Abstract
Cachexia is characterized by inexorable muscle wasting that significantly affects patient prognosis and increases mortality. Therefore, understanding the molecular basis of this muscle wasting is of significant importance. Recent work showed that components of the forkhead box O (FoxO) pathway are increased in skeletal muscle during cachexia. In the current study, we tested the physiological significance of FoxO activation in the progression of muscle atrophy associated with cachexia. FoxO-DNA binding dependent transcription was blocked in the muscles of mice through injection of a dominant negative (DN) FoxO expression plasmid prior to inoculation with Lewis lung carcinoma cells or the induction of sepsis. Expression of DN FoxO inhibited the increased mRNA levels of atrogin-1, MuRF1, cathepsin L, and/or Bnip3 and inhibited muscle fiber atrophy during cancer cachexia and sepsis. Interestingly, during control conditions, expression of DN FoxO decreased myostatin expression, increased MyoD expression and satellite cell proliferation, and induced fiber hypertrophy, which required de novo protein synthesis. Collectively, these data show that FoxO-DNA binding-dependent transcription is necessary for normal muscle fiber atrophy during cancer cachexia and sepsis, and further suggest that basal levels of FoxO play an important role during normal conditions to depress satellite cell activation and limit muscle growth.—Reed, S. A., Sandesara, P. B., Senf, S. F., Judge, A. R. Inhibition of FoxO transcriptional activity prevents muscle fiber atrophy during cachexia and induces hypertrophy.
Keywords: cancer, sepsis, satellite cells, myostatin, protein synthesis
Cachexia is a complex metabolic syndrome characterized by the loss of muscle and fat mass that is unresponsive to nutritional supplementation alone (1). It is commonly associated with a variety of diseases, including cancer, sepsis, heart failure, chronic obstructive pulmonary disease, and HIV/AIDs (2–7). Strikingly, cachexia affects up to 80% of patients with cancer (4), significantly affects patient prognosis (with the severity of the cachexia being inversely related to patient survival time; ref. 8), significantly affects recovery from cancer (3, 9, 10), and accounts for over a quarter of cancer-related deaths (11). Similarly, cachexia associated with sepsis significantly prolongs the patient's stay in the intensive care unit and negatively affects prognosis (2). Therefore, understanding the cellular mechanisms that are necessary for the loss of skeletal muscle mass during cachexia is highly significant from a clinical perspective.
Although the molecular regulation of muscle wasting during cachexia is multifactorial and may depend on the underlying disease state, glucocorticoids and proinflammatory cytokines have been identified as important mediators of muscle atrophy during a variety of cachectic conditions, including sepsis and cancer. Indeed, in patients with sepsis, the plasma levels of lipopolysaccharides (LPS) and glucocorticoids are increased (12, 13), and in patients with lung or gastroesophageal cancers, serum levels of the cytokines tumor necrosis factor α (TNF-α), interleukin (IL)-1, IL-6 and IL-8 are elevated (14, 15). Notably, various studies using rodent models of experimental sepsis or cancer cachexia demonstrate that glucocorticoids (16), TNF-α (17, 18), IL-6 (19), and interferon γ (IFNγ; ref. 20) are necessary for the associated muscle wasting.
More recently, the atrophic signaling pathways activated during sepsis and cancer cachexia, as well as following LPS, glucocorticoid, and cytokine treatment, have been explored in skeletal muscle. One pathway commonly increased during these physiological and experimental models of cachexia is the forkhead box O (FoxO) signaling pathway (21–26). There are 3 FoxO family members present in skeletal muscle (FoxO1, FoxO3a, and FoxO4), and the importance of this family of transcription factors in the regulation of skeletal muscle mass has been clearly demonstrated in a number of genetic studies. Muscle-specific overexpression of FoxO1 or FoxO3a is sufficient to cause skeletal muscle atrophy in vivo (24, 27), and inhibition of FoxO transcriptional activity by expression of a dominant negative (DN) FoxO prevents at least half of disuse-induced muscle fiber atrophy in vivo (28, 29) and attenuates myotube atrophy induced by the glucocorticoid dexamethasone (dex; ref. 24).
The regulation of muscle mass by FoxO transcription factors is presumably due to their regulation of muscle atrophy-related genes. Indeed, 4 genes whose expression levels are commonly increased during multiple models of skeletal muscle atrophy, including cancer and sepsis, are atrogin-1/MAFbx, MuRF1, cathepsin L, and Bnip3 (23, 30–41), and there is evidence to suggest each are FoxO target genes. In this regard, overexpression of FoxO3a is sufficient to activate an atrogin-1 and a MuRF1 promoter reporter and increase atrogin-1 mRNA levels in skeletal muscle (24, 28, 42). This is significant, since atrogin-1 and MuRF1 are required for normal muscle wasting, at least during denervation (34). FoxO3a overexpression is also sufficient to increase Bnip3 expression in HEK293 cells, while inhibition of FoxO transcriptional activity decreases Bnip3 expression (32). In skeletal muscle, Bnip3 is sufficient to induce autophagosome formation and mediates FoxO3a-induced autophagosome formation (32). Expression of cathepsin L, a lysosomal protease, is increased in skeletal muscles overexpressing FoxO1 (27), and FoxO1 is required for the fasting-induced increase in cathepsin L (30). Although these atrophy-related FoxO target genes are concomitantly increased with various markers of FoxO signaling during cachectic conditions, the requirement of FoxO for the transcriptional activation of these specific atrophy genes and in the progression of muscle fiber atrophy during any cachectic condition is currently unknown. Therefore, the objective of the current work was to inhibit FoxO-dependent transcription in whole skeletal muscle during two distinct cachectic conditions, cancer and sepsis, to determine the extent to which FoxO regulates atrophy gene transcription and skeletal muscle fiber atrophy during cachexia.
MATERIALS AND METHODS
Animals
Male C57BL/6 mice (8 wk of age on arrival) from Charles River Laboratories (Wilmington, MA, USA) were used for all experiments. Animals were housed at the University of Florida in a temperature-controlled facility on a 12-h light-dark cycle. Standard diet and water were provided ad libitum. All protocols and procedures were approved by the University of Florida's institutional animal care and use committee.
Expression and reporter plasmids
The DN FoxO cDNA was a kind gift of Dr. Paul Coffer (University Medical Center, Utrecht, The Netherlands). This cDNA consists of only the DNA-binding domain of FoxO3a (aa 141–266), which allows it to bind to DNA but not activate transcription. Therefore, as a DN construct, the DN FoxO binds to FoxO binding sites in target genes and prevents their activation by endogenous FoxO. Because of the significant homology between the FoxO-DNA binding domains of the FoxO factors, this construct also has the potential to repress FoxO1- and FoxO4-dependent transcription (24, 43). The DN FoxO-DsRed fusion plasmid was constructed by us and has been previously described (29). DsRed2-c1 (Clontech, Palo Alto, CA, USA) was used as a control plasmid. The DAF-16 (FoxO) reporter plasmid has been previously described and used (44). The CAGA12-luciferase reporter, a kind gift of Dr. Peter Ten Dijke (Leiden University Medical Centre, Leiden, The Netherlands), encodes 12 repeats of the Smad-binding site (CAGA) and has previously been used and described (45). pRL-TK-Renilla was purchased from Promega (Madison, WI, USA).
Plasmid injection and electroporation
In vivo transfection of plasmid DNA into skeletal muscle has been detailed previously (28). Briefly, the soleus and tibialis anterior (TA) muscles were isolated and injected with 7.5 or 20 μl (soleus and TA, respectively) of 1× PBS containing the plasmid constructs. Following injection, electric pulses were delivered using an electric pulse generator (Electro Squareporator ECM 830; BTX, Hawthorne, NY, USA) by placing needle electrodes on either side of the muscle. Five pulses were delivered in 200-ms interpulse intervals, each with an effective intensity of 75 V/cm and 20-ms duration. These electroporation parameters do not induce muscle damage in our hands (unpublished data) or others (46, 47). The soleus received 5 μg of total plasmid DNA, and the TA received 20 μg plasmid DNA/injection. The TA and soleus in each leg were injected with the same plasmids. Animals were allowed to recover for 5 d before the induction of sepsis or inoculation with Lewis lung carcinoma (LLC) cells. In experiments measuring satellite cell proliferation, animals received an intraperitoneal injection with 50 mg/kg bromodeoxyuridine (BrdU, Sigma Aldrich, St. Louis, MO, USA) daily for 4 d starting 1 wk after plasmid electrotransfer. In experiments to inhibit protein synthesis, cycloheximide (Sigma Aldrich; 15 mg/kg) was injected subcutaneously daily for 6 d starting on d 4 following plasmid injection.
Cecal ligation and puncture
Sepsis was induced via cecal ligation and puncture (CLP). Mice were anesthetized with isoflurane, and a 1- to 2-cm incision was made down the midline of the abdomen. The cecum was removed from the abdominal cavity, and the distal end was ligated with 3.0-gauge sterile silk suture. The ligated cecal cavity was perforated through and through with a 28-gauge needle. The intestines were then carefully returned to the abdominal cavity, and sutures were used to secure the incision in the peritoneal wall and skin. Control animals underwent a sham surgery, during which the cecum was exposed but not subjected to ligation or puncture. All mice were administered 1 ml saline subcutaneously and allowed to recover from anesthesia. The TA and soleus muscles were harvested 4 d following CLP for measurement of FoxO-dependent reporter activity and gene expression and 7 d following CLP for muscle fiber atrophy.
LLC cachexia
LLC cells were obtained from the National Cancer Institute Tumor Repository (Frederick, MD, USA). Cells were cultured in α-MEM (Invitrogen, Carlsbad, CA, USA) supplemented with 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine at 37°C in a 5% CO2 humidified atmosphere. To induce cancer cachexia, animals were injected subcutaneously in the flank with 5 × 106 cells suspended in PBS. The TA and soleus muscles were harvested ∼14 d after LLC injection, when maximal tumor diameter reached 1.5 cm.
Muscle preparation and analysis
The soleus and TA muscles harvested for mRNA expression and other biochemical analysis were rapidly frozen in liquid nitrogen and stored at −80°C. Muscles removed for histological measurements were embedded in freezing medium in a tissue-embedding cassette at a standardized length prior to freezing in liquid-nitrogen-cooled isopentane, and stored at −80°C.
Reporter plasmid analysis
Whole muscle was homogenized in 1× passive lysis buffer (Promega) and subsequently centrifuged for 20 min at 5000 g at 4°C. Supernatant (10 μl) was added to luciferase reagent (100 μl), and total muscle luciferase activity was measured on a Modulus single-tube multimode reader (Turner Biosystems; Promega).
RNA isolation, cDNA synthesis, and qRT-PCR
Muscles were homogenized in TRIzol reagent, and RNA was isolated as described previously (28). RNA (1 μg) was reverse transcribed to cDNA using Ambion's RETROscript first-strand synthesis kit (Ambion, Austin, TX, USA). cDNA was used as a template for real-time PCR using the primers listed below and a 7300 real-time PCR system (Applied Biosystems, Foster City, CA, USA). TaqMan probe-based chemistry was used to detect PCR products, and quantification was performed using a relative standard curve. Primers used were FoxO1 (GenBank accession no. NM_019739.3), FoxO3a (GenBank NM_019740.2), FoxO4 (GenBank NM_018789.1), atrogin-1 (GenBank NM_026346.2), MuRF-1 (GenBank NM_001039048.2), Bnip3 (GenBank NM_009761.3), cathepsin-L (GenBank NM_009984.3), Pax7 (GenBank NM_011039.2), MyoD (GenBank NM_010866.2), myostatin (GenBank NM_010834.2), and 18S (GenBank X03205.1). All primers were purchased from Applied Biosystems. Expression of 18S was used for normalization, as no differences were identified between any groups.
Histology
To determine cross-sectional area, 10-μm sections were taken from the midbelly of muscles using a Microm HM 550 cryostat (Microm International, Walldorf, Germany). Sections were fixed for 20 min in 4% formaldehyde, followed by washing in PBS. To visualize cell membranes, sections were incubated with Alexa Fluor 350-conjugated wheat germ agglutinin (Invitrogen) for 2 h. Images were captured using a Leica DM5000B microscope (Leica Microsystems, Wetzlar, Germany). To measure cross-sectional area, transfected fibers (i.e., those expressing DN FoxO-DsRed or DsRed) were traced and measured using the Leica application suite, version 3.5.0 (Leica Microsystems). The cross-sectional area of ∼100 transfected fibers was measured from each muscle. Given that the DN FoxO-DsRed fusion protein localizes to the nucleus (29) and that not every cross-section dissects a nucleus for each muscle fiber, only fibers with visible fluorescence were measured.
Immunohistochemistry
Muscle cross-sections were collected as described above and rehydrated in PBS prior to antigen retrieval with citrate buffer (1.8 mM citric acid and 8.2 mM sodium citrate, pH 6.0). Sections were washed twice with PBS. Endogenous peroxidase activity was blocked with 0.3% hydrogen peroxide for 5 min, followed by a PBS wash. Sections were incubated with primary antibody (anti-BrdU, Roche Diagnostics, Indianapolis, IN, USA; anti-laminin, Sigma Aldrich; or anti-dystrophin, Thermo Scientific Lab Vision, Fremont, CA, USA) overnight at 4°C and then washed extensively with PBS. For immuofluorescent detection, sections were incubated for 1 h with secondary antibodies (Alexa Fluor 594 anti-mouse IgG and Alexa Fluor 488 anti-rabbit IgG; Invitrogen) prior to extensive washing with PBS and visualization. Images were captured using a Leica DM5000B microscope. BrdU-positive nuclei were counted in 5 ×20 fields for each muscle and averaged. In the laminin-stained sections, only BrdU-positive nuclei within the basal lamina of muscle fibers were counted, since those outside the basal lamina are nonmuscle nuclei. In the dystrophin-stained sections, only BrdU-positive nuclei under dystrophin were counted, to identify satellite cells that proliferated and fused with muscle fibers to become myonuclei. Again, BrdU-positive nuclei were counted in 5 ×20 fields for each muscle and averaged.
Cell culture and transfection
C2C12 myoblasts (American Type Culture Collection, Rockville, MD, USA) were cultured on 0.1% gelatin in high-glucose DMEM (Invitrogen) containing 10% FBS (Invitrogen), 100 U/ml penicillin, 100 μg/ml streptomycin, and 0.1 mg/ml gentamicin at 37°C in a 5% CO2 humidified atmosphere. In the reporter experiments, myoblasts were transfected with the CAGA12-luciferase reporter plasmid and the pRL-TK Renilla luciferase reporter plasmid, plus an empty vector or DN FoxO expression plasmid at ∼80% confluence using FuGend HD transfection reagent (Promega). In the myotube-diameter experiments, myoblasts were transfected with either DsRed or DN FoxO-DsRed. After 16 h, the cells were placed in differentiation medium (2% horse serum in DMEM) to induce differentiation into myotubes. In the reporter experiments, after 3 d, the myotubes were either harvested or treated with vehicle or 100 ng/ml of recombinant myostatin (R&D Systems, Minneapolis, MN, USA) for 12 h and then harvested. Cells were harvested in passive lysis buffer (Promega), and luciferase activity was determined by normalizing firefly luciferase activity to pRL-TK Renilla luciferase activity using a dual-luciferase reporter assay (Promega). In the myostatin myotube-diameter experiments, C2C12 cells were differentiated for 24 h and then treated with vehicle or 100 ng/ml of recombinant myostatin for 96 h, replenishing myostatin every 24 h. In the rapamycin myotube-diameter experiments, C2C12 cells were differentiated for 3 d and then treated with either vehicle or 30 nM of rapamycin (EMD Chemicals, San Diego, CA, USA) for 72 h, replenishing rapamycin every 24 h. In each myotube diameter experiment, myotubes were fixed in 4% paraformaldehyde, and images were captured using a Leica DM5000B microscope. To measure myotube diameter, ∼100 transfected myotubes/treatment were analyzed in ×10 fields using Leica Application Suite 3.5.0. Three diameter measurements were made along the length of each myotube, and the average of these measurements was considered as one value.
Statistical analysis
All data were analyzed using either a 2-way ANOVA followed by Bonferroni post hoc comparisons or a Student's t test (GraphPad Software, San Diego, CA, USA). All data are expressed as means ± se, and significance was established at P ≤ 0.05.
RESULTS
During cachectic conditions, atrophy is more pronounced in glycolytic muscles than in oxidative muscles (48–50). We therefore selected to study both the glycolytic TA and the oxidative soleus muscles in the current study to detect any differences in FoxO-DNA binding-dependent transcription between these two muscle types during cachexia.
FoxO gene expression and transcriptional activity in skeletal muscle during cachexia
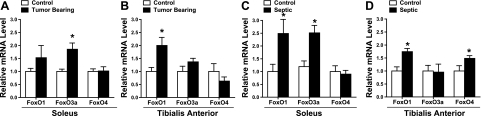
To demonstrate, in our hands, that FoxO signaling is increased in skeletal muscle during cachexia, we first measured FoxO mRNA levels in the soleus and TA muscles of mice during cancer and sepsis. In the soleus of tumor-bearing animals, FoxO3a mRNA levels were increased 1.8-fold compared to controls (Fig. 1A), but FoxO1 mRNA was unchanged. However, in the TA of tumor-bearing mice, FoxO1 mRNA was increased 2-fold compared to controls (Fig. 1B), but FoxO3a remained unchanged. There were no changes in FoxO4 mRNA levels in either muscle of tumor-bearing mice. During sepsis, FoxO1 mRNA levels were significantly increased 2.5-fold in the soleus and 1.7-fold in the TA muscles compared to controls (Fig. 1C, D). Interestingly, in the soleus muscle, FoxO3a mRNA levels were increased 2.5-fold, but FoxO4 mRNA was unchanged, while in the TA, FoxO4 mRNA levels were increased 1.5-fold, but FoxO3a mRNA was unchanged. Combined, these findings show that during cancer and sepsis, FoxO3a mRNA levels are increased in the soleus, and FoxO1 mRNA levels are increased in the TA.
Figure 1.
Cachexia increases the mRNA expression of FoxO family members. Relative mRNA levels of FoxO1, FoxO3a, and FoxO4 in soleus (A, C) and TA muscle (B, D) at 14 d after LLC cell injection (A, B) and after 4 d of sepsis (C, D). Bars represent means ± se for 6 muscles/group. *P < 0.05 vs. control.
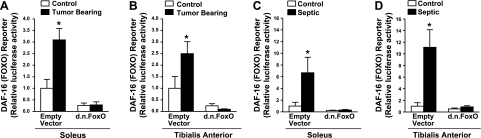
We next determined the extent to which FoxO transcriptional activity is increased in skeletal muscle during cachexia. In these experiments, a FoxO-dependent reporter plasmid plus a control or DN FoxO expression plasmid was injected and electrotransfered into the TA and soleus muscles prior to 4 d of sepsis or ∼14 d of tumor bearing. In tumor-bearing mice, FoxO transcriptional activity was increased 3.1-fold in the soleus and 2.5-fold in the TA (Fig. 2A, B), and during sepsis FoxO transcriptional activity increased almost 7-fold in the soleus and 11-fold in the TA (Fig. 2C, D). These findings demonstrate that the magnitude of increase in FoxO activation is similar in the soleus and TA of tumor-bearing mice, but during sepsis, it is more pronounced in the TA than the soleus. Since, in subsequent experiments, we aimed to determine whether an increase in FoxO-dependent transcription is required for the increased expression of specific atrophy-related genes and muscle fiber atrophy during cachexia, we expressed DN FoxO in a subset of mice during each experimental condition to demonstrate that this construct represses FoxO transcriptional activity during cachexia. Indeed, the increase in FoxO transcriptional activity in both the soleus and TA muscles was completely abolished in muscles from tumor-bearing and septic mice injected with the DN FoxO plasmid (Fig. 2). Basal FoxO transcriptional activity was also decreased by overexpression of DN FoxO.
Figure 2.
Cachexia increases FoxO transcriptional activity. FoxO-dependent luciferase reporter activity from the soleus (A, C) and TA muscles (B, D) of control and LLC tumor-bearing mice (A, B) and control and septic mice (C, D). Muscles were injected with an empty vector or DN FoxO plasmid and harvested ∼14 d after LLC cell injection or after 4 d of sepsis. Bars represent means ± se for 6 muscles/group. *P < 0.05 vs. control empty vector.
Increased FoxO transcriptional activity is required for the increased transcription of specific atrophy-related genes in skeletal muscle during cachexia
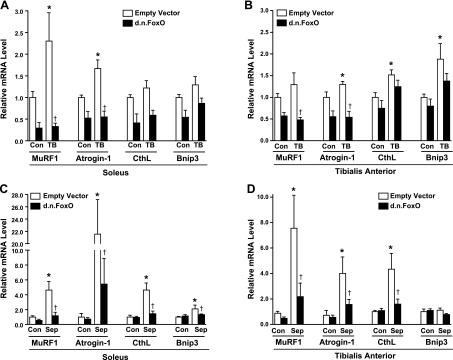
Given our finding that FoxO transcriptional activity is significantly increased in skeletal muscle during both cancer and sepsis, and the knowledge that FoxO can regulate the transcription of atrogin-1 (24, 51), MuRF1 (51, 52), cathepsin L (30), and Bnip3 (53), each of which is increased in skeletal muscle during cachexia (31, 54, 55), we next used qRT-PCR to determine the requirement of FoxO-DNA binding for their increased transcription during cachexia. As shown in Fig. 3A, MuRF1 and atrogin-1 mRNA levels were significantly increased in the control-injected soleus muscle of tumor-bearing mice by 2.3-fold and 1.7-fold, respectively. The increase in each of these genes during cancer was abolished in soleus muscles injected with the DN FoxO. Neither cathepsin L nor Bnip3 was significantly increased in the soleus muscle of tumor-bearing mice. In the TA muscle, cancer cachexia induced significant increases in the mRNA levels of atrogin-1 (1.3-fold), cathepsin L (1.5-fold), and Bnip3 (1.9-fold). The increase in atrogin-1 was abolished in TA muscles from LLC tumor-bearing mice expressing DN FoxO. However, although the increased expression of cathepsin L and Bnip3 was inhibited by 52 and 57%, respectively, in TA muscles from tumor-bearing mice expressing DN FoxO compared to muscles injected with empty vector, neither reached statistical significance (Fig. 3B). MuRF1 mRNA levels were increased 1.4-fold in the TA of tumor-bearing mice, but this did not reach statistical significance.
Figure 3.
FoxO transcriptional activation is required for the cachexia-induced increase in the mRNA levels of selected atrophy-related genes. Relative mRNA levels of MuRF1, atrogin-1, cathepsin L, and Bnip3 from soleus (A, C) and TA muscles (B, D) injected with an empty vector or DN FoxO plasmid and harvested after ∼14 d from control and LLC tumor-bearing mice (A, B) or after 4 d from control and septic mice (C, D). Bars represent means ± se for 6 muscles/group. *P < 0.05 vs. control (Con) empty vector; †P < 0.05 vs. tumor-bearing (TB) or septic (Sep) empty vector.
The sepsis-induced increases in the mRNA levels of MuRF1 (4.6-fold), atrogin-1 (21-fold), cathepsin L (4.6-fold), and Bnip3 (2.1-fold) in the soleus muscle were significantly inhibited in muscles injected with DN FoxO (Fig. 3C). In the TA muscle, the sepsis-induced increases in the mRNA levels of MuRF1 (7.5-fold), atrogin-1 (4-fold), and cathepsin L (4.3-fold) were inhibited by DN FoxO (Fig. 3D). There was no change in Bnip3 expression in the TA during sepsis. Combined, these gene data show that atrogin-1, MuRF1, cathepsin L, and/or Bnip3 are FoxO targets during cachexia. The data also suggest that changes in gene transcription of these atrophy-related genes are more similar between different muscles within a cachectic condition than between the same muscles in different cachectic conditions.
Inhibition of FoxO transcriptional activity prevents muscle fiber atrophy during cachexia
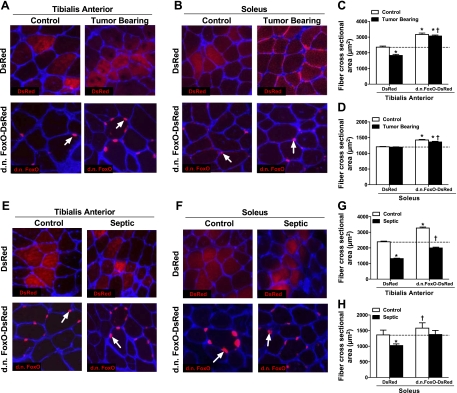
Since FoxO transcriptional activity is increased in the skeletal muscle of cachectic mice and is necessary for the increased transcription of several atrophy-related genes, we next aimed to determine whether FoxO transcriptional activation is necessary for muscle fiber atrophy during cancer and sepsis. The TA and soleus muscles of each hindlimb were injected with DsRed or DN FoxO-DsRed 5 d prior to the induction of sepsis or inoculation with LLC cells. After 7 d of sepsis or control conditions, and ∼14 d of tumor-bearing or control conditions, muscles were harvested, and the cross-sectional areas (CSAs) of fibers expressing either DsRed or DN FoxO-DsRed were measured and compared. In TA muscles expressing DsRed, mice bearing the LLC tumor showed a 22% decrease in the CSA compared to control mice (Fig. 4C; control, 2362±66.7 μm2; tumor bearing, 1833±70.82 μm2, P<0.0001). However, cancer-induced TA muscle fiber atrophy was abolished in DN FoxO-DsRed-expressing fibers (control, 3172±89 μm2; tumor bearing, 3069±67.5 μm2). In fact, the CSA of DN FoxO-DsRed-expressing fibers in the TA of both control and tumor-bearing mice was significantly larger than the CSA of DsRed-expressing fibers in the TA of control mice (23 and 25% larger, respectively; P<0.0001). Although LLC inoculation did not induce soleus muscle fiber atrophy (Fig. 4D; control, 1209±8.6 μm2; tumor bearing, 1195±8.9 μm2), the CSA of DN FoxO-DsRed-expressing fibers was increased in both control and tumor-bearing mice compared to DsRed-transfected fibers in control mice (17.7 and 12.5% increase compared to control, respectively; P<0.0001).
Figure 4.
FoxO transcriptional activation is necessary for cachexia-induced muscle fiber atrophy. A, B, E, F) Representative cross sections taken from the TA (A, E) and soleus muscles (B, F) of control and ∼14 d LLC tumor-bearing mice (A, B), and control and 7-d septic mice (E, F), each transfected with either DsRed or DN FoxO-DsRed. Sections were incubated with wheat germ agglutinin to allow for visualization of muscle fibers and the CSA of transfected fibers (fibers expressing either DsRed or DN FoxO-DsRed) were measured and compared. Arrows indicate representative DN FoxO-DsRed expression, which we have previously shown localizes to the nucleus. C, D) Mean TA (C) and soleus (D) fiber CSA from control and LLC tumor bearing mice transfected with either DsRed or DN FoxO-DsRed in panels A and B, respectively. G, H) Mean TA (G) and soleus (H) fiber CSA from control and septic mice transfected with either DsRed or DN FoxO-DsRed in panels E and F, respectively. Bars represent means ± se for 6 muscles/group. *P < 0.05 vs. control DsRed; †P < 0.05 vs. tumor-bearing or septic DsRed.
Seven days of sepsis caused a 52 and 25% decrease in the CSA of DsRed expressing fibers in TA and soleus muscles, respectively, compared to DsRed-expressing fibers in control mice (TA control, 2389±42.72 μm2; TA sepsis, 1311±29.28 μm2; soleus control, 1360±153.5 μm2; soleus sepsis, 1022±52.08 μm2; Fig. 4G, H). Expression of DN FoxO-DsRed attenuated sepsis-induced muscle fiber atrophy in the TA by 68% and completely prevented muscle fiber atrophy in the solei of septic animals. Again, expression of DN FoxO-DsRed in the muscles of control animals resulted in a 24 and a 16% increase in muscle fiber CSA in the TA and soleus muscles, respectively, compared to DsRed-transfected fibers in control mice.
Inhibition of FoxO transcriptional activity induces muscle fiber hypertrophy
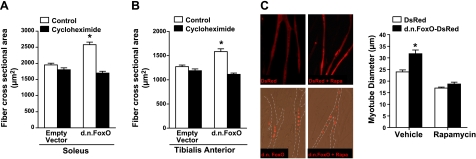
Combined, the above findings demonstrate that inhibition of FoxO signaling during cancer and sepsis not only inhibits muscle fiber atrophy during cachexia but also causes significant muscle fiber hypertrophy in normal control muscle. This is in agreement with our recent findings in rat solei transfected with DN FoxO that also showed fiber hypertrophy (29) and suggests that FoxO plays an important role in limiting muscle growth during control, nonpathophysiological conditions. To determine the extent to which this hypertrophy requires de novo protein synthesis, we transfected the TA and soleus muscles of control mice with DsRed or DN FoxO-DsRed and 4 d later began daily injections with cycloheximide, a global protein synthesis inhibitor, for a period of 6 d. CSA was measured in transfected fibers from mice injected with vehicle only or with cycloheximide. In the TA and soleus muscles of mice injected with vehicle only, the CSA of fibers expressing DN FoxO-DsRed was increased by 28 and 27%, respectively, compared to DsRed-expressing fibers (Fig. 5A, B). However, treatment with cycloheximide prevented the increase in fiber CSA in DN FoxO-DsRed-expressing fibers in both the TA and the soleus. These findings indicate that hypertrophy resulting from inhibition of FoxO transcriptional activity requires de novo protein synthesis.
Figure 5.
Inhibition of FoxO activity induces myofiber hypertrophy via increased de novo protein synthesis. A, B) Soleus and TA muscles were transfected with an empty vector or a DN FoxO expression plasmid; 4 d later, mice were injected with cycloheximide or vehicle for 6 d. Myofiber hypertrophy in soleus (A) and TA (B) muscle fibers expressing DN FoxO in vehicle-injected mice was prevented in mice injected with the protein synthesis inhibitor cycloheximide. C) Representative images and mean myotube diameter of C2C12 myotubes transfected (as myoblasts) with either DsRed or DN FoxO-DsRed and treated with vehicle or rapamycin (Rapa) for 72 h. *P < 0.05 vs. control empty vector or vehicle DsRed.
Because the Akt-mTOR signaling pathway is a key regulator of protein synthesis and muscle hypertrophy (reviewed in ref. 56), we next determined the requirement of mTOR for DN FoxO-induced hypertrophy. In these experiments, C2C12 myoblasts were transfected with either DsRed or DN FoxO-DsRed, differentiated for 3 d, and then treated with either vehicle or rapamycin, to inhibit mTOR, for 3 d. The diameter of myotubes transfected with DN FoxO-DsRed was 33% larger than the diameter of myotubes transfected with DsRed. Rapamycin treatment caused a 29% decrease in the diameter of myotubes transfected with the control plasmid (DsRed), and DN FoxO was unable to increase the diameter of myotubes treated with rapamycin (Fig. 5C). These findings suggest that mTOR is required for the DN FoxO-induced increase in myotube diameter.
Inhibition of FoxO transcriptional activity increases satellite cell proliferation and fusion into myofibers
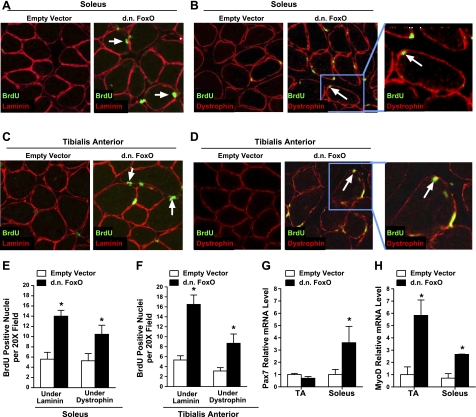
In addition to increased protein synthesis, an increase in muscle fiber size may also involve the activation and proliferation of satellite cells, which fuse into existing myofibers. We therefore tested the extent to which inhibition of FoxO transcriptional activity, via expression of DN FoxO, increased satellite cell proliferation. To do this, soleus and TA muscles were transfected with an empty vector or DN FoxO plasmid 7 d prior to daily injections of BrdU, a synthetic thymidine analog that is incorporated into cellular DNA during mitosis. Since muscle nuclei, but not satellite cell nuclei, are postmitotic, BrdU expression inside the basal lamina of muscle fibers is a result of satellite cell proliferation. Following 4 d of BrdU injections, muscles were harvested and sectioned, and muscle cross sections were incubated with anti-BrdU and anti-laminin antibodies to detect the total number of BrdU-positive nuclei inside the basal lamina (Fig. 6A, C). In each section, the average number of BrdU-positive nuclei under the basal lamina in TA and soleus muscles injected with an empty vector was ∼5. This increased to ∼16 and ∼14 in TA and soleus muscles, respectively, expressing DN FoxO (Fig. 6E, F). These findings suggest that inhibition of FoxO transcriptional activity increases satellite cell proliferation. We subsequently determined the extent to which the increased numbers of proliferating satellite cells in the DN FoxO-expressing muscles fuse with existing myofibers. To do this, we incubated muscle cross sections with anti-BrdU and anti-dystrophin antibodies to detect the number of BrdU-positive nuclei under dystrophin (Fig. 6B, D). In each section, the average number of BrdU-positive nuclei under dystrophin in TA and soleus muscles expressing DN FoxO was ∼8 and ∼10, respectively (Fig. 6E, F). This indicates that inhibition of FoxO transcriptional activity causes an increase in both proliferation and fusion of satellite cells with myofibers.
Figure 6.
Inhibition of FoxO activity activates muscle satellite cells. Soleus and TA muscles were transfected with an empty vector or DN FoxO; 7 d later, the mice were injected with bromodeoxyuridine (BrdU) for 4 d. A–D) Representative cross sections of the soleus (A, B) and TA (C, D), showing BrdU incorporation visualized by immunohistochemistry. Total number of BrdU-positive nuclei under the basal lamina (A, C) and under dystrophin (B, D) were counted in each muscle. Arrows indicate representative BrdU-positive nuclei under the basal lamina or dystrophin. E, F) Mean numbers of BrdU-positive nuclei per ×20 field under laminin and dystrophin in the soleus (E) and TA (F). G, H) Relative mRNA level of Pax7 (G) and MyoD (H) in TA and soleus muscles transfected with an empty vector or DN FoxO. Bars represent means ± se for 6 muscles/group. *P < 0.05 vs. respective empty vector group.
To further support these findings, we measured Pax7 and MyoD mRNA levels in whole muscle transfected with a control plasmid or the DN FoxO expression plasmid. Pax7 is expressed in quiescent and proliferating satellite cells and then down-regulates as the satellite cells commit to myogenic differentiation (57, 58). The expression level of the myogenic regulatory factor MyoD is increased in active and proliferating satellite cells and promotes progression to terminal differentiation (59–61). In the TA muscle, overexpression of DN FoxO had no effect on Pax7 transcription but increased MyoD mRNA levels almost 6-fold, whereas in the soleus muscle, DN FoxO increased Pax7 transcription 3.6-fold and MyoD mRNA 2.6-fold (Fig. 6G, H). These findings suggest that specification of satellite cells to the myogenic lineage in muscles expressing DN FoxO may be progressing more rapidly in the TA compared to the soleus.
Inhibition of FoxO transcriptional activity decreases activity of the myostatin/Smad pathway
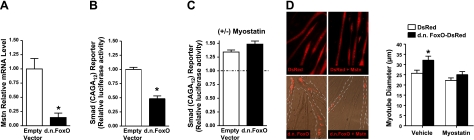
While there are several factors that may regulate satellite cell proliferation, the hormone myostatin has received considerable attention in recent years due to its ability to regulate both satellite cell proliferation and muscle fiber size. In this regard, myostatin inhibits satellite cell activation and proliferation and decreases muscle mass (62–64). Because overexpression of DN FoxO increases satellite cell proliferation and muscle fiber size, we next questioned whether DN FoxO regulates myostatin expression in whole muscle. Indeed, myostatin mRNA levels were decreased by 85% in muscles expressing DN FoxO (Fig. 7A), demonstrating that repression of FoxO transcriptional activity decreases myostatin transcription. This suggests that under normal physiological conditions, basal levels of FoxO transcriptional activity regulate myostatin expression. To further support that FoxO regulates myostatin signaling, we next determined whether inhibition of FoxO-DNA binding-dependent transcription also decreases the activity of Smads, the transcription factor family downstream of myostatin (65). C2C12 myoblasts were cotransfected with a Smad-dependent luciferase reporter plus either an empty vector or DN FoxO plasmid and differentiated for 3 d. Expression of DN FoxO decreased basal Smad transcriptional activity by 52% (Fig. 7B). To determine whether this decrease in Smad transcriptional activity by DN FoxO requires a decrease in myostatin expression, we subsequently tested whether DN FoxO could still repress the Smad reporter in the presence of recombinant myostatin. Treatment with myostatin reversed the ability of DN FoxO to repress Smad reporter activity (Fig. 7C), suggesting that the inhibition of Smad transcriptional activity by DN FoxO occurs through decreasing myostatin levels. We next determined whether a decrease in myostatin is required for the increase in the diameter of C2C12 myotubes expressing DN FoxO. The 30% increase in the diameter of myotubes transfected with DN FoxO-DsRed was abolished in DN FoxO-DsRed-expressing myotubes treated with recombinant myostatin (Fig. 7D). These combined findings indicate that inhibition of FoxO transcriptional activity decreases myostatin/Smad signaling, which is required for DN FoxO-induced hypertrophy.
Figure 7.
Inhibition of FoxO activity decreases activity of the myostatin/Smad pathway. A) Relative mRNA level of myostatin in TA muscle injected and electroporated with an empty vector or DN FoxO. B, C) Smad-dependent reporter activity in C2C12 cells transfected with an empty vector or DN FoxO and differentiated for 3 d and then harvested (B) or treated with vehicle or recombinant myostatin for 12 h and then harvested (C). Data in C are normalized to their respective vehicle-treated group, and are therefore expressed as with or without (±) myostatin. D) Representative images and mean myotube diameter of C2C12 myotubes transfected (as myoblasts) with either DsRed or DN FoxO-DsRed and treated with vehicle or myostatin for 96 h. *P < 0.05 vs. control empty vector or vehicle.
DISCUSSION
Various markers of the FoxO signaling pathway are increased in skeletal muscle during a variety of cachectic conditions, including sepsis (21, 23), cancer (25, 31), and heart failure (66). However, to our knowledge, the current study is the first to show an increase in FoxO transcriptional activity during any cachectic condition and determine the physiological significance of this increase on the progression of the muscle atrophy program. The findings in the present study demonstrate that FoxO transcriptional activity is increased in two metabolically distinct skeletal muscles during both cancer and sepsis, and is required for the increased transcription of several atrophy-related genes and the associated muscle fiber atrophy. Furthermore, inhibition of FoxO transcriptional activity in control muscles increases satellite cell proliferation and fusion with myofibers, increases MyoD transcription, inhibits myostatin transcription and Smad transcriptional activity, and causes significant skeletal muscle fiber hypertrophy that requires de novo protein synthesis.
Our findings of increased FoxO signaling in skeletal muscle during cachexia are consistent with the findings of others (21, 23, 25, 31, 66). During sepsis, FoxO1 and FoxO3a mRNA levels are increased in the skeletal muscle of rats as early as 8 h after the induction of sepsis (23) and remain increased after 24 h (67). Further, the ratio of phosphorylated to total FoxO1 protein is decreased and nuclear FoxO1 protein and FoxO1 DNA binding are increased 16 h after the induction of sepsis (23). While substantially fewer data exist on FoxO signaling in skeletal muscle during cancer cachexia, the ratio of phosphorylated FoxO3a to total FoxO3a is significantly lower in the muscles of human cachectic patients with pancreatic carcinoma compared to noncachectic controls (26), and FoxO1 mRNA is increased in the skeletal muscles of Yoshida ascites hepatoma-bearing rats (31) and in the muscles of sarcoma-180 (S-180) ascites tumor-bearing mice (25). Further, direct evidence supporting a role for FoxO in regulating muscle mass during cancer was demonstrated by treatment of S-180 tumor-bearing mice with a FoxO1 RNA oligonucleotide. This treatment knocked down FoxO1 mRNA expression by 75% and increased muscle mass in tumor-bearing mice by 32% compared to mock-treated tumor-bearing mice (25). Although the magnitude of atrophy attenuation was not reported in this previous study, the findings demonstrate that FoxO1 is necessary for normal muscle wasting during cancer cachexia.
In the current study, we found that muscle fiber atrophy in response to both cancer and sepsis was greater in the TA muscle than the soleus. This finding was not surprising, since cachexia is known to cause more pronounced atrophy in glycolytic muscles than in oxidative muscles when compared at the same time point (48, 50, 68). However, interestingly, the magnitude of increase in atrophy-related genes was actually higher in the soleus than in the TA. Since the transcriptional changes that lead to the induction of atrophy-related genes precede muscle fiber atrophy, we hypothesize that the changes in atrophy gene expression in the glycolytic TA may have peaked prior to the time point at which we measured them. In contrast, the atrophy program may take longer to develop in the soleus, and the time point at which we measured atrophy-related genes may be closer to their peak increase in the soleus. This may explain the lesser magnitude of atrophy in the soleus when compared to the TA at the time point measured. In support of this hypothesis, in the current study, at 4 d after the induction of sepsis, the increase in atrogin-1 mRNA levels was 5-fold higher in the soleus (21-fold) compared to the TA (4-fold), yet it has previously been shown that at 16 h after the induction of sepsis in rats, the increase in atrogin-1 mRNA levels is ∼3-fold higher in the glycolytic extensor digitorum longus (EDL) muscle (15-fold) compared to the soleus (5-fold) (37). Moreover, a >25-fold increase in MuRF1 mRNA and a 15-fold increase in atrogin-1 mRNA were reported in the glycolytic EDL muscle 16 h after the induction of sepsis (37), whereas in the current study, we show a much more modest 2.5-fold increase in MuRF1 mRNA and a 4-fold increase in atrogin-1 mRNA in the glycolytic TA muscle at 4 d after the induction of sepsis. On the basis of these combined findings, we hypothesize that if the cachectic conditions were carried out for a longer duration, the soleus would continue to atrophy, while the atrophy in the TA might begin to plateau. Clearly, time-course experiments would need to be conducted to test this hypothesis.
Notably, expression of DN FoxO significantly attenuated the increased mRNA levels of MuRF1, atrogin-1, cathepsin L, and Bnip3 in the soleus and/or TA muscles during cachexia. This finding demonstrates that an increase in FoxO transcriptional activity is required for the normal increase in these atrophy genes, which has been shown previously for atrogin-1 and MuRF1 in whole muscle during disuse (51) and in C2C12 myotubes treated with dex (24). However, to our knowledge, the work presented here is the first to show this in whole muscle during cachexia. The promoter regions of atrogin-1, MuRF1, cathepsin L, and Bnip3 each have putative FoxO-binding elements (FBEs). Therefore, the attenuated increase in these genes during cachexia in muscles expressing DN FoxO is presumably due to DN FoxO binding to these FBEs and blocking gene transcription by endogenous FoxO.
During sepsis, the inhibition of atrophy in muscle fibers expressing DN FoxO demonstrates that FoxO transcriptional activation is necessary (directly and/or indirectly) for the normal muscle fiber atrophy during sepsis. In addition, since overexpression of DN FoxO resulted in complete inhibition of fiber atrophy in the soleus, compared to 68% attenuation in the TA, these findings further suggest that additional atrophic signaling pathways that are activated in skeletal muscle during sepsis perhaps play a more predominant role in causing fiber atrophy in the glycolytic TA than the oxidative soleus, at least up to the time point that fiber atrophy was measured. Indeed, other transcription factors that are implicated in the muscle atrophy program, including nuclear factor-κB (NF-κB), activator protein-1 (AP-1), and CCAAT/enhancer binding protein (C/EBP), are increased in the glycolytic EDL muscle of septic rats (69, 70), whereas no data exist to show their up-regulation in any oxidative muscle during sepsis.
Surprisingly, overexpression of DN FoxO in the TA muscle of LLC tumor-bearing mice not only abolished the fiber atrophy, but induced hypertrophy to a magnitude directly comparable to control muscles overexpressing DN FoxO. This suggests that DN FoxO induces muscle fiber hypertrophy and that this completely overrides the muscle atrophy program in mice bearing the LLC tumor. However, this was not the case in the TA muscle during sepsis, since fiber size still decreased by 32% in fibers expressing DN FoxO. Therefore, in contrast to tumor-bearing conditions, the hypertrophy effect of DN FoxO was overridden by the atrophy program during sepsis.
The hypertrophy in muscle fibers overexpressing DN FoxO during control conditions indicates a role for FoxO in regulating basal muscle mass. This hypertrophy required de novo protein synthesis, since treatment with the global protein synthesis inhibitor cycloheximide abolished the DN FoxO-induced muscle fiber hypertrophy. Although measurement of specific protein synthesis signaling cascades in the presence of DN FoxO was beyond the scope of the current study, there is evidence that an increase in FoxO is sufficient to repress the Akt/mTOR signaling cascade. Under permissive conditions, active Akt promotes the formation of the mTOR complex 1 (mTORC1), which, in turn, phosphorylates and activates S6K1 and inhibits eIF4E-binding protein (4EBP1) promoting protein synthesis (reviewed in ref. 71). Expression of a constitutively active (CA) FoxO1 decreases mTORC1 activity in Rat1a embryonic fibroblasts, while knockdown of FoxO1 in mouse embryonic fibroblasts increases mTORC1 activity and S6K1 phosphorylation (72). Similarly, in C2C12 cells, expression of CA FoxO1 promotes degradation of raptor and mTOR, two components of mTORC1, as well as S6K1 (73). In the present study, we found that treatment of C2C12 myotubes with rapamycin, which inhibits mTORC1 (74), prevents the DN FoxO-induced increase in myotube diameter. This demonstrates that DN FoxO-induced hypertrophy requires active signaling through the mTORC1 protein complex.
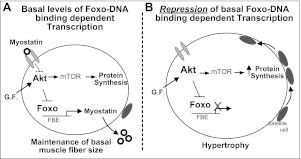
In addition to determining the requirement of de novo protein synthesis for DN FoxO-induced hypertrophy, we also investigated the involvement of muscle satellite cells. Satellite cells are activated by growth or regenerative stimuli, proliferate, and then incorporate into existing myofibers. This addition of myonuclei may serve to maintain the nuclear/cytosol ratio, or the myonuclear domain, during myofiber hypertrophy. We found that muscles expressing DN FoxO exhibited an increased number of BrdU-positive nuclei inside the basal lamina and inside dystrophin, indicative of satellite cell proliferation, and fusion with myofibers. This could be a secondary response to the hypertrophy caused by DN FoxO to maintain the myonuclear domain. Alternatively, inhibition of FoxO transcriptional activity in myofibers could provide a signal to adjoining satellite cells to proliferate. One potential intermediate protein in this signaling process could be myostatin. In adult muscle, myostatin is believed to maintain satellite cells in a quiescent state, and during periods of growth or regeneration, inhibition of myostatin allows satellite cells to reenter the cell cycle and proliferate (62). Given that DN FoxO potently inhibited myostatin mRNA levels in the current study, this could explain the increased satellite cell proliferation. This decrease in myostatin expression in DN FoxO-expressing muscle is in close agreement with previous work showing that FoxO1 knockdown in both C2C12 cells and whole skeletal muscle decreases myostatin expression (25). There are 5 putative FBEs in the mouse myostatin promoter, and, at least in C2C12 myotubes, FoxO1 binding to these FBEs increases myostatin transcription (75). Furthermore, mutation of the most proximal of these FBEs in the myostatin promoter decreases basal promoter activity ∼60%, and basal myostatin promoter activity is decreased ∼80% in a truncated myostatin promoter reporter that lacks all 5 FBEs (75). This suggests that basal levels of FoxO transcription factors bind to and maintain basal levels of myostatin transcriptional activity, which is further supported by the current study. In addition to potentially explaining the increase in satellite cell proliferation, inhibition of myostatin and inhibition of Smads also leads to significant skeletal muscle hypertrophy (63, 65). Indeed, in the current study, we found that a decrease in myostatin is required for the DN FoxO-induced myotube hypertrophy. Given that this DN FoxO-induced hypertrophy also requires mTOR, it is possible that DN FoxO decreases myostatin expression, which, in turn, increases mTOR signaling and protein synthesis, resulting in muscle hypertrophy. In support of this speculation, it has been shown that decreases in myostatin signaling cause an up-regulation of mTOR signaling, increase protein synthesis, and induce muscle hypertrophy (76, 77). Therefore, the inhibition of myostatin by DN FoxO could explain both the increased satellite cell proliferation and the muscle fiber hypertrophy (Fig. 8).
Figure 8.
Proposed mechanisms for Foxo-DNA binding-dependent transcription in the maintenance of muscle fiber size. A) During normal conditions, maintenance of muscle fiber size is controlled through balancing protein synthesis and protein degradation. Growth factor (G.F.) stimulation and myostatin-mediated repression contribute to basal levels of Akt signaling and protein synthesis, mediated through mTOR. Foxo signaling, while predominantly inactive under normal conditions, regulates the transcription of myostatin, as well as genes involved in protein degradation, which at basal levels, helps to balance protein synthesis and maintain muscle fiber size. In addition, satellite cells are maintained in a quiescent state during normal conditions, which is mediated, in part, through myostatin. B) In the current study, we demonstrate that muscle hypertrophy accompanied by satellite cell proliferation and fusion into muscle fibers is induced following repression of basal levels of Foxo-DNA binding-dependent transcription. This hypertrophy required active mTOR signaling and de novo protein synthesis. Because the hypertrophy induced via repression of Foxo also required a decrease in myostatin, which is known to increase satellite cell proliferation, mTOR signaling, and protein synthesis, as well as cause hypertrophy, our data support a model in which the basal activity of Foxo maintains muscle size through regulating myostatin transcription.
In further support of the proposal that inhibition of FoxO transcriptional activity in myofibers signals to adjoining satellite cells to proliferate, we found that whole muscles expressing DN FoxO showed increased mRNA levels of MyoD. Although this increase in MyoD could be transcribed from myonuclear DNA, since we show an increase in satellite cell proliferation in DN FoxO-injected muscles, it is likely derived from satellite cell DNA, since this myogenic transcription factor is increased in active and proliferating satellite cells (59). Our finding that DN FoxO increases MyoD mRNA is in agreement with Liu et al. (25), who reported that knockdown of FoxO1 increases mRNA levels of MyoD in whole muscle, and with Kitamura et al. (78), who found an increase in MyoD expression in skeletal-muscle-specific FoxO1-knockout mice.
In summary, FoxO transcriptional activity is increased in both the TA and the soleus muscles of LLC tumor-bearing mice and septic mice, which is required for the increased transcription of atrophy-related genes and muscle fiber atrophy during these cachectic conditions. Furthermore, inhibition of basal FoxO activity in normal muscle decreases myostatin expression and Smad transcriptional activity, increases MyoD expression, increases satellite cell proliferation and fusion into myofibers, and causes muscle fiber hypertrophy, which requires de novo protein synthesis. These findings highlight the central role of FoxO-dependent transcription in regulating multiple variables important to the maintenance of muscle mass during both pathophysiological and physiological conditions.
Acknowledgments
This work was supported by Bankhead Coley Cancer Research Program grant 09BN-09, and U.S. National Institute of Arthritis and Musculoskeletal and Skin Diseases grants R03AR056418 and R01AR060209 (to A.R.J.). S.M.S. is supported by U.S. National Institute of Child Health and Human Development grant T32-HD-043730.
Footnotes
- BrdU
- bromodeoxyuridine
- CLP
- cecal ligation and puncture
- CSA
- cross-sectional area
- dex
- dexamethasone
- DN
- dominant negative
- EDL
- extensor digitorum longus
- FBE
- FoxO-binding element
- FoxO
- forkhead box O
- LLC
- Lewis lung carcinoma
- S-180
- sarcoma-180
- TA
- tibialis anterior
REFERENCES
- 1. Evans W. J., Morley J. E., Argiles J., Bales C., Baracos V., Guttridge D., Jatoi A., Kalantar-Zadeh K., Lochs H., Mantovani G., Marks D., Mitch W. E., Muscaritoli M., Najand A., Ponikowski P., Rossi Fanelli F., Schambelan M., Schols A., Schuster M., Thomas D., Wolfe R., Anker S. D. (2008) Cachexia: a new definition. Clin. Nutr. 27, 793–799 [DOI] [PubMed] [Google Scholar]
- 2. Hasselgren P. O., Menconi M. J., Fareed M. U., Yang H., Wei W., Evenson A. (2005) Novel aspects on the regulation of muscle wasting in sepsis. Int. J. Biochem. Cell Biol. 37, 2156–2168 [DOI] [PubMed] [Google Scholar]
- 3. Vigano A., Bruera E., Jhangri G. S., Newman S. C., Fields A. L., Suarez-Almazor M. E. (2000) Clinical survival predictors in patients with advanced cancer. Arch. Intern. Med. 160, 861–868 [DOI] [PubMed] [Google Scholar]
- 4. Fearon K. C., Voss A. C., Hustead D. S. (2006) Definition of cancer cachexia: effect of weight loss, reduced food intake, and systemic inflammation on functional status and prognosis. Am. J. Clin. Nutr. 83, 1345–1350 [DOI] [PubMed] [Google Scholar]
- 5. Anker S. D., Ponikowski P., Varney S., Chua T. P., Clark A. L., Webb-Peploe K. M., Harrington D., Kox W. J., Poole-Wilson P. A., Coats A. J. (1997) Wasting as independent risk factor for mortality in chronic heart failure. Lancet 349, 1050–1053 [DOI] [PubMed] [Google Scholar]
- 6. Wang A. Y., Sea M. M., Tang N., Sanderson J. E., Lui S. F., Li P. K., Woo J. (2004) Resting energy expenditure and subsequent mortality risk in peritoneal dialysis patients. J. Am. Soc. Nephrol. 15, 3134–3143 [DOI] [PubMed] [Google Scholar]
- 7. Kotler D. P., Tierney A. R., Wang J., Pierson R. N., Jr. (1989) Magnitude of body-cell-mass depletion and the timing of death from wasting in AIDS. Am. J. Clin. Nutr. 50, 444–447 [DOI] [PubMed] [Google Scholar]
- 8. Harvey K. B., Bothe A., Jr., Blackburn G. L. (1979) Nutritional assessment and patient outcome during oncological therapy. Cancer. 43, 2065–2069 [DOI] [PubMed] [Google Scholar]
- 9. Ross P. J., Ashley S., Norton A., Priest K., Waters J. S., Eisen T., Smith I. E., O'Brien M. E. (2004) Do patients with weight loss have a worse outcome when undergoing chemotherapy for lung cancers? Br. J. Cancer 90, 1905–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dewys W. D., Begg C., Lavin P. T., Band P. R., Bennett J. M., Bertino J. R., Cohen M. H., Douglass H. O., Jr., Engstrom P. F., Ezdinli E. Z., Horton J., Johnson G. J., Moertel C. G., Oken M. M., Perlia C., Rosenbaum C., Silverstein M. N., Skeel R. T., Sponzo R. W., Tormey D. C. (1980) Prognostic effect of weight loss prior to chemotherapy in cancer patients. Eastern Cooperative Oncology Group. Am. J. Med. 69, 491–497 [DOI] [PubMed] [Google Scholar]
- 11. Fearon K. C. (2008) Cancer cachexia: developing multimodal therapy for a multidimensional problem. Eur. J. Cancer 44, 1124–1132 [DOI] [PubMed] [Google Scholar]
- 12. Vaughan G. M., Becker R. A., Allen J. P., Goodwin C. W., Jr., Pruitt B. A., Jr., Mason A. D., Jr. (1982) Cortisol and corticotrophin in burned patients. J. Trauma 22, 263–273 [DOI] [PubMed] [Google Scholar]
- 13. Opal S. M. (2010) Endotoxins and other sepsis triggers. Contrib. Nephrol. 167, 14–24 [DOI] [PubMed] [Google Scholar]
- 14. Fortunati N., Manti R., Birocco N., Pugliese M., Brignardello E., Ciuffreda L., Catalano M. G., Aragno M., Boccuzzi G. (2007) Pro-inflammatory cytokines and oxidative stress/antioxidant parameters characterize the bio-humoral profile of early cachexia in lung cancer patients. Oncol. Rep. 18, 1521–1527 [PubMed] [Google Scholar]
- 15. Krzystek-Korpacka M., Matusiewicz M., Diakowska D., Grabowski K., Blachut K., Kustrzeba-Wojcicka I., Banas T. (2007) Impact of weight loss on circulating IL-1, IL-6, IL-8, TNF-alpha, VEGF-A, VEGF-C and midkine in gastroesophageal cancer patients. Clin. Biochem. 40, 1353–1360 [DOI] [PubMed] [Google Scholar]
- 16. Hall-Angeras M., Angeras U., Zamir O., Hasselgren P. O., Fischer J. E. (1990) Interaction between corticosterone and tumor necrosis factor stimulated protein breakdown in rat skeletal muscle, similar to sepsis. Surgery 108, 460–466 [PubMed] [Google Scholar]
- 17. Costelli P., Carbo N., Tessitore L., Bagby G. J., Lopez-Soriano F. J., Argiles J. M., Baccino F. M. (1993) Tumor necrosis factor-alpha mediates changes in tissue protein turnover in a rat cancer cachexia model. J. Clin. Invest. 92, 2783–2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Llovera M., Carbo N., Garcia-Martinez C., Costelli P., Tessitore L., Baccino F. M., Agell N., Bagby G. J., Lopez-Soriano F. J., Argiles J. M. (1996) Anti-TNF treatment reverts increased muscle ubiquitin gene expression in tumour-bearing rats. Biochem. Biophys. Res. Commun. 221, 653–655 [DOI] [PubMed] [Google Scholar]
- 19. Strassmann G., Fong M., Kenney J. S., Jacob C. O. (1992) Evidence for the involvement of interleukin 6 in experimental cancer cachexia. J. Clin. Invest. 89, 1681–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Matthys P., Heremans H., Opdenakker G., Billiau A. (1991) Anti-interferon-gamma antibody treatment, growth of Lewis lung tumours in mice and tumour-associated cachexia. Eur. J. Cancer 27, 182–187 [DOI] [PubMed] [Google Scholar]
- 21. Crossland H., Constantin-Teodosiu D., Gardiner S. M., Constantin D., Greenhaff P. L. (2008) A potential role for Akt/FOXO signalling in both protein loss and the impairment of muscle carbohydrate oxidation during sepsis in rodent skeletal muscle. J. Physiol. 586, 5589–5600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moylan J. S., Smith J. D., Chambers M. A., McLoughlin T. J., Reid M. B. (2008) TNF induction of atrogin-1/MAFbx mRNA depends on Foxo4 expression but not AKT-Foxo1/3 signaling. Am. J. Physiol. Cell Physiol. 295, C986–C993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Smith I. J., Alamdari N., O'Neal P., Gonnella P., Aversa Z., Hasselgren P. O. (2010) Sepsis increases the expression and activity of the transcription factor forkhead box O 1 (FOXO1) in skeletal muscle by a glucocorticoid-dependent mechanism. Int. J. Biochem. Cell Biol. 42, 701–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sandri M., Sandri C., Gilbert A., Skurk C., Calabria E., Picard A., Walsh K., Schiaffino S., Lecker S. H., Goldberg A. L. (2004) Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117, 399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu C. M., Yang Z., Liu C. W., Wang R., Tien P., Dale R., Sun L. Q. (2007) Effect of RNA oligonucleotide targeting Foxo-1 on muscle growth in normal and cancer cachexia mice. Cancer Gene Ther. 14, 945–952 [DOI] [PubMed] [Google Scholar]
- 26. Schmitt T. L., Martignoni M. E., Bachmann J., Fechtner K., Friess H., Kinscherf R., Hildebrandt W. (2007) Activity of the Akt-dependent anabolic and catabolic pathways in muscle and liver samples in cancer-related cachexia. J. Mol. Med. 85, 647–654 [DOI] [PubMed] [Google Scholar]
- 27. Kamei Y., Miura S., Suzuki M., Kai Y., Mizukami J., Taniguchi T., Mochida K., Hata T., Matsuda J., Aburatani H., Nishino I., Ezaki O. (2004) Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated Type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. J. Biol. Chem. 279, 41114–41123 [DOI] [PubMed] [Google Scholar]
- 28. Senf S. M., Dodd S. L., Judge A. R. (2010) FOXO signaling is required for disuse muscle atrophy and is directly regulated by Hsp70. Am. J. Physiol. Cell Physiol. 298, C38–C45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reed S. A., Senf S. M., Cornwell E. W., Kandarian S. C., Judge A. R. (2011) Inhibition of IκB kinase alpha (IKKα) or IKKβ (IKKβ) plus forkhead box O (Foxo) abolishes skeletal muscle atrophy. Biochem. Biophys. Res. Commun. 405, 491–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yamazaki Y., Kamei Y., Sugita S., Akaike F., Kanai S., Miura S., Hirata Y., Troen B. R., Kitamura T., Nishino I., Suganami T., Ezaki O., Ogawa Y. (2010) The cathepsin L gene is a direct target of FOXO1 in skeletal muscle. Biochem. J. 427, 171–178 [DOI] [PubMed] [Google Scholar]
- 31. Lecker S. H., Jagoe R. T., Gilbert A., Gomes M., Baracos V., Bailey J., Price S. R., Mitch W. E., Goldberg A. L. (2004) Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 18, 39–51 [DOI] [PubMed] [Google Scholar]
- 32. Mammucari C., Milan G., Romanello V., Masiero E., Rudolf R., Del Piccolo P., Burden S. J., Di Lisi R., Sandri C., Zhao J., Goldberg A. L., Schiaffino S., Sandri M. (2007) FoxO3 controls autophagy in skeletal muscle in vivo. Cell. Metab. 6, 458–471 [DOI] [PubMed] [Google Scholar]
- 33. Stevenson E. J., Giresi P. G., Koncarevic A., Kandarian S. C. (2003) Global analysis of gene expression patterns during disuse atrophy in rat skeletal muscle. J. Physiol. 551, 33–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bodine S. C., Latres E., Baumhueter S., Lai V. K., Nunez L., Clarke B. A., Poueymirou W. T., Panaro F. J., Na E., Dharmarajan K., Pan Z. Q., Valenzuela D. M., DeChiara T. M., Stitt T. N., Yancopoulos G. D., Glass D. J. (2001) Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294, 1704–1708 [DOI] [PubMed] [Google Scholar]
- 35. Fareed M. U., Evenson A. R., Wei W., Menconi M., Poylin V., Petkova V., Pignol B., Hasselgren P. O. (2006) Treatment of rats with calpain inhibitors prevents sepsis-induced muscle proteolysis independent of atrogin-1/MAFbx and MuRF1 expression. Am. J. Physiol. 290, R1589–R1597 [DOI] [PubMed] [Google Scholar]
- 36. Frost R. A., Nystrom G. J., Jefferson L. S., Lang C. H. (2007) Hormone, cytokine, and nutritional regulation of sepsis-induced increases in atrogin-1 and MuRF1 in skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 292, E501–E512 [DOI] [PubMed] [Google Scholar]
- 37. Menconi M. J., Arany Z. P., Alamdari N., Aversa Z., Gonnella P., O'Neal P., Smith I. J., Tizio S., Hasselgren P. O. (2010) Sepsis and glucocorticoids downregulate the expression of the nuclear cofactor PGC-1β in skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 299, E533–E543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Poylin V., Fareed M. U., O'Neal P., Alamdari N., Reilly N., Menconi M., Hasselgren P. O. (2008) The NF-κB inhibitor curcumin blocks sepsis-induced muscle proteolysis. Mediators Inflamm. 2008, 317851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wray C. J., Mammen J. M., Hershko D. D., Hasselgren P. O. (2003) Sepsis upregulates the gene expression of multiple ubiquitin ligases in skeletal muscle. Int. J. Biochem. Cell Biol. 35, 698–705 [DOI] [PubMed] [Google Scholar]
- 40. Murton A. J., Alamdari N., Gardiner S. M., Constantin-Teodosiu D., Layfield R., Bennett T., Greenhaff P. L. (2009) Effects of endotoxaemia on protein metabolism in rat fast-twitch skeletal muscle and myocardium. PLoS One 4, e6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Alamdari N., Smith I. J., Aversa Z., Hasselgren P. O. (2010) Sepsis and glucocorticoids upregulate p300 and downregulate HDAC6 expression and activity in skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 299, R509–R520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lee S. W., Dai G., Hu Z., Wang X., Du J., Mitch W. E. (2004) Regulation of muscle protein degradation: coordinated control of apoptotic and ubiquitin-proteasome systems by phosphatidylinositol 3 kinase. J. Am. Soc. Nephrol. 15, 1537–1545 [DOI] [PubMed] [Google Scholar]
- 43. Dijkers P. F., Birkenkamp K. U., Lam E. W., Thomas N. S., Lammers J. W., Koenderman L., Coffer P. J. (2002) FKHR-L1 can act as a critical effector of cell death induced by cytokine withdrawal: protein kinase B-enhanced cell survival through maintenance of mitochondrial integrity. J. Cell Biol. 156, 531–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Doppler H., Storz P., Li J., Comb M. J., Toker A. (2005) A phosphorylation state-specific antibody recognizes Hsp27, a novel substrate of protein kinase D. J. Biol. Chem. 280, 15013–15019 [DOI] [PubMed] [Google Scholar]
- 45. Piek E., Westermark U., Kastemar M., Heldin C. H., van Zoelen E. J., Nister M., Ten Dijke P. (1999) Expression of transforming-growth-factor (TGF)-beta receptors and Smad proteins in glioblastoma cell lines with distinct responses to TGF-beta1. Int. J. Cancer 80, 756–763 [DOI] [PubMed] [Google Scholar]
- 46. Schertzer J. D., Lynch G. S. (2008) Plasmid-based gene transfer in mouse skeletal muscle by electroporation. Methods Mol. Biol. 433, 115–125 [DOI] [PubMed] [Google Scholar]
- 47. Schertzer J. D., Plant D. R., Lynch G. S. (2006) Optimizing plasmid-based gene transfer for investigating skeletal muscle structure and function. Mol. Ther. 13, 795–803 [DOI] [PubMed] [Google Scholar]
- 48. Li P., Waters R. E., Redfern S. I., Zhang M., Mao L., Annex B. H., Yan Z. (2007) Oxidative phenotype protects myofibers from pathological insults induced by chronic heart failure in mice. Am. J. Pathol. 170, 599–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Acharyya S., Butchbach M. E., Sahenk Z., Wang H., Saji M., Carathers M., Ringel M. D., Skipworth R. J., Fearon K. C., Hollingsworth M. A., Muscarella P., Burghes A. H., Rafael-Fortney J. A., Guttridge D. C. (2005) Dystrophin glycoprotein complex dysfunction: a regulatory link between muscular dystrophy and cancer cachexia. Cancer Cell 8, 421–432 [DOI] [PubMed] [Google Scholar]
- 50. Minnaard R., Drost M. R., Wagenmakers A. J., van Kranenburg G. P., Kuipers H., Hesselink M. K. (2005) Skeletal muscle wasting and contractile performance in septic rats. Muscle Nerve 31, 339–348 [DOI] [PubMed] [Google Scholar]
- 51. Senf S. M., Dodd S. L., McClung J. M., Judge A. R. (2008) Hsp70 overexpression inhibits NF-κB and Foxo3a transcriptional activities and prevents skeletal muscle atrophy. FASEB J. 22, 3836–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Waddell D. S., Baehr L. M., van den Brandt J., Johnsen S. A., Reichardt H. M., Furlow J. D., Bodine S. C. (2008) The glucocorticoid receptor and FOXO1 synergistically activate the skeletal muscle atrophy-associated MuRF1 gene. Am. J. Physiol. Endocrinol. Metab. 295, E785–E797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhao J., Brault J. J., Schild A., Cao P., Sandri M., Schiaffino S., Lecker S. H., Goldberg A. L. (2007) FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell. Metab. 6, 472–483 [DOI] [PubMed] [Google Scholar]
- 54. Acharyya S., Ladner K. J., Nelsen L. L., Damrauer J., Reiser P. J., Swoap S., Guttridge D. C. (2004) Cancer cachexia is regulated by selective targeting of skeletal muscle gene products. J. Clin. Invest. 114, 370–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Asp M. L., Tian M., Wendel A. A., Belury M. A. (2010) Evidence for the contribution of insulin resistance to the development of cachexia in tumor-bearing mice. Int. J. Cancer 126, 756–763 [DOI] [PubMed] [Google Scholar]
- 56. Favier F. B., Benoit H., Freyssenet D. (2008) Cellular and molecular events controlling skeletal muscle mass in response to altered use. Pflügers Arch. 456, 587–600 [DOI] [PubMed] [Google Scholar]
- 57. Zammit P. S., Golding J. P., Nagata Y., Hudon V., Partridge T. A., Beauchamp J. R. (2004) Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J. Cell Biol. 166, 347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Seale P., Sabourin L. A., Girgis-Gabardo A., Mansouri A., Gruss P., Rudnicki M. A. (2000) Pax7 is required for the specification of myogenic satellite cells. Cell 102, 777–786 [DOI] [PubMed] [Google Scholar]
- 59. Cornelison D. D., Wold B. J. (1997) Single-cell analysis of regulatory gene expression in quiescent and activated mouse skeletal muscle satellite cells. Dev. Biol. 191, 270–283 [DOI] [PubMed] [Google Scholar]
- 60. Yablonka-Reuveni Z., Rivera A. J. (1994) Temporal expression of regulatory and structural muscle proteins during myogenesis of satellite cells on isolated adult rat fibers. Dev. Biol. 164, 588–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Charge S. B., Rudnicki M. A. (2004) Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 84, 209–238 [DOI] [PubMed] [Google Scholar]
- 62. Lee S. J. (2004) Regulation of muscle mass by myostatin. Annu. Rev. Cell Dev. Biol. 20, 61–86 [DOI] [PubMed] [Google Scholar]
- 63. McPherron A. C., Lawler A. M., Lee S. J. (1997) Regulation of skeletal muscle mass in mice by a new TGF-β superfamily member. Nature 387, 83–90 [DOI] [PubMed] [Google Scholar]
- 64. Durieux A. C., Amirouche A., Banzet S., Koulmann N., Bonnefoy R., Pasdeloup M., Mouret C., Bigard X., Peinnequin A., Freyssenet D. (2007) Ectopic expression of myostatin induces atrophy of adult skeletal muscle by decreasing muscle gene expression. Endocrinology 148, 3140–3147 [DOI] [PubMed] [Google Scholar]
- 65. Sartori R., Milan G., Patron M., Mammucari C., Blaauw B., Abraham R., Sandri M. (2009) Smad2 and 3 transcription factors control muscle mass in adulthood. Am. J. Physiol. Cell Physiol. 296, C1248–C1257 [DOI] [PubMed] [Google Scholar]
- 66. Schulze P. C., Fang J., Kassik K. A., Gannon J., Cupesi M., MacGillivray C., Lee R. T., Rosenthal N. (2005) Transgenic overexpression of locally acting insulin-like growth factor-1 inhibits ubiquitin-mediated muscle atrophy in chronic left-ventricular dysfunction. Circ. Res. 97, 418–426 [DOI] [PubMed] [Google Scholar]
- 67. Nystrom G. J., Lang C. H. (2008) Sepsis and AMPK activation by AICAR differentially regulate FoxO-1, -3 and -4 mRNA in striated muscle. Int. J. Clin. Exp. Med. 1, 50–63 [PMC free article] [PubMed] [Google Scholar]
- 68. Hasselgren P. O., James J. H., Benson D. W., Hall-Angeras M., Angeras U., Hiyama D. T., Li S., Fischer J. E. (1989) Total and myofibrillar protein breakdown in different types of rat skeletal muscle: effects of sepsis and regulation by insulin. Metabolism 38, 634–640 [DOI] [PubMed] [Google Scholar]
- 69. Penner C. G., Gang G., Wray C., Fischer J. E., Hasselgren P. O. (2001) The transcription factors NF-κB and AP-1 are differentially regulated in skeletal muscle during sepsis. Biochem. Biophys. Res. Commun. 281, 1331–1336 [DOI] [PubMed] [Google Scholar]
- 70. Penner G., Gang G., Sun X., Wray C., Hasselgren P. O. (2002) C/EBP DNA-binding activity is upregulated by a glucocorticoid-dependent mechanism in septic muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 282, R439–R444 [DOI] [PubMed] [Google Scholar]
- 71. Hay N., Sonenberg N. (2004) Upstream and downstream of mTOR. Genes Dev. 18, 1926–1945 [DOI] [PubMed] [Google Scholar]
- 72. Chen C. C., Jeon S. M., Bhaskar P. T., Nogueira V., Sundararajan D., Tonic I., Park Y., Hay N. (2010) FoxOs inhibit mTORC1 and activate Akt by inducing the expression of Sestrin3 and Rictor. Dev. Cell. 18, 592–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wu A. L., Kim J. H., Zhang C., Unterman T. G., Chen J. (2008) Forkhead box protein O1 negatively regulates skeletal myocyte differentiation through degradation of mammalian target of rapamycin pathway components. Endocrinology 149, 1407–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Feldman M. E., Apsel B., Uotila A., Loewith R., Knight Z. A., Ruggero D., Shokat K. M. (2009) Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 7, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Allen D. L., Unterman T. G. (2007) Regulation of myostatin expression and myoblast differentiation by FoxO and SMAD transcription factors. Am. J. Physiol. Cell Physiol. 292, C188–C199 [DOI] [PubMed] [Google Scholar]
- 76. Lipina C., Kendall H., McPherron A. C., Taylor P. M., Hundal H. S. (2010) Mechanisms involved in the enhancement of mammalian target of rapamycin signalling and hypertrophy in skeletal muscle of myostatin-deficient mice. FEBS Lett. 584, 2403–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Welle S., Mehta S., Burgess K. (2011) Effect of postdevelopmental myostatin depletion on myofibrillar protein metabolism. Am. J. Physiol. Endocrinol. Metab. 300, E993–E1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kitamura T., Kitamura Y. I., Funahashi Y., Shawber C. J., Castrillon D. H., Kollipara R., DePinho R. A., Kitajewski J., Accili D. (2007) A Foxo/Notch pathway controls myogenic differentiation and fiber type specification. J. Clin. Invest. 117, 2477–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]