Abstract
Hybrid rice has greatly contributed to the global increase of rice productivity. A major component that facilitated the development of hybrids was a mutant showing photoperiod-sensitive male sterility (PSMS) with its fertility regulated by day length. Transcriptome studies have shown that large portions of the eukaryotic genomic sequences are transcribed to long noncoding RNAs (lncRNAs). However, the potential roles for only a few lncRNAs have been brought to light at present. Thus, great efforts have to be invested to understand the biological functions of lncRNAs. Here we show that a lncRNA of 1,236 bases in length, referred to as long-day–specific male-fertility–associated RNA (LDMAR), regulates PSMS in rice. We found that sufficient amount of the LDMAR transcript is required for normal pollen development of plants grown under long-day conditions. A spontaneous mutation causing a single nucleotide polymorphism (SNP) between the wild-type and mutant altered the secondary structure of LDMAR. This change brought about increased methylation in the putative promoter region of LDMAR, which reduced the transcription of LDMAR specifically under long-day conditions, resulting in premature programmed cell death (PCD) in developing anthers, thus causing PSMS. Thus, a lncRNA could directly exert a major effect on a trait like a structure gene, and a SNP could alter the function of a lncRNA similar to amino acid substitution in structural genes. Molecular elucidating of PSMS has important implications for understanding molecular mechanisms of photoperiod regulation of many biological processes and also for developing male sterile germplasms for hybrid crop breeding.
Keywords: epigenetics, Oryza sativa
Photoperiod regulates a large array of processes underlying growth and development of plants (1). It has been known for centuries that flowering of plants is regulated by day length (2). In recent years, many genes for photoperiod-controlled flowering have been identified in a number of plants, leading to characterization of pathways in flowering regulation (3–9). However, photoperiod regulation of other developmental processes has been studied in far less detail than flowering. It has been reported in several plants that male fertility is also sensitive to photoperiod; a critical day length is required for normal pollen development (10–15). However, nothing is known about the molecular control of the photoperiod regulation of male fertility, despite the tremendous progress in recent years in genetic and molecular understanding of male reproductive development in many plant species (16–22).
Hybrid rice has greatly contributed to the global increase of rice productivity (23, 24). A spontaneous mutant exhibiting photoperiod-sensitive male sterility (PSMS) was found in 1973 in a japonica rice (Oryza sativa ssp. japonica) variety Nongken 58 (referred to as 58N) in Hubei Province, China (12). Intensive studies of the mutant (58S) established that its pollen fertility is regulated by day length; it is completely sterile when grown under long-day conditions, whereas pollen fertility varies when it is grown under short-day conditions (SI Appendix, Fig. S1); and the critical stage for photoperiod induction comes between secondary branch differentiation and microsporogenesis during panicle development, suggesting that photoperiod regulation of PSMS is independent of photoperiod flowering (25–27). For hybrid rice breeding, PSMS rice can be used to propagate itself under short days, and to produce hybrid seeds by interplanting it with normal fertile lines under long-day conditions. Moreover, PSMS rice has a broad spectrum of restoration; all of the normal rice varieties tested can restore the fertility of F1 hybrids (28). Two-line hybrids developed using this PSMS germplasm have occupied millions of hectares of rice fields mostly in China for more than a decade (29). PSMS germplasms have also been found and explored in several other crops (15, 30–32).
Transcriptome studies have shown that large portions of the eukaryotic genomic sequences are transcribed to long noncoding RNAs (lncRNAs). However, the potential roles for only a few lncRNAs have been brought to light at present (33–39).
Here we show that a locus regulating PSMS in rice encodes a lncRNA. A SNP between 58N and 58S in this lncRNA caused epigenetic modifications, which reduced expression of this lncRNA, resulting in male sterility under long-day conditions.
Results
Mapping of the pms3 Locus to a 12-kb Region.
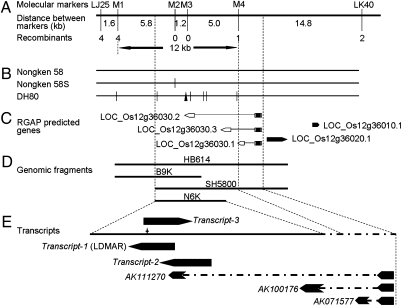
It was determined previously that pms3 on chromosome 12 was the original mutation that changed 58N to 58S (40–44). Using an F2 population of 7,000 plants from a cross between 58S and DH80, a doubled haploid line derived from a cross between 58S and 1514 (a normal japonica variety), which were examined for fertility under natural field long-day conditions in Wuhan, the pms3 locus was delimited to a 28.4-kb DNA fragment between two molecular markers LJ25 and LK40 (45) (Fig. 1A). We sequenced this 28.4-kb region and found seven polymorphic sites between 58N and DH80 and a SNP resulting from substituting guanine (G) in 58N with cytosine (C) in 58S (Fig. 1B). We developed four additional molecular markers, which resolved pms3 to a 12-kb region between M1 and M4 (Fig. 1A).
Fig. 1.
Mapping and cloning of the pms3. (A) Genetic and physical maps of pms3. (B) Polymorphisms identified by comparative sequencing of a 28.4-kb mapping region in 58S, DH80, and 58N. The triangle represents an insertion, and vertical thin bars indicate single nucleotide substitutions relative to 58N. (C) Predicted genes in the pms3 region according to the Rice Genome Annotation Project database (http://rice.plantbiology.msu.edu/). (D) Genomic fragments used for making constructs for transformation. (E) Transcripts in both strands detected in the target region. The black arrows represent exons and the dotted lines represent introns. The quadrangular legend represents the single SNP between 58N and 58S.
We assayed the SNP genotypes of 52 rice accessions representing diverse germplasms, including both wild and cultivated species. All these accessions had the G nucleotide at this position, indicating that this C substitution was probably unique in 58S (SI Appendix, Table S1).
Although there were three predicted genes in the 28.4-kb region (LOC_12g36010, LOC_12g36020, and LOC_12g36030), the targeted 12-kb region contained only part of LOC_12g36030 (Fig. 1C). LOC_12g36030 encodes three alternatively spliced transcripts supported by full-length cDNAs, AK071577, AK100176, and AK111270 (http://cdna01.dna.affrc.go.jp/cDNA/), each of which had two exons. The ORF was in the first exon that was shared among all three transcripts, whereas the second exon was specific to each transcript.
Complexity of Transcripts in the pms3 Region.
We prepared four constructs containing overlapping genomic DNA fragments covering the 12-kb region by digesting clone 117H5 from a BAC library of 58N constructed in our laboratory (Fig. 1D) and subcloning into the vector pCAMBIA1301 (SI Appendix, Fig. S2A). We transformed the constructs into 58S. We also overexpressed the three alternative transcripts (AK071577, AK100176, and AK111270) in both 58N and 58S (SI Appendix, Fig. S2A). At least 15 independent T0 plants were produced for each of the construct/genotype combinations. Surprisingly, no statistically significant difference was detected in spikelet fertility between the transgene-positive and -negative plants in the T0 and/or T1 progenies under long-day conditions in any of the construct/genotype combinations.
Because the targeted 12-kb region contained only part of LOC_12g36030 (Fig. 1 D and E), we speculated that there might be other transcripts from this region that regulate PSMS. To check this possibility, we designed 24 pairs of oligonucleotide primers to cover the genomic region ∼3 kb in length surrounding the SNP site (SI Appendix, Fig. S3A). Three unique transcripts were identified from the two strands (Fig. 1E and SI Appendix, Fig. S3 B–D), which we named Transcript-1 to -3. RT-PCR showed that none of the three transcripts had an intron. Transcript-1 and -2 were transcribed from the sense strand relative to AK111270. Transcript-1 was 1,236 bases in length, and its 5′ end had an 110-base overlap with the 3′ end of AK111270. The 1,268-base sequence of Transcript-2 was located upstream of Transcript-1 and overlapped with the second exon of AK111270 in its entire length. Transcript-3, 1,088 bases in length, was transcribed from the complementary strand, overlapping with Transcript-1 by 688 bases. Thus, this region had a complex transcription pattern producing multiple partial overlapping transcripts. This complexity may also explain why transformation with the genomic fragments could not recover male fertility.
Expression Patterns of the Transcripts.
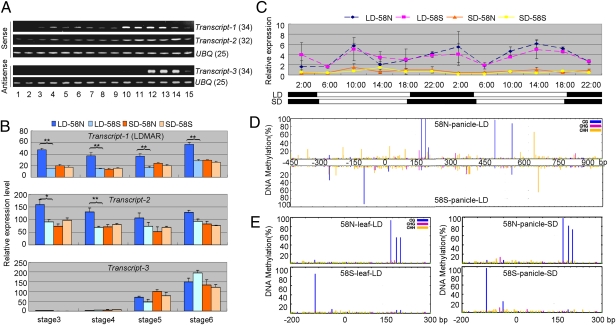
Using a strand-specific RT-PCR method, we assayed the expression profiles of these transcripts in 58N and 58S using RNA from 15 tissues spanning the entire life cycle of the rice plant (Fig. 2A). Transcript-1 was detected in almost all of the tissues, but clearly higher in young panicles. Transcript-2 was constitutively expressed and slightly higher in young panicles. Transcript-3 was specifically expressed in young panicles from pollen mother cell-formation stage to pollen mitosis stage and hardly detectable in vegetative organs. The levels of all three transcripts were extremely low in all of the tissues assayed.
Fig. 2.
Expression analysis of transcripts identified in the pms3 region. (A) Expression profile of transcripts in 58N (tissue description in SI Appendix, Table S2). Numbers after gene names represent the number of cycles in RT-PCR amplification. (B) Comparative expression analysis of the transcripts between 58N and 58S under both long-day (LD) and short-day (SD) conditions in young panicles at four developing stages: stage3, secondary branch primordium differentiation; stage4, pistil/stamen primordium differentiation; stage5, pollen mother cell formation; and stage6, pollen meiosis. All three transcripts were quantified using a common check. Significant differences detected between LD-58N and LD-58S are indicated by **P < 0.01 or *P < 0.05, respectively. Data are means ± SEM (n = 4). (C) Diurnal expression patterns of Transcript-1 under LD and SD conditions in 58N and 58S. The black bars indicate the dark period, and the white bars indicate the light period. The numbers on the Top of the bars indicate hours of the day. Data are means ± SEM (n = 4). (D) DNA methylation levels (%) of the 1.5-kb genome region, including 450-bp putative promoter and the entire LDMAR region (1,050 bp), from young panicles of 58N and 58S grown under LD conditions. (E) DNA methylation levels (%) of the 500-bp genome region (−200 bp to +300 bp of LDMAR) from leaves of plants grown under LD conditions and young panicles of plants under SD conditions, for 58N and 58S.
RNA in situ hybridization showed that, similar to RT-PCR results, Transcript-1 was detectable in leaf and sheath and produced intensive signals in microspore mother cells and vacuolated pollen cells (SI Appendix, Fig. S4 A–E). Transcript-2 was detectable in all tissues with signals more intense than Transcript-1 (SI Appendix, Fig. S4 F–J). The signal of Transcript-3 was only detectable in anthers at vacuolated pollen cell stage (SI Appendix, Fig. S4 K–O).
Comparative analysis in young panicles (Fig. 2B) showed that the expression level of Transcript-1 in 58N under long-day conditions was significantly higher than the other three treatments: 58N under short-day and 58S under both long-day and short-day conditions in all four developmental stages. The levels of Transcript-1 in 58S were nearly the same under long-day and short-day conditions. The expression of Transcript-2 in 58N under long days was also significantly higher than the other three treatments in the two early stages, but the difference diminished at later stages. The expression of Transcript-3 increased gradually with panicle development but there was no significant difference among the four treatments at any stage.
We analyzed the diurnal expression pattern of Transcript-1 in 58N and 58S in leaves at pistil/stamen primordium differentiation stage at 4-h intervals for a total of 48 h under both long-day and short-day conditions (Fig. 2C). Generally, the expression of Transcript-1 was much higher under long days than short days in both 58N and 58S. There seemed no identifiable rhythm expression pattern. Unlike in panicles, however, the abundance of this transcript in leaves was similar in 58N and 58S, under both long-day and short-day conditions.
Validation of Transcript-1 as the Candidate for pms3.
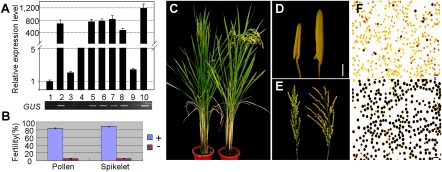
We took Transcript-1 as the best candidate and overexpressed it in 58S with the construct Actin:NT1 (osβ-actin1-58N-Transcript-1) containing the full-length sequence of Transcript-1 from 58N driven by the rice β-actin1 promoter (SI Appendix, Fig. S2 A and B), which produced 26 independent T0 plants. Transgene-positive T0 plants showed a significant increase in spikelet fertility under natural long days compared with the negative plants (Table 1), without obvious morphological difference (SI Appendix, Table S3). The results were confirmed by further examination of three T1 families randomly selected from T0 plants with single-copy transgene, which showed perfect cosegregation between the transgene and pollen fertility. All positive transgenic plants produced normal pollen grains and were fertile, whereas negative plants produced smaller anthers with collapsed pollen (Fig. 3). We thus named Tanscript-1 as pms3.
Table 1.
Fertility of plants overexpressing various constructs of Transcript-1 (LDMAR)
| Transgene | Genotype | No. of plants | Pollen fertility, % | Spikelet fertility, % |
| Actin:NT1 (T0) | Positive | 19 | 48.7 ± 6.72 | |
| Negative | 7 | 2.9 ± 1.00 | ||
| t | 6.75 | |||
| P | 0.0000 | |||
| Actin:ST1 (T0) | Positive | 14 | 46.6 ± 5.51 | |
| Negative | 5 | 3.2 ± 1.82 | ||
| t | 7.48 | |||
| P | 0.0000 | |||
| Actin:NT1 (T1) from three families | Positive | 58 | 83.7 ± 1.03 | 88.0 ± 1.00 |
| Negative | 26 | 5.1 ± 0.74 | 5.6 ± 0.83 | |
| t | 61.67 | 63.13 | ||
| P | 0.0000 | 0.0000 | ||
| Actin:ST1 (T1) from three families | Positive | 58 | 80.8 ± 1.08 | 79.3 ± 1.85 |
| Negative | 25 | 4.3 ± 0.99 | 4.7 ± 0.94 | |
| t | 52.16 | 35.96 | ||
| P | 0.0000 | 0.0000 | ||
| Actin:NT1-mORF (T0) | Positive | 14 | 43.6 ± 8.18 | |
| Negative | 8 | 2.9 ± 1.49 | ||
| t | 3.70 | |||
| P | 0.0007 | |||
| Actin:NT1-ORF (T0) | Positive | 17 | 7.4 ± 1.97 | |
| Negative | 1 | 1.3 | ||
| t | 0.72 | |||
| P | 0.2300 | |||
| Actin:NT1-△SNP (T0) | Positive | 15 | 7.6 ± 2.84 | |
| Negative | 2 | 0.4 ± 0.36 | ||
| t | 0.91 | |||
| P | 0.1891 | |||
| Actin:NT1-△110bp (T0) | Positive | 17 | 40.7 ± 7.47 | |
| Negative | 4 | 4.8 ± 4.65 | ||
| t | 2.27 | |||
| P | 0.0177 |
All data are given as mean ± SEM. P values were obtained by t tests between positive and negative plants.
Fig. 3.
The effect of overexpressing Transcript-1 (LDMAR) on male fertility. (A) Expression level of Transcript-1 (Top) in transgene-positive and -negative plants (Bottom) in a T1 family. Data are means ± SEM (n = 3). (B) Fertility scores of transgene-positive and -negative plants in three T1 families. There are 58 positive and 26 negative segregants from three T1 families. Data are means ± SEM. (C) Whole plants of transgene-negative (Left) and -positive (Right) segregants from T1 family. (D) Anther of transgene-negative (Left) and -positive (Right) plants. (Scale bars, 0.5 mm.) (E) Panicles of transgene-negative (Left, mostly sterile spikelets) and -positive (Right, mostly fertile) plants. (F) Pollen grains of transgene-negative (Top, sterile) and -positive (Bottom, fertile) plants. (Scale bars, 50 μm.)
Pms3 Likely Encodes a lncRNA.
We used the ExPASy translate tool to predict the translated protein of Transcript-1, which suggested a short ORF of 73 amino acids, with no homology to any known proteins. To investigate whether this transcript indeed encodes a protein, we changed the predicted start codon ATG to CTG, and overexpressed it (Actin:NT1-mORF, SI Appendix, Fig. S2 A and B) in 58S, resulting in 22 independent T0 plants. Similar to the plants overexpressing Transcript-1, pollen fertility of the transgene-positive plants under natural long days was much higher than that of the negative plants (Table 1). We also prepared a construct overexpressing only the predicted ORF (Actin:NT1-ORF, SI Appendix, Fig. S2 A and B); transformation of this construct in 58S produced 18 independent T0 plants. No fertility recovery was observed in any of the 17 positive plants (Table 1). These results suggested that Transcript-1 may not encode a protein but a lncRNA, which is referred to as long-day–specific male-fertility–associated RNA (LDMAR).
Functional Importance of the Sequences Surrounding the SNP Site.
The SNP between 58N and 58S was located at the 789-bp site of LDMAR (SI Appendix, Fig. S2B). To investigate how the SNP influenced the function of LDMAR, we prepared three constructs, Actin:ST1 containing the full-length sequence of LDMAR from 58S, Actin:NT1-△110bp in which the 110 bp at the 5′ end was deleted, and Actin:NT1-△SNP in which a 52-bp region containing the SNP site was deleted (SI Appendix, Fig. S2B). These constructs were individually transformed into 58S. Examination of transgenic plants (Table 1) showed that Actin:ST1 could recover the spikelet fertility under long days just like LDMAR from 58N, suggesting that sufficient quantity of LDMAR could recover the fertility in 58S despite the presence of the SNP. The 110-bp deletion in the 5′ end did not affect the function of the transcript in fertility recovery. By contrast, the construct deleting the SNP-containing 52-bp sequence failed to recover fertility, indicating that the 52-bp sequence was in a region critical for the function of LDMAR for fertility recovery.
Different Methylation Patterns in the pms3 Region Between 58N and 58S.
To understand why LDMAR was expressed differently in 58N and 58S, we performed bisulfite sequencing of DNA methylation in the 1.5-kb region of this transcript, including the 450-bp putative promoter and the 1,050-bp transcribed region, using DNA from young panicles at pistil/stamen primordium differentiation stage of plants grown under natural long-day conditions. There was a contrasting pattern of DNA methylation between the putative promoter and transcribed region of LDMAR (Fig. 2D). In the promoter region, the methylation level of CG contexts in 58S was much higher than that in 58N, whereas in the transcribed region the methylation level of CG contexts in 58N was much higher than in 58S. Similar difference in methylation patterns between 58S and 58N was observed using leaf tissues of plants grown under long days and panicle tissues of plants under short days (Fig. 2E). Thus, the DNA methylation status was not tissue or day-length specific.
Recent reports showed that DNA methylation is always correlated with small RNA (46–48). We visualized the RNA secondary structure of LDMAR to find a possible stem-loop structure, which is usually involved in genes encoding microRNAs (http://www.tbi.univie.ac.at/RNA/man/RNAfold.html). A 145-base fragment including the SNP site was predicted to form a stem loop, in which the SNP site was located in the stem region and the G→C change of the SNP influenced the stem-loop structure (SI Appendix, Figs. S2B and S5A). Several small RNAs were also predicted, which may be encoded by this region (SI Appendix, Fig. S5B). We tried to verify these predicted small RNAs. Three small RNAs could be detected by a stem-loop qRT-PCR method but not by small RNA gel blot analysis (SI Appendix, Fig. S5C). The average Ct values of the three small RNAs studied were larger than 32 (SI Appendix, Fig. S5D), whereas Ct values of most known miRNAs are less than 25 (49, 50), which might explain why small RNA gel blot analysis could not detect the signal. These results suggest a likely involvement of small RNAs in regulation of PSMS.
Abnormal Degeneration of Tapetum in Anthers of 58S.
We examined cross-sections of anthers from 58N and 58S plants grown under natural long-day conditions. No obvious differences were detected between 58N and 58S before sporogenous cell stage. Visible abnormalities in anther development of 58S were observed soon after the formation of sporogenous cells and thereafter, featured by condensed, deformed, and vacuolated cytoplasm of tapetal cells and microspore mother cells and deformed, crumpled, and empty microspores, as well as delayed degeneration of tapetal cells (SI Appendix, Fig. S6 A–L). A terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay of anthers at different developmental stages showed that programmed cell death (PCD) occurred earlier in 58S than in 58N (SI Appendix, Fig. S7 A–H).
Discussion
On the basis of the results obtained in this study, we proposed a model to explain the genetic and molecular basis of the PSMS, centered on the role of LDMAR (SI Appendix, Fig. S8). A sufficient amount of LDMAR is required for male fertility under long-day conditions. The spontaneous G→C mutation causing a SNP between 58N and 58S altered the RNA secondary structure of this region. This alteration eventually brought about heritable increased methylation in the promoter region of LDMAR, thus reducing the level of transcription specifically under long-day conditions. Such a reduced level of LDMAR results in premature PCD in anther development under long days, thus causing male sterility. However, several questions remain to be answered for complete understanding of PSMS including: (i) the biochemical mechanism by which LDMAR regulates pollen development; (ii) the reason why LDMAR is needed in large quantity in long-days but not in short-days; and (iii) the relation of pms3 to pms1, the other locus for PSMS identified in genetic analysis of the same cross (42–44, 51), in regulating PSMS.
Elucidation of the genetic and molecular bases of PSMS, although still far from completion at this stage, provides important implications for the studies of other photoperiod-regulated processes. It indicates that each of the processes may have a distinct genetic and molecular control at least at the lower level of regulatory hierarchy of photoperiodism. Therefore, the processes have to be investigated individually to understand them. We also speculate that at higher levels, such as the perception of day length, time keeping, and photoreceptors, among other things, these processes should be subjected to the same regulatory machineries as photoperiod flowering. Thus, the example provided by this study may expand the norm of studies on photoperiodism in plants.
Analyses of transcriptomes have revealed surprising complexity of lncRNAs that are often overlapping with, or interspersed in multiple coding and noncoding transcripts (52, 53). They are classified into different types on the basis of their genomic proximity to protein-coding genes, including overlapping, cis-antisense, bidirectional, and intronic ncRNAs (54). In this study, we detected three transcripts (LDMAR, Transcript-2 and Transcript-3) that were transcribed from the same locus; all of them are likely lncRNAs as none of them was predicted to encode a protein. LDMAR and Transcript-2 were transcribed from the same strand and overlapped with each other. Transcript-3 was transcribed from the complementary strand of LDMAR, thus they are mutually cis-antisene. We also showed that all of them had tissue-specific expression patterns, implying that they are involved in specific development processes. This was in agreement with recent findings that the expression of many lncRNAs is restricted to particular developmental contexts (54). Besides associating LDMAR with PSMS, the lncRNAs identified here provide additional opportunities to reveal the roles of lncRNAs in plants.
Two-line hybrids enabled by PSMS rice represent a significant advance in rice breeding. The mechanistic understanding of PSMS and the DNA polymorphism identified in the pms3 region may greatly facilitate the development of new PSMS lines in rice breeding programs.
Methods
Plant Materials and Field Growth Condition.
58N and 1514 are japonica rice varieties. 58S is a spontaneous mutant from 58N and DH80 is a doubled haploid line derived from a cross between 58S and 1514. The field growth conditions of rice plants are described in SI Appendix, SI Methods.
Generation of Constructs and Transformation.
Gnomic fragments were cut from the BAC clone 117H5 of 58N and cloned into the vector pCAMBIA1301 (SI Appendix, Fig. S2A). To generate overexpression constructs of LDMAR (SI Appendix, Fig. S2A), the full-length transcribed sequence was amplified from the panicles of 58N and 58S by RT-PCR using primers OSNPF and OSNPR (SI Appendix, Table S4). The PCR products were digested with BamHI and PstI and inserted into pCAMBIA1301A under the control of the rice β-actin1 promoter (55). Other overexpression constructs were generated by similar strategy. All of these constructs were individually transformed into the Agrobacterium strain EHA105. Transformation was conducted according to a published protocol (56).
RNA Quantification.
The diurnal expression patterns of the transcripts were carried out using strand-specific RT-PCR, in which 1 μg total RNA was reverse transcribed using SuperScript III reverse transcriptase (Invitrogen) with the mixture of gene-specific primers (for sense: SNP-F5, P-R1, and UBQR1; for antisense: pms3-F9 and UBQR1). We used primers SNP-F5 and SNP-R1 for amplifying the transcript of Transcript-1, P-R1 and pms3-F8 for Transcript-2, and pms3-F9 and pms3-R9 for Transcript-3 (SI Appendix, Table S4). Quantitative RT-PCR was performed on an Applied Biosystems 7500 Real-Time PCR system according to the manufacturer's instructions. The measurements were obtained using the relative quantification method (57). The entire life cycle expression profile of these transcripts was carried out using semiquantitative RT-PCR. PCR products were separated in 2% (wt/vol) agarose gels and normalized against the rice ubiquitin (UBQ, LOC_Os03g13170) gene.
Bisulfite Sequencing.
Genomic DNA from young panicles and leaves was bisulfite treated using the EpiTect Bisulfite kit (Qiagen). The candidate regions were amplified using bisulfite primers (SI Appendix, Table S4) designed using MethPrimer software and cloned into the pGEM-T vector. Each fragment was sequenced in at least 30 clones. Sequence analysis was performed by the Web-based tool, Kismeth.
More methods and data collection are provided in SI Appendix, SI Methods. DNA sequences of the two alleles of LDMAR have been deposited in GenBank under accession nos. JQ317784 (Nongken 58) and JQ317785 (Nongken 58S).
Supplementary Material
Acknowledgments
We thank D. Zhou for his suggestions and critically reading the manuscript. This work was supported by 863 Project Grant 2012AA100103 and National Natural Science Foundation of China Grant 30921091.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences of the two alleles of LDMAR reported in this paper have been deposited in the GenBank database [accession nos. JQ317784 (Nongken 58) and JQ317785 (Nongken 58S)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1121374109/-/DCSupplemental.
References
- 1.Thomas B, Vince-Prue D. Photoperiodism in Plants. 2nd Ed. San Diego: Academic; 1997. [Google Scholar]
- 2.Henfrey A. The Vegetation of Europe, Its Conditions and Causes. London: J Van Voorst; 1852. [Google Scholar]
- 3.Albani MC, Coupland G. Comparative analysis of flowering in annual and perennial plants. Curr Top Dev Biol. 2010;91:323–348. doi: 10.1016/S0070-2153(10)91011-9. [DOI] [PubMed] [Google Scholar]
- 4.Tsuji H, Taoka K, Shimamoto K. Regulation of flowering in rice: Two florigen genes, a complex gene network, and natural variation. Curr Opin Plant Biol. 2011;14:45–52. doi: 10.1016/j.pbi.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 5.Jung C, Müller AE. Flowering time control and applications in plant breeding. Trends Plant Sci. 2009;14:563–573. doi: 10.1016/j.tplants.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Kobayashi Y, Weigel D. Move on up, it's time for change—mobile signals controlling photoperiod-dependent flowering. Genes Dev. 2007;21:2371–2384. doi: 10.1101/gad.1589007. [DOI] [PubMed] [Google Scholar]
- 7.Böhlenius H, et al. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science. 2006;312:1040–1043. doi: 10.1126/science.1126038. [DOI] [PubMed] [Google Scholar]
- 8.Xue W, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40:761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- 9.Wu C, et al. RID1, encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice. Proc Natl Acad Sci USA. 2008;105:12915–12920. doi: 10.1073/pnas.0806019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batch JJ, Morgan DG. Male sterility induced in barley by photoperiod. Nature. 1974;250:165. [Google Scholar]
- 11.Fisher JE. The transformation of stamens to ovaries and of ovaries to inflorescences in T. aestivum L. under short day treatment. Bot Gaz. 1972;133:78–85. [Google Scholar]
- 12.Shi M. The discovery and preliminary studies of the photoperiod-sensitive recessive male sterile rice (Oryza sativa L. subsp. japonica) Sci Agric Sin. 1985;2:44–48. [Google Scholar]
- 13.Moss GI, Heslop-Harson J. Photoperiod and pollen sterility in maize. Ann Bot (Lond) 1968;32:833–846. [Google Scholar]
- 14.Knox RB, Heslop-Harrison J. Control of pollen fertility through the agency of the light regime in the grass Dichanthium aristatum. Phyton. 1966;11:3–4. [Google Scholar]
- 15.Virmani SS, Ilyas MA. Environment-sensitive genic male sterility (EGMS) in crops. Adv Agron. 2001;72:139–195. [Google Scholar]
- 16.Ma H. Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu Rev Plant Biol. 2005;56:393–434. doi: 10.1146/annurev.arplant.55.031903.141717. [DOI] [PubMed] [Google Scholar]
- 17.Wilson ZA, Zhang DB. From Arabidopsis to rice: Pathways in pollen development. J Exp Bot. 2009;60:1479–1492. doi: 10.1093/jxb/erp095. [DOI] [PubMed] [Google Scholar]
- 18.Ge X, Chang F, Ma H. Signaling and transcriptional control of reproductive development in Arabidopsis. Curr Biol. 2010;20:R988–R997. doi: 10.1016/j.cub.2010.09.040. [DOI] [PubMed] [Google Scholar]
- 19.Ariizumi T, Toriyama K. Genetic regulation of sporopollenin synthesis and pollen exine development. Annu Rev Plant Biol. 2011;62:437–460. doi: 10.1146/annurev-arplant-042809-112312. [DOI] [PubMed] [Google Scholar]
- 20.Li N, et al. The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell. 2006;18:2999–3014. doi: 10.1105/tpc.106.044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, et al. Rice APOPTOSIS INHIBITOR5 coupled with two DEAD-box adenosine 5′-triphosphate-dependent RNA helicases regulates tapetum degeneration. Plant Cell. 2011;23:1416–1434. doi: 10.1105/tpc.110.082636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vizcay-Barrena G, Wilson ZA. Altered tapetal PCD and pollen wall development in the Arabidopsis ms1 mutant. J Exp Bot. 2006;57:2709–2717. doi: 10.1093/jxb/erl032. [DOI] [PubMed] [Google Scholar]
- 23.Yuan L. Hybrid rice technology for food security in the world. Crop Res. 2004;18:185–186. [Google Scholar]
- 24.Khush GS. Green revolution: The way forward. Nat Rev Genet. 2001;2:815–822. doi: 10.1038/35093585. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Z, Yuan S. The influence of photoperiod on pollenfertility change of Hubei photoperiod-sensitive genic male sterile rice. Chin J Rice Sci. 1987;1:137–143. [Google Scholar]
- 26.Yuan S, Zhang Z, Xu C. Studies on the critical stage of fertility change induced by light and its phase development in HPGMR. Acta Agron Sin. 1988;14:7–12. [Google Scholar]
- 27.Yuan S, et al. Two photoperiodic-reactions in photoperiod-sensitive genic male-sterile rice. Crop Sci. 1993;33:651–660. [Google Scholar]
- 28.Li Z. Advances in studies and applications of photoperiod sensitive genic male sterility (PGMS) in rice. In: Li Z, Jin Z, Lu X, Zhu Y, editors. Studies on the Photoperiod Sensitive Genic Male Sterility in Rice and Utilization in Breeding. Wuhan, China: Hubei Science and Technology; 1995. pp. 1–42. [Google Scholar]
- 29.Yang S, Cheng B, Shen W, Xia J. Progress of application and breeding on two-line hybrid rice in China. Hybrid Rice. 2009;24:5–9. [Google Scholar]
- 30.Smith MB, Horner HT, Palmer RG. Temperature and photoperiod effects on sterility in a cytoplasmicmale-sterile soybean. Crop Sci. 2001;41:702–704. [Google Scholar]
- 31.Yang G, Fu T, Yang X. Studies on the ecotypical male stetile line of Brassica napus L. I. inheritance of the ecotypical male sterile line. Acta Agron Sin. 1995;21:129–135. [Google Scholar]
- 32.Guo RX, Sun DF, Tan ZB, Rong DF, Li CD. Two recessive genes controlling thermophotoperiod-sensitive male sterility in wheat. Theor Appl Genet. 2006;112:1271–1276. doi: 10.1007/s00122-006-0228-z. [DOI] [PubMed] [Google Scholar]
- 33.Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143:390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pandey RR, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 36.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 38.Swiezewski S, Liu F, Magusin A, Dean C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature. 2009;462:799–802. doi: 10.1038/nature08618. [DOI] [PubMed] [Google Scholar]
- 39.Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- 40.Zhang D, et al. Chromosomal location of the photoperiod sensitive male genic sterile gene in Nongken 58S. J Huazhong Agric Univ. 1990;9:407–419. [Google Scholar]
- 41.Zhang Q, et al. Using bulked extremes and recessive class to map genes for photoperiod-sensitive genic male sterility in rice. Proc Natl Acad Sci USA. 1994;91:8675–8679. doi: 10.1073/pnas.91.18.8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mei M, Dai X, Xu C, Zhang Q. Mapping and genetic analysis of the genes for photoperiod-sensitive genic sterility in rice using the original mutant Nongken 58S. Crop Sci. 1999;39:1711–1715. [Google Scholar]
- 43.Mei M, et al. pms3 is the locus causing the original photoperiod-sensitive male sterility mutation of ‘Nongken 58S’. Sci China C Life Sci. 1999;42:316–322. doi: 10.1007/BF03183609. [DOI] [PubMed] [Google Scholar]
- 44.Li X, Lu Q, Wang F, Xu C, Zhang Q. Separation of the two-locus inheritance of photoperiod-sensitive genic male sterility in rice and precisely mapping the pms3 locus. Euphytica. 2001;119:343–348. [Google Scholar]
- 45.Lu Q, Li XH, Guo D, Xu CG, Zhang Q. Localization of pms3, a gene for photoperiod-sensitive genic male sterility, to a 28.4-kb DNA fragment. Mol Genet Genomics. 2005;273:507–511. doi: 10.1007/s00438-005-1155-4. [DOI] [PubMed] [Google Scholar]
- 46.Zhang H, Zhu J. RNA-directed DNA methylation. Curr Opin Plant Biol. 2011;14:142–147. doi: 10.1016/j.pbi.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu L, et al. DNA methylation mediated by a microRNA pathway. Mol Cell. 2010;38:465–475. doi: 10.1016/j.molcel.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 48.Matzke M, Kanno T, Huettel B, Daxinger L, Matzke AJ. Targets of RNA-directed DNA methylation. Curr Opin Plant Biol. 2007;10:512–519. doi: 10.1016/j.pbi.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 49.Chen C, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen J, Xie K, Xiong L. Global expression profiling of rice microRNAs by one-tube stem-loop reverse transcription quantitative PCR revealed important roles of microRNAs in abiotic stress responses. Mol Genet Genomics. 2010;284:477–488. doi: 10.1007/s00438-010-0581-0. [DOI] [PubMed] [Google Scholar]
- 51.Wang F, Mei M, Xu C, Zhang Q. pms1 is not the locus relevant to fertility difference between the photoperiod-sensitive male sterile rice Nongken 58S and normal rice Nongken58. Acta Bot Sin. 1997;39:922–925. [Google Scholar]
- 52.Carninci P, et al. FANTOM Consortium. RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 53.Kapranov P, et al. Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays. Genome Res. 2005;15:987–997. doi: 10.1101/gr.3455305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 55.Xiao B, Huang Y, Tang N, Xiong L. Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor Appl Genet. 2007;115:35–46. doi: 10.1007/s00122-007-0538-9. [DOI] [PubMed] [Google Scholar]
- 56.Lin YJ, Zhang Q. Optimising the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell Rep. 2005;23:540–547. doi: 10.1007/s00299-004-0843-6. [DOI] [PubMed] [Google Scholar]
- 57.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.