absence of autoproteolysis sites on α-helices (SI Appendix, Fig. S2). Although the selection for autoproteolysis at the terminus of a β-strand likely reflects diverse evolutionary pressures, it also suggests that an intimate connection exists between the conformational states of these proteins’ active site residues and the autoproteolytic chemistry they share.
Fig. 3.
Representative autoproteolytic proteins from distinct folds. Autoproteolysis sites (black arrows) separate the N-terminal domain (silver) from the C-terminal domain (red). In each system, the autoproteolysis site is found at the terminus of a β-strand. The Ntn hydrolases (A) and the D/O hydrolases (D) share similar αββα architecture, but the connectivity between the secondary structure elements is different, indicating their relationship through convergent evolution. Protein Data Bank ID: (A) 2A8I, hTaspase1; (B) 2Q5X, hNup98; (C) 2ACM, Muc1 SEA; (D) 3S3U, ThnT. For clarity, a single monomer of hTaspase1 and ThnT is shown.
Two Conformations of the Scissile Bond are Crystallographically Observed.
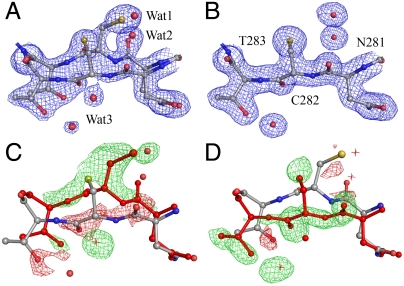
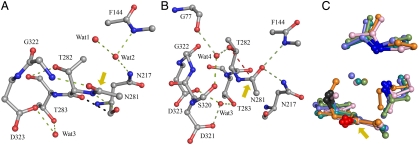
Despite the low rmsd, 0.11 Å2, between the subunits (designated π1 and π2) of ThnT T282C, the electron density flanking the scissile bond (N↑CT) in π1 and π2 contain significant differences. This segment of π1 was readily modeled with the nucleophilic thiol present on the re-face of the scissile bond (state B, Fig. 4, B). Attempts to model the N↑CT segment of π2 with a single conformer consistently produced low-quality maps. Relying on the electron-rich sulfur atom of C282 to guide placement of the side chain, we assigned a major conformer with the thiol on the si-face of the scissile bond (state A, Fig. 4, A). Inspection of the resulting Fo - Fc map showed the remaining electron density precisely matched state B. Determination of a 55∶45 ratio of the A and B states in π2 required including the partial occupancy of Wat1 and Wat2 in state B, which occupy the same space as the thiol and scissile carbonyl of state A (Fig. 4). During model building, C282 in state A consistently refined to a disallowed region of ϕ-ψ space (87°, 114°), suggesting that specific structural features must favor what is otherwise a highly strained conformation.
Fig. 4.
Dual occupancy at the active site residues N281, C282, and T283. (A) The π2 subunit with states A and B modeled with a 55∶45 ratio, 2mFo - DFc electron density map (blue) calculated at 1.0 σ. (B) The π1 subunit with a single conformer, state B, modeled. (C) Modeling state B alone into π2 produces poor Fo - Fc electron density at 4.0 σ with negative (red) and positive (green) density corresponding to the omitted conformer (red residues, water as asterisks). (D) Modeling state A alone into π2 clearly shows that the missing density may be satisfied by the partial occupancy of state B, identified in π1.
The origin of the differences between each subunit may be related to the asymmetry of their crystalline environments, as indicated by B-factor analysis. The average B factor of the atoms modeled in π1, 17.2 Å2, is 25% higher than the π2 atoms, 13.7 Å2. This asymmetry is distributed broadly throughout the subunit and may reflect significant differences in the packing forces across the asymmetric unit (SI Appendix, Fig. S3). The mechanism through which state A in π2 is preferentially stabilized is not apparent. Residues L319–D321, which have crystal contacts, are adjacent to the active site and also populate two conformations in π2, suggesting that long-range conformational coupling may affect the ThnT active site. The existing crystallographic data, however, cannot determine a causal relationship between multiple conformational states.
Autoproteolysis Proceeds Through a Si-Face Attack.
Each state defined by the structure of ThnT T282C implicates a different face of the scissile bond for the initial nucleophilic attack. Based on the chemical demands of the N-O acyl shift, we established a procedure to identify a potential cleavage-competent state. A reactive rotamer is defined by its ability to reach a Bürgi-Dunitz (B-D) trajectory of 105 ± 5° (34). Inspection of the active site must show this reactive rotamer is sterically allowed. For systems displaying the RRE, the reactive rotamer will be favored by Thr relative to Ser. In order to estimate the population of a given rotamer, we employed a backbone-dependent rotamer library. (35). Lastly, we note that crystal structures of disabled autoprocessing sites should be viewed generously, because mutational inactivation and crystal packing may alter the active site geometry.
In state B of ThnT, the B-D trajectory may be reached from the g+ rotamer of T282, however, this forces a strong steric clash with the carbonyl oxygen of the scissile amide (SI Appendix, Fig. S4). A single viable rotamer for T282 can be rationalized, with the hydroxyl anti to the scissile bond (Fig. 5A). Therefore, we conclude state B could be populated by ThnT, but could not effect the N-O acyl shift. In state A, the g- rotamer of T282 can reach the B-D trajectory for attack into the si-face of the scissile bond (Fig. 5B), requiring bond angle compression that is also necessary for formation of the oxazolidine intermediate. In this conformation, the γ-methyl is 2.7 Å from the carbonyls of A142 and G320. These steric clashes could be ameliorated by small structural adjustments. Finally, state A favors the g- rotamer of Thr more than Ser by a factor of 1.6, as indicated by the backbone-dependent rotamer library. The capacity of Ser to hydrogen bond to the carbonyls of A142 or G320 in the nonreactive anti-rotamer, which Thr cannot populate, may further shift the rotamer distribution to account for the observed 4.3-fold rate deceleration observed with the T282S mutant.
Fig. 5.
Active and inactive states of ThnT. (A) Model of the inactive state B of ThnT. Hydrogen bonds are shown as green dashes. The 2.7-Å steric clash between T282 and N217 is shown with black dashes. A yellow arrow indicates the scissile bond. (B) Model of the cleavage-competent state of ThnT where the steric clash and twisted amide of T282 have been removed. The direction of attack is indicated with red dashes. (C) Alignment of T282C-state B with precursor Ntn hydrolases (2X1C, teal; 2IWM, pink; 1OQZ, slate; 1KEH, green) using the two oxyanion hole nitrogen atoms, oxygen of the scissile bond, and Cβ atoms (blue, red, and gray, respectively). This alignment reveals significant structural similarity between each active site and the occupation of the oxyanion hole with a crystallographic water molecule, corresponding to Wat2 in (A). For these Ntn hydrolases, a hidden rearrangement that displaces the bound water and places the scissile carbonyl in the oxyanion would activate the system for autoproteolysis.
Consequently, state A satisfies the known constraints for ThnT autoproteolysis and may represent a cleavage-competent state. Assignment of a si-face attack is corroborated by the presence of an oxyanion hole, comprised of the δNH2 of N217 and the amide NH of F144, with optimal distances and angles for polarization of the scissile bond and stabilization of the oxazolidine intermediate. The mutation N217D disrupts autoproteolysis, consistent with a mechanism utilizing this oxyanion hole (SI Appendix, Fig. S1). Hence, application of simple constraints based on the underlying chemistry unambiguously identifies state A, the net minor conformer of ThnT, as the cleavage-competent state.
Factors Promoting Formation of the Cleavage-Competent State.
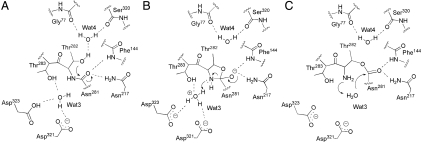
ThnT promotes the formation of a reactive conformation by several distinct means. In the inactive state B, the carbonyl of T282 has a 2.7-Å steric clash with N217 and a severely strained ω angle of 159° (Fig. 5). Formation of state A removes the clash and relaxes the ω angle to 177°. Concurrently T282 moves into a “forbidden” region of the Ramachandran plot, accompanied by the expulsion of Wat1 and polarization of the γOH by its hydrogen bond to Wat4. The group undergoing the most significant change is the scissile bond itself, which rotates 90° into the oxyanion hole, replacing Wat2. This peptide flip maintains a strained ω angle of 172° and exchanges the hydrogen bond partner of the amide nitrogen from the carbonyl oxygen of N217 to Wat3, which shifts into hydrogen bond distance with the amide nitrogen of the scissile bond. An oxyanion hole is a common feature of enzyme active sites where polarization increases the electrophilicity of reactive carbonyls and stabilizes tetrahedral intermediates. Polarization, however, also raises the barrier to rotation of amide bonds and we cannot speculate on the energy necessary to maintain the torsional strain present on the scissile bond of ThnT. In sum, preferential hydrogen bonding, relief of torsional strain, removal of steric clashes, and liberation of solvent may combine to compensate for formation of a highly strained, chemically reactive state.
A Mechanism for ThnT Autoproteolysis.
Two proton transfers accompany the N-O acyl shift: deprotonation of the nucleophile and protonation of the amine leaving group. In all Ntn and D/O enzymes, maturation yields an N-terminal amine that functions as the general base during catalysis (31). However, this group is not available prior to (thio)ester formation. The lack of a strongly activated nucleophile may contribute to the unexplained observation that the kobs of self-cleavage is orders of magnitude slower than the kcat of substrate cleavage in mature Ntn hydrolases (21, 26, 36). Wat4 is uniquely oriented in state A to act as a proton acceptor for T282 by its hydrogen bonds with the backbone carbonyls of S320 and G322. We hypothesize that this water molecule acts as the base for the reaction (Fig. 6). Once activated, the nucleophile may then follow the B-D trajectory for attack into the scissile bond.
Fig. 6.
Proposed autoproteolytic mechanism of ThnT. (A) The precleavage conformation shown in Fig. 5B. Dashed lines represent hydrogen bonds critical to activation of the system. A fourth hydrogen bond exists for Wat4, enabling it to shuttle the labile proton on T282 into solvent. (B) The amine leaving group is protonated by Wat3 and expelled, generating a planar ester. (C) The nucleophilic Wat3 is activated by the nascent amine and hydrolyzes the ester.
Protonation of the leaving group can occur on the amide prior to nucleophilic attack or on the 2° amine of the oxazolidine intermediate. As discussed above, the Muc1 SEA domain promotes autoproteolysis by applying torsional strain to the scissile bond. This strain raises the pKa of the amide nitrogen sufficiently for protonation at neutral pH, prior to nucleophilic attack (11, 37), and was suggested to be a common mechanism for autoprocessing. We note that a grossly strained scissile bond has yet to be observed in a structure of an uncleaved autoproteolytic protein. More importantly, the basicity of the amide nitrogen is lowered by the polarizing effect of the oxyanion hole provided in the Ntn and D/O systems. Consequently, we propose that nucleophilic attack into the scissile bond precedes protonation of the nitrogen for ThnT and its relatives (Fig. 6).
There is a single candidate water molecule, Wat3, for protonation of the oxazolidine intermediate. In state A, this water is tightly coordinated by the side chains of T283, D321, and D323. The conservative mutations D321N and D323N completely eliminate self-cleavage, demonstrating the importance of these residues (SI Appendix, Fig. S1). The close proximity of their carboxylate groups, 4.3 Å, suggests that one of these residues may have an elevated pKa, allowing it to act as a general acid. Although D321 is closer to Wat3, D323 is conserved across the D/O superfamily, suggesting it is the acidic group. Water-mediated proton transfer from D323 to the secondary amine would then allow for collapse of the oxazolidine, generating a planar ester intermediate. The nascent amine of T282 then activates Wat3 to hydrolyze the ester, completing autoproteolysis (Fig. 6).
Two Distinct Mechanistic Pathways for Cis-Autoproteolysis with Thr.
The autoactivation of many Thr-utilizing Ntn hydrolases has been investigated with X-ray crystallography. We applied our criteria for identification of a cleavage-competent state and found that several of these structures cannot support the N-O acyl shift. The proteasome β-subunit (PS) is the only Ntn hydrolase reported to initiate autoproteolysis through a si-face attack. Hence, it shares the same intrinsic geometric constraints for the N-O acyl shift as ThnT. The structure of uncleaved PS was solved using Thr → Ala inactivation, which may have altered its active site geometry due to the removal of the γOH, whose interactions are better preserved in the Thr → Cys structure of ThnT reported here. A model of PS activation that brings the observed ϕ-ψ angles of the nucleophile closer to those found for ThnT would significantly increase the probability of populating the reactive rotamer (SI Appendix, Table S1). GA is the most intensely studied cis-autoproteolytic system, where the current model for self-cleavage contains features from two different inactivated structures (28, 38). The geometry captured by mutation of the nucleophile, T152C, is almost identical to the inactive state of ThnT. As was shown for ThnT, population of a reactive rotamer in this state causes a preclusive steric clash and, therefore, cannot support the N-O acyl shift without prior structural rearrangement. The inactive W11F GA structure retains the Thr nucleophile and contains an active site geometry that does allow formation of the reactive rotamer. This conformation indicates autoproteolysis occurs through a re-face attack from the same g- rotamer proposed for ThnT and PS.
Our analysis of uncleaved structures for γ-glutamyltranspeptidase (20, 39), l-asparaginase EcAIII (40, 41), Taspase1 (24), and GA (28) indicates that they have crystallized in inactive states (SI Appendix, Table S1). For gGT and EcaIII, the active site geometry was previously hypothesized to represent an active state that is comparable to state B of ThnT T282C and GA T152C. Because this state is now firmly established as inactive and has been shown to rearrange to a conformation giving rise to either re-face or si-face attack, additional information is needed before a mechanistic pathway may be proposed for these proteins. However, it is notable that each of the plausible mechanisms for autoproteolysis with Thr proceeds from the g- rotamer of the nucleophile. This empirical result could have been anticipated by the RRE because this is the only rotamer that places the γ-methyl in the favorable anti position. We believe that autoproteolysis initiated from the g- rotamer is a general feature of Thr-utilizing cis-autoproteolytic systems.
Evidence for Hidden Conformational Rearrangement Preceding Ntn Hydrolase Maturation.
Fewer data exist for autoproteolysis with a Ser or Cys nucleophile. Mechanistic analysis is more difficult because these residues lack the added constraint of a γ-methyl. Nonetheless, the structurally characterized systems, PA and CA, display an active site that bears several striking similarities to the inactive state of ThnT (17, 18, 42). The protein backbone flanking the scissile bond is comparable to the inactive conformation identified in state B of ThnT. Also, the oxyanion hole is occupied by a water molecule corresponding to Wat2 of ThnT (Fig. 5C). The proposed mechanism for these enzymes invokes the bound water molecule to activate the nucleophile by acting as a proton acceptor. The oxyanion hole, however, polarizes this water to act as a proton donor. To rectify the perplexing mechanistic implications suggested by these structures, we hypothesize a peptide flip places the reactive carbonyl into the conserved oxyanion hole, expels the “bound” water and positions the nucleophile for si-face attack. Our hypothesis is further supported by observations in CA, where the inactive F177P variant compresses the active site and expels the bound water (43). Instead of disrupting self-cleavage by removing an inappropriately polarized water molecule, the observed compression may prevent a peptide flip necessary for formation of a cleavage-competent state.
Lastly, a large planar deviation in the ω angle of the nucleophilic residue has been observed for PA, ThnT, and GA (28, 38). It was proposed that relief of this strain upon ester formation would contribute up to 5 kcal/mol to drive the N-O acyl shift. However, for ThnT, this strain is relieved upon formation of the active state, supporting an alternative hypothesis: Conformational strain drives autoprocessing by destabilizing nonproductive states.
Conclusions
Autoprocessing is a unique and challenging branch of enzymology. Because chemistry is directly encoded in the folded-state ensemble, nowhere is the link between structure and function so clearly defined. We attribute the accelerated rate of autoproteolysis with a Thr nucleophile to the RRE, providing a basis for the evolutionary selection of this residue. In the crystal structure of T282C, we observed two conformations at the active site and identified the net minor conformer as a cleavage-competent state. By extending our mechanism-based analysis to previously reported uncleaved structures, we have shown why many are unlikely to promote the requisite chemistry. This deficiency may be addressed by invoking a hidden conformational rearrangement within the proenzyme active site. For CA and PA, evidence suggests that this rearrangement is comparable to that observed for ThnT. Although great strides have been made in understanding autoprocessing, an important lesson emerges from this investigation: X-ray crystallography of mutationally inactivated precursors is not necessarily a reliable technique for the identification of reactive states.
Methods
Expression Vector Construction.
Construction of a codon-optimized expression vector has been described previously (19). Mutant genes were made using overlap extension PCR with primers T282S-F: 5′- GCCACCCTCAACAGCACGCTCGCCGTG -3′ and T282S-R: 5′- CACGGCGAGCGTGCTGTTGAGGGTGGC -3′ (Sigma-Genosys), digested with NdeI and HindIII, and ligated into pET28b(+) (Novagen) to create N-6His-tag fusions. All mutations were sequence-verified and transformed into Escherichia coli Rosetta2(DE3)-competent cells (Novagen).
Protein Expression and Purification.
All ThnT mutants were expressed in E. coli Rosetta2(DE3) cells as previously reported (19). Eluent from the Ni-nitrilotriacetate column was desalted into either 10 mM potassium phosphate, pH 7.5 with 10% glycerol (for kinetic experiments) or 20 mM potassium phosphate, pH 7.5 with 5% glycerol (for crystallography), passed through 700 µL Q-Sepharose resin (Sigma-Aldrich), and concentrated in 10 K molecular weight cutoff Amicon filtration devices (Bio-Rad).
Autoproteolysis Assays.
Purified protein was diluted to 1 μg/μL into 100 mM citric acid/phosphate buffer, pH, 7.5, 100 mM KCl and 10% glycerol, and preincubated to 37 °C for 5 min. For assays extending beyond 12 h, 100 mM Tris pH 7.5 was used instead of 100 mM citric acid/phosphate buffer. Discrete time points were collected by quenching an aliquot of protein in 5× SDS-PAGE loading buffer and heated at 98 °C for 1 min. Samples were run on a 15% SDS-PAGE gel and stained with Coomassie blue. Densitometry was performed using ImageJ software and the fraction of uncleaved protein fit to a single exponential (Eq. 1) in Prism (GraphPad) (SI Appendix, Fig S5):
| [1] |
Crystallization and Data Collection.
Automated crystal screening was performed by the University of Maryland Crystallography Core using an OryxNano (Douglas Instruments) with 7.7 μg/μL T282C and solutions from Qiagen. Crystallization was repeated using sitting-drop vapor diffusion against a 1-mL reservoir solution of 0.45–0.65 M sodium acetate and 12–14% PEG 3350. Crystals were cryoprotected by immersion into 0.5 M sodium acetate, 17% PEG3350, and 20% glycerol prior to flash-freezing in N2. Crystals of T282C for multiwavelength anomalous dispersion phasing experiments were derivatized with a 20-min soak in cryoprotection buffer supplemented with 200 μM ethylmercuric phosphate. Diffraction data were collected on BL 12-2 at the Stanford Synchrotron Light Source (SI Appendix, Table S3).
Structure Solution and Refinement.
Phasing was performed using the multiple wavelength anomalous diffraction method with ethylmercuric-derivatized protein and then applied to nonderivatized T282C structure factors using direct Fourier synthesis. Data were processed in Collaborative Computational Project Number 4 using SHELX/C/D/E, followed by solvent flattening and histogram matching with DM and automated model building with wARP/ARP (44–47). Iterative rounds of manual model building were performed in Coot using REFMAC5 for refinement (48). Atomic coordinates were deposited in the Research Collaboratory for Structural Bioinformatics Data Bank (Protein Data bank ID 3S3U).
Supplementary Material
Acknowledgments.
We thank Prof. Edvin Pozharskiy and the University of Maryland Crystallography Core for assistance in crystal screening and data collection. Densitometry was performed in the laboratory of Prof. Karen G. Fleming. This work was supported by a National Institutes of Health (NIH) Research Grant R01 AI001437 (to C.A.T.) and NIH GM61017 (to J.F.S.), and the Dimitri V. d’Arbeloff Postdoctoral Fellowship (to N.T.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3S3U).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113633109/-/DCSupplemental.
References
- 1.Perler FB, Xu MQ, Paulus H. Protein splicing and autoproteolysis mechanisms. Curr Opin Chem Biol. 1997;1:292–299. doi: 10.1016/s1367-5931(97)80065-8. [DOI] [PubMed] [Google Scholar]
- 2.Macao B, Johansson DGA, Hansson GC, Hard T. Autoproteolysis coupled to protein folding in the SEA domain of the membrane-bound MUC1 mucin. Nat Struct Mol Biol. 2006;13:71–76. doi: 10.1038/nsmb1035. [DOI] [PubMed] [Google Scholar]
- 3.Rosenblum JS, Blobel G. Autoproteolysis in nucleoporin biogenesis. Proc Natl Acad Sci USA. 1999;96:11370–11375. doi: 10.1073/pnas.96.20.11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paulus H. Protein splicing and related forms of protein autoprocessing. Annu Rev Biochem. 2000;69:447–496. doi: 10.1146/annurev.biochem.69.1.447. [DOI] [PubMed] [Google Scholar]
- 5.Lin HH, et al. Autocatalytic cleavage of the EMR2 receptor occurs at a conserved G protein-coupled receptor proteolytic site motif. J Biol Chem. 2004;279:31823–31832. doi: 10.1074/jbc.M402974200. [DOI] [PubMed] [Google Scholar]
- 6.Tinel A, et al. Autoproteolysis of PIDD marks the bifurcation between pro-death caspase-2 and pro-survival NF-kappa B pathway. EMBO J. 2007;26:197–208. doi: 10.1038/sj.emboj.7601473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ditzel L, et al. Conformational constraints for protein self-cleavage in the proteasome. J Mol Biol. 1998;279:1187–1191. doi: 10.1006/jmbi.1998.1818. [DOI] [PubMed] [Google Scholar]
- 8.Volpato G, Rodrigues RC, Fernandez-Lafuente R. Use of enzymes in the production of semi-synthetic penicillins and cephalosporins: Drawbacks and perspectives. Curr Med Chem. 2010;17:3855–3873. doi: 10.2174/092986710793205435. [DOI] [PubMed] [Google Scholar]
- 9.Dodson G, Wlodawer A. Catalytic triads and their relatives. Trends Biochem Sci. 1998;23:347–352. doi: 10.1016/s0968-0004(98)01254-7. [DOI] [PubMed] [Google Scholar]
- 10.Radzicka A, Wolfenden R. Rates of uncatalyzed peptide bond hydrolysis in neutral solution and the transition state affinities of proteases. J Am Chem Soc. 1996;118:6105–6109. [Google Scholar]
- 11.Sandberg A, Johansson DGA, Macao B, Hard T. SEA domain autoproteolysis accelerated by conformational strain: Energetic aspects. J Mol Biol. 2008;377:1117–1129. doi: 10.1016/j.jmb.2008.01.051. [DOI] [PubMed] [Google Scholar]
- 12.Johansson DGA, Macao B, Sandberg A, Hard T. SEA domain autoproteolysis accelerated by conformational strain: Mechanistic aspects. J Mol Biol. 2008;377:1130–1143. doi: 10.1016/j.jmb.2008.01.050. [DOI] [PubMed] [Google Scholar]
- 13.Smith JL, et al. Structure of the allosteric regulatory enzyme of purine biosynthesis. Science. 1994;264:1427–1433. doi: 10.1126/science.8197456. [DOI] [PubMed] [Google Scholar]
- 14.Cheng H, Grishin NV. DOM-fold: A structure with crossing loops found in DmpA, ornithine acetyltransferase, and molybdenum cofactor-binding domain. Prot Sci. 2005;14:1902–1910. doi: 10.1110/ps.051364905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hewitt L, et al. Structure of a slow processing precursor penicillin acylase from Escherichia coli reveals the linker peptide blocking the active-site cleft. J Mol Biol. 2000;302:887–898. doi: 10.1006/jmbi.2000.4105. [DOI] [PubMed] [Google Scholar]
- 16.Kershaw NJ, et al. ORF6 from the clavulanic acid gene cluster of Streptomyces clavuligerus has ornithine acetyltransferase activity. Eur J Biochem. 2002;269:2052–2059. doi: 10.1046/j.1432-1033.2002.02853.x. [DOI] [PubMed] [Google Scholar]
- 17.Kim Y, Kim S, Earnest TN, Hol WGJ. Precursor structure of cephalosporin acylase—insights into autoproteolytic activation in a new N-terminal hydrolase family. J Biol Chem. 2002;277:2823–2829. doi: 10.1074/jbc.M108888200. [DOI] [PubMed] [Google Scholar]
- 18.Kim JK, et al. Crystal structures of glutaryl 7-aminocephalosporanic acid acylase: Insight into autoproteolytic activation. Biochemistry. 2003;42:4084–4093. doi: 10.1021/bi027181x. [DOI] [PubMed] [Google Scholar]
- 19.Freeman MF, McIshos KA, Bodner MJ, Li RF, Townsend CA. Four enzymes define the incorporation of coenzyme A in thienamycin biosynthesis. Proc Natl Acad Sci USA. 2008;105:11128–11133. doi: 10.1073/pnas.0804500105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki H, Kumagai H. Autocatalytic processing of gamma-glutamyltranspeptidase. J Biol Chem. 2002;277:43536–43543. doi: 10.1074/jbc.M207680200. [DOI] [PubMed] [Google Scholar]
- 21.Boanca G, Sand A, Barycki JJ. Uncoupling the enzymatic and autoprocessing activities of Helicobacter pylori gamma-glutamyltranspeptidase. J Biol Chem. 2006;281:19029–19037. doi: 10.1074/jbc.M603381200. [DOI] [PubMed] [Google Scholar]
- 22.Guan CD, et al. Activation of glycosylasparaginase—formation of active N-terminal threonine by intramolecular autoproteolysis. J Biol Chem. 1996;271:1732–1737. doi: 10.1074/jbc.271.3.1732. [DOI] [PubMed] [Google Scholar]
- 23.Marc F, Weigel P, Legrain C, Glansdorff N, Sakanyan V. An invariant threonine is involved in self-catalyzed cleavage of the precursor protein for ornithine acetyltransferase. J Biol Chem. 2001;276:25404–25410. doi: 10.1074/jbc.M100392200. [DOI] [PubMed] [Google Scholar]
- 24.Khan JA, Dunn BM, Tong L. Crystal structure of human Taspase1, a crucial protease regulating the function of MLL. Structure. 2005;13:1443–1452. doi: 10.1016/j.str.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 25.Seemuller E, Lupas A, Baumeister W. Autocatalytic processing of the 20S proteasome. Nature. 1996;382:468–470. doi: 10.1038/382468a0. [DOI] [PubMed] [Google Scholar]
- 26.Guan C, et al. Characterization and functional analysis of the cis-autoproteolysis active center of glycosylasparaginase. J Biol Chem. 1998;273:9695–9702. doi: 10.1074/jbc.273.16.9695. [DOI] [PubMed] [Google Scholar]
- 27.Saarela J, Oinonen C, Jalanko A, Rouvinen J, Peltonen L. Autoproteolytic activation of human aspartylglucosaminidase. Biochem J. 2004;378:363–371. doi: 10.1042/BJ20031496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu QA, Buckley D, Guan CD, Guo HC. Structural insights into the mechanism of intramolecular proteolysis. Cell. 1999;98:651–661. doi: 10.1016/s0092-8674(00)80052-5. [DOI] [PubMed] [Google Scholar]
- 29.Jung ME, Piizzi G. gem-Disubstituent effect: Theoretical basis and synthetic applications. Chem Rev. 2005;105:1735–1766. doi: 10.1021/cr940337h. [DOI] [PubMed] [Google Scholar]
- 30.Powers JC, Asgian JL, Ekici OD, James KE. Irreversible inhibitors of serine, cysteine, and threonine proteases. Chem Rev. 2002;102:4639–4750. doi: 10.1021/cr010182v. [DOI] [PubMed] [Google Scholar]
- 31.Ekici OD, Paetzel M, Dalbey RE. Unconventional serine proteases: Variations on the catalytic Ser/His/Asp triad configuration. Prot Sci. 2008;17:2023–2037. doi: 10.1110/ps.035436.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shao Y, Xu MQ, Paulus H. Protein splicing: Evidence for an N-O acyl rearrangement as the initial step in the splicing process. Biochemistry. 1996;35:3810–3815. doi: 10.1021/bi952592h. [DOI] [PubMed] [Google Scholar]
- 33.Sun Y, Guo HC. Structural constraints on autoprocessing of the human nucleoporin Nup98. Prot Sci. 2008;17:494–505. doi: 10.1110/ps.073311808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burgi HB, Dunitz JD, Shefter E. Geometrical reaction coordinates 2. Nucleophilic addition to a carbonyl group. J Am Chem Soc. 1973;95:5065–5067. [Google Scholar]
- 35.Shapovalov MV, Dunbrack RL., Jr A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Structure. 2011;19:844–858. doi: 10.1016/j.str.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y, Guan C, Aronson NN. Site-directed mutagenesis of essential residues involved in the mechanism of bacterial glycosylasparaginase. J Biol Chem. 1998;273:9688–9694. doi: 10.1074/jbc.273.16.9688. [DOI] [PubMed] [Google Scholar]
- 37.Johansson DGA, et al. Protein autoproteolysis: Conformational strain linked to the rate of peptide cleavage by the pH dependence of the N → O acyl shift reaction. J Am Chem Soc. 2009;131:9475–9477. doi: 10.1021/ja9010817. [DOI] [PubMed] [Google Scholar]
- 38.Wang YM, Guo HC. Crystallographic snapshot of glycosylasparaginase precursor poised for autoprocessing. J Mol Biol. 2010;403:120–130. doi: 10.1016/j.jmb.2010.08.038. [DOI] [PubMed] [Google Scholar]
- 39.Okada T, Suzuki H, Wada K, Kumagai H, Fukuyama K. Crystal structure of the gamma-glutamyltranspeptidase precursor protein from Escherichia coli—structural changes autocatalytic processing and implications for the maturation mechanism. J Biol Chem. 2007;282:2433–2439. doi: 10.1074/jbc.M607490200. [DOI] [PubMed] [Google Scholar]
- 40.Michalska K, Hernandez-Santoyo A, Jaskolski M. The mechanism of autocatalytic activation of plant-type L-asparaginases. J Biol Chem. 2008;283:13388–13397. doi: 10.1074/jbc.M800746200. [DOI] [PubMed] [Google Scholar]
- 41.Michalska K, Borek D, Hernandez-Santoyo A, Jaskolski M. Crystal packing of plant-type L-asparaginase from Escherichia coli. Acta Crystallogr D Biol Crystallogr. 2008;64:309–320. doi: 10.1107/S0907444907068072. [DOI] [PubMed] [Google Scholar]
- 42.Bokhove M, et al. Structures of an isopenicillin N converting Ntn-hydrolase reveal different catalytic roles for the active site residues of precursor and mature enzyme. Structure. 2010;18:301–308. doi: 10.1016/j.str.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 43.Yoon J, et al. A bound water molecule is crucial in initiating autocatalytic precursor activation in an N-terminal hydrolase. J Biol Chem. 2004;279:341–347. doi: 10.1074/jbc.M309281200. [DOI] [PubMed] [Google Scholar]
- 44.Bailey S. The CCP4 suite—programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 45.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 46.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 47.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.