Abstract
Our objective was to test whether or not plastids and mitochondria, the two DNA-containing organelles, move between cells in plants. As our experimental approach, we grafted two different species of tobacco, Nicotiana tabacum and Nicotiana sylvestris. Grafting triggers formation of new cell-to-cell contacts, creating an opportunity to detect cell-to-cell organelle movement between the genetically distinct plants. We initiated tissue culture from sliced graft junctions and selected for clonal lines in which gentamycin resistance encoded in the N. tabacum nucleus was combined with spectinomycin resistance encoded in N. sylvestris plastids. Here, we present evidence for cell-to-cell movement of the entire 161-kb plastid genome in these plants, most likely in intact plastids. We also found that the related mitochondria were absent, suggesting independent movement of the two DNA-containing organelles. Acquisition of plastids from neighboring cells provides a mechanism by which cells may be repopulated with functioning organelles. Our finding supports the universality of intercellular organelle trafficking and may enable development of future biotechnological applications.
Keywords: plasmodesmata, horizontal gene transfer, plastid transformation, chloroplast biotechnology
Plant cells have three DNA-containing cellular compartments: the nucleus, plastids, and mitochondria. The plastid and mitochondrial genomes (ptDNA and mtDNA) in all plant species have been massively reduced relative to their prokaryotic ancestors through the evolutionary process of intracellular gene transfer. The 155-kb plastid and 430-kb mitochondrial genomes of Nicotiana tabacum encode only 112 (1) and 60 (2) genes, respectively. Experimental reconstruction of this evolutionary process in the laboratory revealed that plastid-to-nucleus gene transfer occurs at a surprisingly high frequency (3). The recent demonstration of the exchange of genetic material between cells in plant tissue grafts reconstructed the evolutionary process of intercellular gene transfer (4).
Intercellular movement of mitochondria in mammalian cells was found to be a basic biological process and involved in tissue repair (5). In coculture, donor cells extend cytoplasmic projections toward target cells and mitochondria stream from cell to cell (6–8). This transfer was shown to result in replacement of diseased mitochondrial genomes with mtDNA from the donor cells (5).
However, there is no report yet on the intercellular movement of DNA-containing organelles, plastids, and mitochondria, between plant cells. In contrast to animal cells, plant cells have a rigid cell wall. However, plant cells are connected by sophisticated intercellular channels (plasmodesmata), which actively and passively regulate cell-to-cell movement of nutrients, hormones, and information macromolecules, including transcription factors, phloem proteins, mRNA, and sRNAs (9, 10).
Our objective was to determine whether chloroplasts or mitochondria could move from cell to cell in plants. To test this hypothesis, we grafted two different species of tobacco with genetic markers in their plastids, mitochondria and nuclei. Grafting triggers formation of new cell-to-cell connections (11) that creates an opportunity for cell-to-cell movement of organelles. Here, we report evidence supporting the movement of plastids (ptDNA) between cells in graft tissue. However, the related (nonselected) mitochondria were absent in the same plants, suggesting independent transfer of plastids through the graft junction. We discuss acquisition of plastids from neighboring cells as a potential mechanism to repopulate cells with functional organelles and the possibilities of cell-to-cell movement of plastids for biotechnological applications.
Results
Experimental Design.
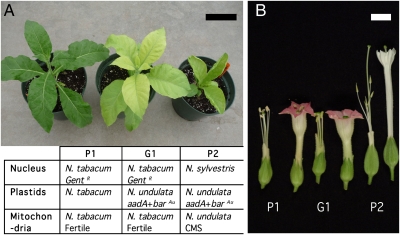
Because of the difficulty to directly observe rare intercellular organelle movement, we chose graft partners with distinct nuclear and organellar genomes to test for cell-to-cell transfer of plastids and mitochondria in graft junctions (Fig. 1). We grafted two species of tobacco, N. tabacum (partner P1) with a selectable transgenic nuclear gentamycin resistance gene and Nicotiana sylvestris (partner P2) with plastids carrying a selectable spectinomycin resistance (aadA) gene and the aurea young leaf color phenotype (barau gene). The N. sylvestris partner carried the plastids and mitochondria of a third species, N. undulata, providing a large number of organellar DNA markers. The P1 partner with the N. tabacum nucleus was fertile and the P2 partner with the N. sylvestris nucleus cytoplasmic male sterile (CMS) (Fig. 1B), a trait controlled by mitochondria (12). The grafted plants were grown in culture for ten days (Fig. 2A) and sections of the graft junctions were selected for the gentamycin and spectinomycin resistance traits carried by the P1 nucleus and in P2 plastids, respectively (Fig. 2B). Of 30 graft junctions, a total of 3 plastid graft transmission (PGT) events (G1, G3, and G4) were recovered. The plants regenerated from the graft junction displayed the leaf morphology, growth habit, and pink flowers associated with the selected N. tabacum nucleus but the aurea leaf color of the P2 partner, a plastid trait (Fig. 1 A and B).
Fig. 1.
Phenotypes of the graft partners and the G1 graft transfer plant. (A) Partner P1 is N. tabacum (2N = 48) with a nuclear gentamycin resistance transgene and wild-type N. tabacum plastids and mitochondria. Partner P2 has a wild-type N. sylvestris (2N = 24) nuclear genome, N. undulata plastids with aadA transgenes for spectinomycin selection and the aurea young leaf color phenotype (barau gene), and N. undulata mitochondria that confer cytoplasmic male sterility (CMS-92). Shown is also the G1 plant and its markers. (Black scale bar: 10 cm.) (B) Flower morphology of the P1 and P2 partners and G1 PGT plant. (White scale bar: 1 cm.)
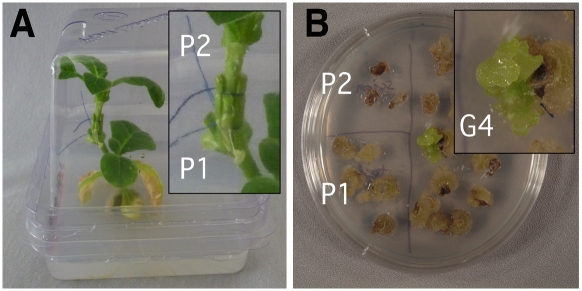
Fig. 2.
Identification of plastid graft transfer events. (A) Grafted plant. Note that the P2 scion shown here is green because the expression of the barau gene is restricted to fast-growing tissue and is sensitive to environmental conditions. (B) Selection in cultures of 1–2-mm graft sections for gentamycin and spectinomycin resistance. On the left are stem sections from above (P2) and below (P1) the graft and on the right from the graft region. Note a green, proliferating callus that yielded the G4 PGT plants.
No Exchange of Chromosomes in the PGT Plants.
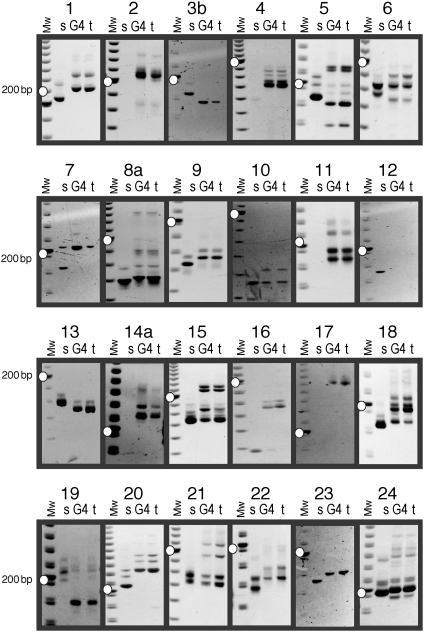
To investigate the contribution of nuclear genetic material to the PGT plants, we examined 24 simple sequence repeat (SSR) (or microsatellite) polymorphic DNA markers previously mapped to each of the N. tabacum chromosomes (13). These markers distinguished N. tabacum from N. sylvestris ecotype TW137 and indicated the presence of the chromosomes of the N. tabacum P1 partner that carried the selectable nuclear gene without contribution from the nonselected P2 N. sylvestris nucleus (Fig. 3). The presence of chromosomal markers from one partner excluded chimera formation as the source of double resistance of the G1, G3, and G4 PGT plants. However, we cannot exclude limited transfer of chromosome fragments that remained undetected in the study.
Fig. 3.
SSR markers confirm N. tabacum chromosomes in the G4 plant by testing each of the 24 chromosomes (numbered 1–24). Lanes are marked with s, G4, and t for the P2, G4, and P1 plants, respectively (see legend of Fig. 1). Some markers do not amplify the N. sylvestris template (13). White dots indicate the 200-bp fragment of the 20-bp molecular-mass ladder.
Mitochondria Remain Associated with the Selected Nucleus.
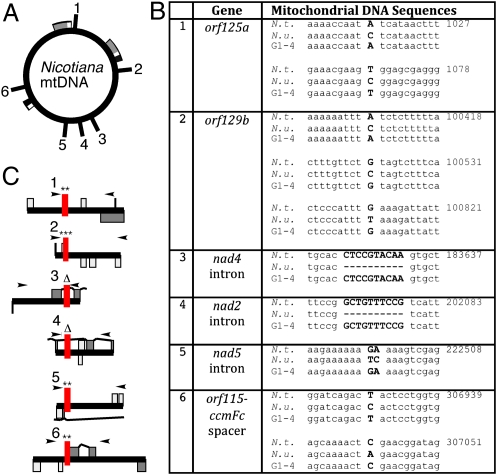
The graft partners carried distinct mitochondrial genomes determining the flower type (Fig. 1B). The P1 partner with the N. tabacum nucleus had normal anthers and produced fertile pollen, whereas the P2 partner with the N. sylvestris nucleus had stigmatoid anthers, a phenotype controlled by mitochondria. The G1, G3, and G4 PGT plants were male fertile and lacked the stigmatoid anthers of the CMS P2 partner. In line with the flower morphology, the CMS92 mtDNA markers were absent in the G1, G3, and G4 plants. To determine the source of the mitochondrial genome in the PGT plants, we identified six SNP and insertion/deletion markers that are suitable to distinguish the N. undulata CMS92 mtDNA (Fig. 4) from the fertile N. tabacum mtDNA (2). Sanger sequencing of PCR fragments indicated that the G1, G3, and G4 plants have the mitochondrial genome of the nuclear donor (Fig. 4). Thus, we did not find evidence for the transfer of mitochondrial DNA in the PGT plants. Given the tendency of mitochondria for fusion (14) and mtDNA for recombination (12, 15), should mtDNA be transferred, we would expect to find at least chimeric mtDNA. The absence of nonselected mitochondrial DNA suggests limited organelle transfer, rather than large-scale mixing of the two cytoplasms at the graft junction. Although we did not find evidence for the cotransfer of mtDNA with plastids in the lines tested, it is possible that mtDNA transfer could be detected with selection in a larger PGT plant population.
Fig. 4.
Identification of the source of mtDNA in the PGT plants. (A) Schematic representation of the tobacco mtDNA master circle with the position of polymorphic regions marked. Repeated regions are marked with boxes. (B) Mitochondrial DNA sequence polymorphisms. (C) Map position of polymorphic sites relative to the sequencing primers and gene features.
PGT Plants Contain the Entire Selected Plastid Genome.
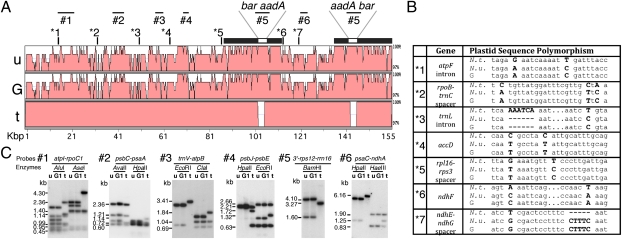
Dual selection for the nucleus- and plastid-encoded antibiotic resistances ensured that the PGT plants would carry both transgenes. The N. tabacum-specific SSR markers in the G1, G3, and G4 plants indicated the presence of the P1 chromosomes alone in the PGT plants. However, the presence of the plastid markers did not distinguish between a transformation-like process that involves incorporation of ptDNA fragments and intercellular movement of plastids implied by the transfer of complete plastid genomes, either of which is compatible with the earlier report (4). To determine how much of the P2 ptDNA is present in the G1, G3, and G4 plants, we first examined markers distant from the transgenes by probing total cellular DNA on blots. Southern probing of the six previously identified RFLP markers (Fig. 5C) and PCR analyses (Fig. 5B) suggested the presence of the entire plastid genome of the P2 partner and that the PGT plants carried a uniform population of P2 transplastomes. To exhaust the search for a contribution to the PGT plastid genomes from the nonselected P1 plastome, we performed next-generation sequencing of the plastid genomes of the P1 and P2 partners and the G1, G3, and G4 PGT plants. We report here that the sequence of the 160,743-nt transplastomes in the P2 partner and in three PGT plants are identical (GenBank accession no. JN563930). The P2 and PGT plastid genomes are larger than the 155,863-nt wild-type N. undulata plastid genome (GenBank accession no. JN563929) because the transplastomes also contain the spectinomycin resistance (aadA) and the aurea barau transgenes. We also sequenced the plastid genome in partner P1 that carries the wild-type N. tabacum ptDNA of cv. Petit Havana. We have found that the sequence of cv. Petit Havana ptDNA is identical to the cv. Bright Yellow sequence deposited in GenBank (GenBank Accession number Z00044). However, the N. undulata ptDNA differs from the N. tabacum cv. Petit Havana ptDNA by 805 SNPs, 52 insertions, and 61 deletions and the transgene cassettes. Differences between the plastid genomes are depicted on the mVISTA identity plots shown in Fig. 5A and Fig. S1: the 500-bp sliding window in Fig. 5A gives an overview; and the 100-bp sliding window in Fig. S1 provides more precise information about the location of SNPs and insertions/deletions. Importantly, we observed all of these polymorphic loci, with an average density of 200 bp/SNP (170 bp/polymorphism) in the plastid genome of the three graft transmission plants indicating the transfer of intact ptDNA from the P2 graft partner. We also tested transmission of the plastid-encoded spectinomycin resistance in reciprocal backcrosses with the G1 PGT plant. When the G1 plant was the mother and the wild type the father, each of the 208 seedlings was resistant, whereas when the G1 plant was the father and the wild type the mother, each of the 318 seedlings was spectinomycin sensitive. Thus, spectinomycin resistance exhibited uniform, maternal inheritance, as expected for a homoplastomic N. tabacum, a species with strict maternal plastid inheritance (16, 17).
Fig. 5.
Identification of the N. undulata plastids in the PGT plants. (A) Identity plots of the plastid genomes of the transplastomic P2 partner carrying N. undulata ptDNA (u) with the aadA and barau transgenes (GenBank accession no. JN563930); the G1, G3, and G4 (G) PGT plants; and the P1 partner with N. tabacum ptDNA (t) (GenBank accession no. Z00044) aligned with the mVISTA program using a 500-bp sliding window. Shown above the map are the positions of the DNA probes (#1 through #6) and DNA polymorphisms (*1 through *7). (B) Plastid DNA sequence polymorphisms. For map position, see Fig. 5A. (C) DNA gel blot to identify RFLP markers in ptDNA. For probes see Fig. 5A.
Discussion
Cell-to-Cell Migration of Plastids.
Here, we report cell-to-cell movement of entire plastid genomes. We considered two possible mechanisms for the transfer of genome-size ptDNA: the intercellular transport of extraorganellar (“naked”) DNA or the ptDNA traveling within an intact organelle. Selection for movement of ptDNA to the nucleus led to the discovery that incorporation of kilobase-size ptDNA fragments is frequent and that the source most probably is degraded organellar genomes (18–20). Movement of entire genomes may require more protection than the fragments. Better protection could be provided if the extraorganellar ptDNA would be encapsulated in membrane-bound vesicles that are shed from fragmented chloroplast stromules (21), although ptDNA is normally absent from stromules (22). Because of the need for capacity for translation, plastids cannot be created de novo from membranes and DNA (23). Thus, if “naked” ptDNA is transferred, an invading plastome would need to enter an existing plastid with transcription and translation machinery and displace the existing plastome by a transformation-like process to explain our observations. However, a transformation-like process would yield mosaic genomes if different genomes were present, because plastid genomes within an organelle undergo frequent recombination (24–26). The absence of chimeric genomes in the PGT plants makes it unlikely that naked DNA transfer is the mechanism of intercellular ptDNA transfer.
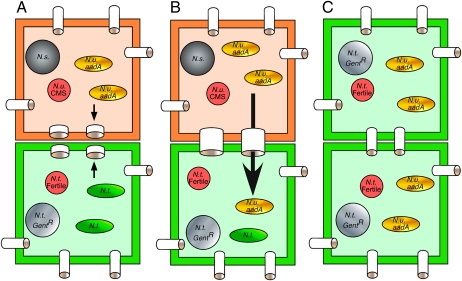
More likely, vehicles of cell-to-cell movement of entire plastid genomes could be the organelles themselves. The avenue for the movement of intact organelles could be damage to cell walls that allows for some mixing of cytoplasms in the graft junctions. A more likely mechanism would be the transfer of proplastids via newly formed connections between cells that are well documented at graft junctions (11). The size of proplastids, ∼1 μm, is well above the size exclusion limit of plasmodesmata normally defined by molecular mass. However, the size exclusion limit changes during development and depends on tissue type (9, 27). We speculate that the new openings, formed by thinning of opposing cell walls at the site of future plasmodesmata, permit intercellular movement of proplastids. Our preferred model of intercellular plastid transfer in graft junctions is shown in Fig. 6.
Fig. 6.
Model for cell-to-cell movement of plastids via initial cytoplasmic connection in graft junctions. (A) Cells at graft junction reconnect by plasmodesmata. Arrows point to sites where opposite parts of the contact walls are synchronously thinned (11). These are future sites of plasmodesmata. Proplastids (ovals), mitochondria (small circles), and nuclei (large circles) are identified in scion and rootstock. Ns, N. sylvestris; Nt, N. tabacum; Nu, N. undulata. (B) Proplastid is transferred via initial cytoplasmic connection. (C) Transferred spectinomycin-resistant plastid takes over on selective medium. Note that the cells derive from the bottom cell in Fig. 6B.
The Role of Cell-to-Cell Movement of Plastids.
The capacity of a plant cell to acquire organelles from a neighboring cell is a basic biological process. Acquisition of plastids from neighboring cells may be important because once the ribosomes are lost, translation cannot be restored, because some of the ribosomal proteins are encoded in the plastid genome and their translation is dependent on plastid ribosomes (23). Therefore, during certain stages of development, including dedifferentiation associated with forming new connections in grafted tissues (11), plants cells may allow intercellular transport of organelles. In this regard it is intriguing to note that the redox state of plastids regulates symplastic permeability and that ectopic expression of the proplastid-targeted GAT1 protein increased plasmodesmal size exclusion limit (28). The functional state of mitochondria also regulates the size exclusion limit of intercellular trafficking (29) and reprogramming of diseased mammalian cells was associated with acquisition of functional mitochondria (5, 7). The discovery of intercellular movement of plastids supports the universality of intercellular organelle trafficking and calls for testing the biological significance of this process in plants.
Horizontal Gene Transfer and Cell-to-Cell Movement of Organelles.
The formation of interspecific cytoplasmic connections and exchange of genetic material has also been reported between parasitic flowering plants and their hosts. All interspecific secondary plasmodesmata have been localized in thinned-wall areas at the contact between host and parasite, which corresponds to the observations on graft unions (11). Although horizontal gene transfer (HGT) in plant mitochondrial genomes is rampant when a parasitic flowering plant is involved as a donor or recipient, it very rarely occurs in plastids (30, 31). Cell-to-cell movement of plant mitochondria and the observed massive mitochondrial fusion (14) would provide an efficient mechanism for evolutionary gene transfer. In contrast, plastid fusion has been rarely observed under experimental conditions (25, 32, 33), explaining the scarcity of HGT. The cell-to-cell movement of entire plastids is restricted to closely related species, because plastid-nucleus incompatibility prevents incorporation of entire unmodified ptDNA in a distantly related host (34, 35). However, fragments of the incoming plastid genome may find their way into the nucleus and mitochondria of the host.
Applications in Plastid Genetics and Biotechnology.
Because in most species both plastids and mitochondria are maternally inherited, they cannot be separated by crossing. Thus far protoplast fusion has been the only option to obtain new combinations of plastids and mitochondria (12). The result is intercellular transfer of parental plastids, but formation of recombinant mitochondrial genomes. The protocol we report here enables combination of parental plastids and nonrecombinant mitochondria by PGT, a significant improvement over the protoplast-based process that yields recombinant mitochondria.
An additional application of PGT could be rapid introgression of transformed plastids into commercial cultivars. Plastid transformation is a powerful tool for biotechnological applications because the transgenes that are integrated into the plastid genome are expressed at high levels, can be clustered in operons, and are not subject to silencing (36, 37). Currently, the option is to transform the plastids in permissive cultivars then introduce them into commercial lines by repeated backcrossing using the commercial cultivar as a recurrent pollen parent. Based on the findings in this report, backcrossing can be replaced in the future by graft transfer of the transformed plastids, instantly yielding a substitution line with transgenic plastids combined with the valuable commercial nuclear genome.
Materials and Methods
Partner P1 (Nt-pHC19) has an allotetraploid N. tabacum cv. Petit Havana (2N = 48) nucleus with the aacC1 transgene for gentamycin resistance and wild-type N. tabacum plastid and mitochondrial genomes (38). Partner P2 (Ns-pCK2-6W2) has a wild-type diploid N. sylvestris TW137 (2N = 24) nuclear genome, N. undulata plastids with aadA transgenes for spectinomycin selection and the aurea young leaf color phenotype (barau gene), and N. undulata (CMS-92) mitochondria that confer cytoplasmic male sterility (39). For grafting, the plants were grown aseptically on a medium containing Murashige and Skoog salts and 3% sucrose (40). Grafting was carried out reciprocally in equal numbers. Plants were regenerated from the graft junctions on RMOP shoot regeneration media supplemented with 500 mg/L spectinomycin and 100 mg/L gentamycin (40). Southern probing for ptDNA polymorphisms was carried out using six previously identified polymorphic regions (16). Organellar DNA was amplified using total cellular DNA as a template (41) and appropriate PCR primers (Tables S1 and S2). Primer design for ptDNA was based on GenBank accession nos. Z00044 and JN563929; for mtDNA, primer design was based on GenBank accession no. BA000042. The P1, P2, and G1 plastid genomes were amplified in 34 PCRs using primers listed in Table S3. DNA sequence was determined on an Illumina Genome Analyzer II using 80-bp paired-end (500-bp insert) library. Total leaf DNA fragments of P1, P2, G1, G3, and G4 plants were also analyzed on a SOLiD 5500xl sequencer (Applied Biosystems) using 76-nt reads. Reference guided assembly of the ptDNA was essentially carried out as described elsewhere (42). Nuclear SSR markers (13) were amplified using primers listed in Table S4.
Note Added in Proof
Stegemann et al. report similar findings in this issue of PNAS (43).
Supplementary Material
Acknowledgments
G.T. was supported by a Waksman Institute of Microbiology Predoctoral Fellowship and a teaching assistantship from the Department of Genetics, Rutgers University. We thank Drs. Ramsey Lewis and Hyunsook Moon, North Carolina State University, for unpublished information on the N. sylvestris SSR markers.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JN563929 and JN563930).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1114297109/-/DCSupplemental.
References
- 1.Shimada H, Sugiura M. Fine structural features of the chloroplast genome: Comparison of the sequenced chloroplast genomes. Nucleic Acids Res. 1991;19:983–995. doi: 10.1093/nar/19.5.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sugiyama Y, et al. The complete nucleotide sequence and multipartite organization of the tobacco mitochondrial genome: Comparative analysis of mitochondrial genomes in higher plants. Mol Genet Genomics. 2005;272:603–615. doi: 10.1007/s00438-004-1075-8. [DOI] [PubMed] [Google Scholar]
- 3.Bock R, Timmis JN. Reconstructing evolution: Gene transfer from plastids to the nucleus. Bioessays. 2008;30:556–566. doi: 10.1002/bies.20761. [DOI] [PubMed] [Google Scholar]
- 4.Stegemann S, Bock R. Exchange of genetic material between cells in plant tissue grafts. Science. 2009;324:649–651. doi: 10.1126/science.1170397. [DOI] [PubMed] [Google Scholar]
- 5.Spees JL, Olson SD, Whitney MJ, Prockop DJ. Mitochondrial transfer between cells can rescue aerobic respiration. Proc Natl Acad Sci USA. 2006;103:1283–1288. doi: 10.1073/pnas.0510511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Onfelt B, et al. Structurally distinct membrane nanotubes between human macrophages support long-distance vesicular traffic or surfing of bacteria. J Immunol. 2006;177:8476–8483. doi: 10.4049/jimmunol.177.12.8476. [DOI] [PubMed] [Google Scholar]
- 7.Acquistapace A, et al. Human mesenchymal stem cells reprogram adult cardiomyocytes toward a progenitor-like state through partial cell fusion and mitochondria transfer. Stem Cells. 2011;29:812–824. doi: 10.1002/stem.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koyanagi M, Brandes RP, Haendeler J, Zeiher AM, Dimmeler S. Cell-to-cell connection of endothelial progenitor cells with cardiac myocytes by nanotubes: A novel mechanism for cell fate changes? Circ Res. 2005;96:1039–1041. doi: 10.1161/01.RES.0000168650.23479.0c. [DOI] [PubMed] [Google Scholar]
- 9.Lucas WJ, Ham BK, Kim JY. Plasmodesmata - Bridging the gap between neighboring plant cells. Trends Cell Biol. 2009;19:495–503. doi: 10.1016/j.tcb.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Molnar A, et al. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science. 2010;328:872–875. doi: 10.1126/science.1187959. [DOI] [PubMed] [Google Scholar]
- 11.Ehlers K, Kollmann R. Primary and secondary plasmodesmata: Structure, origin, and functioning. Protoplasma. 2001;216:1–30. doi: 10.1007/BF02680127. [DOI] [PubMed] [Google Scholar]
- 12.Gillman JD, Bentolila S, Hanson MR. In: Cytoplasmic male sterility in Petunia. Petunia. Gerats T, Strommer J, editors. New York: Springer; 2009. pp. 107–129. [Google Scholar]
- 13.Moon HS, Nicholson JS, Lewis RS. Use of transferable Nicotiana tabacum L. microsatellite markers for investigating genetic diversity in the genus Nicotiana. Genome. 2008;51:547–559. doi: 10.1139/G08-039. [DOI] [PubMed] [Google Scholar]
- 14.Sheahan MB, McCurdy DW, Rose RJ. Mitochondria as a connected population: Ensuring continuity of the mitochondrial genome during plant cell dedifferentiation through massive mitochondrial fusion. Plant J. 2005;44:744–755. doi: 10.1111/j.1365-313X.2005.02561.x. [DOI] [PubMed] [Google Scholar]
- 15.Boeshore ML, Hanson MR, Izhar S. A variant mitochondrial DNA arrangement specific to Petunia stable sterile somatic hybrids. Plant Mol Biol. 1985;4:125–132. doi: 10.1007/BF02418759. [DOI] [PubMed] [Google Scholar]
- 16.Svab Z, Maliga P. Exceptional transmission of plastids and mitochondria from the transplastomic pollen parent and its impact on transgene containment. Proc Natl Acad Sci USA. 2007;104:7003–7008. doi: 10.1073/pnas.0700063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruf S, Karcher D, Bock R. Determining the transgene containment level provided by chloroplast transformation. Proc Natl Acad Sci USA. 2007;104:6998–7002. doi: 10.1073/pnas.0700008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang CY, Ayliffe MA, Timmis JN. Direct measurement of the transfer rate of chloroplast DNA into the nucleus. Nature. 2003;422:72–76. doi: 10.1038/nature01435. [DOI] [PubMed] [Google Scholar]
- 19.Stegemann S, Hartmann S, Ruf S, Bock R. High-frequency gene transfer from the chloroplast genome to the nucleus. Proc Natl Acad Sci USA. 2003;100:8828–8833. doi: 10.1073/pnas.1430924100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheppard AE, et al. Transfer of plastid DNA to the nucleus is elevated during male gametogenesis in tobacco. Plant Physiol. 2008;148:328–336. doi: 10.1104/pp.108.119107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanson MR, Sattarzadeh A. Stromules: Recent insights into a long neglected feature of plastid morphology and function. Plant Physiol. 2011;155:1486–1492. doi: 10.1104/pp.110.170852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newell CA, et al. Exclusion of plastid nucleoids and ribosomes from stromules in tobacco and Arabidopsis. Plant J. 2012;69:399–410. doi: 10.1111/j.1365-313X.2011.04798.x. [DOI] [PubMed] [Google Scholar]
- 23.Zubko MK, Day A. Stable albinism induced without mutagenesis: A model for ribosome-free plastid inheritance. Plant J. 1998;15:265–271. doi: 10.1046/j.1365-313x.1998.00195.x. [DOI] [PubMed] [Google Scholar]
- 24.Palmer JD. Chloroplast DNA exists in two orientations. Nature. 1983;301:92–93. [Google Scholar]
- 25.Medgyesy P, Fejes E, Maliga P. Interspecific chloroplast recombination in a Nicotiana somatic hybrid. Proc Natl Acad Sci USA. 1985;82:6960–6964. doi: 10.1073/pnas.82.20.6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fejes E, Engler D, Maliga P. Extensive homologous chloroplast DNA recombination in the pt14 Nicotiana somatic hybrid. Theor Appl Genet. 1990;79:28–32. doi: 10.1007/BF00223782. [DOI] [PubMed] [Google Scholar]
- 27.Burch-Smith TM, Stonebloom S, Xu M, Zambryski PC. Plasmodesmata during development: Re-examination of the importance of primary, secondary, and branched plasmodesmata structure versus function. Protoplasma. 2011;248:61–74. doi: 10.1007/s00709-010-0252-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benitez-Alfonso Y, et al. Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc Natl Acad Sci USA. 2009;106:3615–3620. doi: 10.1073/pnas.0808717106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stonebloom S, et al. Loss of the plant DEAD-box protein ISE1 leads to defective mitochondria and increased cell-to-cell transport via plasmodesmata. Proc Natl Acad Sci USA. 2009;106:17229–17234. doi: 10.1073/pnas.0909229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- 31.Bock R. The give-and-take of DNA: Horizontal gene transfer in plants. Trends Plant Sci. 2010;15:11–22. doi: 10.1016/j.tplants.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 32.Baldev A, et al. Recombination between chloroplast genomes of Trachystoma ballii and Brassica juncea following protoplast fusion. Mol Gen Genet. 1998;260:357–361. doi: 10.1007/s004380050904. [DOI] [PubMed] [Google Scholar]
- 33.Than ND, Medgyesy P. Limited chloroplast gene transfer via recombination overcomes plastome-genome incompatibility between Nicotiana tabacum and Solanum tuberosum. Plant Mol Biol. 1989;12:87–93. doi: 10.1007/BF00017450. [DOI] [PubMed] [Google Scholar]
- 34.Bock R, Kössel H, Maliga P. Introduction of a heterologous editing site into the tobacco plastid genome: The lack of RNA editing leads to a mutant phenotype. EMBO J. 1994;13:4623–4628. doi: 10.1002/j.1460-2075.1994.tb06784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitz-Linneweber C, et al. Pigment deficiency in nightshade/tobacco cybrids is caused by the failure to edit the plastid ATPase alpha-subunit mRNA. Plant Cell. 2005;17:1815–1828. doi: 10.1105/tpc.105.032474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maliga P, Bock R. Plastid biotechnology: Food, fuel, and medicine for the 21st century. Plant Physiol. 2011;155:1501–1510. doi: 10.1104/pp.110.170969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cardi T, Lenzi P, Maliga P. Chloroplasts as expression platforms for plant-produced vaccines. Expert Rev Vaccines. 2010;9:893–911. doi: 10.1586/erv.10.78. [DOI] [PubMed] [Google Scholar]
- 38.Carrer H, Staub JM, Maliga P. Gentamycin resistance in Nicotiana conferred by AAC(3)-I, a narrow substrate specificity acetyl transferase. Plant Mol Biol. 1990;17:301–303. doi: 10.1007/BF00039510. [DOI] [PubMed] [Google Scholar]
- 39.Maliga P, Svab Z. Engineering the plastid genome of Nicotiana sylvestris, a diploid model species for plastid genetics. In: Birchler JJ, editor. Plant Chromosome Engineering: Methods and Protocols. Vol 701. New York: Springer; 2011. pp. 37–50. [DOI] [PubMed] [Google Scholar]
- 40.Lutz KA, Svab Z, Maliga P. Construction of marker-free transplastomic tobacco using the Cre-loxP site-specific recombination system. Nat Protoc. 2006;1:900–910. doi: 10.1038/nprot.2006.118. [DOI] [PubMed] [Google Scholar]
- 41.Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cronn R, et al. Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic Acids Res. 2008;36:e122. doi: 10.1093/nar/gkn502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stegemann S, Keuthe M, Greiner S, Bock R. Horizontal transfer of chloroplast genomes between plant species. Proc Natl Acad Sci USA. 2012;109:2434–2438. doi: 10.1073/pnas.1114076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.