Abstract
Methanol is a large volume industrial chemical and widely used solvent and fuel additive. Methanol’s well known toxicity and use in a wide spectrum of applications has raised long-standing environmental issues over its safety, including its carcinogenicity. Methanol has not been listed as a carcinogen by any regulatory agency; however, there are debates about its carcinogenic potential. Formaldehyde, a metabolite of methanol, has been proposed to be responsible for the carcinogenesis of methanol. Formaldehyde is a known carcinogen and actively targets DNA and protein, causing diverse DNA and protein damage. However, formaldehyde-induced DNA adducts arising from the metabolism of methanol have not been reported previously, largely due to the absence of suitable DNA biomarkers and the inability to differentiate what was due to methanol compared with the substantial background of endogenous formaldehyde. Recently, we developed a unique approach combining highly sensitive liquid chromatography-mass spectrometry methods and exposure to stable isotope labeled chemicals to simultaneously quantify formaldehyde-specific endogenous and exogenous DNA adducts. In this study, rats were exposed daily to 500 or 2000 mg/kg [13CD4]-methanol by gavage for 5 days. Our data demonstrate that labeled formaldehyde arising from [13CD4]-methanol induced hydroxymethyl DNA adducts in multiple tissues in a dose-dependent manner. The results also demonstrated that the number of exogenous DNA adducts was lower than the number of endogenous hydroxymethyl DNA adducts in all tissues of rats administered 500 mg/kg per day for 5 days, a lethal dose to humans, even after incorporating an average factor of 4 for reduced metabolism due to isotope effects of deuterium-labeled methanol into account.
Keywords: DNA adducts, genetic toxicology, formaldehyde, methanol, hydroxymethyl adducts, mass spectrometry
Methanol is a high volume industrial chemical that is widely used as a solvent, fuel, or fuel additive and a starting reagent for the synthesis of other important industrial chemicals, such as formaldehyde (Kavet and Nauss, 1990). It is well documented that acute exposure to high doses of methanol can result in metabolic acidosis, ocular toxicity, and even death in humans (Burkhart, 1997; Liesivuori and Savolainen, 1991; Martin-Amat et al., 1978; Agency for Toxic Substances and Disease Registry, 1993). These toxicities, coupled with wide spread human exposure has raised long-standing public concerns over its safety, including its developmental toxicity and carcinogenesis. Although developmental toxicity has been demonstrated in rodents (Rogers and Mole, 1997; Rogers et al., 2004), there remain debates regarding the potential carcinogenicity of methanol.
Methanol currently is not classified as a carcinogen by any regulatory body or the International Agency for Research on Cancer (Cruzan, 2009). No epidemiology studies have been reported for methanol-exposed cohorts. Several animal cancer bioassays have been conducted and widely discussed in the literature (New Energy Development Organization [NEDO], 1985a,b, 1987; Soffritti et al., 2002). The Japanese NEDO performed inhalation carcinogenicity studies in rats, mice, and monkeys in 1982–1984, and no positive findings were identified. Another study in Sprague-Dawley rats was conducted by the European Ramazzini Foundation (ERF) (Soffritti et al., 2002). They exposed rats to 0, 500, 5000, and 20,000 ppm of methanol in drinking water for 2 years. Significantly increased incidences for hemolymphoreticular tumors and ear duct carcinomas were reported for animals in the highest dose group (Soffritti et al., 2002). However, their use of conventionally maintained animals, high incidence of lung pathology, and potential misidentification of lung lesions have raised serious concerns about the conclusions drawn from this study (Cruzan, 2009; Schoeb et al., 2009b; Schoeb and McConnell, 2011b).
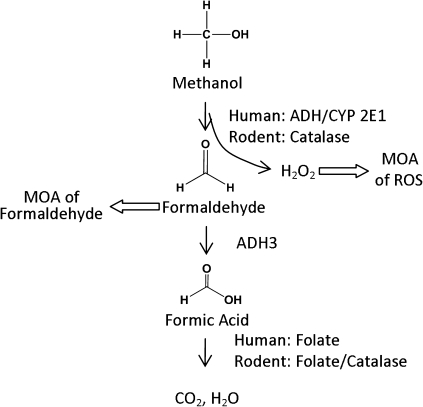
Despite the inconsistent results across the studies and the lack of substantial evidence for the mode of action (MOA) of methanol’s carcinogenesis, two possible mechanisms were proposed (Cruzan, 2009; Sweeting et al., 2010), as shown in Figure 1. The most prevalent hypothesis is that methanol’s metabolism to formaldehyde is responsible for the carcinogenicity of methanol. Methanol is metabolized to formaldehyde in rodents and humans with the participation of different enzyme systems (Clary, 2003; Harris et al., 2003; Johlin et al., 1987). Humans primarily use alcohol dehydrogenase (ADH) and cytochrome P450 2E1 (CYP2E1) to metabolize methanol, whereas rodents use catalase to metabolize methanol to formaldehyde. Another hypothesis is that oxidative DNA damage from reactive oxygen species is generated during the metabolism of methanol by catalase (Cederbaum and Qureshi, 1982).
FIG. 1.
Metabolism of methanol and two proposed MOAs for the carcinogenicity of methanol, adapted from Sweeting et al. (2010).
Formaldehyde is a known carcinogen, causing squamous cell carcinoma in nasal tissue of rats and nasopharyngeal cancer in humans (Kerns et al., 1983; Swenberg et al., 1980). Formaldehyde is a very reactive compound that targets DNA and proteins, causing diverse DNA and protein damage. However, the induction of formaldehyde-induced DNA damage following methanol exposure has not been successfully investigated (Wang et al., 2008), which is largely due to the lack of suitable DNA biomarkers and the interference of substantial endogenous background adducts of formaldehyde. It is well documented that both methanol and its metabolite, formaldehyde, are endogenous metabolites in animals and humans.
Our laboratory has recently developed highly sensitive liquid chromatography-mass spectrometry (LC-MS/MS) methods to analyze formaldehyde-specific DNA biomarkers (Lu et al., 2010a, 2011). Coupled with the use of stable isotope labeled–formaldehyde for exposure, the endogenous and exogenous formaldehyde–induced DNA adducts could be differentiated and accurately measured for the first time. In our previous studies, we have clearly demonstrated that exogenous formaldehyde induced N2-hydroxymethyl-dG adducts, but not N6-hydroxymethyl-dA adducts, in nasal epithelium of rats and monkeys exposed by inhalation to [13CD2]-formaldehyde, whereas endogenous N2-hydroxymethyl-dG and N6-hydroxymethyl-dA were readily detected in all tissues examined (Lu et al., 2010a).
Here, we used a similar approach by exposing rats orally to [13CD4]-methanol through gavage for 5 days, followed by the analysis of DNA adducts using our highly sensitive LC-MS/MS selected reaction monitoring (SRM) methods with a limit of detection of 20–240 amol on column. This study was specifically designed to examine whether or not methanol-derived formaldehyde causes DNA damage in rats. Furthermore, we were interested in evaluating the individual contribution of endogenous formaldehyde and methanol-derived formaldehyde. Using our unique approach, DNA adducts originating from both endogenous and exogenous formaldehyde were quantified simultaneously in rats exposed to different doses of [13CD4]-methanol. We have demonstrated that [13CD4]-methanol–derived formaldehyde DNA adducts were induced in multiple tissues in a dose-dependent manner. Moreover, ratios of exogenous versus endogenous formaldehyde DNA adducts clearly show that endogenous DNA adducts are present in larger amounts than are exogenous DNA adducts in rats treated with 500 or 2000 mg/kg daily for 5 days.
MATERIALS AND METHODS
Chemicals and materials.
Deoxyguanosine, deoxyadenosine, potassium phosphate, Tris-HCl, MgCl2, formic acid, NaCNBH3, methanol, acetonitrile, phosphates, and high-performance liquid chromatography (HPLC) grade water were all purchased from Sigma (St Louis, MO). Twenty percent of formaldehyde in water was procured from Tousimis (Rockville, MD). DNase I, alkaline phosphatase, and phosphodiesterases were purchased from Sigma. [13C10 15N5]-dG was ordered from Cambridge Isotope Lab (Cambridge, MA). N2-CH3-dG and N6-Me-dA was obtained from Berry & Associates (Dexter, MI). All chemicals were used as received unless otherwise stated.
Preparation of internal standards.
A 10mM [13C10 15N5]-dG and [15N5]-dA were mixed with 100mM formaldehyde in phosphate buffer (pH = 7.2) overnight at 37°C. The reaction mixture was separated by HPLC using a 150 × 2.5 mm C18 T3 analytical column. N2-Hydroxymethyl-dG and N6-hydroxymethyl-dA were collected, followed by incubation with 50mM NaCNBH3 (pH = 7.1) overnight at 37°C. The resultant solution was further separated using HPLC. [13C10 15N5]-N2-CH3-dG and [15N5]-N6-CH3-dA eluted at 27.2 and 32.6 min on a 150 × 2.5 mm T3 column. The concentration of [13C10 15N5]-N2-CH3-dG and [15N5]-N6-CH3-dA was determined by HPLC with corresponding unlabeled N2-CH3-dG and N6-Me-dA as references. The conversion rate from hydroxymethyl to methyl groups was ∼65 to 85%.
Exposure experiment.
Eight Sprague-Dawley rats were exposed to [13CD4]-methanol at 500 or 2000 mg/kg/day by gavage for 5 days. Control animals received saline. Liver, lung, kidney, spleen, bone marrow, thymus, brain, and white blood cells (WBC) were collected 6 h following the last dosing, flash frozen, and stored at −80°C until analysis.
DNA isolation, treatment, and digestion.
DNA was isolated from tissues of rats using a NucleoBond DNA Isolation Kit (Bethlehem, PA) according to the manufacturer’s instruction. The isolated DNA was dissolved in water, quantified by UV spectroscopy, and frozen at −80°C for later analysis. About 200 μg of DNA was incubated with 50mM NaCNBH3 at 37°C for 6 h in phosphate buffer (pH = 7.2). Then, DNA was treated with DNase I (200U) for 10 min in the digestion buffer (80mM Tris-HCl and 20mM MgCl2, pH = 7.2), followed by the addition of 25 μl of alkaline phosphatase and 25 μl of phosphodiesterases for an additional hour. Enzymes and undigested DNA were removed by a Millipore Microcon YM-10 spin column, and the resultant solution was separated by HPLC to collect the fractions containing the corresponding DNA adducts.
High performance liquid chromatography.
The purification of formaldehyde-DNA adducts was carried out on an Agilent 1200 series HPLC system equipped with a diode-array detector (Santa Clara, CA). Analytes were separated by reverse phase chromatography using a 150 × 4.6 mm C18 T3 analytical column from Waters (Milford, MA). The mobile phases were 10mM ammonium acetate with 0.1% acetic acid (A) and methanol (B). The gradient was used as follows, at a flow rate of 1 ml/min and monitored at 254 nm: 0 min, 5% solvent B; 5 min, 5% solvent B; 10 min, 8% solvent B; 20 min, 10% solvent B; 30 min, 15% solvent B; 45 min, 30% solvent B; 45.1 min, 5% solvent B; and 60 min, 5% solvent B. N2-Me-dG and N6-Me-dA eluted at 26.5 and 31.7 min on the column in this system, respectively.
Liquid chromatography-mass spectrometry.
LC-MS analyses were performed on a triple quadrupole mass spectrometer TSQ-Quantum Ultra (Thermo Electron, Waltham, MA) operating in SRM mode to detect and quantify hydroxymethyl-DNA adducts. The mass spectrometer was interfaced with a nano-Ultra Performance Liquid Chromatography system from Waters. Mobile phases were comprised of water with 0.1% Acetic Acid (A) or Acetonitrile with 0.1% Acetic Acid (B). Both capillary and nano-flow rates were used to quantify formaldehyde-DNA adducts. For the capillary method, a linear gradient was run from 2 to 60% B over 10 min, at 40 μl/min. The electrospray ionization (ESI) source was set as follows: spray voltage, 4.0 kV; capillary temperature, 300°C; sheath gas pressure, 40 arbitrary units (au); and auxiliary gas pressure, 10 au. For the nano method, analytes were first retained on a trap column with a flow rate of 5 μl/min of 2% mobile phase B, followed by transfer to the analytical column with an initial starting condition of 2% B at 0.6 μl/min for 1 min followed by a linear gradient to 60% B over 14 min. The column was then cleaned at 80% B for 1.5 min followed by a reequilibration for an additional 7.5 min. The analytes were introduced to the MS using positive mode ESI with a source voltage of 2200 V and no additional gasses. The ion transfer tube was held at 325°C and skimmer offset set to zero. Scan speed was set at 0.1 s, scan width at 1.0 m/z, and peak widths for Q1 and Q3 at 0.3 and 0.5 m/z, respectively. Collision energy was set at 17 eV with Argon as the collision gas set at 1.5 au. N2-HOCH2-dG was quantified as N2-CH3-dG after reduction using the transition of m/z 282.2→m/z 166.1, and N2-HO13CD2-dG was quantified as N2-13CD2H-dG with the transition of m/z 285.2→m/z 169.1. Two additional transitions, including m/z 284.2→m/z 168.1 and m/z 283.2→m/z 167.1, were also monitored in case H-D exchange occurred. Similarly, five transitions, including m/z 266.2→m/z 150.1, m/z 269.2→m/z 153.1, m/z 271.2→m/z 150.1, m/z 268.2→152.1, and m/z 267.2→m/z 151.1, were used to detect endogenous, exogenous, internal standard, and possible products after H-D exchange for dA adducts, respectively. The calibration curves for quantitation were obtained using the integrated peak area and amount of injected analytical standard and internal standard.
Statistical analysis.
Data represent mean ± SD. Unpaired Student’s t-tests were performed with the sample size ranging from 4 to 6 for adduct analysis. Differences were considered statistically significant if p < 0.05.
RESULTS
Method and Validation
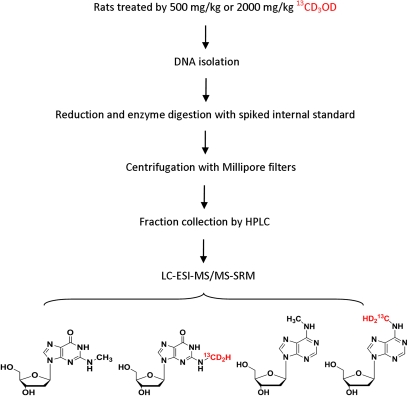
Figure 2 illustrates the outline of the experiment design and analytical approach for the analysis of hydroxymethyl-DNA adducts after their reduction. The method has been described in detail previously. Transitions to monitor potential H-D exchange are extremely important to make peak assignments and accurate quantitation. Therefore, the mass spectrometer was set up to monitor the following transitions: m/z 282.2→m/z 166.1 for N2-CH3-dG; m/z 285.2→m/z 169.1 for N2-13CD2H-dG, and m/z 297.2→m/z 176.1 for internal standard [13C1015N5]-N2-CH3-dG. Two additional transitions, including m/z 283.2→m/z 167.1 and m/z 284.2→m/z 168.1, were used to monitor adducts that may arise after potential H-D exchange. Likewise, for the detection of dA adducts, the mass spectrometer was set up to monitor the following transitions: m/z 266.2→m/z 150.1 for N6-CH3-dA; m/z 269.2→m/z 153.1 for N6-13CD2H-dA; m/z 271.2→m/z 150.1 for internal standard [15N5]-N6-CH3-dA; and m/z 268.2→m/z 152.1 and m/z 267.2→m/z 151.1 for products after H-D exchange.
FIG. 2.
Schematic representation of experiment design and analytical approach for formaldehyde-DNA adducts from endogenous source and [13CD4]-methanol.
The typical calibration curves used for the quantitative measurements of Me-dG and Me-dA adducts, respectively, which have good linearity within three orders of magnitude, are given in Supplementary figure S1. The good precision and accuracy of this assay was validated by spiking known amounts of analytical standards into liver DNA, as shown in Supplementary table S1.
Establishment of Criteria for the Assignment of Adduct Peaks
Hydrogen-deuterium exchange is a well-documented phenomenon for deuterium-labeled compounds. Therefore, potential H-D exchange could occur during exposure, which has been confirmed by a cell culture study using [13CD2]-formaldehyde for exposure (Lu et al., 2011). Therefore, it is critical to establish criteria to make peak assignments and accurate quantitation. For dG adducts, if one deuterium atom was lost, the signal would shift to m/z = 284.2. If two deuterium atoms were lost, the signal of exogenous peak could further move to m/z = 283.2. However, the peak at m/z = 283.2 could be also from the normal isotope distribution of the endogenous peak at m/z = 282.2. Therefore, the ratio of peak283→167 versus peak282→166 needed to be checked to determine if the peak283 is only the consequence of natural abundance of isotopes.
Figure 3 gives the natural abundance of different isotope peaks for spiked analytical standards, as reveled by multiple SRM scans. Me-dG analytical standard could give two isotope peaks at m/z = 284 and 283, whereas Me-dA analytical standard only shows a single isotope peak at m/z = 267. Using analytical standards (n = 20) and control DNA samples (n = 40), the peak ratios were determined to be 8.3 ± 0.6% (peak283/peak282) and 6.5 ± 1.3% (peak284/peak283) for Me-dG adducts. The peak ratio was determined to be 8.4 ± 0.5% (only peak267 and peak266 were observed) for dA adducts. Therefore, 9 and 8% were selected as threshold values to justify the formation of exogenous dG adducts, whereas 9% was used for exogenous dA adducts if H-D exchange occurred. The specific decision rules are illustrated in Supplementary figure S2. Briefly, if a peak at 285 is detected, it could be unequivocally assigned to exogenous dG adducts. If the peak at 285 is absent and peaks at 284 and 283 are detected, the peak ratios are used to make assignments. Similarly, the peaks at 269 and 268 unambiguously identify the formation of exogenous dA adducts, whereas the ratio of 267/266 is used for the identification of adducts if only the peak at 267 is observed. Likewise, the quantitation of DNA adducts also needs to consider the contributions of natural isotope abundance and H-D exchange.
FIG. 3.
Contribution of natural abundance of isotope peaks for 100 fmol N2-methyl-dG (A) and 40 fmol N6-methyl-dA analytical standards injected on column (B).
Formation of N2-Hydroxymethyl-dG DNA Adducts From Endogenous Sources and [13CD4]-Methanol
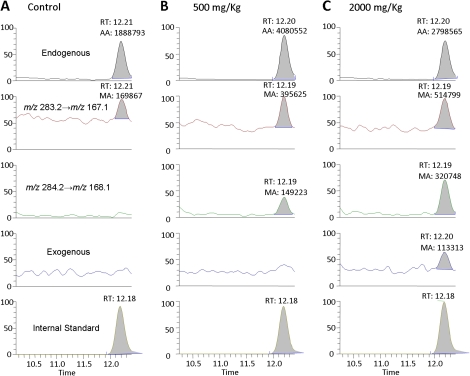
Figure 4 shows the LC-ESI-MS/MS SRM chromatograms of N2-Me-dG DNA adducts in spleen of rats exposed to [13CD4]-methanol for 5 days. The peak corresponding to the specific transition of m/z 282.2→m/z 166.1 and the same retention time with [13C1015N5]-N2-CH3-dG internal standard unambiguously identified endogenous formaldehyde–induced N2-HOCH2-dG, as shown by the peak at 12.2 min in the top panel of Figure 4. The signal corresponding to the transition of m/z 285.2→m/z 169.1 coeluted with the internal standard, which is attributed to N2-HO13CD2-dG arising from exogenous [13CD4]-methanol–derived formaldehyde, as shown in Figure 4C. In addition to these three peaks, two more transitions, including m/z 284.2→m/z 168.1 and m/z 283.2→m/z 167.1, were also monitored to examine whether significant H-D exchange occurred during exposure. There is no peak at m/z 285.2 in the 500 mg/kg dose group, as shown in Figure 4B. However, the peak ratio of 284/283 (∼38%) is significantly higher than 8%, the threshold for the identification of adducts. This clearly demonstrated the formation of [13CD4]-methanol–derived DNA adducts and significant H-D exchange during 5-days exposure. Likewise, the peak ratios of 285/284, 284/283, and 283/282 are 35, 62, and 18% for rats treated with 2000 mg/kg methanol, as shown in Figure 4C. These numbers are all much higher than the cutoff values arising from natural isotope abundance, further supporting the formation of exogenous DNA damage from [13CD4]-methanol–derived formaldehyde and the occurrence of H-D exchange during exposure.
FIG. 4.
Typical LC-ESI-MS/MS SRM chromatograms of N2-Me-dG DNA adducts in spleen of rats exposed to 0 (A), 500 (B) or 2000 (C) mg/kg for 5 days.
Formation of N6-Hydroxymethyl-dA From Endogenous Sources and [13CD4]-Methanol
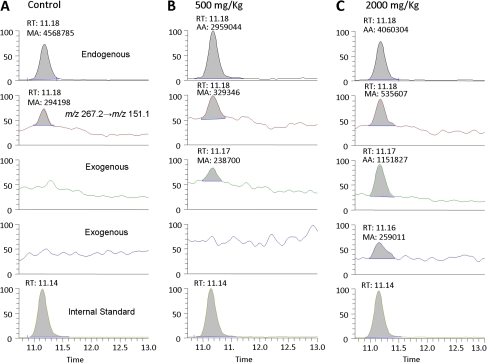
Figure 5 shows the LC-ESI-MS/MS SRM chromatogram of N6-methyl-dA in bone marrow DNA of rats exposed to [13CD4]-methanol for 5 days. The peak corresponding to the specific transition of m/z 266.2→m/z 150.1 and the same retention time with [15N5]-N6-CH3-dA internal standard unequivocally identified endogenous formaldehyde–induced N6-HOCH2-dA, as shown by the peak at 11.1 min in the top panel of Figure 5. The signal corresponding to the transition of m/z 269.2→m/z 153.1 are generally attributed to N6-HO13CD2-dA arising from exogenous [13CD4]-methanol–derived formaldehyde. However, as shown in Figure 5B, no peak was observed for the transition at m/z 269.2→m/z 153.1. The peak at m/z 268 was able to identify the formation of exogenous dA adducts, as we discussed above. Moreover, the ratio of peak268→152 versus peak267→151 was 72%, which further demonstrated this peak was primarily from exogenous exposure instead of the consequence of natural isotope distribution (∼8 to 9%). Similarly, as the consequence of H-D exchange, the peak ratios of 269/268, 268/267, and 267/266 in the sample from the 2000 mg/kg group were 22, 215, and 13%, separately, which are all above the natural abundance of isotope peaks (Fig. 5C).
FIG. 5.
Typical LC-ESI-MS/MS SRM chromatograms of N6-Me-dA DNA adducts in bone marrow of rats exposed to 0 (A), 500 (B) or 2000 (C) mg/kg for 5 days.
Numbers of Hydroxymethyl-DNA Adducts in Rats Exposed to [13CD4]-Methanol
Table 1 summarizes the number of dG adducts in the tissues examined. [13CD4]-methanol–derived exogenous dG adducts were detected in all tissues except brain. It should be noticed that these adduct numbers and others were directly calculated from our mass spectrometry data without any adjustment based on the isotope effect of deuterium-labeled methanol on metabolism, unless otherwise stated. The potential influence of isotope effects on adduct formation and corresponding adjustment are described in the “Discussion” and “Conclusion” sections. The numbers of exogenous N2-hydroxymethyl-dG adducts induced by [13CD4]-methanol were shown to be 0.08 ± 0.08, 0.13 ± 0.04, 0.12 ± 0.04, 0.19 ± 0.12, 0.37 ± 0.08, 0.16 ± 0.06, and 0.09 ± 0.03 adducts/107 dG in liver, lung, kidney, spleen, bone marrow, thymus, and WBC of rats treated with 500 mg/kg/day [13CD4]-methanol, respectively. The exogenous adduct numbers were determined to be 0.41 ± 0.14, 0.22 ± 0.06, 0.39 ± 0.09, 0.90 ± 0.26, 1.42 ± 0.29, 0.42 ± 0.03, and 0.19 ± 0.02 adducts/107 dG in liver, lung, kidney, spleen, bone marrow, thymus, and WBC of 2000 mg/kg/day [13CD4]-methanol–treated rats. Clearly, methanol induced the formation of exogenous dG adducts in a dose-dependent manner. Interestingly, endogenous dG adducts also significantly increased in liver, lung, kidney, spleen, and thymus compared with control, whereas no increase in endogenous dA adducts was observed following exposure.
TABLE 1.
N2-Hydroxymethyl-dG Adducts in Rats Exposed to 0, 500, or 2000 mg/kg Methanol for 5 Days
| Dose | Liver | Lung | Kidney | Spleen | Bone marrow | Thymus | WBC | Brain |
| Control | ||||||||
| Endogenous | 4.35 ± 1.01 | 4.55 ± 1.93 | 4.31 ± 2.4 | 3.70 ± 1.34 | 2.99 ± 0.56 | 2.55 ± 0.37 | 3.32 ± 0.45 | 6.69 ± 2.91 |
| Exogenous | nd | nd | nd | nd | nd | nd | nd | nd |
| 500 mg/kg/day | ||||||||
| Endogenous | 5.66 ± 0.52* | 7.24 ± 1.95 | 8.48 ± 1.50* | 5.85 ± 1.12* | 2.99 ± 0.73 | 3.49 ± 0.12* | 3.65 ± 0.43 | 7.95 ± 2.37 |
| Exogenous | 0.08 ± 0.08 | 0.13 ± 0.04 | 0.12 ± 0.04 | 0.19 ± 0.12 | 0.37 ± 0.08 | 0.16 ± 0.06 | 0.09 ± 0.03 | nd |
| 2000 mg/kg/day | ||||||||
| Endogenous | 8.14 ± 2.03* | 10.32 ± 1.83* | 7.86 ± 2.14* | 4.89 ± 0.69* | 3.34 ± 0.49 | 3.73 ± 0.17* | 3.92 ± 0.25 | 10.38 ± 4.84 |
| Exogenous | 0.41 ± 0.14 | 0.22 ± 0.06 | 0.39 ± 0.09 | 0.90 ± 0.26 | 1.42 ± 0.29 | 0.42 ± 0.03 | 0.19 ± 0.02 | nd |
Note. nd, nondetectable.
*Significantly different from control (p < 0.05).
In contrast, [13CD4]-methanol–derived exogenous dA adducts were only observed in bone marrow and kidney, as listed in Table 2. The number of exogenous N6-hydroxymethyl-dA adducts induced by [13CD4]-methanol were 0.10 ± 0.08 and 0.29 ± 0.09 adducts/107 dA in bone marrow of rats in 500 or 2000 mg/kg/day groups, respectively. The amount of exogenous dA adducts in kidney was detectable but not quantifiable by our capillary LC method. Therefore, a more sensitive nano-LC method was used to quantify adducts in the different regions of kidney, as discussed below. Typically, the numbers of exogenous dA adducts in kidney were ∼0.1 and ∼0.6 adducts/107 dA in rats treated with 500, or 2000 mg/kg [13CD4]-methanol for 5 days. The observation that exogenous dA adducts only formed in bone marrow and kidney may suggest fundamental differences in the formation of dA versus dG adducts, which were found in most examined tissues. In addition, both endogenous dG and dA adducts were readily detected across all tissues analyzed.
TABLE 2.
N6-Hydroxymethyl-dA Adducts in Rats Exposed to 500 or 2000 mg/kg Methanol for 5 Days
| Dose | Liver | Lung | Kidney | Spleen | Bone marrow | Thymus | WBC | Brain |
| Control | ||||||||
| Endogenous | 1.40 ± 0.51 | 1.85 ± 0.19 | 1.19 ± 0.45 | 1.68 ± 0.25 | 1.65 ± 0.85 | 1.05 ± 0.25 | 2.53 ± 0.69 | 2.00 ± 1.59 |
| Exogenous | nd | nd | nd | nd | nd | nd | nd | nd |
| 500 mg/kg/day | ||||||||
| Endogenous | 1.48 ± 0.29 | 1.72 ± 0.28 | 1.33 ± 0.65 | 1.75 ± 0.49 | 1.07 ± 0.37 | 1.24 ± 0.16 | 1.43 ± 0.32 | 1.86 ± 0.61 |
| Exogenous | nd | n.d. | nd | nd | 0.10 ± 0.08 | nd | nd | nd |
| 2000 mg/kg/day | ||||||||
| Endogenous | 1.89 ± 0.44 | 1.75 ± 0.23 | 1.00 ± 0.16 | 1.82 ± 0.54 | 1.01 ± 0.16 | 1.66 ± 0.51 | 1.71 ± 0.30 | 2.08 ± 1.04 |
| Exogenous | nd | nd | Detectablea | nd | 0.29 ± 0.09 | nd | nd | nd |
Note. nd, nondetectable.
Detectable but not quantifiable by our capillary LC method.
Dose-Response of Exogenous DNA Adducts
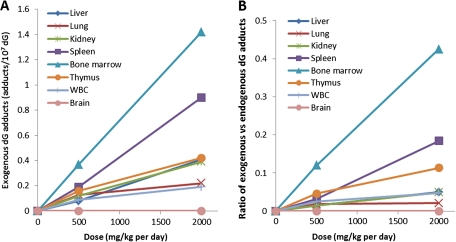
Figure 6A illustrates the exogenous adduct numbers for dG adducts in tissues of rats exposed to 500 mg/kg, or 2000 mg/kg methanol for 5 days. The exogenous dG adducts ranged from 0.08 to 0.37 adducts/107 dG for 500 mg/kg treated rats, whereas 2000 mg/kg methanol exposures induced 0.19–1.42 exogenous adducts/107 dG in different tissues. Bone marrow had the highest amounts of exogenous dG adducts, followed by spleen of rats exposed to methanol. The formation of exogenous dG adducts was less than linear, suggesting saturation of metabolism in lung, kidney, and thymus. An approximately linear dose-response was observed for other organs, as illustrated in Figure 6A.
FIG. 6.
The number of exogenous N2-hydroxymethyl-dG adducts in different tissues (A) and the ratios of exogenous versus endogenous dG adducts (B).
Figure 6B shows the ratios of exogenous versus endogenous dG adducts in multiple tissues, which was calculated using the average adduct numbers of the same dose groups. The ratios give relatively larger values for bone marrow, spleen, and thymus, whereas the numbers for liver, lung, kidney, and WBC are typically 1–3-fold lower. Among all the tissues, the ratios in bone marrow were the largest in both 500 and 2000 mg/kg/day exposure groups. However, exogenous adducts were still less than endogenous adducts in all organs evaluated, even in rats treated with 2000 mg/kg daily for 5 days.
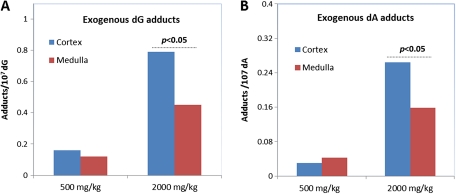
Adduct Formation in Different Regions of Kidney
As methanol is partially excreted through the kidney at high doses, exogenous DNA adducts were analyzed to evaluate adduct distribution within the kidney. Figure 7 illustrates the difference for the formation of exogenous adducts at the different locations of kidney. There was no statistically significant difference between cortex and medulla for either exogenous dG or dA adducts in rats exposed to 500 mg/kg methanol for 5 days (Fig. 7A). However, more exogenous dG and dA adducts formed in cortex than medulla when rats were exposed to 2000 mg/kg methanol for 5 days (p < 0.05, Fig. 7B). This interesting observation suggests differential exposure, metabolism, or excretion of methanol in kidney as the dose increases.
FIG. 7.
Formation of exogenous hydroxymethyl-dG (A) and -dA adducts (B) in different regions of kidney.
DISCUSSION AND CONCLUSIONS
In this study, we have applied our highly sensitive LC-ESI-MS/MS SRM methods to detect hydroxymethyl-DNA adducts in rats exposed to 500 mg/kg, or 2000 mg/kg [13CD4]-methanol for 5 days. Both exogenous and endogenous DNA adducts were quantified simultaneously in the same samples. Our results show exogenous hydroxymethyl-dG adducts formed in multiple tissues we examined. In addition, we have demonstrated that [13CD4]-methanol induced exogenous dA adducts in kidney and bone marrow. Moreover, the ratios of exogenous versus endogenous DNA adducts indicate that endogenous hydroxymethyl-DNA adducts are greater than those generated from [13CD4]-methanol.
Methanol currently is not listed as a carcinogen by the International Agency for Research on Cancer, the National Toxicology Program (NTP), or the U.S. Environmental Protection Agency (EPA). Likewise, there have been no evaluations of cancer in human epidemiology studies. Discussions on the potential carcinogenicity of methanol are largely focused on several animal cancer bioassays, including NEDO and ERF studies (NEDO, 1985a,b, 1987; Soffritti et al., 2002). However, conflicting results have been reported from these long-term rodent cancer bioassays. Thus, the carcinogenicity of methanol remains unclassifiable.
The Japanese NEDO conducted a series of bioassays in rats, mice, and monkeys following inhalation exposure (NEDO, 1985a,b, 1987). Among these studies, a 24-month carcinogenicity bioassay was conducted, in which 52 F344 rats/sex/group were exposed to 0, 10, 100, or 1000 ppm methanol vapor. In the original reports by NEDO, no statistical significance on cancer induction was reported due to exposure to methanol. However, recently, EPA reanalyzed the results and identified potential treatment-related changes in the lungs of male rats and the adrenal medulla of female rats in the more detailed translation of the original report. Specifically, the observed incidence rate for pulmonary adenoma/adenocarcinoma in high-dose males of 13.5% (7/52) was significantly elevated (p < 0.05) over the concurrent control rate of 2% (1/52) and historical control rates of 2.5 ± 2.6% (n = 1054) and 3.84 ± 2.94% (n = 1199) reported by NTP for the pre-1995 control F344 male rats fed NIH-07 diet and post-1994 control F344 male rats fed NTP-2000 diet, respectively. Also, the incidence of pheochromocytomas in female rats exhibited a significant dose-response trend (p < 0.05).
The ERF conducted a drinking water study by Soffritti et al. (2002) exposing 100 male and 100 female Sprague-Dawley rats to 0, 500, 5000, or 20000 ppm (or 0, 53.2, 524, and 1780 mg/kg day for males and 0, 66.0, 624.1, and 2177 mg/kg day for females) methanol in drinking water for 2 years. The rats were kept for their lifetime (up to 153 weeks). Compared with the control group, increased incidences of hemolymphoreticular tumors as well as ear duct carcinomas were reported. Specifically, the incidence of hemolymphoreticular tumors (lymphomas/leukemias) was 28 and 40% for female and male rats treated with 2177 and 1780 mg/kg methanol for 2 years, which were higher than the incidence of 13.3 and 20.6% in female historical controls (2274 rats) and male historical controls (2265 rats), respectively. In addition, a significant increase in the high-dose male ear duct carcinomas was also reported. However, this study has been criticized due to the ERF’s refusal to have it peer reviewed (Schoeb et al., 2009a,b), and the finding that the majority of hemolymphoreticular cancers were found in the lungs. A recent review by the NTP had major differences in diagnoses and raised serious concerns regarding the misdiagnosis of lung infections as hemolymphoreticular cancers (Schoeb and McConnell, 2011a,b).
Although available data on methanol carcinogenicity are highly controversial and there are no consistent carcinogenic responses between the NEDO and ERF studies, two MOAs have been proposed for the carcinogenicity of methanol. Formaldehyde is considered a mutagenic metabolite of methanol and is a known carcinogen. Both genotoxicity and cytotoxicity are key events in formaldehyde’s carcinogenicity. Another possible MOA is oxidative stress induced by hydrogen peroxide arising from metabolism of methanol by catalase. However, recent studies have shown that a 6-h methanol exposure (2000 mg/kg ip) in mice, rabbits and monkeys did not cause increased oxidative DNA damage in lung, liver, kidney, bone marrow, or spleen in any of these species (McCallum et al., 2011b). Chronic treatment of mice with 2000 mg/kg daily for 15 days also did not result in increased amounts of oxidative DNA adducts in these tissues (McCallum et al., 2011a).
In this study, we have clearly demonstrated that oral exposure to methanol induces exogenous DNA adducts at multiple tissues in rats treated by 500, or 2000 mg/kg daily for 5 days. The mean number of exogenous dG adducts was 0.08–0.37 and 0.19–1.42 adducts/107 dG for 500 and 2000 mg/kg groups, respectively. Interestingly, the number of endogenous dG adducts also increased 1.3- to 2.3-fold, whereas no increase in endogenous dA adducts was observed. Increased endogenous dG adducts could be the consequence of saturation of formaldehyde detoxification pathways. In addition, the marked contrast between endogenous dG and dA adducts may also reflect the involvement of different repair systems. This study provided the first evidence that the metabolite of methanol, formaldehyde, can cause DNA damage at multiple organs. Since the formation of DNA adducts is a potential key event during carcinogenesis, it is important to keep the dose-response for both exogenous and endogenous DNA adducts from formaldehyde in perspective. Considerable amounts of endogenous DNA damage are always present in cells, and this background damage, like its exogenous counterpart, would be expected to be involved in the induction of cancer. Therefore, it is more informative to analyze the contribution of exogenous exposure in the context of its corresponding endogenous background, as we have demonstrated in a recent perspective on the relationship between endogenous and exogenous DNA adducts in risk assessment (Swenberg et al., 2011). In this study, as shown in Figure 6, the number of exogenous DNA adducts are less than endogenous DNA adducts in tissues we analyzed. Bone marrow had the highest ratio of exogenous versus endogenous dG adducts, with 0.12 and 0.43 for 500 and 2000 mg/kg groups, respectively. Placing this in a more general perspective, if a rat took the mean dose of methanol from dietary intake of aspartame (∼0.7 mg/kg in humans), the ratio would be 0.00016 for bone marrow, i.e., 1 exogenous adduct out of ∼6000 identical endogenous adducts.
Furthermore, we treated rats with 500, or 2000 mg/kg daily for 5 days, which modeled the conditions used in the ERF cancer bioassay (Soffritti et al., 2002). These doses are very high and actually lethal to humans. Under normal physiological conditions, methanol is metabolized to formaldehyde by ADH, followed by rapid conversion to formic acid by formaldehyde dehydrogenase and eventually carbon dioxide and water by a folate-dependent dehydrogenase. Humans have limited folate, which results in the accumulation of formic acid after exposure to high doses of methanol. In contrast, rodents have much higher capacity of folate, and metabolism of methanol can occur through both catalase- and folate-dependent pathways. Thus, they have much higher tolerance for high amounts of methanol by efficiently preventing the formic acid accumulation. However, even under such high concentrations of exposure, the endogenous DNA adducts were higher than corresponding exogenous adducts in all tissues we analyzed.
This study shows that exogenous DNA adducts are formed in a dose-dependent manner. The doses used in our study, 500, or 2000 mg/kg, are below and above the documented saturation dose (600 mg/kg) for rats (NEDO, 1987). The numbers of exogenous dG DNA adducts in liver, lung, kidney, spleen, bone marrow, thymus, and WBC of animals treated with 2000 mg/kg were 5.1, 1.69, 3.25, 4.74, 3.84, 2.63, and 2.11 times higher than those in the 500 mg/kg dose group. The exogenous dA adducts in bone marrow and kidney of rats exposed to 2000 mg/kg methanol were also several fold higher than those exposed to 500 mg/kg methanol. These data clearly demonstrate that higher doses of exposure lead to increased numbers of exogenous DNA damage, although it has been reported that the conversion of methanol to formaldehyde in rats is saturated at doses above 600 mg/kg.
Unlike formaldehyde inhalation exposures, we detected both exogenous dG and dA adducts in select tissues in this study, whereas only exogenous dG could be found following inhalation exposure. This clear difference supports our hypothesis that exogenous dG and dA adducts are formed through different mechanisms. Specifically, we hypothesize that the difference could be a consequence of their different involvement in the formation of DNA-protein cross-links (DPC)- or DNA-protein interaction and suggest that exogenous dG adducts are actually formed through DPC intermediates. Our previous study demonstrated that dG actively forms DPC through cross-linking with lysine and cysteine, whereas dA only cross-linked with cysteine and histidine in much lower amounts (Lu et al., 2010b). Lysine-dG cross-links are the primary DPC formed; however, they are very unstable (Lu et al., 2010b). Decomposition of these labile DNA-protein cross-links may facilitate the formation of dG monoadducts. However, this may not occur with dA because it is much less involved in the formation of DPC. Rather, exogenous dA adducts could be the consequence of direct reaction between DNA and formaldehyde generated by metabolism of other compounds. Direct inhalation exposure only leads to the formation of exogenous dG adducts, whereas exogenous dA adducts appear to only be formed by formaldehyde generated from the metabolism of other formaldehyde-generating compounds. This difference is of importance when evaluating the relevance of available DNA adduct data for the risk assessment of formaldehyde through inhalation. As we demonstrated previously, exogenous dA adducts cannot be induced by direct inhalation exposure (Lu et al., 2010a) but do form through the metabolism of other chemicals like methanol. Therefore, these results further cast a question on the relevance of dA adducts in risk assessment of formaldehyde inhalation exposures other than that endogenous dA adducts would also be expected to increase mutations when DNA replication is increased by formaldehyde’s cytotoxicity.
In contrast to our previous formaldehyde inhalation studies, exogenous DNA damage was found in multiple tissues, such as liver, lung, spleen, thymus, bone marrow, kidney, and WBC following methanol exposure. Thus, this study could be viewed as a general positive control and further validation of the methodology used in previous studies (Lu et al., 2010a, 2011; Moeller et al., 2011). If the exogenous DNA adducts were present in distant tissues, our highly sensitive method would readily measure them, as demonstrated in this study. The huge difference of tissue distribution of formaldehyde-DNA adducts between formaldehyde inhalation exposure and methanol oral exposure highlights the importance of exposure route on the formation of DNA adducts. Formaldehyde is a very reactive compound, and its effects are generally localized within nasal and upper respiratory passages after inhalation. Inhaled formaldehyde does not appear to reach distant sites in an active form that causes DNA damage (Lu et al., 2010a; Moeller et al., 2011). However, methanol is rapidly distributed to all organs and metabolized thereafter once it is absorbed following exposure. This is consistent with the systematic tissue distribution of exogenous DNA adducts discovered in this study.
Another interesting finding was that exogenous dA adducts were only detected in kidney and bone marrow. Bone marrow seems to be a unique organ, with the highest exogenous/endogenous dG adduct ratio among all the tissues we analyzed. As we discussed above, exogenous dA adducts may only be formed by formaldehyde generated from metabolism of related chemicals. Therefore, higher amounts of exogenous dA adducts might suggest greater availability of intact methanol or increased metabolic activities, which is partially supported by the fact that methanol is excreted by kidney after exposure to high doses of methanol. Certainly, more studies are needed to elucidate potential mechanisms leading to this intriguing observation.
In addition, it should be noted that there are large species differences in the metabolism of methanol (Sweeting et al., 2010, 2011). In humans, only ADH is involved, whereas catalase is the primary enzyme responsible for the metabolic activation of methanol in rodents. Moreover, the utilization of folate is much higher in rodents, which prevents the accumulation of formic acid. Since the tissue distribution of hydroxymethyl-DNA adducts is largely controlled by metabolic activity, the distribution spectrum of DNA adducts might be quite different between humans and rodents after exposure to methanol. Furthermore, although we detected exogenous DNA adducts in multiple tissues of rats, the weight of evidence for the genotoxicity of methanol is negative, as most previous studies have been negative in multiple test systems.
An important issue that may influence data interpretation arises from the rate difference between unlabeled and deuterium-labeled methanol during metabolism. Previous studies have shown that deuterium-labeled methanol has a slower conversion or reaction rate (Brooks and Shore, 1971; Kraus and Simon, 2011; Shea et al., 1983; Zhang et al., 2006), resulting in a significant isotope effect. However, the values of isotope effects are inconsistent among studies, ranging from 1.7 to 8.4 depending on the experimental systems and methodologies (Brooks and Shore, 1971; Kraus and Simon, 2011; Shea et al., 1983; Zhang et al., 2006). No experiments have been conducted exactly the same as ours. Therefore, we used an isotope effect of 4, the mean of reported values from several available studies, to estimate the potential impact of deuterium on the metabolism of methanol and subsequent adduct formation. Thus, the exogenous dG adduct numbers and ratios of exogenous versus endogenous dG adducts would increase approximately fourfold. Specifically, the ratio of exogenous versus endogenous dG adducts would be 0.056, 0.072, 0.056, 0.13, 0.48, 0.18, and 0.1 in liver, lung, kidney, spleen, thymus, bone marrow, and WBC of rats exposed to 500 mg/kg per day for 5 days, respectively. Likewise, the ratios of exogenous versus endogenous dG adducts in rats exposed to 2000 mg/kg would increase to 0.20, 0.084, 0.20, 0.74, 1.70, 0.45, and 0.19 in liver, lung, kidney, spleen, thymus, bone marrow, and WBC. However, it should be noted that 500 mg/kg dose is already a lethal dose to humans (estimated to be 300–1000 mg/kg). Even at a dose lethal to humans that far exceeds environmentally relevant concentrations, exogenous dG adducts arising from methanol are still less than endogenous ones, if one takes an isotope effect of 4 into account. Nevertheless, the impact of isotope effects on exogenous adduct numbers and the ratio of exogenous versus endogenous adducts need to be considered when applying our data for quantitative risk assessment of methanol.
Finally, one fact to point out is that methanol and its metabolite, formaldehyde, are both endogenous compounds. Therefore, a substantial endogenous formaldehyde and methanol background is always present and cannot be simply ignored. This ever present background could and should be considered for the risk assessment of such compounds, especially at low doses relevant to human exposure. Two critical questions should be asked when dealing with the risk assessment for chemicals that form DNA adducts identical to endogenous DNA adducts. What is the quantitative contribution of exogenous exposure to endogenous-driven biological responses? Which exposure, exogenous or endogenous, is driving mutagenesis or carcinogenesis at low exposures? Knowledge of this issue should allow science to better address a critical question, such as, what is the safe exposure level of a chemical when substantial amounts of the same chemical are always present in cells and tissues?
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
This research was supported in part by National Institutes of Health (P30-ES10126 and P42-ES05948 to J.A.S.) and the Methanol Institute. B.C.M. received support from T32-ES007126.
Supplementary Material
References
- Brooks RL, Shore JD. Effect of substrate structure on the rate of the catalytic step in the liver alcohol dehydrogenase mechanism. Biochemistry. 1971;10:3855–3858. doi: 10.1021/bi00797a009. [DOI] [PubMed] [Google Scholar]
- Burkhart K. Methanol and ethylene glycol toxicity. J. Toxicol. Clin. Toxicol. 1997;35:149–150. doi: 10.3109/15563659709001185. [DOI] [PubMed] [Google Scholar]
- Cederbaum AI, Qureshi A. Role of catalase and hydroxyl radicals in the oxidation of methanol by rat liver microsomes. Biochem. Pharmacol. 1982;31:329–335. doi: 10.1016/0006-2952(82)90179-4. [DOI] [PubMed] [Google Scholar]
- Clary JJ. Methanol, is it a developmental risk to humans? Regul. Toxicol. Pharmacol. 2003;37:83–91. doi: 10.1016/s0273-2300(02)00031-4. [DOI] [PubMed] [Google Scholar]
- Cruzan G. Assessment of the cancer potential of methanol. Crit. Rev. Toxicol. 2009;39:347–363. doi: 10.1080/10408440802475199. [DOI] [PubMed] [Google Scholar]
- Harris C, Wang SW, Lauchu JJ, Hansen JM. Methanol metabolism and embryotoxicity in rat and mouse conceptuses: Comparisons of alcohol dehydrogenase (ADH1), formaldehyde dehydrogenase (ADH3), and catalase. Reprod. Toxicol. 2003;17:349–357. doi: 10.1016/s0890-6238(03)00013-3. [DOI] [PubMed] [Google Scholar]
- Johlin FC, Fortman CS, Nghiem DD, Tephly TR. Studies on the role of folic acid and folate-dependent enzymes in human methanol poisoning. Mol. Pharmacol. 1987;31:557–561. [PubMed] [Google Scholar]
- Kavet R, Nauss KM. The toxicity of inhaled methanol vapors. Crit. Rev. Toxicol. 1990;21:21–50. doi: 10.3109/10408449009089872. [DOI] [PubMed] [Google Scholar]
- Kerns WD, Pavkov KL, Donofrio DJ, Gralla EJ, Swenberg JA. Carcinogenicity of formaldehyde in rats and mice after long-term inhalation exposure. Cancer Res. 1983;43:4382–4392. [PubMed] [Google Scholar]
- Kraus A, Simon H. Stereochemistry and mechanism of the methanol oxidase from Candida boidinii (author's transl) Hoppe Seylers Z. Physiol. Chem. 2011;356:1477–1484. [PubMed] [Google Scholar]
- Liesivuori J, Savolainen H. Methanol and formic acid toxicity: Biochemical mechanisms. Pharmacol. Toxicol. 1991;69:157–163. doi: 10.1111/j.1600-0773.1991.tb01290.x. [DOI] [PubMed] [Google Scholar]
- Lu K, Collins LB, Ru H, Bermudez E, Swenberg JA. Distribution of DNA adducts caused by inhaled formaldehyde is consistent with induction of nasal carcinoma but not leukemia. Toxicol. Sci. 2010a;116:441–451. doi: 10.1093/toxsci/kfq061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Moeller B, Doyle-Eisele M, McDonald J, Swenberg JA. Molecular dosimetry of N2-hydroxymethyl-dG DNA adducts in rats exposed to formaldehyde. Chem. Res. Toxicol. 2011;24:159–161. doi: 10.1021/tx1003886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Ye W, Zhou L, Collins LB, Chen X, Gold A, Ball LM, Swenberg JA. Structural characterization of formaldehyde-induced cross-links between amino acids and deoxynucleosides and their oligomers. J. Am. Chem. Soc. 2010b;132:3388–3399. doi: 10.1021/ja908282f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Amat G, McMartin KE, Hayreh SS, Hayreh MS, Tephly TR. Methanol poisoning: Ocular toxicity produced by formate. Toxicol. Appl. Pharmacol. 1978;45:201–208. doi: 10.1016/0041-008x(78)90040-6. [DOI] [PubMed] [Google Scholar]
- McCallum GP, Siu M, Ondovcik SL, Sweeting JN, Wells PG. Methanol exposure does not lead to accumulation of oxidative DNA damage in bone marrow and spleen of mice, rabbits or primates. Mol. Carcinog. 2011a;50:163–172. doi: 10.1002/mc.20701. [DOI] [PubMed] [Google Scholar]
- McCallum GP, Siu M, Sweeting JN, Wells PG. Methanol exposure does not produce oxidatively damaged DNA in lung, liver or kidney of adult mice, rabbits or primates. Toxicol. Appl. Pharmacol. 2011b;250:147–153. doi: 10.1016/j.taap.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Methanol toxicity. Agency for Toxic Substances and Disease Registry. Am. Fam. Physician. 1993;47:163–171. [PubMed] [Google Scholar]
- Moeller BC, Lu K, Doyle-Eisele M, McDonald J, Gigliotti A, Swenberg JA. Determination of N2-hydroxymethyl-dG adducts in the nasal epithelium and bone marrow of nonhuman primates following 13CD2-formaldehyde inhalation exposure. Chem. Res. Toxicol. 2011;24:162–164. doi: 10.1021/tx1004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New Energy Development Organization (NEDO) 1985a. 18 Month Inhalation Carcinogenicity Study on Methanol in B6C3F1 Mice. Tokyo, Japan: Mitsubishi Kasei Institute for Toxicological and Environmental Sciences; [Google Scholar]
- New Energy Development Organization (NEDO) 1985b. 24 Month Inhalation Carcinogenicity Study on Methanol in Fischer Rats. Tokyo, Japan: Mitsubishi Kasei Institute for Toxicological and Environmental Sciences; [Google Scholar]
- New Energy Development Organization (NEDO) 1987. Toxicological Research of Methanol as a Fuel for Power Station. Tokyo, Japan: Mitsubishi Kasei Institute for Toxicological and Environmental Sciences; pp. 1–297. [Google Scholar]
- Rogers JM, Brannen KC, Barbee BD, Zucker RM, Degitz SJ. Methanol exposure during gastrulation causes holoprosencephaly, facial dysgenesis, and cervical vertebral malformations in C57BL/6J mice. Birth Defects Res. B Dev. Reprod. Toxicol. 2004;71:80–88. doi: 10.1002/bdrb.20003. [DOI] [PubMed] [Google Scholar]
- Rogers JM, Mole ML. Critical periods of sensitivity to the developmental toxicity of inhaled methanol in the CD-1 mouse. Teratology. 1997;55:364–372. doi: 10.1002/(SICI)1096-9926(199706)55:6<364::AID-TERA2>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Schoeb TR, McConnell EE. Commentary: Further comments on Mycoplasma pulmonis and lymphoma in bioassays of rats. Vet. Pathol. 2011a;48:420–426. doi: 10.1177/0300985810377183. [DOI] [PubMed] [Google Scholar]
- Schoeb TR, McConnell EE. Mycoplasma pulmonis and lymphoma in a methanol bioassay. Vet. Pathol. 2011b;48:903–905. doi: 10.1177/0300985811404713. [DOI] [PubMed] [Google Scholar]
- Schoeb TR, McConnell EE, Juliana MM, Davis JK, Davidson MK, Lindsey JR. Mycoplasma pulmonis and lymphoma. Environ. Mol. Mutagen. 2009a;50:1–3. doi: 10.1002/em.20465. [DOI] [PubMed] [Google Scholar]
- Schoeb TR, McConnell EE, Juliana MM, Davis JK, Davidson MK, Lindsey JR. Mycoplasma pulmonis and lymphoma in bioassays in rats. Vet. Pathol. 2009b;46:952–959. doi: 10.1354/vp.08-VP-0240-S-COM. [DOI] [PubMed] [Google Scholar]
- Shea JP, Nelson SD, Ford GP. MNDO calculation of kinetic isotope effects in model cytochrome P-450 oxidations. J. Am. Chem. Soc. 1983;105:5451–5454. [Google Scholar]
- Soffritti M, Belpoggi F, Cevolani D, Guarino M, Padovani M, Maltoni C. Results of long-term experimental studies on the carcinogenicity of methyl alcohol and ethyl alcohol in rats. Ann. N. Y. Acad. Sci. 2002;982:46–69. doi: 10.1111/j.1749-6632.2002.tb04924.x. [DOI] [PubMed] [Google Scholar]
- Swenberg J A, Kerns W D, Mitchell R I, Gralla E J, Pavkov K L. Induction of squamous cell carcinomas of the rat nasal cavity by inhalation exposure to formaldehyde vapor. Cancer Res. 1980;40:3398–3402. [PubMed] [Google Scholar]
- Swenberg JA, Lu K, Moeller BC, Gao L, Upton PB, Nakamura J, Starr TB. Endogenous versus exogenous DNA adducts: their role in carcinogenesis, epidemiology, and risk assessment. Toxicol. Sci. 2011;120(Suppl. 1):S130–S145. doi: 10.1093/toxsci/kfq371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeting JN, Siu M, McCallum GP, Miller L, Wells PG. Species differences in methanol and formic acid pharmacokinetics in mice, rabbits and primates. Toxicol. Appl. Pharmacol. 2010;247:28–35. doi: 10.1016/j.taap.2010.05.009. [DOI] [PubMed] [Google Scholar]
- Sweeting JN, Siu M, Wiley MJ, Wells PG. Species- and strain-dependent teratogenicity of methanol in rabbits and mice. Reprod. Toxicol. 2011;31:50–58. doi: 10.1016/j.reprotox.2010.09.014. [DOI] [PubMed] [Google Scholar]
- Wang M, Cheng G, Villalta PW, Kassie F, Hecht SS. Analysis of Formaldehdye-DNA Adducts in Leukocyte DNA by Liquid Chromatography-Electrospray Ionization-Tandem Mass Spectrometry: Application to Rats Treated with Aspartame. 2008. In Proceedings of the 99th Annual Meeting of the American Association for Cancer Research; 12–16 Apr 2008, San Diego, CA, Abstract no. 2250. AACR, Philadelphia, PA. [Google Scholar]
- Zhang R, Nagraj N, Lansakara-P DSP, Hager LP, Newcomb M. Kinetics of two-electron oxidations by the compound I derivative of chloroperoxidase, a model for cytochrome P450 oxidants. Org. Lett. 2006;8:2731–2734. doi: 10.1021/ol060762k. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.