Abstract
The protein Vezf1 plays multiple roles important for embryonic development. In Vezf1−/− mouse embryonic stem (mES) cells, our earlier data showed widespread changes in gene-expression profiles, including decreased expression of the full-length active isoform of Dnmt3b methyltransferase and concomitant genome-wide reduction in DNA methylation. Here we show that in HeLaS3 cells there is a strong genome-wide correlation between Vezf1 binding and peaks of elongating Ser2-P RNA polymerase (Pol) ll, reflecting Vezf1-dependent slowing of elongation. In WT mES cells, the elongating form of RNA pol II accumulates near Vezf1 binding sites within the dnmt3b gene and at several other Vezf1 sites, and this accumulation is significantly reduced at these sites in Vezf1−/− mES cells. Depending upon genomic location, Vezf1-mediated Pol II pausing can have different regulatory roles in transcription and splicing. We find examples of genes in which Vezf1 binding sites are located near cassette exons, and in which loss of Vezf1 leads to a change in the relative abundance of alternatively spliced messages. We further show that Vezf1 interacts with Mrg15/Mrgbp, a protein that recognizes H3K36 trimethylation, consistent with the role of histone modifications at alternatively spliced sites.
Transcriptional regulatory factors that recognize long strings of poly dG (“G-strings”) have been identified in a variety of organisms. Early interest in these proteins was related both to the unusual stability and conformational properties of the homopolymer duplexes that are their targets, and to their association with potential regulatory sites in the promoters of the chicken adult β-globin promoter and the sea urchin gene LpS1, which bind the factors β-globin protein 1 (BGP1) and sea urchin G-string binding factor 1, respectively (1, 2). Recently, we have begun to reinvestigate the role of BGP1 as a regulatory factor because of our identification of BGP1 binding sites (3) within the compound insulator element at the 5′ end of the chicken βA-globin locus, where they play an essential role (4).
Previous studies of the expression patterns of the BGP1 mouse homolog, Vezf1, have implicated it in vascular system development (5) and the human homolog DB1 has been identified as a coactivator of human IL-3 expression (6). Our studies have shown that loss of the zinc-finger protein Vezf1 causes genome-wide loss of DNA methylation in mouse ES (mES) cells. This result was found to be because of reduced expression of the full-length isoform of DNA methyltransferase Dnmt3b1, whereas only a mild decrease was seen in the expression of the shorter isoform Dnmt3b6. We also identified two Vezf1 binding sites, one at the 3′ end of the Dnmt3b gene in an intron just downstream of the alternatively spliced exons 22 and 23, and the second site in the 3′ UTR of the gene (7). We suggested that bound Vezf1 might affect the relative abundance of the two splice variants by slowing the elongation rate of RNA polymerase (Pol) II.
During the transcription process, Pol II goes through the phases of initiation, elongation, and termination, all of which are subject to regulation. The transition of Pol II from the initiation to the elongation phase is accompanied by sequential changes in the state of phosphorylation of the Pol II C-terminal domain at the serine (Ser) 5 and 2 residues. Phosphorylated Ser5 marks the polymerase in the early phases of transcription, whereas Ser2 phosphorylation prevails throughout the gene body and especially toward the 3′ end. Postinitiation promoter proximal pausing of Pol II, which involves stalling of Pol II at the 5′ end of the transcription unit, has been observed in Drosophila and mES cells (8–13). The Ser2-phosphorylated, fully elongation-competent form of Pol II is then engaged by RNA processing and termination factors as it transcribes processively toward the poly-adenylation site.
Elongating Pol II also is subject to pausing at downstream sites within transcribed genes. Although it seems plausible that DNA-bound protein factors could interfere with the advance of polymerase and modulate pausing, available evidence suggests that most factors do not have such an effect (14). Only the Myc-associated zinc-finger protein (MAZ) element has been well-characterized as a site that can interfere with Pol II elongation. This element can bind the MAZ protein, but may also function at least in part in the absence of the protein (15). MAZ pause sites were discovered in the sequences between the closely spaced human genes C2 and Factor B, as well as between g11-C4 and the intron of mouse IgM-D. These pause sites were shown to promote efficient transcriptional termination in stably transformed cell lines, thus preventing transcriptional interference (16).
There is abundant evidence for functional coupling between transcription and precursor mRNA (pre-mRNA) processing, including splicing and alternative splicing (17–21). Both processes are regulated by cis-regulatory elements on the pre-mRNA that bind to the cognate trans-acting factors. It has also been shown that change in rate of Pol II elongation can affect inclusion of alternative exons by providing different windows of opportunity in the use of weak and strong splice sites (22–26). This kinetic model for coupling of alternative splicing and transcription was suggested by experiments in which introduction into a transgene of a MAZ4 element resulted in an altered splicing choice in the transcript (14, 27). It is also known that transcription factors that associate with Pol II at the promoters and alter the elongation rate can influence alternative splicing (24, 26). There has, however, been no evidence for Pol II pausing at naturally occurring transcription-factor binding sites (including MAZ4) within genes that could, as a result, affect alternative splicing.
Here we describe a genome-wide ChIP-Seq analysis of Vezf1 binding sites in HeLa S3 cells, and in parallel, a similar survey of Ser2P-Pol II distribution. We find that more than half of Vezf1 binding sites are associated with peaks of elongating Pol II. SiRNA knockdown of Vezf1 in HeLa S3 cells results in the decreased accumulation of Pol II at tested Vezf1 binding sites. Thus, there is a strong correlation between Vezf1 binding and Ser2P-Pol II pausing throughout the genome. By comparing mouse WT and Vezf1−/− ES cells, we also identify several sites in mouse ES cells at which Pol II accumulates in a Vezf1-dependent manner. We show additional instances where alternative splicing patterns are changed in Vezf1−/− cells compared with WT cells. These results suggest that Vezf1 plays an important role in modulating transcription elongation, which in some cases affects the outcome of splicing events.
Results
Paused Elongating Pol II at Genomic Vezf1 Binding Sites.
Our earlier observations of the effects of Vezf1 on gene expression (7) led us to speculate that DNA-bound Vezf1 may alter the rate of chain elongation by RNA Pol II. To test this hypothesis, we investigated genome-wide the relationship between Vezf1 binding and relative abundance of elongating Pol II by ChIP-Seq. Flag-tagged Vezf1 was expressed in HeLa S3 cells (SI Appendix, Fig. S1); cross-linked chromatin was sonicated and immunoprecipitated either with anti-Flag M2 agarose or anti-Pol II (ser2 phosphorylated) antibody. Isolated DNA from both samples was subjected to high-throughput sequencing using the Illumina sequencing system.
To quantify the distribution of Vezf1, we defined discrete binding sites for each ChIP-Seq (Materials and Methods and SI Appendix, Table S1). The coverage profile of Vezf1 binding showed that the majority (60%) of bound sites are localized to coding regions of which 54% are within 1 kb of transcription start sites (TSS) (SI Appendix, Fig. S2A). In addition, 65% of promoters with CpG islands had Vezf1 binding sites, compared with only 7% of promoters without islands (SI Appendix, Fig. S2B). Vezf1 binding sites are known to have intrinsic preferences for poly(C) tracts in the genome (3). Although the Vezf1 peaks in these data are too heterogeneous for a reasonable motif search, assessing their correlation with repeats showed that Vezf1 peaks were enriched (90- to 50-fold) for many GC-rich simple repeats (SI Appendix, Table S2).
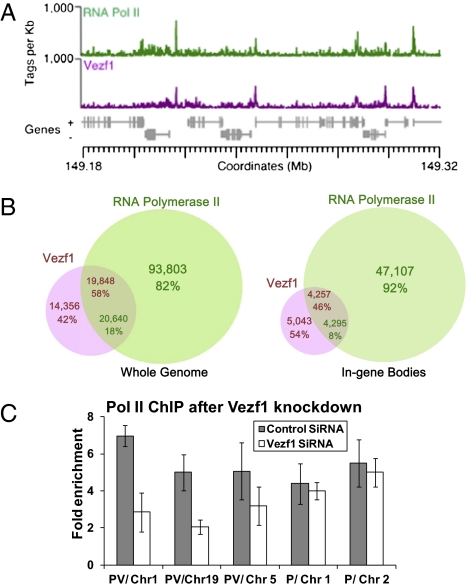
Next, we compared the genome-wide tag distributions for Vezf1, Ser2P-Pol II, and input DNA. We defined discrete enrichment sites for Pol II binding by methods similar to those for Vezf1 (Fig. 1A). The global distributions of Vezf1 and Ser2-Pol II were strikingly similar (Fig. 1A). By comparing the coverage profiles of these bound sites, we showed that the Vezf1 binding profile was very highly correlated to that of Ser2-Pol II (mean chromosomal Spearman's R = 0.573 ± 0.041, expectation = 0.003 ± 0.022, bootstrapped P < 10−5). Overall, 58% of Vezf1 binding sites overlapped with those of Ser2- Pol II-enriched sites (Fig. 1B). We also showed that the average within-gene coverage was very highly correlated (Spearman's R = 0.80, bootstrapped P < 10−5) (SI Appendix, Fig. S3). For an elongating Pol II, accumulation of tags at any position within a gene indicates pausing (i.e., a longer residence time, during elongation). Analysis of the genomic distribution of Vezf1/Ser2-Pol II co-occupied sites showed that 57% are located at 5′ ends of genes (TSS ± 1 kb). To evaluate the significant co-occurrence of the Vezf1/Ser2-Pol II sites within gene bodies but not at the TSS (n = 9,300), we compared the in-gene Ser2-Pol II paused sites (Materials and Methods) to Vezf1 binding sites and found that 46% of intragenic Vezf1 sites overlapped such sites (Fig. 1B), far more than the expected 2.6%.
Fig. 1.
ChIP-Seq of Flag-Vezf1 and Ser2 Phos Pol II in HeLa S3 cells. (A) Vezf1 and Ser2-Pol II ChIP-Seq coverage are highly correlated. The ChIP-Seq tag coverage for each protein is shown across a representative region on chromosome 1. To generate coverage, each tag was extended 150 nt 3′ to the tag start and coverage is calculated in 100-nt nonoverlapping bins. Tag normalized control background (input DNA) has been subtracted and only windows with positive tag counts are shown. (B) Vezf1 binds frequently at Ser2-Pol II binding sites. (Left) whole genome; (Right) in-gene bodies. In-gene binding sites were defined as binding sites that were located in coding regions but that were >500 bp from a TSS. The numbers of binding sites for each sample differ in the intersect because binding site overlaps are not 1:1. (C) Effects of siRNA mediated Vezf1 knockdown in HeLa S3 cells. HeLa S3 cells were treated with siRNA to knock down the expression of endogenous Vezf1. Cross-linked chromatin was immunoprecipitated using anti Ser2Phos Pol II antibody followed by qPCR using Taqman probes. In the bar diagram, PV indicates where Pol II peaks overlap with Vezf1 binding sites and P indicates control sites with only Pol II peaks. The significance of the Pol II decrease is indicated by the t test-derived P values, which are 0.001, 0.05, and 0.1 for Chr1, -19, and -5, respectively.
To investigate whether Pol II accumulation at Vezf1 sites was modulated by Vezf1 binding in HeLa S3 cells, we performed siRNA knock down of Vezf1 (SI Appendix, Fig. S1) and examined changes in Pol II accumulation by ChIP-quantitative PCR (qPCR) at five randomly picked Pol II binding sites, three with Vezf1 binding sites and two without (SI Appendix, Fig. S4A). As shown in Fig. 1C, there was a decrease in Pol II binding at all three Vezf1 binding sites, whereas there was no change at control sites. There was no change in the binding of Pol II in the region upstream of the Vezf1 binding sites, thus confirming the Vezf1-mediated pausing of Pol II at the tested sites (SI Appendix, Fig. S4B).
There is a curious predominance of Vezf1/Pol II at the 5′ ends of genes, which is likely to be associated with a distinct role for Pol II pausing at such sites. We checked for alternative transcription events at these promoters. Intriguingly, Pol II/Vezf1 peaks were more frequently associated with alternative promoters (62%; defined in University of California at Santa Cruz AltEvents table) than with those that were not (45%; P = 10−105, one-sided binomial test), suggesting a possible role for Vezf1-mediated polymerase pausing in facilitating such events. We investigated the possible role of the co-occupancy of Pol II/Vezf1 in promoter choice. Vezf1 was knocked down in HeLaS3 cells and the relative change in the occupancy by Ser2P-Pol II at the canonical promoter, and the alternate promoter of four randomly selected genes was measured. Although we noticed a decrease in the level of Ser2P-Pol II at the promoters of all of the tested genes, we did not see any change in the Ser2P-Pol II level at the alternate promoters (SI Appendix, Fig S5A). Thus, although Vezf1 induces pausing at promoter proximal sites, its specific role in transcriptional regulation at these sites remains to be determined.
Another distinct role for Vezf1 is associated with its activity within the chicken β-globin insulator, where it is essential for barrier insulator function (see Discussion). The β-globin insulator contains, in addition to Vezf1, a binding site for the CTCF insulator protein (28). We find a very significant overlap between Vezf1 sites (non-TSS) and CTCF sites, with 20% of Vezf1 sites present within 1 kb of a CTCF binding site (SI Appendix, Fig. S5B). This finding supports the genome-wide role of Vezf1 in insulator barrier function.
Role of Pol II Pausing at Vezf1 Binding Sites.
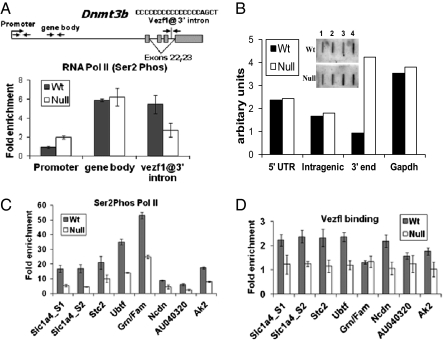
A strong Vezf1 binding site is present at a 3′ intron of the dnmt3b gene; we investigated the possible role of paused Pol II at this Vezf1 binding site. Using an antibody specific to Ser2P-Pol II, we performed ChIP experiments on cross-linked chromatin from WT and Vezf1−/− ES cells (Fig. 2A). The data show a greater accumulation of Ser2P-Pol II at Vezf1 binding sites in WT ES cells than in Vezf1−/− ES cells. The co-occupancy of Vezf1 and Pol II at Vezf1 binding sites was confirmed by a sequential ChIP analysis for the two proteins (SI Appendix, Fig. S6A).
Fig. 2.
ChIP followed by qPCR using Taqman probes. (A) Illustration of the Dnmt3b gene shows the position of probes (arrows). Boxes represent exons; alternatively spliced exons 22 and 23 are indicated. (A, C, and D) Cross-linked chromatin from WT and Vezf1−/− (null) ES cells was immunoprecipitated with anti-Ser2 phosphorylated Pol II antibody (ab5095) or polyclonal anti-Vezf1 antibody. For C and D, the positions of primer/probes for qPCR are marked by black arrows in SI Appendix, Fig. S4. (B) Nuclear run on assay using nuclei purified from WT and Vezf1−/− (null) ES cells. The probes to detect the 5′ UTR (lane 1), intragenic region (lane 2) and 3′ (lane 3) end transcripts of the dnmt3b gene were slot-blotted and hybridized with total RNA purified after the run-on reaction. Gapdh is used as a control for signal intensity (lane 4). Bar diagram shows the quantification of the intensity of bands using a PhosphoImager.
To investigate the effect of Pol II pausing on Dnmt3b transcription, we performed nuclear run-on assays using nuclei purified from WT and Vezf1−/− ES cells. Probes specific to the 5′ UTR, an intragenic region and the 3′ end of the dnmt3b gene, were used. As shown in Fig. 2B, there is no significant difference in the transcript level between WT and Vezf1−/− ES cells at the promoter region or the intragenic region. In contrast, at the 3′ end the band intensity detected by the probe 1-kb downstream of the Vezf1 binding site is higher in Vezf1−/− ES cells than in WT cells, suggesting that Vezf1 binding affects transcription elongation by inducing pausing of Pol II near Vezf1 binding sites. We also confirmed the pausing of RNA Pol II in vitro at Vezf1 binding sites, taking advantage of an S1 nuclease protection assay developed earlier to show RNA Pol II pausing at MAZ binding sites (15) (SI Appendix, Fig. S6B).
To examine further the relationship between Vezf1 and Pol II pausing, we carried out a genome-wide ChIP-Seq survey of Ser2P-Pol II binding in WT and Vezf1−/− ES cells. We had found that direct immunoprecipitation by antibodies to Vezf1, although satisfactory for ChIP-qPCR studies at individual sites, did not give sufficiently robust signals for a genome-wide study. We therefore chose individual potential Vezf1 binding sites (poly-GC rich), where peaks of enrichment of Pol II were higher in WT cells compared with the Vezf1−/− ES cells (SI Appendix, Fig. S7A). The qPCR analysis at these sites showed approximately twofold higher enrichment of Pol II in the WT compared with the null cells (Fig. 2C). To test if this change correlated to loss of Vezf1 binding at these sites, we performed ChIP using polyclonal anti-Vezf1 antibody. As shown in Fig. 2D, seven of eight sites at which Ser2P-Pol II was depleted in the null cells were adjacent to sites occupied by Vezf1 in WT ES cells. To address the biological importance of Pol II pausing in the above-studied genes, we measured gene expression by qRT-PCR analysis; Vezf1−/− cells showed decreased expression of four of these six associated genes (SI Appendix, Fig. S7B). The above data are consistent with the genome-wide correlation between Vezf1 binding and pausing of elongating Pol II, and suggests a regulatory role in gene expression.
Pol II Pausing and Regulation of Alternative Splicing.
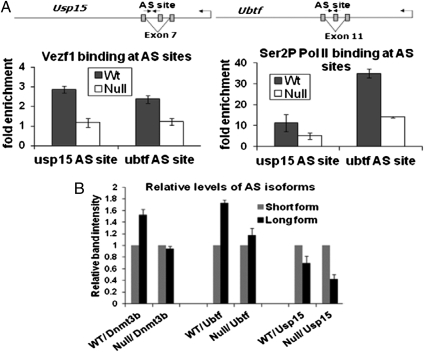
Regulation of alternative splicing takes place through several mechanisms (29). One of these mechanisms involves reducing the elongation rate of Pol II downstream of weak exons, thus giving enough time for inclusion of these exons in the spliced transcript. We had observed earlier (7) that the absence of Vezf1 results in alteration in the relative abundance of dnmt3b splice variants, and that Vezf1−/− ES cells expressing ectopic Vezf1 show partial rescue of Dnmt3b expression and of DNA methylation. This behavior strongly suggests a role of Vezf1 mediated Pol II pausing in regulating alternative splicing of Dnmt3b. Although Vezf1 should directly affect expression at many sites genome-wide, its effect on Dnmt3b, and therefore on DNA methylation, assigns it an equally important role in a complex circuit regulating global gene expression. To investigate whether modulation of elongation by Vezf1 had an effect on alternative splicing at genes other than dnmt3b (7), we tested two genes, usp15 and ubtf, which have two alternatively spliced forms coexpressed in mES cells (SI Appendix, Fig. S8). ChIP analysis of each gene reveals both Vezf1 binding and accumulation of Ser2P-Pol II at the sites of alternative splicing (Fig. 3A). By semiquantitative PCR analysis, we determine that as with Dnmt3b, there is a decrease in the expression of the long isoforms of usp15 and ubtf in Vezf1−/− ES cells, thus changing the relative ratio of the two isoforms (Fig. 3B) and supporting this role of Vezf1-mediated Pol II pausing at alternative splicing sites.
Fig. 3.
Role of Vezf1 mediated Pol II pausing in alternative splicing. (A) Illustration of Usp15 and Ubtf genes showing the cassette exons. Arrows indicate the position of the primers around the Vezf1 binding sites used for qPCR. Cross-linked chromatin from WT and Vezf1−/− ES cells were immunoprecipitated with anti-Vezf1 antibody (rabbit polyclonal) or anti-Ser2Phos Pol II antibody. AS, alternative splicing. (B) One-step RT-PCR was performed with total RNA from WT and Vezf1−/− cells. The primers flanking the alternatively spliced exons were used to amplify isoforms in the same reaction, which then were separated by PAGE (SI Appendix, Fig. S8). The relative density of bands on the gel was measured. The data represents the average calculated from two independent experiments. The abundances of long and short isoforms are represented as relative intensities.
Given these results, we wanted to determine, from the genome-wide studies in HeLaS3, the frequency of Vezf1/Ser2P-Pol II cobinding sites downstream of cassette exons. We excluded cassette exons that were within 1 kb of the 5′ end of a gene to prevent bias arising from broad peaks near the transcription start site. Eleven percent of intragenic cooccupied sites were present within 5 kb downstream of cassette exons and about 3% of cassette exons contained a co-occupied binding site in the region 5 kb downstream (SI Appendix, Fig. S9). These data suggest that Vezf1-mediated Pol II pausing may represent a significant regulatory mechanism for alternative splicing.
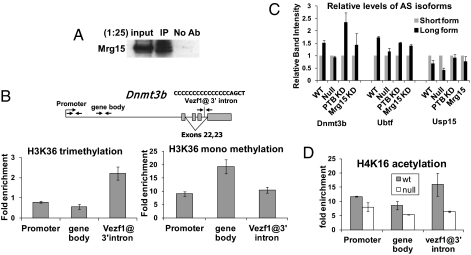
We showed above that Pol II accumulation at Vezf1 binding sites decreased in mouse Vezf1−/− ES cells. To understand the mechanism by which Vezf1 affects transcription elongation, we searched for factors that might be recruited by Vezf1. Coimmunoprecipitation of V5-tagged Vezf1 from ES nuclear extracts followed by mass spectrometry revealed several interacting partners (SI Appendix, Fig. S10). Among other chromatin-associated factors, we identified both Mrg15, which recognizes H3K36 trimethylation (30), and Mrg binding protein (Mrgbp). We confirmed the interaction of Vezf1 with Mrg15 by Western blot, following its coimmunoprecipitation from nuclear extracts with anti-Vezf1 antibody (Fig. 4A). We measured H3K36 tri- and monomethylation at sites in the Dnmt3b gene, including the 3′ Vezf1 site. In agreement with published data (31), we see the highest accumulations of H3K36 trimethylation in the alternatively spliced region (Fig. 4B). Differential inclusion of exons is regulated in some cases by polypyrimidine tract binding protein (Ptb), and previous data (31) showed a subset of Ptb-dependent alternative splicing events was mediated by Mrg15. We tested the effect of siRNA mediated knockdown of Ptb and Mrg15 on alternative splicing of dnmt3b, usp15, and ubtf transcripts. Fig. 4C and SI Appendix, Fig. S11 show that in dnmt3b and usp15, Ptb knockdown causes a substantial increase in the ratio of long form to short form transcript, the opposite of the effect of Vezf1 knockout. Alternative splicing of ubtf transcript did not show a clear Ptb dependence. Knockdown of Mrg15, however, resulted in smaller changes in the ratios of alternatively spliced forms and without a clear bias, indicating a less direct role in the above splicing events.
Fig. 4.
Role of Vezf1 interacting factors and histone modifications. (A) Vezf1 interacting proteins were coimmunoprecipitated from ES cell nuclear extract using anti-Vezf1 anibody followed by detection on Western blot using antibody against Mrg15. (B and D) ChIP-qPCR. Cross-linked chromatin from indicated mES cells was immunoprecipitated with antibody against (B) H3K36 tri- and monomethylation or (D) against H4K16 acetylation. (C) The splicing isoforms, L (long) and S (short) of dnmt3b, ubtf, and usp15 were amplified by RT-PCR and separated by PAGE. The density of specific bands from the gel was quantified as in Fig. 3B using the PhospoImager Image Quant TL.
Mrg15 is known to be a component of both the Tip60 histone acetyltransferase (HAT) and histone deacetylases complexes (32–34), and Mrgbp is a subunit specific to the Tip60 HAT complex (35), which has an established role in transcriptional elongation (ref. 36 and references therein). This complex catalyzes acetylation of sites on histones H3 and H4 (37). In agreement with a role of Vezf1 in recruiting these modifications through one or both of these proteins, we observed a higher accumulation of H4K16 acetylation in the region of alternative splicing of dnmt3b, which is decreased in Vezf1−/− cells (Fig. 4D). A decrease in H4K16 acetylation was also seen in usp15 and ubtf alternative splice sites in Vezf1−/− cells (SI Appendix, Fig. S11C). H4K16 acetylation is associated with lower levels of chromatin compaction (38). Our data are consistent with a model in which binding of the Mrg15/Mrgbp complex, recruited to chromatin around Vezf1 binding sites, is stabilized by high levels of H3K36 trimethylation. This process, in turn, could recruit higher levels of H4K16 acetylation, facilitating the release of paused Pol II and completion of the transcription cycle. Thus, at the sites of alternative splicing, Vezf1 could play a dual role in stimulating Pol II pausing and recruiting chromatin modifications that facilitate its release from the paused state.
Discussion
We found in an earlier study (7) that absence of Vezf1 leads to loss of DNA methylation at many sites in the genome, and that this could be attributed to a decrease in levels of the RNA splice variant that codes for the active form of the methyltransferase Dnmt3b. In cells expressing Vezf1, the protein is bound at two downstream sites, one within a 3′ intron. This finding led us to suggest that at this intronic site Vezf1 modulates the movement of elongating RNA pol II along the gene.
We have shown here, through a genome-wide analysis, that the majority of sites occupied by the transcription factor Vezf1 in HeLa S3 cells are associated genome-wide with peaks of elongating Pol II (Fig. 1A). We point out that these changes are observed against a background of elongating Pol II, which is distributed over expressed genes. It is possible that Vezf1 and other sites enriched in poly G/C DNA could have an intrinsic ability, independent of Vezf1 binding, to partially slow elongation. Although such effects have been reported for T7 bacteriophage RNA polymerase (39), and could perhaps affect transcription rates by Pol II, our data make clear that binding of Vezf1 regulates accumulation of Ser2-Pol II.
The Vezf1/Ser2P-Pol II binding sites associated with genes may be divided into two classes: those around the TSS and those within gene bodies. Our initial interest in the regulatory effects of Vezf1 on Dnmt3b expression led us to focus on downstream sites within genes, and to an examination of the role of Vezf1 in modulating splicing choices, particularly at Vezf1/Pol II sites where Vezf1 and elongating pol II are bound near alternatively spliced exons. There is accumulating evidence that splicing and alternative splicing occur cotranscriptionally (refs. 19, 20, and 40, and references therein). The first evidence that Vezf1 mediates splicing choice came from our earlier study of the Dnmt3b gene (7), in which we observed a change in the relative abundance of two splice variants in mES cells lacking Vezf1 (SI Appendix, Fig. S8C). Here we have shown (Fig. 3B) that similar effects can be observed at other genes where Vezf1 and elongating Pol II are bound near alternatively spliced exons. This finding suggests that Vezf1 has that function genome-wide. We have observed a very high enrichment of Vezf1 binding sites in GC-rich simple repeats. Earlier studies have emphasized the role of simple repeats in alternative splicing through the activity of specific RNA binding proteins (41). It is plausible that these sequences have a synergistic effect on exon inclusion through both RNA- and DNA-mediated mechanisms.
Several recent studies have highlighted the important role of promoter-associated transcription factors (24, 26) and chromatin structure in alternative splicing (40, 41). We have shown here that Mrg15, which recognizes H3K36 trimethylation (30), and Mrgbp, copurify with Vezf1 (Fig. 4A). Mrg15 and Mrgbp are components of the Tip60 HAT complex, which by acetylating H4K16 at these Pol II pausing sites could facilitate Pol II release and prevent the premature termination of transcription.
The properties of Vezf1 may allow it to play more than one role within the nucleus. At other intragenic sites, Vezf1 may regulate transcription by modulating transcriptional elongation, or equally possible, the cofactors with which it is associated may lead to site-specific changes in histone modifications. Our original interest in Vezf1 function arose because of its activity as a boundary element within the chicken β-globin insulator. Our observation that there are many sites in the genome occupied by Vezf1 that are adjacent to CTCF-occupied sites strongly suggests that Vezf1 plays a role in insulator function genome-wide. Deletion of any one of three Vezf1 binding sites within the insulator destroys its ability to protect against heterochromatic silencing (3). We have shown that the heterochromatic region upstream of the chicken β-globin insulator is maintained by dicer-dependent mechanisms involving low levels of transcription from the region (42). It may be that at the insulator and similar sites, bound Vezf1, by blocking RNA Pol II, prevents the extension into active chromatin of low-level transcription from adjacent heterochromatic regions, and that this protects against silencing.
Materials and Methods
Comparison of Ser2-Pol II Coverage in WT and Vezf1−/− mES Cells.
The genome-wide coverage of Ser2-Pol II was calculated in the WT and Vezf1−/− genetic backgrounds. Genomic loci with higher Ser2-Pol II binding in WT were identified by comparing the number of tags in WT and Vezf1−/− for each 50-nt window. A binomial test was used to assess significance and a Bonferroni-corrected P value threshold of 10−3 used. Adjacent sites ± 50 bp were merged and several such sites were chosen for qPCR analysis to assess Vezf1 binding.
Binding Site Identification from ChIP-Seq Data.
Statistically overrepresented sites were identified by sliding a 50-nt nonoverlapping window across the genome. A binomial test was used at each site to assess if the tag count was significantly enriched with respect to the control (input). Windows with no tags in the control were not considered. Adjacent enriched windows were merged and the maximum P value assigned. We assessed the false-discovery rate (FDR) for this method by switching the control and treatment data and identifying enriched regions in the control. We selected a P value threshold such that the FDR for each experiment was less than 10%. Thus, a log10 (P value) of 1.5 was used as a cutoff, yielding FDRs of 7.0% for the Vezf1 sample and 2.9% for Ser2-Pol II. To assess the correlation between replicates and between Vezf1 and Ser2-Pol II binding for each chromosome (SI Appendix, Fig. S12), a vector was generated from the coverage in enriched regions for each replicate dataset for each protein. The Spearman rank correlation was then calculated between the coverage vectors for each chromosome for each dataset. Loci with zero coverage in the first sample were discarded for each comparison. To assess the expected random correlation, we permuted the strengths of the first sample in each comparison.
On a gene-by-gene basis, Ser2-Pol II binding sites that had a magnitude greater than the mean in-gene coverage + 1 SD were identified and termed “in-gene” paused sites. This normalization helps to distinguish paused Ser2-Pol II from the background transcription level and is particularly useful for distinguishing Ser2-Pol II accumulations in highly expressed genes. The expected overlap between Vezf1 in-gene sites and paused Ser2-Pol II sites was calculated by calculating the Vezf1 site overlap with randomly distributed Ser2-Pol II sites. For this analysis, the Ser2-Pol II sites were randomly distributed in transcribed, nongap regions of the genome, at least 500 nt from a TSS. Ten thousand iterations were performed.
Supplementary Material
Acknowledgments
We thank the members of our laboratory for their advice and help; Dr. Nick Proudfoot for providing pMLPIII-MAZ constructs for in-vitro transcription assays; and Dr. Ann Dean for providing antibodies. This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. GSE34785).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1121538109/-/DCSupplemental.
References
- 1.Clark SP, Lewis CD, Felsenfeld G. Properties of BGP1, a poly(dG)-binding protein from chicken erythrocytes. Nucleic Acids Res. 1990;18:5119–5126. doi: 10.1093/nar/18.17.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hapgood J, Patterton D. Purification of an oligo(dG).oligo(dC)-binding sea urchin nuclear protein, suGF1: A family of G-string factors involved in gene regulation during development. Mol Cell Biol. 1994;14:1402–1409. doi: 10.1128/mcb.14.2.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dickson J, et al. VEZF1 elements mediate protection from DNA methylation. PLoS Genet. 2010;6:e1000804. doi: 10.1371/journal.pgen.1000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Recillas-Targa F, et al. Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc Natl Acad Sci USA. 2002;99:6883–6888. doi: 10.1073/pnas.102179399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiong JW, Leahy A, Lee HH, Stuhlmann H. Vezf1: A Zn finger transcription factor restricted to endothelial cells and their precursors. Dev Biol. 1999;206:123–141. doi: 10.1006/dbio.1998.9144. [DOI] [PubMed] [Google Scholar]
- 6.Koyano-Nakagawa N, Nishida J, Baldwin D, Arai K, Yokota T. Molecular cloning of a novel human cDNA encoding a zinc finger protein that binds to the interleukin-3 promoter. Mol Cell Biol. 1994;14:5099–5107. doi: 10.1128/mcb.14.8.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gowher H, Stuhlmann H, Felsenfeld G. Vezf1 regulates genomic DNA methylation through its effects on expression of DNA methyltransferase Dnmt3b. Genes Dev. 2008;22:2075–2084. doi: 10.1101/gad.1658408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lis JT. Imaging Drosophila gene activation and polymerase pausing in vivo. Nature. 2007;450:198–202. doi: 10.1038/nature06324. [DOI] [PubMed] [Google Scholar]
- 11.O'Brien T, Lis JT. RNA polymerase II pauses at the 5′ end of the transcriptionally induced Drosophila hsp70 gene. Mol Cell Biol. 1991;11:5285–5290. doi: 10.1128/mcb.11.10.5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilchrist DA, et al. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts GC, Gooding C, Mak HY, Proudfoot NJ, Smith CW. Co-transcriptional commitment to alternative splice site selection. Nucleic Acids Res. 1998;26:5568–5572. doi: 10.1093/nar/26.24.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yonaha M, Proudfoot NJ. Specific transcriptional pausing activates polyadenylation in a coupled in vitro system. Mol Cell. 1999;3:593–600. doi: 10.1016/s1097-2765(00)80352-4. [DOI] [PubMed] [Google Scholar]
- 16.Ashfield R, et al. MAZ-dependent termination between closely spaced human complement genes. EMBO J. 1994;13:5656–5667. doi: 10.1002/j.1460-2075.1994.tb06904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alexander RD, Innocente SA, Barrass JD, Beggs JD. Splicing-dependent RNA polymerase pausing in yeast. Mol Cell. 2010;40:582–593. doi: 10.1016/j.molcel.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrillo Oesterreich F, Preibisch S, Neugebauer KM. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol Cell. 2010;40:571–581. doi: 10.1016/j.molcel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 19.Kornblihtt AR. Coupling transcription and alternative splicing. Adv Exp Med Biol. 2007;623:175–189. doi: 10.1007/978-0-387-77374-2_11. [DOI] [PubMed] [Google Scholar]
- 20.Perales R, Bentley D. “Cotranscriptionality”: The transcription elongation complex as a nexus for nuclear transactions. Mol Cell. 2009;36:178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Auboeuf D, Batsché E, Dutertre M, Muchardt C, O'Malley BW. Coregulators: Transducing signal from transcription to alternative splicing. Trends Endocrinol Metab. 2007;18:122–129. doi: 10.1016/j.tem.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 22.de la Mata M, et al. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 23.Howe KJ, Kane CM, Ares M., Jr Perturbation of transcription elongation influences the fidelity of internal exon inclusion in Saccharomyces cerevisiae. RNA. 2003;9:993–1006. doi: 10.1261/rna.5390803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadener S, et al. Antagonistic effects of T-Ag and VP16 reveal a role for RNA pol II elongation on alternative splicing. EMBO J. 2001;20:5759–5768. doi: 10.1093/emboj/20.20.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kadener S, Fededa JP, Rosbash M, Kornblihtt AR. Regulation of alternative splicing by a transcriptional enhancer through RNA pol II elongation. Proc Natl Acad Sci USA. 2002;99:8185–8190. doi: 10.1073/pnas.122246099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nogues G, Kadener S, Cramer P, Bentley D, Kornblihtt AR. Transcriptional activators differ in their abilities to control alternative splicing. J Biol Chem. 2002;277:43110–43114. doi: 10.1074/jbc.M208418200. [DOI] [PubMed] [Google Scholar]
- 27.Robson-Dixon ND, Garcia-Blanco MA. MAZ elements alter transcription elongation and silencing of the fibroblast growth factor receptor 2 exon IIIb. J Biol Chem. 2004;279:29075–29084. doi: 10.1074/jbc.M312747200. [DOI] [PubMed] [Google Scholar]
- 28.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 29.Chen M, Manley JL. Mechanisms of alternative splicing regulation: Insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang P, et al. Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucleic Acids Res. 2006;34:6621–6628. doi: 10.1093/nar/gkl989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luco RF, et al. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yochum GS, Ayer DE. Role for the mortality factors MORF4, MRGX, and MRG15 in transcriptional repression via associations with Pf1, mSin3A, and Transducin-Like Enhancer of Split. Mol Cell Biol. 2002;22:7868–7876. doi: 10.1128/MCB.22.22.7868-7876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai Y, et al. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J Biol Chem. 2003;278:42733–42736. doi: 10.1074/jbc.C300389200. [DOI] [PubMed] [Google Scholar]
- 34.Doyon Y, Côté J. The highly conserved and multifunctional NuA4 HAT complex. Curr Opin Genet Dev. 2004;14:147–154. doi: 10.1016/j.gde.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 35.Hayakawa T, et al. RBP2 is an MRG15 complex component and down-regulates intragenic histone H3 lysine 4 methylation. Genes Cells. 2007;12:811–826. doi: 10.1111/j.1365-2443.2007.01089.x. [DOI] [PubMed] [Google Scholar]
- 36.Wang Z, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ikura T, et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 38.Shogren-Knaak M, et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 39.Belotserkovskii BP, et al. Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proc Natl Acad Sci USA. 2010;107:12816–12821. doi: 10.1073/pnas.1007580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 41.Hui J, Bindereif A. Alternative pre-mRNA splicing in the human system: Unexpected role of repetitive sequences as regulatory elements. Biol Chem. 2005;386:1265–1271. doi: 10.1515/BC.2005.143. [DOI] [PubMed] [Google Scholar]
- 42.Giles KE, Ghirlando R, Felsenfeld G. Maintenance of a constitutive heterochromatin domain in vertebrates by a Dicer-dependent mechanism. Nat Cell Biol. 2010;12(1):94–99; sup pp 91–96. doi: 10.1038/ncb2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.