Abstract
Syncytins are envelope protein genes of retroviral origin that have been captured for a function in placentation. Two such genes have already been identified in simians, two distinct, unrelated genes have been identified in Muridae, and a fifth gene has been identified in the rabbit. Here, we searched for similar genes in the Laurasiatheria clade, which diverged from Euarchontoglires—primates, rodents, and lagomorphs—shortly after mammalian radiation (100 Mya). In silico search for envelope protein genes with full-coding capacity within the dog and cat genomes identified several candidate genes, with one common to both species that displayed placenta-specific expression, which was revealed by RT-PCR analysis of a large panel of tissues. This gene belongs to a degenerate endogenous retroviral element, with precise proviral integration at a site common to dog and cat. Cloning of the gene for an ex vivo pseudotype assay showed fusogenicity on both dog and cat cells. In situ hybridization on placenta sections from both species showed specific expression at the level of the invasive fetal villi within the placental junctional zone, where trophoblast cells fuse into a syncytiotrophoblast layer to form the maternofetal interface. Finally, we show that the gene is conserved among a series of 26 Carnivora representatives, with evidence for purifying selection and conservation of fusogenic activity. The gene is not found in the Pholidota order and, therefore, it was captured before Carnivora radiation, between 60 and 85 Mya. This gene is the oldest syncytin gene identified to date, and it is the first in a new major clade of eutherian mammals.
Keywords: endogenous retrovirus, gene capture, cell–cell fusion, endotheliochorial placenta, evolution
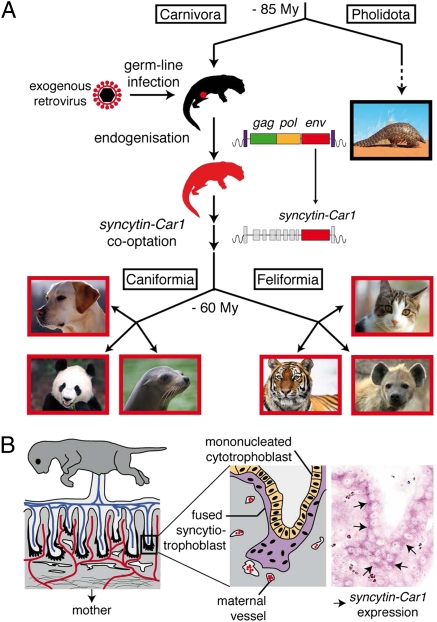
Previous studies have identified envelope protein (env) genes of retroviral origin that have been independently captured by their host for a function in placentation and that have been named syncytins. In simians, syncytin-1 (1–4) and syncytin-2 (5, 6) entered the primate genome 25 and >40 Mya, respectively, and retained their coding capacity in all of the subsequent lineages. They display placenta-specific expression and are fusogenic in ex vivo fusion assays, and one of them (syncytin-2) displays immunosuppressive activity (7). A pair of env genes from endogenous retroviruses (ERVs) was then identified in the mouse, named syncytin-A and -B, and they share closely related functional properties, although they have a completely distinct origin, showing a divergent sequence and a different genomic location compared with the primate syncytins (8). Finally, a syncytin gene was identified in a third order of mammals, namely in the lagomorphs, with the rabbit syncytin-Ory1, which again, was unrelated to the simian and murine genes but most probably shares with them a similar physiological function (9). Recently, we have unambiguously shown by the generation of syncytin-A KO mice that these genes are indeed essential for placentation, with a lack of cell–cell fusion observed in vivo at the level of the syncytiotrophoblast interhaemal layer of the mutant placenta; this lack results in impaired maternofetal exchanges and death of the embryos at midgestation (10). Furthermore, in the sheep, ERV-derived env genes, clearly distinct from the syncytins, were shown to be involved in periimplantation placental morphogenesis through loss of function experiments (11). Therefore, it seems that, on several occasions in the course of mammalian evolution, env genes from endogenous retroviruses have been co-opted by their host to participate in the formation of the placenta.
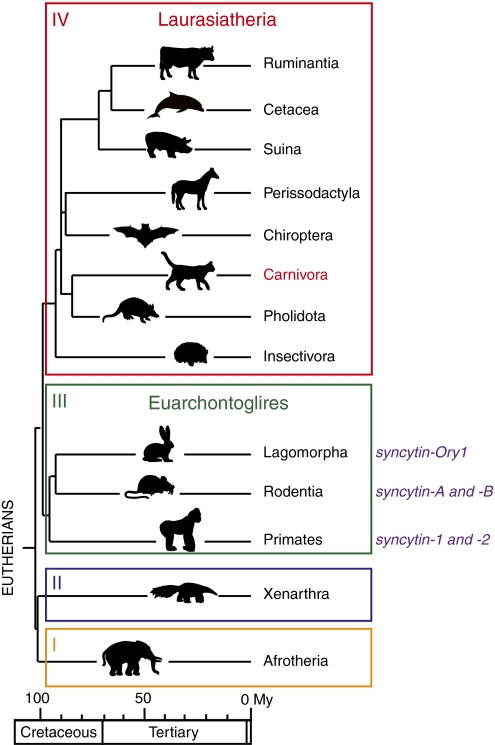
Interestingly, this stochastic acquisition of genes of exogenous origin might be related to the unexpectedly large diversity observed in placental structures and the physiology of placentation among eutherian mammals (12–15). Three main types of placenta have been identified involving different interfaces between the mother and fetal tissues. In the simplest one, the epitheliochorial placenta, both tissues are simply apposed; in the most intricate one, the hemochorial placenta, the placental tissue is in close contact with the mother's blood. The third type, the endotheliochorial placenta, displays intermediate characteristics. It turns out that the primates, rodents, and lagomorphs in which syncytin genes have been identified all belong to the same clade of eutherian mammals, the Euarchontoglires, which diverged from a second major clade of eutherian mammals, the Laurasiatheria, ∼100 Mya (Fig. 1) (16). The latter comprises a large number of species belonging to the orders Cetartiodactyla (Ruminantia, Cetacea, Suina, etc.), Perissodactyla, Chiroptera, Insectivora, Pholidota, and Carnivora that display various types of placenta. To determine whether syncytin capture is a general process also found in this anciently diverged clade of mammals and whether syncytins can be found in animals with a nonhemochorial placentation, we searched for such genes in Carnivora. Carnivora was selected, because the genome from three representatives of this order had been sequenced, namely the dog, the cat, and more recently, the giant panda (17–20). Moreover, at least for the former two representatives, placenta can be easily recovered at various stages of gestation. In addition, placentation in Carnivora is of the endotheliochorial type (13, 15).
Fig. 1.
Phylogeny of Eutherians and previously identified syncytin genes. Eutherians can be grouped into four major clades: Afrotheria (I), Xenarthra (II), Euarchontoglires (III), and Laurasiatheria (IV). Adapted from ref. 16. Branch length is proportional to time (in million years), and the orders where syncytins have been identified to date are indicated with the corresponding gene name.
An in silico search for full-length env genes with an uninterrupted ORF within the dog and cat genomes identified several candidate genes, which were tested for specific expression in the placenta by quantitative RT-PCR on RNA isolated from a large panel of tissues. This test resulted in the identification of a placenta-specific env gene belonging to an endogenous retrovirus present in the dog and cat genomes that is also found in the giant panda. This env gene is distinct from previously identified syncytins, and it corresponds to an independent retrovirus capture by a common ancestor of Carnivora. Functional characterization of the identified placenta-expressed env gene, by cloning into a CMV-driven expression vector and transient transfection experiments, showed its fusogenic activity in an ex vivo infectivity assay with viral pseudotypes. Finally, in situ hybridization of dog and cat placenta sections revealed specific expression of this gene at the level of the syncytial junctional zone where the invading placental tissue contacts the maternal myometrium, consistent with a role in the formation of the maternofetal interface. The gene was found at the same genomic position in the 26 Carnivora species that we further investigated, with evidence for purifying selection. This gene that we accordingly named syncytin-Car1 should, therefore, have entered the Carnivora order before its radiation (i.e., >60 Mya) (16, 21), and as such, it is the oldest syncytin found to date that has been conserved in a functional state for a physiological role in placentation.
The identification of a syncytin gene within a new major clade of mammals, the Laurasiatheria, strongly supports the notion that, on several occasions, retroviral infections have resulted in the independent capture of genes that have been positively selected for a convergent physiological role. It also shows that genomic and genetic analyses cannot solely rely on the notion that mammalian genes have evolved through a series of mutations/recombinations from primitive genes common to most living species, but they have to take into account stochastic de novo acquisition of exogenous genes of parasitic origin, possibly responsible for dramatic evolutionary transitions. Transition from egg-laying animals to placental mammals might be one illustrative example of such a genomic “coup de force.”
Results
In Silico Search for Retroviral env Genes Within the Dog (Canis lupus familiaris) and Cat (Felis catus) Genomes.
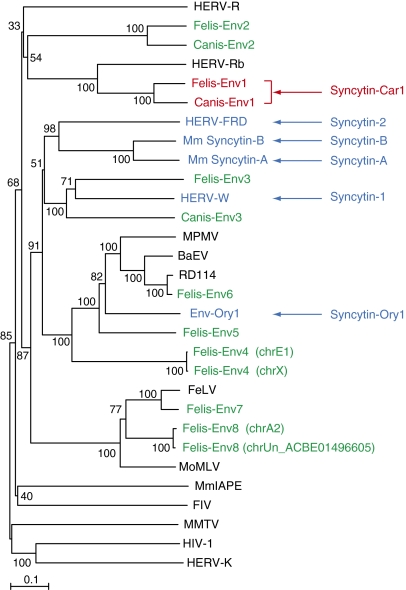
To identify putative env-derived syncytin genes in Carnivora, we made use of the available dog (Canis lupus familiaris) and cat (Felis catus) genome sequences and the method that we previously devised to screen the human, mouse, and rabbit genomes for such genes (8, 9, 22). Basically, it makes use of the CKS17 consensus motif that is associated with the immunosuppressive domain of retroviral envelope proteins and designed to match the majority of env genes of exogenous and endogenous origin (22). Dog and cat sequences were screened with a slightly modified CKS17 motif using the BioMotif program, and only coding sequences longer than 1 kb (from start to stop codon) were considered (Methods). Accordingly, 3 and 10 coding sequences were identified in the dog and cat genomes, respectively. For the cat genes, two pairs of sequences displayed high identity, and all of the identified coding sequences within the dog and cat genomes could finally be grouped into 11 families that we named canis-env1 to -env3 and felis-env1 to -env8, respectively (Figs. 2 and 3). A phylogenetic tree constructed with these sequences and including a series of retroviral env genes as well as the known syncytins (Fig. 2) showed that the identified genes are distinct from the previously identified syncytins, with very short branch lengths observed for both the paired felis-env4 and felis-env8 sequences.
Fig. 2.
Retroviral envelope protein-based phylogenetic tree with the identified Canis- and Felis-Env protein candidates. The tree was determined by the neighbor-joining method using envelope amino acid sequences from murine and human endogenous retroviruses and a series of infectious retroviruses. The horizontal branch length is proportional to the percentage of amino acid substitutions from the node (scale bar on the left), and the percent bootstrap values obtained from 1,000 replicates are indicated at the nodes. The two pairs of Felis-Env proteins that were grouped into single families of elements (Felis-Env4 and Felis-Env8) are distinguished by indication of their chromosomal position. BaEV, baboon endogenous virus; FeLV, feline leukemia virus; FIV, feline immunodeficiency virus; HERV, human ERV; MoMLV, moloney murine leukemia virus; MmIAPE, Mus musculus intracisternal A-type particle with an envelope gene; MMTV, murine mammary tumor virus; MPMV, Mason–Pfizer monkey virus; RD114, feline endogenous type-C retrovirus.
Fig. 3.
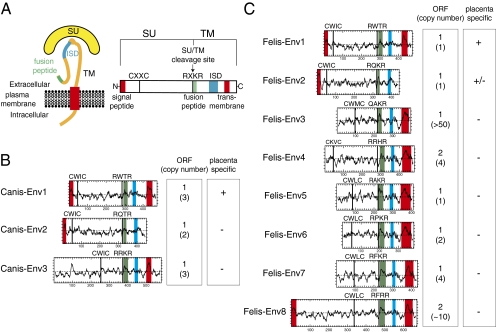
Structure of a canonical retroviral envelope protein and characterization of the identified dog and cat candidates. (A) Schematic representation of a retroviral envelope protein with the SU and TM subunits delineated and the furin cleavage site (consensus: R/K-X-R/K-R) between the two subunits together with the C-X-X-C domain involved in SU–TM interaction indicated; the hydrophobic signal peptide (red), fusion peptide (green), transmembrane domain (red), and putative immunosuppressive domain (ISD; blue) are also indicated. (B and C) Characterization of the dog and cat candidate envelope proteins, respectively. (Left) The hydrophobicity profile for each candidate is shown with the canonical structural features listed in A positioned when present (same color code). (Right) Number of full-length coding sequences (ORF) of env genes within each family of element and total number of genomic copies (in parenthesis) together with indication for specific placental expression (details in Fig. 4).
In silico analysis of the identified sequences (Fig. 3) also revealed that they possess at least part of the characteristic features of retroviral envelope proteins (23, 24), with a putative furin cleavage site (consensus: R/K-X-R/K-R) delineating the surface (SU) and transmembrane (TM) subunits and a C-X-X-C motif corresponding to a binding domain between the two subunits. Hydrophobicity plots identify a hydrophobic transmembrane domain within the TM subunits, a sequence required for anchoring of the envelope protein within the plasma membrane and membrane fusion, with the exception of Canis-Env2 and Felis-Env2 because of the presence of a premature stop codon. A putative hydrophobic fusion peptide is also identified at the N terminus of the TM subunits as well as a canonical immunosuppressive domain (ISD), which was expected from the screening procedure. In some cases (e.g., Canis-Env1 and -Env2 and Felis-Env1, -Env2, and -Env8), a clear signal peptide is predicted by the Phobius program (http://phobius.sbc.su.se/) at the N terminus of the sequences.
Finally, a BLAST search among the corresponding databases revealed (Fig. 3) that canis-env1 to -env3 are present at a low copy number (one to three copies, with only one copy with a full-length ORF in each case), with a similar situation for most of the cat sequences (one to four copies). For felis-env3 and -env8, a much larger number of copies can be identified (up to 50 copies for felis-env3) but again, with only one or two containing a complete ORF in each family of elements.
Identification of Placenta-Specific env Genes.
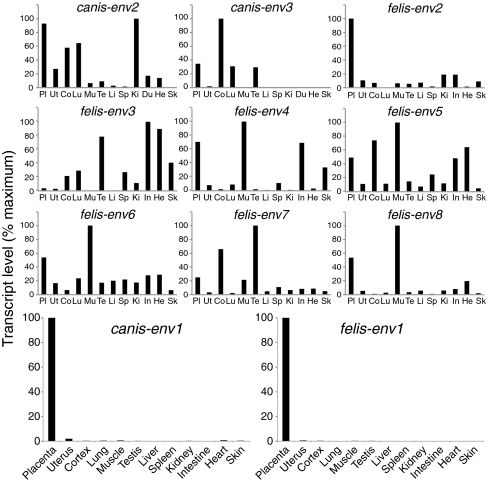
Quantitative RT-PCR (qRT-PCR) analysis of transcript levels for each candidate env gene was then performed using primers that were designed to be specific for the ORF-containing sequences within each family of elements (Methods). Accordingly, as shown in Figs. 3 and 4, only canis-env1 and felis-env1, and to a much lesser extent, felis-env2, showed the characteristic property of a syncytin gene, with high-level expression in the placenta and limited expression in other tissues. Expression is indeed severely restricted to the placenta for canis-env1 and felis-env1, with <2% of the placental level found in only a few other organs (e.g., uterus, cortex, and heart). Felis-env2 displays up to 20% of the placenta expression level in several tissues, whereas the other candidate genes are found to be expressed at higher levels in nonplacental than placental tissues. Altogether, in silico analyses combined with qRT-PCR assays for the dog and cat retroviral env genes clearly identify canis-env1 and felis-env1 as putative syncytin genes. Felis-env2, despite its pattern of expression and its close relationship with canis-env2 (76% identity in amino acid sequences; see also Fig. 2), was not considered further in the present study, because it lacks the transmembrane domain essential for fusogenic activity.
Fig. 4.
Real-time qRT-PCR analysis of the candidate env gene transcripts from dog and cat. Transcript levels are expressed as percent of maximum and were normalized relative to the amount of a control gene (PPIA) (Methods) RNA. The two placenta-specific canis-env1 and felis-env1 genes are displayed in enlarged panels in the fourth row; the same tissues (abbreviated and displayed in the same order) for the nine other env gene candidates are shown. Values for the placenta and uterus are the means of at least five samples for both dog and cat; RNAs of other tissues are from Zyagen.
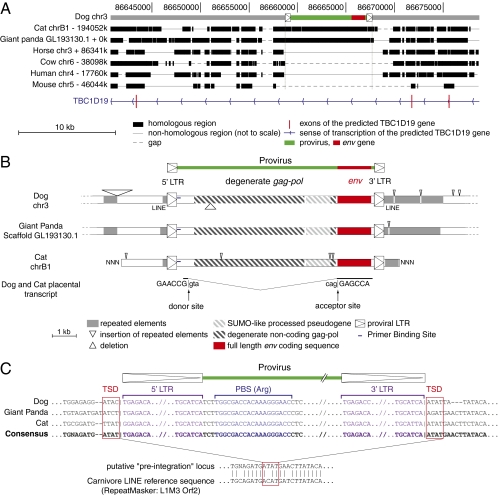
As already suggested by the relatively short branch length separating canis-env1 and felis-env1 (Fig. 2), these two genes are closely related, with 78% identity of their amino acid sequences. Furthermore, a BLAST search using either sequence against the third available Carnivora genome, namely the genome of the giant panda bear (Ailuropoda melanoleuca), identified a homologous env gene sequence within this species that we consequently named ailur-env1. As illustrated in Fig. 5, all three genes are homologous (80% and 78% amino acid sequence identity between ailur-env1 and canis- and felis-env1, respectively), with all of the canonical sites and domains characteristic for a retroviral envelope protein being highly conserved. Interestingly, a search using the Comparative Genomic tool of the University of California Santa Cruz (UCSC) Genome Browser (http://genome.ucsc.edu) showed that canis-env1, felis-env1, and ailur-env1 are positioned at the same syntenic genomic locus: chromosome 3 for the dog, chromosome B1 for the cat, and scaffold GL193130.1 for the giant panda (Fig. 6A). Of note, although intrinsically not fusogenic because of a premature stop codon before the transmembrane domain, canis-env2, felis-env2, and a homologous gene in the giant panda ailur-env2 (80% and 73% amino acid sequence identity between ailur-env2 and canis-env2 and felis-env2, respectively) are also positioned at a syntenic genomic locus in these three species.
Fig. 5.
Primary sequences and alignment of Canis-Env1, Felis-Env1, and Ailur-Env1. Primary amino acid sequence and characteristic structural features of the homologous Env protein from dog, cat, and giant panda. Same color code and abbreviations as in Fig. 3; asterisks indicate amino acid identity, and colons indicate amino acid similarity.
Fig. 6.
Characterization of the dog, cat, and giant panda env1-containing endogenous retrovirus and its integration site. (A) Evidence for orthology of the dog, cat, and panda sequences. The canis-env1–containing provirus (shown in green with its LTRs schematized by boxed triangles and the env gene shown in red) was used as a reference, and synteny between the dog, cat, giant panda, horse, cow, human, and mouse genomes was determined with the Comparative Genomic tool of the UCSC Genome Browser (http://genome.ucsc.edu/); the position of exons (vertical red lines) of the resident TBC1D19 gene and the sense of transcription (blue arrows) are indicated. Homologous regions are shown as black boxes, nonhomologous regions are shown as thin lines (not to scale), and gaps are shown as dotted lines. (B) Structure of the corresponding proviruses and integration sites. Homologous regions common to all three sequences are aligned, with the insertions and deletions represented by triangles placed above and below each sequence, respectively; repeated mobile elements (dark gray) as identified by the RepeatMasker website program are positioned. The proviral LTRs, the degenerate gag-pol gene, and the env gene coding sequence are indicated together with the SUMO-like processed pseudogene inserted within the provirus (the symbols used are given below the panel). Splice sites for the env subgenomic transcript as determined by RT-PCR from dog and cat placental RNA are indicated together with the donor and acceptor site sequences. (C) Sequence alignment of proviral and flanking sequences: evidence for retroviral integration. Characteristic sequences are shown, including LTRs (purple with TG … CA borders), primer binding site (blue) for an arginine tRNA, and target site duplication (red boxes). Sequence alignment of the reconstituted, putative preintegration site with the Carnivore LINE L1M3 Orf2 (prototypic reference sequence from the RepeatMasker website) provides evidence for precise proviral integration into a preexisting ancestral LINE.
Orthologous Candidate syncytin Genes of Retroviral Origin.
Close examination of the canis-env1, felis-env1, and ailur-env1 genes shows that they are part of a proviral structure with degenerate but still identifiable flanking LTRs and internal gag and pol gene sequences (Fig. 6B). A primer binding site sequence can also be identified downstream to the 5′ LTR, which is commonly observed for retroviruses (23, 25, 26), where it is used for priming reverse transcription, and found in the present case to be complementary to the Carnivora Arginine tRNA (Fig. 6C). The retroviral origin of the env1 genes is also supported by the occurrence of a retroviral-like mode of generation of the subgenomic env transcripts, with a putative donor splice site located close to the 5′ LTR and an acceptor site 5′ to env. These sites were predicted by the NetGene2 program (http://www.cbs.dtu.dk/services/NetGene2) and confirmed by RT-PCR analysis—and sequencing—of the env1 transcripts from the dog and cat placenta using appropriate primers (Methods and Fig. 6B). Close inspection of the flanking sequences also unambiguously shows that insertion of the ancestral provirus occurred within a LINE retroelement and was of the retroviral type, with evidence for a target site duplication of 4 nt (as shown on the consensus sequence derived from the three loci), a common feature of retroviruses (23, 25, 26) (Fig. 6C).
As shown in Fig. 6A, the dog, cat, and giant panda proviruses are integrated within an intron of the putative TBC1D19 gene in the antisense orientation as commonly observed for endogenous retroviruses (25, 26) (Fig. 6A). Moreover, high sequence homology of the sequences flanking the proviruses (pairwise percentage of identity comprised between 78% and 82% over 1 kb of the 5′ flanking region and between 59% and 67% over 1 kb of the 3′ flanking region for the three proviruses) as well as the presence of a SUMO-like pseudogene inserted at the same position within all three proviruses (Fig. 6B) confirm that they are strict orthologous copies of the same gene in the dog, cat, and giant panda genomes. Therefore, the ancestral provirus integrated within the genome of Carnivora before the dog–cat–giant panda divergence. A PCR assay with the forward primer located within the provirus at the 5′ end of the env gene and the reverse primer located downstream of the provirus in the 3′ flanking sequence resulted in a single amplification product with a conserved size, confirming orthology of the genes in all three species (see below). Accordingly, the Carnivora gene corresponding to these three orthologous genes will now be referred to as env-Car1, whereas canis-env-Car1, ailur-env-Car1, and felis-env-Car1 will refer to the dog, giant panda, and cat representatives, respectively.
Env-Car1 Is a Fusogenic Retroviral Envelope Protein.
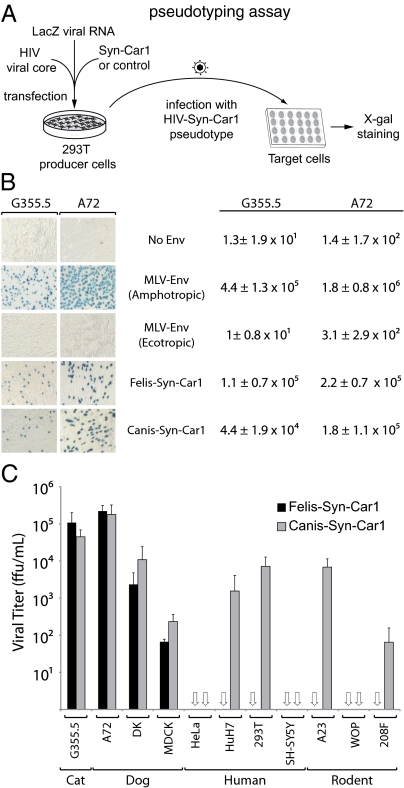
The functionality of Env-Car1 as an ancestral, retrovirally derived fusogenic envelope protein was assayed ex vivo as previously described for other syncytins (6, 9). Basically, it was tested whether this envelope protein added in trans could render a recombinant retrovirus—deprived of its own native env gene—able to mediate infection of test target cells (Fig. 7A, scheme). To do so, the amplification products of canis-env1 and felis-env1 were cloned into a CMV promoter-containing expression vector (Methods), and plasmids with a full-length env gene ORF 100% identical to the genomic sequences were assayed. As expected for an envelope protein of retroviral origin, both Canis-Env1 and Felis-Env1 can produce infectious particles. As illustrated in Fig. 7B, pseudotypes generated in human 293T cells with an HIV core are able to infect the neuroblastoma-derived G355.5 cat cell line and the epithelial kidney-derived A-72 dog cell line, in both cases with a high infectious titer (4.4 ± 1.9 × 104 focus forming unit [ffu] /mL in G355.5 and 1.8 ± 1.1 × 105 ffu/mL in A-72 for Canis-Env1 and 1.1 ± 0.7 × 105 ffu/mL in G355.5 and 2.2 ± 0.7 × 105 ffu/mL in A-72 for Felis-Env1). Of note, as classically observed for most retroviral envelope proteins, not all cells could be infected with identical efficiency, even among cells from the same animal species (Fig. 7C). This finding is illustrated, for instance, by the embryonic kidney-derived MDCK dog cell line, which shows a close to 100-fold reduction in infectivity of both the Canis-Env1 and Felis-Env1 pseudotypes, possibly associated with poor expression of the cognate receptor for this ancestral retroviral envelope protein on this cell line. Interestingly, it can also be observed that the human HeLa and SH-SY5Y as well as the murine WOP cell lines cannot be infected by either pseudotype, whereas other cell lines—such as human 293T and HuH7 or rodent A23 and 208F—can be infected, at least by Canis-Env1. This finding strongly suggests that a human or rodent receptor—with variable levels of expression depending on the cell type—can substitute to some extent to the carnivore receptor to mediate pseudotype cell entry, although it has to be noted that it seems not to be efficient in mediating Felis-Env1 pseudotype cell entry. This difference could be because of a subtle divergence in the expected parallel coevolution of the captured env-Car1 gene (that can now be named syncytin-Car1) and its cognate—but still unknown—receptor.
Fig. 7.
Syncytin-Car1 is a fusogenic retroviral envelope protein. (A) Schematic representation of the assay for cell infection with Syncytin-Car1–pseudotyped virus particles. Pseudotypes are produced by cotransfection of human 293T cells, with expression vectors for the HIV-1 core, Syncytin-Car1 proteins (or control retroviral envelope proteins or an empty vector), and β-Gal encoded by a LacZ-containing retroviral transcript. Productive cell supernatants are then assayed for infection of the indicated target cells, which are X-gal–stained 3 d postinfection. Syn-Car1: Syncytin-Car1. (B) X-gal–stained target cells (Left) and viral titers (Right) for particles pseudotyped with Felis-Syn-Car1, Canis-Syn-Car1, or control Env proteins from murine leukemia viruses of either the ecotropic (infecting only murine cells) or the amphotropic (infecting both murine and nonmurine cells) subtype. Target cells were cat (G355.5) and dog (A72) cells. Values (focus forming unit per milliliter ± SD) are the means from three independent experiments. (C) Viral titers for particles pseudotyped with Felis-Syn-Car1 or Canis-Syn-Car1 assayed on a panel of target cells from cat (G355.5), dog (A72, MDCK, and DK), human (HeLa, HuH7, 293T, and SH-SY5Y), or rodent (A23, WOP, and 208F). Titers are corrected for the background values of control particles without an envelope protein, and they are the means from at least two independent experiments (focus forming unit per milliliter ± SD; open arrows indicate null values).
In Situ Hybridization on Placenta Sections.
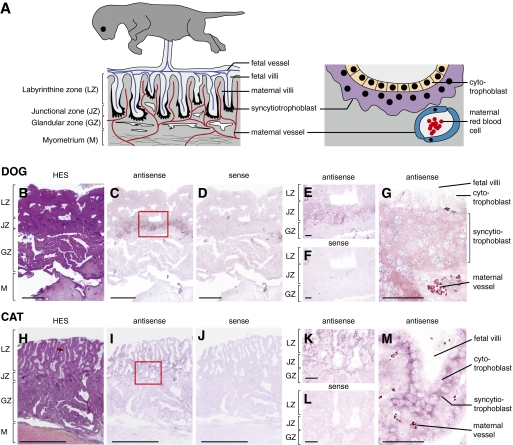
The placenta of Carnivora is of the endotheliochorial type (27–29). As schematized in Fig. 8A, Left, it is composed, from fetus to mother, of the labyrinthine zone where fetal and maternal villi are interspersed, the junctional zone where the fetal villi are invading the maternal tissue, and the maternal uterine glandular zone and myometrium. At the tip of the invasive fetal villi, cytotrophoblast cells migrate and differentiate into a multinucleated syncytial layer, the syncytiotrophoblast, by cell–cell fusion (Fig. 8A, Right). This syncytiotrophoblast is highly invasive and phagocytic: it destroys the maternal epithelium and surrounds the maternal vessels, thus forming the endotheliochorial pattern of the maternofetal interface. At variance with human and mice, the syncytiotrophoblast in Carnivora does not disrupt the maternal vessels.
Fig. 8.
Structure of the Carnivora placenta and in situ hybridization for syncytin-Car1 expression on dog and cat placental sections. (A) Schematic representation of the Carnivora placenta (A Left) with, from mother to fetus, the myometrium (M), the glandular zone (GZ), the junctional zone (JZ), and the labyrinthine zone (LZ). The maternal and fetal vessels are schematized, with the invasive syncytiotrophoblast at the fetomaternal interface colored in black at the tip of the invading fetal villi. An enlarged view (A Right) shows the multinucleated syncytiotrophoblast (purple) generated by fusion from the underlying mononucleated cytotrophoblast cells (yellow) forming the interface between the fetal compartments and the maternal vessels (with the red blood cells schematized). Sections of placenta from dog (B–G) and cat (H–M) at midgestation. (B and H) Hematoxylin–eosin–saffron staining (HES) of the placental section with the four layers indicated. (C–G and I–M) In situ hybridization on serial sections observed at different magnifications using digoxigenin-labeled antisense (C, E, G, I, K, and M) or sense (negative control; D, F, J, and L) riboprobes revealed with an alkaline phosphatase-conjugated antidigoxigenin antibody. (E, F, K, and L) Intermediate magnification of the junctional domain delineated in C and I (red boxed areas). (G and M) Higher magnification with the fetal villi, the labeled syncytiotrophoblast, and the maternal vessels clearly visible. (Scale bars: B–D and H–J, 1 mm; E, F, K, and L, 100 μm; G and M, 50 μm.)
To further assess the physiological relevance of the specific syncytin-Car1 expression in the placenta, which is revealed by the qRT-PCR assays in Fig. 4, in situ hybridization experiments were performed on paraffin sections of dog and cat placentae. Specific digoxigenin-labeled antisense riboprobes were synthesized for the detection of the syncytin-Car1 transcripts from each species as well as the corresponding sense riboprobes to be used as negative controls. As illustrated in Fig. 8, specific labeling was observed only with the antisense probes and not with the control probes. In the dog, syncytin-Car1 expression is restricted to the junctional zone at the tip of the villi in areas where placental cells invade the maternal tissues (Fig. 8 C and E). In the cat, the junctional zone is thinner, and specific labeling is observed on the entire villi (Fig. 8 I and K). Higher magnification images also show that syncytin-Car1 is expressed, for both the dog and cat, at the surface of the fetal villi. Expression is detected at the level of the multinucleated syncytiotrophoblast interface between the fetal mononucleated cytotrophoblasts and the maternal vessels (Fig. 8 G and M). Such a labeling is consistent with a role for the fusogenic Syncytin-Car1 in the formation of the syncytiotrophoblast.
Identification of Syncytin-Car1 in All Carnivora Suborders.
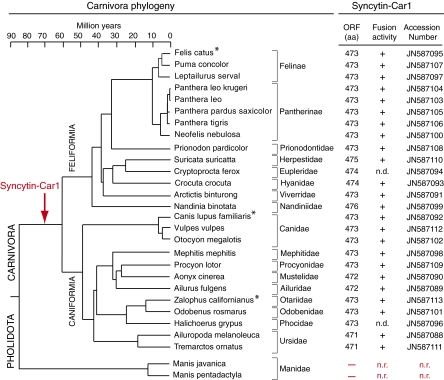
Phylogenetic relationships in the Carnivora order are illustrated in Fig. 9 (adapted from refs. 16, 21, and 30). This order includes two major suborders, the Caniformia and the Feliformia, with the dog and cat as reference species, respectively. Each group comprises a series of families as indicated in Fig. 9. To further characterize syncytin-Car1 and determine its status and evolution in Carnivora, DNA from at least one representative species within each family was recovered to tentatively determine the presence, sequence, and functional properties of the gene. Locus-specific pairs of PCR primers (forward primer in the provirus upstream of syncytin-Car1 and reverse primer downstream of the provirus in the 3′ flanking sequence) (Table S1) were used to amplify genomic DNA from 26 representative species. In all cases, PCR amplification showed a single amplification product with a conserved size, strongly suggesting the presence of the orthologous syncytin-Car1 in all carnivores. This finding was confirmed by sequencing the PCR products (sequences were deposited in GenBank, and accession numbers are listed in Fig. 9), which revealed the presence of a syncytin-Car1 gene encoding a full-length ORF (471–476 aa long) in all of the carnivores tested.
Fig. 9.
Entry date and conservation of syncytin-Car1 in the Carnivora radiation. (Left) Carnivora phylogenetic tree with the Pholidota outgroup indicated. Adapted from refs. 16, 21, and 30. Horizontal branch length is proportional to time (scale bar at the top). The names of the 26 Carnivora species (from both the Feliformia and Caniformia suborders) tested for the presence of the syncytin-Car1 gene are indicated together with the names of their corresponding families. (Right) The length (in amino acids) of the Syncytin-Car1 proteins that were identified for each species and the accession number for the sequences that were deposited in GenBank are indicated; the fusogenic activity for each cloned gene, as determined by the pseudotyping assay described in Fig. 7, is provided (n.d., not determined). For the two Manidae tested (from the Pholidota Order), syncytin-Car1 was not detected (n.r., not relevant), thus dating back acquisition of the gene between 60 and 85 Mya. Asterisks indicate species where placenta-specific expression could be shown (Figs. 4 and 8 for dog [C. lupus familiaris] and cat [F. catus], and Fig. S1 for sea lion [Zalophus californianus]).
The closest outgroup for Carnivora is the Pholidota order (comprising the pangolin species), but no representatives from this group have been sequenced to date. The presence of syncytin-Car1 in this order was, therefore, similarly assayed by PCR with the genomic DNA from two distinct pangolin species (Manis javanica and Manis longicaudata). Both were found negative (Fig. 9). The absence of syncytin-Car1 was confirmed using PCR primers internal to the syncytin-Car1 ORF placed at positions where all previously sequenced Carnivora genes showed a strictly identical nucleotide sequence. Again, PCRs with the pangolin DNA were negative under conditions where DNAs from all of the carnivores in Fig. 9 were positive. Although we cannot formally exclude that pangolin sequences may be too divergent to allow primers annealing and PCR amplification and despite the fact that we could not amplify the empty locus in this species (a rather expected issue taking into consideration that integration in Carnivora was within a LINE mobile element, not necessarily present at the same locus in pangolins), the data strongly suggest that syncytin-Car1 entered the identified locus after the divergence between Carnivora and Pholidota (i.e., 85 Mya). Analyses using the comparative tool from the UCSC Genome browser (http://genome.ucsc.edu/) confirmed the absence of syncytin-Car1 at the syntenic locus of the available horse, cow, human, and mouse genomes. Taking into consideration that the radiation of Carnivora occurred 60 Mya, the presence of a proviral integration at the same locus in all of the carnivores tested indicates that the provirus entered the genome of the common Carnivora ancestor between 60 and 85 Mya.
Purifying Selection and Functional Conservation of syncytin-Car1 in Carnivora.
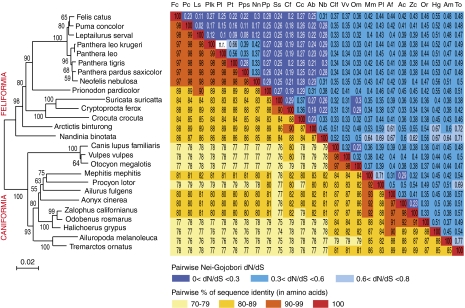
Sequence analysis of the 26 syncytin-Car1 Carnivora genes shows high sequence similarities ranging from 74% to 99% (Fig. 10, Right), which was expected for a bona fide cellular gene. Interestingly, the syncytin-Car1–based phylogenetic tree that can be generated from the aligned sequences using the neighbor-joining method (Fig. 10, Left) is strongly congruent with the Carnivora phylogenetic tree in Fig. 9 (comparison in Fig. S2), with the Feliformia and Caniformia sequences branching into two distinct monophyletic groups. Closer examination only reveals minor differences within these groups, which actually correspond to nodes with low bootstrap values.
Fig. 10.
Sequence conservation and evidence for purifying selection of syncytin-Car1 in Carnivora. (Left) Syncytin-Car1–based phylogenetic tree determined using amino acid alignment of the Syncytin-Car1 proteins identified in Fig. 9 by the neighbor-joining method. The horizontal branch length and scale indicate the percentage of amino acid substitutions. Percent bootstrap values obtained from 1,000 replicates are indicated at the nodes. (Right) Double-entry table for (lower triangle) the pairwise percentage of amino acid sequence identity between the syncytin-Car1 gene among the indicated species and (upper triangle) the pairwise Nei–Gojobori (30) dN/dS; dN/dS between Panthera leo and P. leo krugeri could not be determined (only one nonsynonymous and no synonymous mutation between both species; n.r. not relevant). A color code is provided below the table for both series of values.
To further characterize the conservation/evolution of the syncytin-Car1 gene among Carnivora, analysis of the nonsynonymous to synonymous mutation (dN/dS) ratio between all pairs of species sequences was performed using the method in the work by Nei and Gojobori (31). Accordingly, the entire env gene shows purifying selection between all pairs of species, with dN/dS ratios comprised between 0.02 and 0.77 (Fig. 10, Right). This finding is what is classically found for cellular genes with a physiological role in which nonsynonymous mutations are strongly selected against. However, because syncytin-Car1 has a retroviral origin, it could be asked whether, as observed, for instance, for the HIV env gene (32), some specific domains of the syncytin-Car1 gene could still display some variability (i.e., be under positive selection). To answer this question, we performed a more refined analysis of the sequences using methods to assay site-specific selection (33, 34). These methods use a continuous time Markov chain process, allowing the dN/dS ratio to vary between codons. Such an analysis, using the PAML package (35), provided support for a model (model M8 vs. model M7: χ2 = 12.2, degree of freedom = 2, P value = 2.2e-3) in which most of the codons are under strong purifying selection (1e-4 ≤ dN/dS ≤ 0.82, 77% of the codons), a few codons are under nearly neutral selection (dN/dS = 0.97, 8.5% of the codons), and some codons are under weak positive selection (dN/dS = 1.6, 14.5% of the codons). However, no definite amino acid but one (Fig. S3) and no specific domain can be identified with a dN/dS value significantly higher than unity, suggesting that positive selection, if any, is weak and that syncytin-Car1 is mainly under strong purifying selection. Analyses using the HyPhy package (34) with slightly different site-specific models (random effect likelihood and fixed effect likelihood) or a branch-specific model (GA branch), which allowed the dN/dS value to vary among the branches, lead to similar conclusions (Fig. S3). Syncytin-Car1 genes are, therefore, mainly under strong purifying selection, although one cannot definitely exclude that few events of positive selection may have occurred. In the case of HIV, several domains of the env gene have been shown to be subject to positive selection (e.g., the variable regions of the SU subunit), with mutations in these domains favoring virus escape from the host immune response. Clearly, syncytin-Car1 behaves differently, a result consistent with this gene having acquired a cellular gene status and consequently, having lost the high mutation rate of a replicating retrovirus. Altogether, the data strongly suggest that syncytin-Car1 is now a bona fide cellular gene co-opted by Carnivora for a physiological role in placentation.
To determine whether the strong selective pressure shown for the syncytin-Car1 gene among Carnivora indeed correlates with conservation of its functional properties, an ex vivo assay for its fusogenic activity, which is illustrated in Fig. 7 for the dog and cat representatives, was finally performed. The PCR-amplified syncytin-Car1 genes in Fig. 9 were cloned into the same eukaryotic expression vector, and pseudotypes were similarly assayed using either cat (G355.5 neuroblastoma cells) or dog (A-72 epithelial kidney cells) cells as the target. As indicated in Fig. 9, all of the 24 tested syncytin-Car1 genes were found positive.
Discussion
Here, we have identified syncytin-Car1, the env gene from an endogenous retrovirus that has integrated into the genome from a common ancestor of Carnivora before the radiation of this order more than 60 Mya (16, 21), as the most ancient gene of the syncytin group identified to date. This gene has been maintained as a functional retroviral env gene since that time, being conserved in all of the 26 species tested, which were selected among the known Carnivora families. This gene displays all of the canonical characteristics of a syncytin gene. (i) It exhibits fusogenic activity, because it can functionally replace a present day retroviral env gene within a recombinant infectious retrovirus. (ii) It has been subject to purifying selection in the course of evolution, displaying low rates of nonsynonymous to synonymous substitutions and full conservation of its fusogenic property. (iii) It is specifically expressed in the placenta, which was evidenced by both RT-PCR analyses and in situ hybridization of cat and dog placental tissue sections. The in situ hybridization experiments using syncytin-Car1 sequences as a probe clearly show that expression takes place at the level of the invading fetal villi, consistent with a direct role of this fusogenic syncytin gene in syncytiotrophoblast formation. Syncytin-Car1 adds to the two primate syncytin-1 and -2 genes first identified (1, 2, 5), the two syncytin-A and -B genes later found in Muridae (8), and the syncytin-Ory1 gene recently described in Leporidae (9). Importantly, all six syncytins are unrelated and correspond to independent captures in separate mammalian lineages of genes of retroviral origin. In the case of the syncytin-A gene, KO mice unambiguously showed that it is absolutely required for placentation, with evidence for a defect in syncytiotrophoblast formation causing embryonic lethality in the mutants (10). It, therefore, can be proposed that the other identified syncytins as well as the newly discovered Carnivora syncytin-Car1 are likely to play a similar role in placentation by being involved in syncytiotrophoblast formation. This finding would be consistent with the Primate, Muridae, and Leporidae placentae being of the hemochorial type, where an extended multinucleated syncytial layer forms the interface between maternal blood spaces and fetal vessels (36–38). In the case of Carnivora, placentation is of the endotheliochorial type, and syncytiotrophoblast formation also takes place at the level of the invading fetal trophoblast cells, although in that case, invasion does not result in disruption of the maternal vessels (27–29).
An important outcome of the present investigation is that the discovery of syncytin-Car1 extends the presence of syncytin genes outside the Euarchontoglire clade of placental mammals, where previously identified syncytins had been found, to include members from the highly diverse Laurasiatheria. This clade contains—among others and in addition to Carnivora—the Ruminantia (cow and sheep), the Suina (pig), and the Perissodactyla (horse), and it diverged from the Euarchontoglires 100 Mya (Fig. 1) (16). It, therefore, clearly shows that syncytin gene capture has been a widespread process, which finally turns out to have taken place in several widely separate lineages in the course of eutherian evolution. In this respect, it should be emphasized that, in the course of our search for syncytins in mammals, in every species investigated up to date, at least one such gene could be found. It is, therefore, likely that syncytins are a common feature of placental mammals, and search for their presence in other Laurasiatheria orders, as well as in the two other Xenarthra and Afrotheria clades, is of prime interest. A specific issue should concern (among other Laurasiatheria) the Suina and Perissodactyla, because they display a third type of placentation—the epitheliochorial type—where the fetal and maternal tissues are simply apposed (39–41) without evidence for syncytiotrophoblast formation, thus raising the question of the presence and possible role of syncytins in these species besides cell fusion. In the case of ruminants, an intermediate type of placentation, the synepitheliochorial type, is also observed, with syncytiotrophoblast formation occurring only to a limited extent (42, 43). Accordingly, it is tempting to hypothesize that the large variability in placental structures that can be observed simply results from the diversity of the syncytin genes that have been stochastically captured in the course of mammalian evolution and that parameters, such as the intrinsic level of fusogenicity of the Env proteins, the presence of the receptor required for Env-mediated fusion at the surface of the right neighboring cell, and the specific regulation of syncytin expression in the appropriate tissues, control and finely tune the placentation process. In this respect, the presently identified Carnivora syncytin with its cloning in a large number of carnivore species should help in understanding the subtle differences in placental structures observed among them, especially after the cognate receptor for the Syncytin-Car1 protein will be identified and analyzed for its species-specific interactions.
Methods
Database Screening and Sequence Analyses.
Retroviral env gene sequences were searched by using the BioMotif program (http://www.lpta.univ-montp2.fr/users/menes/bioMotif_html_doc/ref_Run.html) with the degenerate modified CKS17 consensus motif [(L|Y|F|P|W|A|S|M) (Q|N|E|D) N XX (G|A|D|M|V|C) (L|P|I|V) (D|H|N|G|V) X (L|F|P|S|V|T|I) XXXX (G|D|K|E|S) (G|E|S|R|H) X C] as a query and the getorf program from the EMBOSS package (http://emboss.sourceforge.net/apps/cvs/emboss/apps/getorf.html) to identify coding sequences (from start to stop codons) >1 kb. We made use of the available domestic dog and cat genome sequences (7.6× coverage assembly of the C. lupus familiaris genome [UCSC Broad/canFam2, May of 2005] and 2.8× coverage assembly of the F. catus genome [UCSC, catChrV17e, December of 2008], respectively). The identified env coding sequences (Fig. 3) coordinates are listed in SI Methods.
The dog, cat, and giant panda genomes were screened with the identified envelope glycoprotein sequences using the BLAST programs from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST). Multiple alignments of amino acid sequences were carried out by using the Seaview program under ClustalW protocol. Phylogenetic trees were constructed from manually corrected alignments by neighbor-joining with 1,000 replicates using the Seaview program (44).
PAML4 (35) was used to run site-specific selection tests and obtain dN/dS ratios for all syncytin-Car1 sequences. PAML models analyzed assumed no molecular clock (clock = 0) and a single dN/dS for all tree branches (model = 0), and we used likelihood ratio tests to compare the improvement in likelihood for a model (M8), allowing for positive selection compared with a model (M7; NS site = 7–8) that does not. Each analysis ran until convergence (Small_Diff = 0.5e-6), and the control file is available on request. HyPhy (34) was used on the datamonkey webserver (www.datamonkey.org) to run site-specific Random Effect Likelihood (REL) and Fixed Effect Likelihood (FEL), and branch-specific GA-branch selection tests. The best branch-specific model was selected using the Akaike Information Criterion.
The horse genome (Equus caballus, 6.79× coverage, UCSC Broad/equCab2, 2007), the cow genome (Bos taurus, 7.1× coverage, UCSC Baylor 4.0/bosTau4, 2007), the human genome (Homo sapiens, UCSC GRCh37/hg19, 2009), and the mouse genome (Mus musculus, UCSC NCBI37/mm9, 2007) assemblies were also screened for the presence of the identified syncytin-Car1 containing provirus sequence using syntenic genomic regions from the UCSC genome browser (http://genome.ucsc.edu/).
Search for syncytin-Car1 in Other Species.
PCRs were performed on 100 ng genomic DNA using Accuprime Taq DNA Polymerase (Invitrogen) for 40 cycles (30 s at 94 °C, 30 s at 50 °C, and 2 min at 68 °C). All genomic DNAs from carnivores were amplified with primers upstream to the syncytin-Car1 ORF within the provirus and downstream to the 3′ LTR of the provirus within the flanking region. All genes could be amplified with the same primers except for Ailurus fulgens and Mephitis mephitis, where the reverse primer was different (all primers listed in Table S1). Manis longicaudata and Manis javanica genomic DNAs were tentatively amplified with primers conserved in the dog and cat sequences either close to the start and stop codons of the syncytin-Car1 ORF (within the provirus) (Table S1, ORF primers) or external to the provirus (Table S1, locus primers) together with primers internal to the ORF (Table S1, internal primers) and conserved among all sequenced syncytin-Car1 genes. PCR products were directly sequenced without cloning to avoid low-level mutations introduced by PCR.
Biological Samples, RT-PCR, In Situ Hybridization, and Infection Assays.
See SI Methods.
Supplementary Material
Acknowledgments
We thank H. Gachot for the gift of Vulpes vulpes genomic DNA, F. Catzeflis for the gift of Manis longicaudata tissue, R. Potier for blood samples, F.-L. Cosset for the gift of the phCMV-G plasmid and the HuH7 cell line, S. Blaise and J. Richardson for the gift of A72 and DK cell lines, P. Opolon and O. Bawa for their contribution to the histological analyses, and P. Dessen for assistance in the computing work. We thank Christian Lavialle for comments and critical reading of the manuscript. This work was supported by the Centre National de la Recherche Scientifique and a grant from the Ligue Nationale contre Le Cancer (Equipe Labellisée).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 2184.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JN587088–JN587113).
See Author Summary on page 2206.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115346109/-/DCSupplemental.
References
- 1.Mi S, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 2.Blond JL, et al. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol. 2000;74:3321–3329. doi: 10.1128/jvi.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mallet F, et al. The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology. Proc Natl Acad Sci USA. 2004;101:1731–1736. doi: 10.1073/pnas.0305763101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cáceres M, Thomas JW, Thomas JW. NISC Comparative Sequencing Program The gene of retroviral origin Syncytin 1 is specific to hominoids and is inactive in Old World monkeys. J Hered. 2006;97:100–106. doi: 10.1093/jhered/esj011. [DOI] [PubMed] [Google Scholar]
- 5.Blaise S, de Parseval N, Bénit L, Heidmann T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc Natl Acad Sci USA. 2003;100:13013–13018. doi: 10.1073/pnas.2132646100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaise S, Ruggieri A, Dewannieux M, Cosset F-L, Heidmann T. Identification of an envelope protein from the FRD family of human endogenous retroviruses (HERV-FRD) conferring infectivity and functional conservation among simians. J Virol. 2004;78:1050–1054. doi: 10.1128/JVI.78.2.1050-1054.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mangeney M, et al. Placental syncytins: Genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. Proc Natl Acad Sci USA. 2007;104:20534–20539. doi: 10.1073/pnas.0707873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dupressoir A, et al. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc Natl Acad Sci USA. 2005;102:725–730. doi: 10.1073/pnas.0406509102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heidmann O, Vernochet C, Dupressoir A, Heidmann T. Identification of an endogenous retroviral envelope gene with fusogenic activity and placenta-specific expression in the rabbit: A new “syncytin” in a third order of mammals. Retrovirology. 2009 doi: 10.1186/1742-4690-6-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dupressoir A, et al. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc Natl Acad Sci USA. 2009;106:12127–12132. doi: 10.1073/pnas.0902925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunlap KA, et al. Endogenous retroviruses regulate periimplantation placental growth and differentiation. Proc Natl Acad Sci USA. 2006;103:14390–14395. doi: 10.1073/pnas.0603836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leiser R, Kaufmann P. Placental structure: In a comparative aspect. Exp Clin Endocrinol. 1994;102:122–134. doi: 10.1055/s-0029-1211275. [DOI] [PubMed] [Google Scholar]
- 13.Enders AC, Carter AM. What can comparative studies of placental structure tell us?—A review. Placenta. 2004;25(Suppl A):S3–S9. doi: 10.1016/j.placenta.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 14.Carter AM, Enders AC. Comparative aspects of trophoblast development and placentation. Reprod Biol Endocrinol. 2004 doi: 10.1016/j.placenta.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wooding P, Burton G. Placentation Fundamentals. Comparative Placentation: Structures, Functions and Evolution. Berlin: Springer; 2008. pp. 1–46. [Google Scholar]
- 16.Bininda-Emonds ORP, et al. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- 17.Lindblad-Toh K, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 18.Pontius JU, et al. Initial sequence and comparative analysis of the cat genome. Genome Res. 2007;17:1675–1689. doi: 10.1101/gr.6380007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mullikin JC, et al. Light whole genome sequence for SNP discovery across domestic cat breeds. BMC Genomics. 2010 doi: 10.1186/1471-2164-11-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li R, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463:311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eizirik E, et al. Pattern and timing of diversification of the mammalian order Carnivora inferred from multiple nuclear gene sequences. Mol Phylogenet Evol. 2010;56:49–63. doi: 10.1016/j.ympev.2010.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bénit L, Dessen P, Heidmann T. Identification, phylogeny, and evolution of retroviral elements based on their envelope genes. J Virol. 2001;75:11709–11719. doi: 10.1128/JVI.75.23.11709-11719.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vogt VM. Retroviral virions and genomes. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Plainview, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 27–69. [PubMed] [Google Scholar]
- 24.Swanstrom R, Wills JW. Synthesis, assembly, and processing of viral proteins. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Plainview, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 263–333. [PubMed] [Google Scholar]
- 25.Boeke JD, Stoye JP. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Plainview, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 343–435. [PubMed] [Google Scholar]
- 26.Jern P, Coffin JM. Effects of retroviruses on host genome function. Annu Rev Genet. 2008;42:709–732. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- 27.Anderson JW. Ultrastructure of the placenta and fetal membranes of the dog. I. The placental labyrinth. Anat Rec. 1969;165:15–35. doi: 10.1002/ar.1091650103. [DOI] [PubMed] [Google Scholar]
- 28.Leiser R, Koob B. Development and characteristics of placentation in a carnivore, the domestic cat. J Exp Zool. 1993;266:642–656. doi: 10.1002/jez.1402660612. [DOI] [PubMed] [Google Scholar]
- 29.Wooding P, Burton G. Endotheliochorial Placentation: Cat, Dog, Bat. Comparative Placentation: Structures, Functions and Evolution. Berlin: Springer; 2008. pp. 169–184. [Google Scholar]
- 30.Johnson WE, et al. The late Miocene radiation of modern Felidae: A genetic assessment. Science. 2006;311:73–77. doi: 10.1126/science.1122277. [DOI] [PubMed] [Google Scholar]
- 31.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 32.Yoshida I, et al. Change of positive selection pressure on HIV-1 envelope gene inferred by early and recent samples. PLoS One. 2011;6:e18630. doi: 10.1371/journal.pone.0018630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Z, Nielsen R, Goldman N, Pedersen AM. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kosakovsky Pond SL, Frost SD. Not so different after all: A comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 35.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 36.Enders AC. A comparative study of fine structure of trophoblast in several hemochorial placentas. Am J Anat. 1965;116:29–67. doi: 10.1002/aja.1001160103. [DOI] [PubMed] [Google Scholar]
- 37.Georgiades P, Ferguson-Smith AC, Burton GJ. Comparative developmental anatomy of the murine and human definitive placentae. Placenta. 2002;23:3–19. doi: 10.1053/plac.2001.0738. [DOI] [PubMed] [Google Scholar]
- 38.Wooding P, Burton G. Haemochorial Placentation: Mouse, Rabbit, Man, Apes, Monkeys. Comparative Placentation: Structures, Functions and Evolution. Berlin: Springer; 2008. pp. 185–230. [Google Scholar]
- 39.Leiser R, Dantzer V. Structural and functional aspects of porcine placental microvasculature. Anat Embryol (Berl) 1988;177:409–419. doi: 10.1007/BF00304738. [DOI] [PubMed] [Google Scholar]
- 40.Samuel CA, Allen WR, Steven DH. Studies on the equine placenta II. Ultrastructure of the placental barrier. J Reprod Fertil. 1976;48:257–264. doi: 10.1530/jrf.0.0480257. [DOI] [PubMed] [Google Scholar]
- 41.Wooding P, Burton G. Eutheria: Epitheliochorial Placentation Pig and Horse. Comparative Placentation: Structures, Functions and Evolution. Berlin: Springer; 2008. pp. 105–132. [Google Scholar]
- 42.Wooding FB. Current topic: The synepitheliochorial placenta of ruminants: Binucleate cell fusions and hormone production. Placenta. 1992;13:101–113. doi: 10.1016/0143-4004(92)90025-o. [DOI] [PubMed] [Google Scholar]
- 43.Wooding P, Burton G. Synepitheliochorial Placentation: Ruminants (Ewe and Cow). Comparative Placentation: Structures, Functions and Evolution. Berlin: Springer; 2008. pp. 133–168. [Google Scholar]
- 44.Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]