Abstract
Over the past decade, it has become increasingly clear that many tissues have regenerative capabilities. The challenge has been to find the stem cells or progenitors that are responsible for tissue renewal and repair. The revolution in technological advances that permit sophisticated spatial, temporal and kinetic analyses of stem cells has allowed stem cell hunters to ferret out where stem cells live, and to monitor when they come and go from these hiding places.
When cells of mature tissues and organs die, either by natural death or by injury, they must be replenished. This remarkable regenerative feature of adult tissues during tissue homeostasis and in response to injury is a direct result of the activities of stem cells. Adult stem cells have the long-term capacity to self-renew—that is, to divide and create additional stem cells, and also to select a differentiation programme from one or more of a limited number of possible differentiation pathways.
Stem cells are thought to reside within specific anatomical structures or ‘niches’, where they receive signals that maintain them in an undifferentiated state and induce their self-renewal as needed1. The non-stem cell residents of the niche must also be responsive to the needs of the organism so that they can integrate external cues and mobilize the stem cells to exit the niche during homeostatic replacement or on injury. In turn, stem cells are likely to impose signals of their own that also participate in specifying the niche. Identifying the intrinsic and extrinsic signals that balance stem cell proliferation and tissue regeneration is fundamental to understanding cellular diversity and animal survival. This information is also valuable to pinpoint where stem cells reside within tissues and their dynamic actions during regenerative processes. In this review, we will discuss the history and recent advancements made in developing techniques for identifying and monitoring the activity of stem cells within adult tissues.
Identifying stem cells: approaches based on long-term self-renewal and relative quiescence
Given the long-lived nature of stem cells and the long-held hypothesis that they are used sparingly and therefore cycle less frequently when compared with their progeny, putative stem cells were initially identified within a tissue by their ability to retain labelled nucleotides for extended periods of time2. By pulsing mice with 5-bromo-2-deoxyuridine (BrdU) or tritiated thymidine for a sufficient length of time (typically 3–5 days), most proliferative cells within tissues will be labelled. A subsequent long-term chase of the label (typically 2–10 weeks) then allows the highly proliferative cells to dilute the label and the labelled cells that had terminally differentiated to be sloughed from the tissue. The cells that had incorporated label during the pulse, but that divided only a few times if at all, retain the label and map the site(s) of putative long-term slow-cycling stem cells. The technique has been successful in its mission to identify cells that cycle infrequently in the skin and the immune system3–5. In addition, by applying the pulse-chase experiments to epithelial tissues, niches of relatively quiescent cells were identified, including those of the hair follicle bulge6 and the limbus of the eye7.
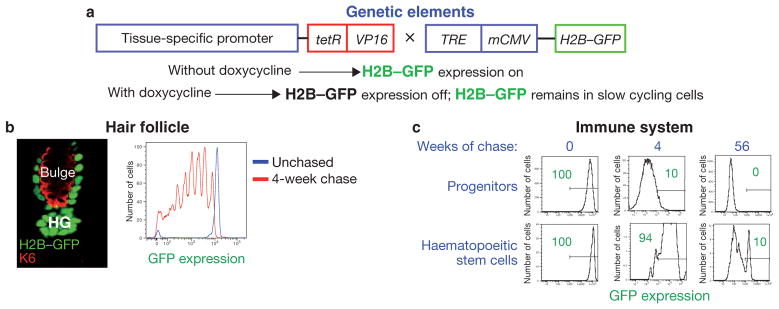
It took many years to demonstrate that label-retaining cells (LRCs) identified in this manner possess the properties of stem cells. Although powerful, this method came with potential challenges. One was the methodology itself, which relies on a stem cell population having at least some actively cycling cells during the nucleotide labelling period. Another was that the cells had to be permeabilized to measure the level of nucleotide retention by fluorescence activated cell sorting (FACS). This precluded the possibility of studying viable stem cells purified on the basis of nucleotide label retention. It wasn’t until years later that these hurdles were overcome by adapting the pulse-chase concept by using the expression of a tissue-specific, tetracycline-inducible histone, tagged with green fluorescent protein (H2B–GFP; Fig. 1a)8.
Figure 1.
Regulated expression of histone H2B–GFP to follow slow-cycling cells within a tissue. (a) Schematic representation of the genetic strategy to mark slow-cycling cells with GFP-labelled histone H2B. Transgenic mice harbouring a tissue-specific promoter driving the tetracycline-repressor (tetR)–VP16 transgene are crossed to transgenic mice expressing a tightly regulated tetracycline-responsive regulatory element (TRE)–mCMV–H2B–GFP element. Without doxycycline, H2B–GFP is uniformly present in the nuclei of all cells within the tissue of interest. With doxycycline, H2B–GFP expression is inhibited, resulting in a dilution of existing fluorescence by 2-fold with each division. By chasing for different time periods in presence of doxycycline, only slow-cycling stem cells or long-lasting, non-dividing, terminally differentiated cells of the tissue will remain labelled. (b) Histology (left) and fluorescence activated cell sorting (FACS; right) analysis of H2B–GFP label retention in the hair follicle when Keratin 5 (K5) is the tissue-specific promoter. Left: at the start of new hair growth, green nuclei, reflective of LRCS, can be detected within the stem cells of the outer layer of the hair follicle bulge (the inner layer is marked by keratin 6; K6), as well as in the hair germ (HG) at the base of the bulge. Right: by analysing GFP expression during a 4-week chase, populations of cells exhibiting a 2-fold reduction in GFP fluorescence, reflective of cell division, can be detected by FACS. (c) FACS analysis of H2B–GFP label retention in the immune system. Haematopoietic progenitors lose H2B–GFP retention after 56 weeks of chase, whereas 10% of stem cells retain label at this time point (green numbers and the lines represent percentage of cells expressing GFP). Images courtesy of Ya-Chieh Hsu, Tudorita Tumbar and Hanno Hock.
In the first application of this kind, slow-cycling H2B–GFP-expressing LRCs at the base of the ‘bulge’ niche of each hair follicle of mouse skin were shown to be activated at the start of a new hair cycle (Fig. 1b). As they exited the niche and proliferated, they diluted out fluorescence by a factor of two with each cell division. And in response to wounding, LRCs from the top of the bulge were mobilized to move upward8. Moreover, with the aid of cell-surface markers, slow-cycling bulge cells could be purified, transcriptionally profiled, cultured and passaged long-term in vitro. Most importantly, when engrafted onto immunocompromised (Nude) mice in vivo, these LRCs were documented to be bona fide multipotent stem cells, able to generate the full set of epithelial tissues found in the skin including the epidermis, hair follicles and sebaceous glands8–10.
In effect, the use of a stable fluorescent histone has transformed the pulse-chase experiment into a lineage tracing method, and it has enabled researchers to monitor not only the slow-cycling stem cells but also their progeny11,12. For some stem cells, such as those of the hair follicle bulge, slow cycling properties emerge in the embryo while the tissue is still undergoing morphogenesis13. With the newfound ability to transcription-ally profile both stem cells and early progeny, the system has become a powerful one for unearthing new features of slow-cycling stem cells.
Haematopoietic stem cells share a number of interesting parallels with hair follicle stem cells, including the ability to cycle slowly. The existence of stem cells within mammalian bone marrow was demonstrated 50 years ago when Till and McCulloch investigated the reconstitution of the haematopoietic system following irradiation2. Judged initially by laborious but effective serial transplantation, it was found that < 1% of bone marrow cells possess long-term regenerative capacity, and most of these cells seemed to be quiescent and in the G0 phase of the cell cycle3,14–16. Despite their quiescence, many of these cells could be labelled when the tissue was exposed to tracer nucleotides for several days17,18, suggesting that they do enter into the cell cycle during the pulse period. When the label was subsequently chased, it was discovered that the majority of haematopoietic stem cells are undergoing a constant, but slow rate of cycling15,16. By contrast, so-called multipotent progenitors divide more frequently, a property that for the haematopoietic system correlates with a loss of stemness.
When the H2B–GFP pulse-chase system was customized for haematopoiesis, new aspects of dormancy and proliferation in haematopoietic stem cells emerged (Fig. 1c)19,20. By labelling during embryogenesis and then chasing postnatally for several months, two different groups uncovered a population of haematopoietic stem cells that retain H2B–GFP label and are induced to proliferate only in response to major injury — either chemically or irradiation-induced. This population would probably not be detected by labelling procedures that initiate the pulse period in adult mice, as these cells divide rarely, if at all, under normal homeostasis.
Overall, these findings raise the possibility that for the haematopoietic system two populations of stem cells exist: a slow-cycling population of long-term haematopoietic stem cells that contributes to homeostasis and a dormant population that is set aside for use in severe injury conditions19,20. Recent studies on the hair follicle suggest that the phenomenon could also apply to this tissue, where the dormant, old stem cell niche from the prior hair cycle no longer contributes to homeostasis, but can be mobilized to proliferate on wounding12.
For the bone marrow, these types of pulse-chase experiments not only illustrated the unique longevity and regenerative properties of haematopoietic cells, but they also gave researchers a handle to mark them in their native niche and further characterize them. Haematopoietic stem cells are now typically distinguished not only by their relative quiescence but also by cell-surface markers (Lin−Sca1+ckit+CD150+CD48−)19–21. Between 20% and 50% of cells purified from the bone marrow in this way exhibit repopulation activity when serially transplanted in vivo.
Are haematopoietic and hair follicle bulge stem cells truly slow cycling or are there other explanations for label retention? A model dating back to the 1970s had posited that stem cells asymmetrically segregate their DNA, keeping the master template DNA strand and passing the less desirable copy strands to the committed daughters22. The similar results obtained when using histone and nucleotide tracers on stem cells argue against this model, as histones and DNA are thought to segregate by different mechanisms during normal cell divisions. Indeed, more recent studies involving multiple nucleotide analogues that mark and track the fates of both the mother and the newly synthesized daughter strands do not favour a master template strand for haematopoietic stem cells or hair follicle stem cells11,16,23. That said, the behaviours of stem cells in murine skeletal muscle and in the Drosophila female germline seem to fit the model24–26, leaving the debate open for some stem cell pools.
Lineage tracing
Not all stem cells divide slowly, and even for the ones that do, approximately only 5–7 divisions of the label-retaining stem cell or of its progeny can be monitored after a pulse-chase before the label has diluted out to the point where it can no longer be traced. Marking a stem cell so that its progeny can be permanently tagged and monitored can be performed by infecting the cell with a replication-defective retrovirus27 or using dyes28. However, to achieve specificity by these methods, the stem cells usually need to be purified first, then marked and returned to the animal’s tissue. This approach works quite effectively for haematopoietic stem cells, which home back to the bone marrow when re-introduced into the animal through tail vein injections29,30. However, most engraftment techniques induce a wound response, which can elicit quite a different behaviour from the stem cells than during normal homeostasis.
Generating a permanent genetic mark in a stem cell in situ has proven to be an effective method for long-term lineage tracing of cells within a tissue. These tools were adapted from site-specific recombination systems of yeast or bacteriophage to organisms containing tissues31,32. A number of techniques are now at hand to genetically mark and locate stem cells within tissues in Drosophila and mice.
The use of heat shock to control the activity of promoters in Drosophila enables cellular descendants of a singly marked cell to be monitored. This strategy has been used to identify stem cells within the Drosophila ovary34 and posterior midgut35. Heat-shock-mediated expression of the yeast FLP (‘flippase’) recombinase33 induces FLP-catalysed site-specific recombination specifically in cycling cells. When recombination is induced between complementary inactive alleles, an active reporter gene is produced that is then expressed in all recombinant cells and their progeny, permanently marking them (for example, the α-tubulin promoter becomes fused to the lacZ gene, the expression of which can be easily monitored by β-galactosidase assays).
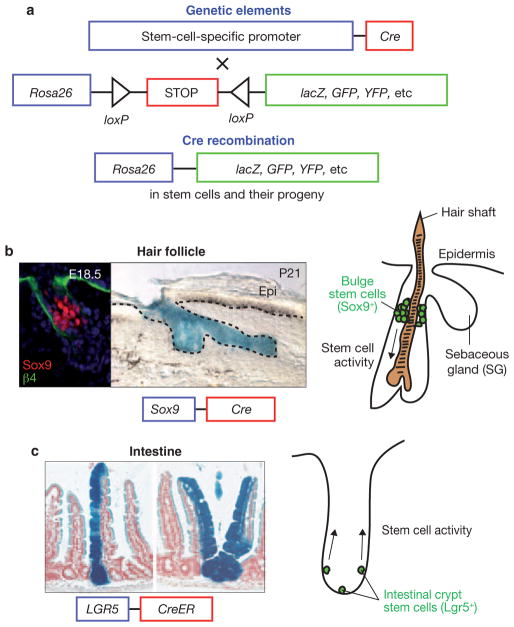
Similar types of genetic technologies have been used in mice to induce recombination within specific cell populations based on promoter expression. The archetypal strategy has been to engineer mice to express an inducible Cre recombinase in the desired cell type of choice, and then mate them to a mouse in which the ubiquitously expressed Rosa26 gene36 has been genetically manipulated so that positioned downstream from the Rosa26 promoter is a stop codon (or secondary gene) flanked by Cre-recombinogenic loxP sites followed by a reporter gene (see Fig. 2 for schematic representation of strategy and examples of its use in lineage tracing). Once the Rosa26 locus has been recombined, reporter activity is faithfully transmitted to all progeny.
Figure 2.
Genetic lineage tracing mediated by Cre recombinase in mammalian tissues. (a) Schematic representation of the genetic strategy to lineage-trace stem cells. The genetic background of one animal is altered such that a specific promoter is used to induce Cre recombination in stem cells. These mice are crossed to a reporter animal harbouring a stop codon flanked by Cre-recombinogenic loxP sites upstream of a reporter gene, such as lacZ or GFP, under the control of the Rosa26 promoter. In mice expressing both genetic elements, Cre recombinase excises the stop codon, such that Rosa26 drives expression of the reporter in stem cells. Once marked in this way, all descendants propagate the expression of the reporter under Rosa26 control. (b) Genetic lineage tracing in the hair follicle using the Sox9 promoter driving expression of Cre recombinase and a Rosa26–lacZ reporter. Sox9 is expressed within the β4-integrin-positive developing hair follicles (left; image of mice at embryonic day (E) 18.5). In mice expressing Cre recombinase under the control of the Sox9 promoter, follicular bulge cells are labelled within 2 days and contribute to multiple follicle lineages after 21 days (middle; Epi, epidermis). These types of studies have demonstrated that the bulge is the bona fide niche for stem cells in the hair follicle. (c) Genetic lineage tracing in the intestine using the Lgr5 promoter driving an inducible Cre recombinase linked to the oestrogen receptor and a Rosa26–lacZ reporter. By inducing nuclear translocation of Cre with tamoxifen, Lgr5-expressing cells give rise to progeny that encompass all cell types in the intestinal crypt, as indicated by blue label (left). Images courtesy of Jonathan Nowak, Ya-Chieh Hsu and Nick Barker.
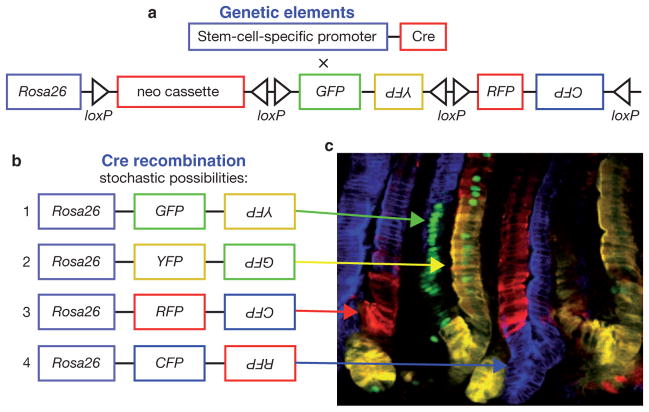
Since its first application in mice32, this theme has seen many variations. The approach has now been used effectively to mark and identify many different types of long-term, self-renewing stem cells. One of the most elaborate examples is the ‘brainbow’ mouse, which was created by applying the Cre/loxP recombination system to create a stochastic choice of expression between up to four different fluorescent proteins (XFPs) from one transgene37 (Fig. 3a,b). When multiple copies of transgenes exist, combinatorial expression can be achieved, providing hundreds of possible fluorescing states for each marked cell within a population. These different colour combinations in labelled cells are achieved through Cre recombination, as stochastic recombination leads to excision and inversion of genetic elements flanked by loxP sites (Fig. 3). Although initially used to directly visualize synaptic interactions and distinguish the neurons converging onto a postsynaptic cell, Brainbow is ideal for stem cell lineage analysis to mark neighbouring stem cells with distinct colours and monitor their progeny37.
Figure 3.
Multicolour Cre-recombinase-mediated reporter for marking stem cells and their progeny. (a) Schematic representation of the genetic strategy to mark stem cells with multiple fluorescent proteins. One animal harbours a transgene encoding a stem-cell-specific promoter driving Cre recombinase expression. These mice are crossed to a reporter animal that, under the control of the ubiquitous Rosa26 promoter, harbour a neomycin resistance gene flanked by Cre-recombinogenic loxP sites, and multiple genes, in sense and antisense orientations, encoding the fluorescent proteins, GFP, RFP, YFP and CFP (green, red, yellow and cyan fluorescent protein, respectively) that are flanked by Cre-recombinogenic loxP and inversion sites. (b) In mice expressing both genetic elements from a, Cre recombinase stochastically excises and inverts at the loxP sites to generate the possible transgenes shown, and allow Rosa26 to drive expression of multiple combinations of fluorescent proteins in stem cells and their progeny. (c) Multicolour lineage tracing in the intestine using the Ah promoter to drive an inducible Cre recombinase. Following induction, Ah-expressing cells are marked with multiple colours in the intestinal crypt based on the stochastic recombinogenic activity of Cre recombinase. Arrows indicate the result of genetic events in b (green, 1; yellow, 2; red, 3; blue, 4). The monoclonal nature of the crypts over time suggests that stem cells stochastically adopt stem cell fates. Image courtesy of Hugo Snippert and Hans Clevers.
Nevertheless, to delineate the behaviour of individual stem cells within a tissue, two major challenges exist for lineage tracing approaches. The first is to engineer mice to express Cre under the control of a promoter that is active in only the desired stem cells. The second is to titrate down the activation of Cre within the stem cell niche such that on average, only one cell per niche is marked. In this utopian situation, the initially marked stem cell will be long-lived and if it does not depart from its niche, it will be recognizable on the basis of its mark and its location. Over time, if it undergoes self-renewal within the niche, a small clone of marked cells will develop that will continue to increase in size over time. By contrast, the progeny are expected to be shorter lived, leaving a trail of transiently labelled cells emanating from the niche, enabling a tracing of where they go and what lineages they differentiate into. This type of analysis was recently employed in the hair follicle, demonstrating that self-renewal and differentiation are separated temporally in follicular stem cells38.
Although this study is an excellent example of single-cell tracing within a tissue, similar analyses have rarely been achieved in other tissues. Most promoters that are active in stem cells are not exclusive to these cells, resulting in a heterogeneous group of stem cells and early progeny that are marked. In addition, long-term quiescent stem cells or stem cells that leave the niche are difficult to reveal with this method. Finally, although many stem cell progeny are replaced rapidly by stem cell activity, others may persist for many months, making it difficult, if not impossible, to distinguish long-lived progeny from stem cells. These caveats aside, lineage tracing has been an effective means of identifying stem cells, particularly when conducted in conjunction with other methods. The method has also been helpful in evaluating the longevity of a particular type of stem cell, its term of residence within the niche, where it goes when it leaves the niche, and what its progeny do during normal homeostasis and/or wound-repair.
One of the most elegant examples using this kind of lineage tracing to track stem cells is that by the Clevers lab, who knocked an EGFP–ires–CreERT2 transgene into the Lgr5 locus. Mice carrying this locus were then crossed to Rosa26–fl–stop–fl–lacZ reporter mice, and the resulting strains were treated with tamoxifen to activate Cre in only a small number of Lgr5-expressing stem cells at the base of the intestinal crypts, which resulted in excision of the floxed stop codon in the Rosa26–lacZ reporter. GFP+ and βgal+ stem cells remained localized, but over time entire discrete crypts were found to be βgal+, but did not express GFP39, suggesting that they were progeny of the Lgr5-expressing cells (Fig. 2c).
The simplest interpretation of these data is that Lgr5 marks the intestinal stem cells and that an individual stem cell can give rise to an individual crypt unit. To test this hypothesis further, Clevers and colleagues recently employed the Brainbow technology. After inducing Cre expression in Lgr5-expressing stem cells, the resulting mix of colours revealed that intestinal stem cells do indeed generate clonality in the small intestine40 (Fig. 3b,c). In the future, applying the Brainbow methodology to interfollicular epidermis and other tissues showing continuous turnover is likely to reveal the dynamics of constantly self-renewing stem cells and their contribution to homeostasis and wound repair.
Lgr5 has turned out to be a broadly expressed marker of stem cells. It is expressed in many other epithelial stem cell niches, including hair follicle, stomach and colon9,41–44. In the hair follicle, Lgr5 is most highly expressed in the slow-cycling CD34+α6+ stem cells of the bulge, but it is also expressed in the activated stem cells and committed progeny of the hair germ and regenerating follicle44. In the resting hair follicle, the expression pattern of Lgr5 mirrors that of a transgene, K15–Cre–PR, that has been used for lineage tracing of hair follicle stem cells12,41. Although perceived discrepancies in the cycling rates of these populations had initially suggested the existence of two populations, this now seems to have been rooted in the timing at which the analyses were conducted and the breadth of the populations tested6,8,9,42. Recent studies demonstrate that the stem cells that are responsible for hair follicle regeneration exist in a quiescent state throughout much of the hair cycle, and are activated only briefly, shortly after the start of each cycle12. Thus, Lgr5 can mark stem cells whether they are fast or slow cycling.
Recently, in a lineage tracing experiment using Lrg5-Cre mice to label the progeny of hair follicle stem cells that have moved away from the bulge, researchers discovered that committed cells that have progressed along the lineage and have lost stemness are nevertheless able to home back to their niche. Interestingly, when back in the niche, these non-stem cells transmit critical inhibitory signals to the stem cells that keep them in a quiescent state even in the presence of positive stimuli and raise the threshold of activators needed to initiate the next round of stem cell activation and hair production12. Whether other stem cells use such mechanisms in controlling tissue homeostasis is not known, but it would seem to be as important for tissue homeostasis and wound repair to instruct stem cells when to stop making tissue as it is to tell them when to start. Moreover, by imposing both stimulatory and inhibitory signals on the stem cell niche, the balance between quiescence and activation of its residents is placed under the dynamic control of the microenvironment.
In tissues without cyclical regeneration or a suitable stem cell promoter, combined expression of markers can sometimes be used for intersectional lineage tracing to trace specific populations of stem cells and their progeny. For example, using a combination of FLP and Cre recombinase technology has allowed the identification of the neural cell types composing the serotonin system based on the intersection of Engrailed and Pet1 expression45. Another method of unbiased clonal sampling is to infect a desired tissue or organ, such as the brain, from a Rosa26-reporter mouse with a Cre-expressing retrovirus. At low dose, the virus randomly infects individual cells within the tissue, and if the cell has the ability to divide at least once to permit integration into the genome, diverse stem cell types can be identified even when there is no prior knowledge of the stem cell46. As these methods evolve, the identification of stem cells in different tissues is bound to continue to expand.
Transitioning from snapshots to movies: monitoring stem cells in action
Despite its many advantages, a single lineage tracing method only offers a snapshot of stem cells and their extended families. By combining and staggering the timing of multiple lineage tracing methods, a temporal series of time lapse snapshots can be strung together to make the beginnings of a movie that will trace stem cells in action12,47. The ideal movie, however, is live imaging of stem cells in their native tissue. Some emerging new technologies suggest that this can be achieved, at least with certain stem cell systems.
The use of microscopy as a means for lineage analysis is an old idea. Direct observation of Caenorhabditis elegans development allowed researchers to map the entire lineage relation of the cells within the organism48. The ability to visualize cells in situ requires an organism that is optically clear and a tissue with sufficiently slow cell divisions to apply conventional time-lapse microscopy approaches. When genetic approaches are then applied so that progenitors are fluorescently marked, confocal time-lapse microscopy permits capturing images over consecutive cell divisions. Recently, such a strategy was used to successfully monitor the cell divisions in the developing brain of live zebrafish embryos49. By imaging each individual cell over a period of 9 hours, researchers were able to unambiguously distinguish those divisions that generated two neural progenitors (symmetric proliferative divisions) from those generating a neuron and a progenitor (asymmetric cell divisions).
When imaging was performed with a Par3–GFP fusion protein of the apical polarity complex, it was clear that asymmetric inheritance of this complex tracks with the progenitor and not the committed neuronal progeny. Although this was established previously in Drosophila neuroblasts, the beautiful study by Clarke and co-workers provided important documentation that asymmetric inheritance of the apical complex is also a powerful determinant of cell fate in the developing vertebrate central nervous system.
Haematopoietic stem cells have also been imaged in their native niche, the bone marrow. Two recent studies have used confocal and two-photon microscopy to follow haematopoietic stem cells in real time during regenerative processes50,51. A variation on this theme has been to combine lineage analysis with live imaging. This approach recently revealed that the previous markers attributable to murine spermatogonial stem cells actually define a heterogeneous population that includes a high proportion of transit amplifying cells, the progeny of stem cells52. This analysis also uncovered signs of spermatogonial cyst fragmentation and fate reversal, and showed that these phenomena increase in response to tissue damage. The study demonstrates the power of combining lineage tracing and live imaging to uncover insights into the physiology of stem cells within their niches.
CONCLUSIONS
The search to understand the regenerative potential within our tissues drives the hunt for stem cells in their native environments. Within recent years, the advancement of label-retention and lineage-tracing methodologies have unearthed the locations of many new stem cell niches and led to our enhanced characterizations of the intrinsic properties of these unique cells and the impact of their niches on their behaviour. Each approach has brought new and refined insights into these biologically fascinating and clinically important residents of adult tissues. Combining these technologies with live imaging to visualize stem cells in action within their native environment is now casting new light on the dynamic processes of stem cells as they exit their niche, migrate, differentiate, and home back again. Many common principles about stem cells have emerged as well as a number of distinct differences. As refinements are made in the tools described in this review, novel insights into the behaviour of these youthful and resourceful cells will most surely be gathered at the dazzling pace that we have now come to expect in the stem cell field.
Acknowledgments
We would like to thank our colleagues in the stem cell field whose ingenuity and creativity have developed these technologies for stem cell biology. In particular, we thank T. Tumbar (Cornell University, USA), H. Hock (Harvard Medical School, Harvard Stem Cell Institute and Cancer Center and Centre for Regenerative Medicine, Harvard University, USA), H. Clevers (Hubrecht Institute, the Netherlands), H. Snippert (Clevers Lab, Hubrecht Institute, the Netherlands), N. Barker (Hubrecht Institute, the Netherlands), Y-C. Hsu (Fuchs lab, Rockefeller University, USA) and J. Nowak (Rockefeller University, USA) for providing images. V.H. is a Pew Scholar in Biomedical Research and is funded by funded by the NIH (4R00AR054775) and the Connecticut Dept. Public Health (09SCAYALE30). E.F. is an HHMI investigator and receives support for her research on the identification and tracking of stem cells from the NIH (R01-AR050452) and New York State.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
Contributor Information
Elaine Fuchs, Howard Hughes Medical Institute, Mammalian Cell Biology and Development, the Rockefeller University, New York, New York 10065, USA.
Valerie Horsley, Email: fuchslb@rockefeller.edu, Department of Molecular, Cellular and Developmental Biology, Yale University, New Haven, Connecticut 06520, USA.
References
- 1.Schofield R. The relationship between the spleen colony-forming cell and the haematopoietic stem cell. Blood Cells. 1978;4:7–25. [PubMed] [Google Scholar]
- 2.Till JE, McCulloch EA. A direct measurement of the radiation sensitivity of normal mouse bone marrow cells. Radiat Res. 1961;14:213–222. [PubMed] [Google Scholar]
- 3.Punzel M, Ho AD. Divisional history and pluripotency of human hematopoietic stem cells. Ann N Y Acad Sci. 2001;938:72–81. doi: 10.1111/j.1749-6632.2001.tb03576.x. discussion 81–82. [DOI] [PubMed] [Google Scholar]
- 4.Potten CS. Keratinocyte stem cells, label-retaining cells and possible genome protection mechanisms. J Investig Dermatol Symp Proc. 2004;9:183–195. doi: 10.1111/j.1087-0024.2004.09305.x. [DOI] [PubMed] [Google Scholar]
- 5.Fuchs E. The tortoise and the hair: slow-cycling cells in the stem cell race. Cell. 2009;137:811–819. doi: 10.1016/j.cell.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cotsarelis G, Sun TT, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990;61:1329–1337. doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- 7.Cotsarelis G, Cheng SZ, Dong G, Sun TT, Lavker RM. Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: implications on epithelial stem cells. Cell. 1989;57:201–209. doi: 10.1016/0092-8674(89)90958-6. [DOI] [PubMed] [Google Scholar]
- 8.Tumbar T, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 10.Claudinot S, Nicolas M, Oshima H, Rochat A, Barrandon Y. Long-term renewal of hair follicles from clonogenic multipotent stem cells. Proc Natl Acad Sci USA. 2005;102:14677–14682. doi: 10.1073/pnas.0507250102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waghmare SK, et al. Quantitative proliferation dynamics and random chromosome segregation of hair follicle stem cells. EMBO J. 2008;27:1309–1320. doi: 10.1038/emboj.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu YC, Pasolli HA, Fuchs E. Dynamics between stem cells, niche, and progeny in the hair follicle. Cell. 2011;144:92–105. doi: 10.1016/j.cell.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nowak JA, Polak L, Pasolli HA, Fuchs E. Hair follicle stem cells are specified and function in early skin morphogenesis. Cell Stem Cell. 2008;3:33–43. doi: 10.1016/j.stem.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 15.Cheshier SH, Morrison SJ, Liao X, Weissman IL. In vivo proliferation and cell cycle kinetics of long-term self-renewing hematopoietic stem cells. Proc Natl Acad Sci USA. 1999;96:3120–3125. doi: 10.1073/pnas.96.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiel MJ, et al. Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature. 2007;449:238–242. doi: 10.1038/nature06115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jetmore A, et al. Homing efficiency, cell cycle kinetics, and survival of quiescent and cycling human CD34(+) cells transplanted into conditioned NOD/SCID recipients. Blood. 2002;99:1585–1593. doi: 10.1182/blood.v99.5.1585. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425:836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- 19.Foudi A, et al. Analysis of histone 2B–GFP retention reveals slowly cycling hematopoietic stem cells. Nat Biotechnol. 2009;27:84–90. doi: 10.1038/nbt.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson A, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 21.Ema H, et al. Adult mouse hematopoietic stem cells: purification and single-cell assays. Nat Protoc. 2006;1:2979–2987. doi: 10.1038/nprot.2006.447. [DOI] [PubMed] [Google Scholar]
- 22.Cairns J. Cancer and the immortal strand hypothesis. Genetics. 2006;174:1069–1072. doi: 10.1534/genetics.104.66886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sotiropoulou PA, Candi A, Blanpain C. The majority of multipotent epidermal stem cells do not protect their genome by asymmetrical chromosome segregation. Stem Cells. 2008;26:2964–2973. doi: 10.1634/stemcells.2008-0634. [DOI] [PubMed] [Google Scholar]
- 24.Shinin V, Gayraud-Morel B, Gomes D, Tajbakhsh S. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nat Cell Biol. 2006;8:677–687. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- 25.Conboy MJ, Karasov AO, Rando TA. High incidence of non-random template strand segregation and asymmetric fate determination in dividing stem cells and their progeny. PLoS Biol. 2007;5:e102. doi: 10.1371/journal.pbio.0050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karpowicz P, et al. The germline stem cells of Drosophila melanogaster partition DNA non-randomly. Eur J Cell Biol. 2009;88:397–408. doi: 10.1016/j.ejcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Price J, Turner D, Cepko C. Lineage analysis in the vertebrate nervous system by retrovirus-mediated gene transfer. Proc Natl Acad Sci USA. 1987;84:156–160. doi: 10.1073/pnas.84.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Axelrod D. Carbocyanine dye orientation in red cell membrane studied by microscopic fluorescence polarization. Biophys J. 1979;26:557–573. doi: 10.1016/S0006-3495(79)85271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dick JE. Retrovirus-mediated gene transfer into hematopoietic stem cells. Ann N Y Acad Sci. 1979;507:242–251. doi: 10.1111/j.1749-6632.1987.tb45805.x. [DOI] [PubMed] [Google Scholar]
- 30.Dzierzak EA, Papayannopoulou T, Mulligan RC. Lineage-specific expression of a human β-globin gene in murine bone marrow transplant recipients reconstituted with retrovirus-transduced stem cells. Nature. 1988;331:35–41. doi: 10.1038/331035a0. [DOI] [PubMed] [Google Scholar]
- 31.Golic KG. Site-specific recombination between homologous chromosomes in Drosophila. Science. 1991;252:958–961. doi: 10.1126/science.2035025. [DOI] [PubMed] [Google Scholar]
- 32.Lakso M, et al. Targeted oncogene activation by site-specific recombination in transgenic mice. Proc Natl Acad Sci USA. 1992;89:6232–6236. doi: 10.1073/pnas.89.14.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harrison DA, Perrimon N. Simple and efficient generation of marked clones in Drosophila. Curr Biol. 1993;3:424–433. doi: 10.1016/0960-9822(93)90349-s. [DOI] [PubMed] [Google Scholar]
- 34.Margolis J, Spradling A. Identification and behavior of epithelial stem cells in the Drosophila ovary. Development. 1995;121:3797–3807. doi: 10.1242/dev.121.11.3797. [DOI] [PubMed] [Google Scholar]
- 35.Ohlstein B, Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2006;439:470–474. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]
- 36.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 37.Livet J, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 38.Zhang YV, Cheong J, Ciapurin N, McDermitt DJ, Tumbar T. Distinct self-renewal and differentiation phases in the niche of infrequently dividing hair follicle stem cells. Cell Stem Cell. 2009;5:267–278. doi: 10.1016/j.stem.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 40.Snippert HJ, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 41.Morris RJ, et al. Capturing and profiling adult hair follicle stem cells. Nat Biotechnol. 2004;22:411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- 42.Jaks V, et al. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet. 2008;40:1291–1299. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- 43.Barker N, et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 44.Greco V, et al. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell. 2009;4:155–169. doi: 10.1016/j.stem.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jensen P, et al. Redefining the serotonergic system by genetic lineage. Nat Neurosci. 2008;11:417–419. doi: 10.1038/nn2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2008;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klein AM, Nakagawa T, Ichikawa R, Yoshida S, Simons BD. Mouse germ line stem cells undergo rapid and stochastic turnover. Cell Stem Cell. 2010;7:214–224. doi: 10.1016/j.stem.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 48.Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 49.Alexandre P, Reugels AM, Barker D, Blanc E, Clarke JD. Neurons derive from the more apical daughter in asymmetric divisions in the zebrafish neural tube. Nat Neurosci. 2010;13:673–679. doi: 10.1038/nn.2547. [DOI] [PubMed] [Google Scholar]
- 50.Celso CL, et al. Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature. 2009;457:92–96. doi: 10.1038/nature07434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie Y, et al. Detection of functional haematopoietic stem cell niche using real-time imaging. Nature. 2009;457:97–101. doi: 10.1038/nature07639. [DOI] [PubMed] [Google Scholar]
- 52.Nakagawa T, Sharma M, Nabeshima Y, Braun RE, Yoshida S. Functional hierarchy and reversibility within the murine spermatogenic stem cell compartment. Science. 2010;328:62–67. doi: 10.1126/science.1182868. [DOI] [PMC free article] [PubMed] [Google Scholar]