Abstract
Major histocompatibility complex (MHC) class I and class II are crucial for the function of the human adaptive immune system. A member of the NLR (nucleotide-binding domain, leucine-rich repeat) protein family, NLRC5, has recently been identified as a transcriptional regulator of MHC class I and related genes. While a ‘master regulator’ of MHC class II genes, CIITA, has long been known, NLRC5 specifically associates with and transactivates the proximal promoters of MHC class I genes. In this study, we analyzed the molecular requirements of NLRC5 nuclear import and transactivation activity. We show that NLRC5-mediated MHC class I gene induction requires an intact nuclear localization signal and nuclear distribution of NLRC5. In addition, we find that the nucleotide-binding domain (NBD) of NLRC5 is critical not only for nuclear translocation but also for the transactivation of MHC class I genes. Changing the cellular localization of NLRC5 is likely to immediately impact MHC class I expression as well as MHC class I-mediated antigen presentation. NLRC5 may thus provide a promising target for the modulation of MHC class I antigen presentation, especially in the setting of transplant medicine.
Keywords: MHC class I, NLR proteins, CITA
1. Introduction
Major histocompatibility complex (MHC) class I and class II molecules play key roles in the activation of the adaptive immune system. MHC class I molecules present peptide antigens of intracellular origin to CD8+ T cells, whereas MHC class II molecules present peptide antigens of extracellular sources to CD4+ T cells [1; 2]. The expression of both constitutive and inducible MHC class II requires the transcriptional co-activator CIITA (MHC class II transactivator) [3]. Although CIITA lacks a DNA binding domain, it can activate the promoters of MHC class II genes by forming a nucleoprotein complex called the MHC-enhanceosome [4; 5] with transcription factors including the trimeric RFX protein, CREB/ATF1 family members, and the NF-Y complex [6; 7].
Although CIITA appears to also transactivate MHC class I genes in vitro [6; 8; 9; 10; 11; 12], CIITA deficiency in both human and animal models results in the impaired expression of MHC class II but not class I genes. [3; 13; 14; 15; 16]. This discrepancy is largely explained by the recent discovery of a transactivator of MHC class I genes, NLRC5 [17; 18]. Similar to CIITA, NLRC5 is IFN-γ-inducible and can shuttle into the nucleus through its nuclear localization signal (NLS). However, NLRC5, or class I transactivator (CITA), specifically associates with and transactivates MHC class I promoters, resulting in the expression of MHC class I and related genes such as β2M [17; 18].
Both CIITA and NLRC5 belong to the NLR or nucleotide-binding domain (NBD), leucine-rich repeat (LRR) family of proteins [19; 20]. Nucleotide binding to the NBD of NLR proteins has been proposed to be critical for the function of these proteins [19; 20]. Indeed, point mutations in the nucleotide-binding (Walker A) motif in the NBD of CIITA or NLRC5 resulted in the failure of MHC gene induction [17; 21]. In the present study, we characterized the role of the Walker A motif in NLRC5 transactivation of MHC class I. We found that disruption of the nucleotide-binding motif prevents nuclear import of NLRC5. Enforced nuclear expression of a mutant lacking the Walker A motif could not, however, restore MHC class I gene induction, suggesting that nucleotide binding is required for both NLRC5 nuclear translocation and target gene transactivation.
2. Materials and methods
2.1. Cell lines and reagents
Human embryonic kidney cells (HEK293T: CRL-11268, ATCC) were cultured in Dulbecco's modified eagle medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and penicillin (100 U/ml)/ streptomycin (100 μg/ml, Gibco). Leptomycin B (LMB) was obtained from LC Laboratories.
2.2. Plasmids
Cloning of human GFP-NLRC5, GFP-CIITA and the NLRC5 import mutants NLS I (RRK133/134/135A) and NLS II (KR121/122A) has been described previously [17]. Combining NLSI and NLSII resulted in the double mutant DM (KR121/122A , RRK132/133/134A). All point mutations were introduced using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). Cloning of the NLRC5 Walker A, B and AB constructs has also been described previously [17]. To rescue the nuclear import defect of the Walker A mutant, SV40 NLS (PKKKRKV) sequences were appended at both ends of NLRC5 by PCR using the following primers: F, 5’-ATATGAATTCGGAGCTCCAAAGAAGAAGCGTAAGGTAGACCCCGTTGGCCTCCAGC TCGG-3’; R, 5’-ATATTCTAGATTATACCTTACGCTTCTTCTTTGGAGCAGTACCCCAAGGGGCCTG-3’ (Walker A2xNLS). Using the same primers, a NLRC5 WT construct containing two SV40 NLSs was constructed and used as a control (WT2xNLS). The MHC class I and class II reporter gene constructs used in reporter gene assays were a kind gift from Dr. P.J. van den Elsen (Leiden University).
2.3.Transfections and generation of stable cell lines
HEK293T cells were transiently transfected using polyethylenimine (1 mg PEI/ ml, pH 7.2, Polysciences, Inc.) at a ratio of 1:3 (DNA:PEI). Medium was changed the following day and cells were analyzed 48 hrs post transfection. To select for stable integration of expression plasmids, 2 mg/ml G418 (Gibco) was added to the culture medium 24 hrs after transfection for 10 days. GFP-positive cells were further enriched by cell sorting using a MoFlo high-speed sorter (Dako).
2.4. Microscopy
HEK293T cells were grown overnight on glass coverslips coated with poly-L-lysine (Sigma-Aldrich). Prior to imaging cells were rinsed with PBS and stained with Hoechst 33342 (1 μg/ml, Invitrogen) to stain the nuclei before fixing with 10% phosphate buffered formalin. Coverslips were mounted onto glass slides using ProLong Gold Antifade Reagent (Invitrogen). Epifluorescence microscopy was performed using a Nikon Eclipse E800 (Nikon Instruments). For confocal microscopy, cells were cultured in a coverslip-mounted 20 mm dish (MatTek) 24 hours prior to imaging. Cells were stained with Hoechst 33342 for 10 min and then washed with PBS just prior to imaging. Images were acquired with the Eclipse Ti spinning disk confocal microscope (Nikon) using the 100X objective (1.40 NA) and the ORCA-ER camera (Hamamatsu). GFP was imaged using the solid phase argon laser at 488nm and a 520/30 band pass filter. The Hoechst 33342 stained nuclei were imaged using the solid phase UV laser at 405 nm and a 475/30 band pass filter. Image analysis was performed using ImageJ (NIH).
2.5. Luciferase assay
HEK293T cells were split into 24-wells and co-transfected with 300 ng of either GFP, GFP-NLRC5, or the corresponding mutant expression plasmids and 100 ng of the indicated luciferase reporter construct. 50 ng per well of Renilla plasmid (pRL-null, Promega) was included to allow for normalization of transfection efficiency. Cells were harvested 48 hrs post transfection, and cell lysates were analyzed using the dual-luciferase system (Promega), according to the manufacturer’s instructions.
2.6. Quantitative real-time PCR analysis
RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. First-strand cDNA was synthesized from 1 μg RNA using the qScript Flex cDNA synthesis kit (Quanta Biosciences), and RNA expression was quantified on the 7300 Real-Time PCR System (Applied Biosystems) using the PerfeCTa SYBR Green SuperMix with ROX (Quanta Biosciences), and analyzed using the 7300 System SDS Software (Applied Biosystems). The primers used for amplification have been described previously [17].
2.7. Western blot analysis
Whole cell extracts were prepared using 1x Cell Lysis Buffer (Cell Signaling) supplemented with 1 mM DTT, and 1 mM PMSF. Following SDS-polyacrylamide gel electrophoresis using 4–12% gradient gels (Invitrogen), proteins were transferred to nitrocellulose membranes (HyBond ECL, Amersham) for 2 hrs at 80V. The following antibodies were used for protein detection: anti-MHC class I heavy chain (3B10.7, a kind gift from Dr. P. Cresswell, Yale University), anti-GFP (JL-8 Clontech), anti-β-actin (I-19, Santa Cruz). Blots were developed using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific), and imaged using the Molecular Imager ChemiDoc XRS+ System (Bio-Rad) or exposed to autoradiography film (Denville). Quantification was performed using ImageQuant (Molecular Dynamics).
2.8. Flow cytometry
To determine HLA class I surface expression, cells were stained with phycoerythrin (PE)-conjugated anti-HLA-A,B,C (W6/32, BioLegend) following fixation with 1% paraformaldehyde. FACS analysis was performed using the FACSCalibur system (BD Biosciences) and analyzed using the FlowJo software (Tree Star).
2.9. Statistical analysis
Data were subjected to Student's t test for analysis of statistical significance and plotted using Prism (GraphPad). Results are given as the mean ± SD. A P-value of < 0.05 was considered to be significant.
3. Results
3.1. Nuclear translocation of NLRC5 is required for the induction of MHC class I genes
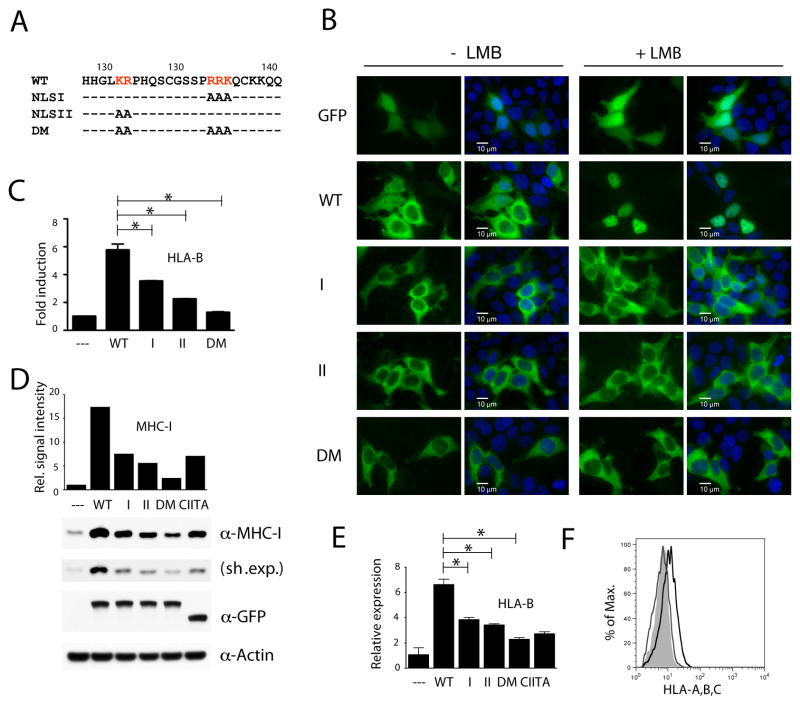
Nuclear translocation of NLRC5 is regulated by a bipartite nuclear localization signal (NLS), which is located between the N-terminal CARD and the central NBD (Fig. 1A). The sequence and position of the NLS is conserved between species and we have previously demonstrated that mutations of either arm of the NLRC5 bipartite NLS result in impaired nuclear import of the protein (Fig. 1B) [17]. In this study we sought to determine if NLRC5-mediated MHC class I gene induction requires nuclear import.
Fig. 1.
NLRC5 requires an intact nuclear localization signal for efficient MHC class I induction.
(A) Sequence of the NLRC5 bipartite NLS in the N-terminus and the indicated mutant versions used in this study. Alanine substitution of the right or left arm of the NLS was used to construct the NLSI and NLSII import mutant expression plasmids. DM: double mutant. (B–F) HEK293T cells were transiently transfected with either expression vectors for GFP, GFP-NLRC5, or GFP-CIITA. (B) Subcellular localization of wild-type and NLS mutant NLRC5 upon LMB treatment (100 nM, for 2 hrs). Scale bar: 10 μm. (C) Reporter gene analysis of the HLA-B promoter. An HLA-B luciferase reporter construct was co-transfected with the indicated plasmids and cell lysates were analyzed 48 hrs post transfection by dual-luciferase assay. Data are a representative of several independent experiments performed in duplicates and are plotted as fold induction with respect to the GFP control vector (--). Error bars represent ± SD. *p < 0.05. (D) Cell extracts were examined for the expression of MHC class I heavy chain by immunoblotting 48 hrs post transfection. Sh. Exp., short exposure. MHC class I signal intensities, obtained by densitometric analysis, were normalized by β-Actin levels (bar diagram). The GFP blot demonstrates equal expression of the NLRC5 and CIITA fusion proteins. (E) RNA samples were isolated and HLA-B transcripts were quantified by qRT-PCR using gene specific primers. (F) Quantitative analysis by flow cytometry using an anti-MHC class I antibody. WT (black line), DM (grey line), empty vector control (shaded histogram).
While the wild-type NLRC5 GFP-fusion protein (WT), which under steady state conditions displays a partial nuclear localization (−LMB), was trapped in the nucleus using the nuclear export inhibitor leptomycin B (+LMB), alanine substitution of either arm of the bipartite NLS (mutants I and II) resulted in a strictly cytosolic localization of the corresponding GFP-fusion proteins (Fig. 1B). Moreover, blocking nuclear export with LMB did not result in nuclear accumulation of the mutants, suggesting that entry in the nucleus is impaired. As can be seen in Fig. 1C–E, this nuclear import defect results in decreased activity of the import mutants I and II as measured by reporter gene assay using an HLA-B promoter construct (Fig. 1C), as well as by quantification of MHC class I protein and HLA-B transcript levels (Fig. 1D,E). While wild-type and mutant NLRC5 fusion proteins were expressed at similar levels (Fig. 1D, GFP blot), disruption of both arms of the bipartite NLS (double mutant, DM) further reduced NLRC5 activity, as can be seen again in the level of HLA-B promoter activity, MHC class I protein and transcript induction (Fig. 1C–E), and in reduced MHC class I surface expression (Fig. 1F). Our findings emphasize the importance of NLRC5 nuclear import for the transactivation of MHC class I genes. It should be noted, however, that even the double import mutant (DM) retains residual activity, which may be due to association of overexpressed proteins with endogenous NLRC5 to form functional protein complexes (Fig. 1C–E).
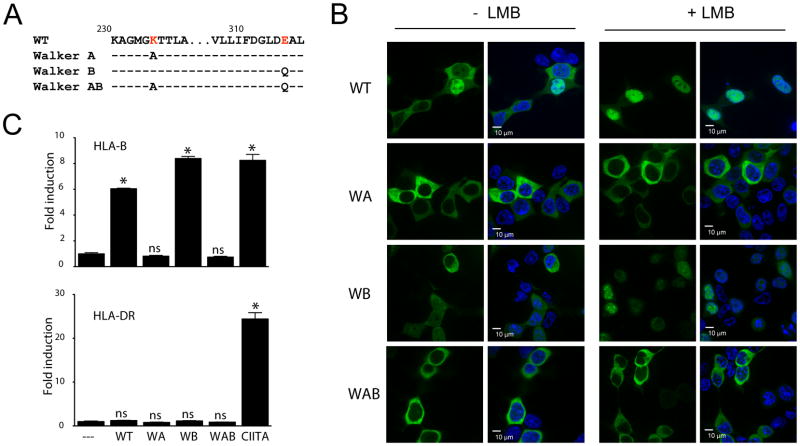
3.2. The nucleotide-binding domain of NLRC5 controls nuclear translocation
To determine whether the NBD also plays a role in controlling the subcellular localization of NLRC5, we generated stable 293T cell lines expressing GFP-fusions of either the wild-type (WT) , the nucleotide-binding deficient Walker A (WA) mutant, the nucleotide hydrolysis-deficient Walker B mutant (WB) or the double mutant combining both alterations (WAB) (Fig. 2A) and imaged by spinning disc confocal microscopy. Disruption of the Walker A motif abolished nuclear import of the GFP-fusion protein, indicating that nucleotide binding is important for NLRC5 activity (Fig. 2B), possibly by promoting a nuclear import-competent conformation. The Walker B mutant behaved similar to the WT protein in both nuclear translocation and reporter gene assays (Fig. 2B,C). Again, LMB treatment resulted in entrapment of NLRC5 WT in the nucleus, while the Walker A and AB mutant remained strictly cytosolic (Fig. 2B). Both the WT and Walker B proteins specifically induced the HLA-B promoter but had no impact on an MHC class II promoter (HLA-DR), while CIITA, used as a positive control, activated the HLA-DR promoter (Fig. 2C). Interestingly, whereas the DM import mutant retained residual activity, the Walker A mutant completely failed to activate the MHC class I promoter (Fig. 1C and Fig. 2C). These data suggest that the activity of the NBD is required not only for nuclear translocation, but may also play a direct role in the transactivation of target genes.
Fig. 2.
An active NBD is required for nuclear translocation of NLRC5.
(A) Sequence of the changes introduced into the Walker A or B motifs of the NBD domain of NLRC5. (B) Subcellular localization of wild-type and NBD mutant NLRC5 upon LMB treatment (100 nM) as observed by spinning disc confocal microscopy. Cell nuclei were stained with Hoechst 33342 (blue). Scale bar: 10 μm. (C) Reporter gene analysis of HLA-B (upper panel) and HLA-DR promoters (lower panel), respectively. Luciferase reporter constructs were co-transfected with the indicated plasmids and cell lysates were analyzed 48 hrs post transfection by dual-luciferase assay. Data were plotted as fold induction with respect to the GFP control vector as the mean ± SD of duplicates. *p < 0.05.
3.3. Transactivation of MHC class I genes requires a functional NBD
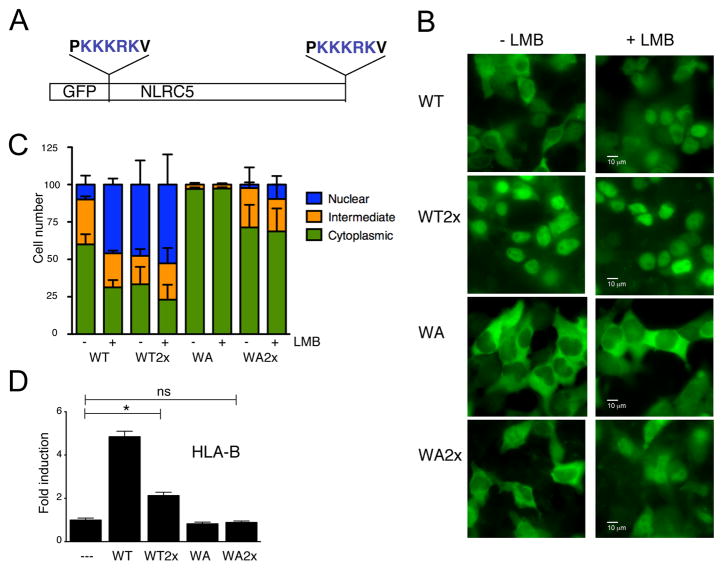
The nuclear import defect of the NLRC5 Walker A mutant makes it impossible to separate the role of the NBD in nuclear import from that in target gene transactivation. To directly examine whether nucleotide binding to the NBD is also required for the transactivation activity of NLRC5, we devised a strategy to overcome the nuclear import deficiency of the Walker A mutant. We appended two SV40 NLS sequences to the N- and C-termini of the NLRC5 coding sequence (Fig. 3A) to restore nuclear translocation of the Walker A mutant. As can be seen in Fig. 3B and 3C, appending two NLS sequences to wild-type NLRC5 resulted in a constitutive nuclear localization of the GFP-fusion protein under steady state conditions (WT-2xNLS). In more than 65% of the cells the WT-2xNLS protein was enriched in the nucleus or found in both nuclear and cytoplamic compartments, and addition of LMB to the medium further increased the percentage of nuclear protein (Fig. 3B, C). Interestingly, while the Walker A mutant is almost exclusively localized to the cytoplasm (97%), the 2xNLS version of the Walker A mutant (WA-2xNLS) was also found in the nucleus and the cytoplasmic distribution of WA-2xNLS was reduced to 71%, almost comparable to that of the wild-type protein (60%), indicating that the nuclear import defect was partially rescued (Fig. 3B,C). However, while adding the NLSs to the Walker A mutant could overcome the impaired nuclear localization, we could not detect any transactivation activity of the protein in a reporter gene assay using an HLA-B promoter luciferase construct (Fig. 3D), or by analyzing HLA-B transcript levels (data not shown). Taken together, our findings suggest that the NBD is critical not only for controlling nuclear transport of NLRC5, but also for its transactivation activity.
Fig. 3.
The NBD is required for transactivation of MHC class I genes.
(A) Schematic representation of NLS-tagged NLRC5. (B–D) HEK293T cells were transiently transfected with the indicated expression vectors. (B) Subcellular localization of wild-type, NBD mutant or NLS tagged NLRC5 upon LMB treatment (100 nM, for 2 hrs) as observed by epifluorescence microscopy. Scale bar: 10 μm. (C) Quantified subcellular localization. Cells were counted in a blind manner as ‘cytosolic’ or ‘nuclear’ if the majority of the GFP signal was detected in the respective compartment, and ‘intermediate’ if the signal intensity in both compartments was comparable. Data was pooled from three independent experiments. Error bars represent ± SD. (D) Reporter gene analysis of the HLA-B promoter. An HLA-B luciferase reporter construct was co-transfectd with the indicated plasmids and cell lysates were analyzed 48 hrs post transfection by dual-luciferase assay. Data are a representative of several independent experiments performed in duplicates and are plotted as fold induction with respect to the GFP control vector (--). Error bars represent ± SD. *p < 0.05.
4. Discussion
We have previously shown that the Walker A mutant of NLRC5 is unable to induce transcription of MHC class I genes [17]. However, the role of the NBD in controlling the subcellular localization and/or function of NLRC5 was not clear. In this study, we have investigated the requirements of NLRC5 nucleocytoplasmic transport. We found that NLRC5-mediated induction of MHC class I genes relies on the nuclear presence of the protein since disruption of the N-terminal NLS resulted in impaired nuclear import as well as in reduced induction of MHC class I genes (Fig. 1). Interestingly, also the Walker A mutant of NLRC5 remained in the cytoplasm even in the presence of the nuclear export inhibitor LMB. This suggests that nucleotide binding to the NBD is required for nuclear import, presumably by facilitating conformational changes or oligomerization of the protein (Fig. 2). The nuclear import defect of the Walker A mutant could be a possible explanation as to why transactivation of MHC class I genes is not observed. Alternatively, to determine if nucleotide binding also plays a more direct role in target gene transactivation, we enforced nuclear localization of the Walker A mutant by appending two additional SV40-derived NLSs. Although NLS tagging partially rescued the impaired nuclear import of the Walker A mutant, no promoter activity of MHC class I gene was detected, suggesting that nucleotide binding is critical for target gene transactivation (Fig. 3). Our findings are reminiscent of what has been previously reported for CIITA, further underlining the similarity of these two MHC transcriptional regulators. Similar to NLRC5, CIITA shuttles between the cytoplasm and the nucleus through complex regulatory mechanisms which, in addition to multiple NLSs, also involves nuclear export signals (NES) and activity of a functional NBD and LRRs [22; 23; 24; 25]. It has previously been shown that GTP binding by the Walker A motif of the NBD is essential for the nuclear localization of CIITA [22]. In addition, consistent with our findings, enforced nuclear localization of a CIITA Walker A mutant did not rescue the transactivation of MHC class II genes [21]. Although it has not been established whether GTP or ATP binds to the NBD of NLRC5, our findings indicate that the NBDs of NLRC5 and CIITA have similar functions in MHC gene induction; NTP binding to the NBD is essential for the nuclear translocation and transactivation activity of both proteins. It has been suggested that nuclear CIITA exists in its GTP-bound form [21]; it is thus tempting to speculate that nuclear NLRC5 may also represent the nucleotide bound form of NLRC5.
Interestingly, the NLRC5 NLS mutants, although displaying a clear cytoplasmic distribution, retained residual transcriptional activity of MHC class I genes (Fig. 1). Similar to CIITA, it has also been demonstrated that NLRC5 can form oligomers through homotypic interactions, although the stoichiometry remains unclear [26]. Therefore, it may be possible that interactions with endogenous protein are responsible for the residual activity of the double NLS mutant, which may enter the nucleus in the form of a heteromeric complex with endogenous NLRC5. A rather unexpected finding is the observation that enforced nuclear localization of WT NLRC5 resulted in a decreased transactivation of MHC class I genes (Fig. 3D). One possibility is that NLS tagging impaired conformational requirements for the transactivation. Alternatively, this finding may be explained by the requirement for an additional cytosolic posttranslational modification that is needed to maintain the transcriptional potency of NLRC5 in the nucleus. It is well known that the activity of transcriptional regulators is controlled by the balance of nuclear to cytoplasmic levels of the corresponding factors. Post-translational modifications such as phosphorylation can often shift the balance from one compartment to the other by impacting import or export kinetics. A prominent example is the STAT (signal transducer and activators of transcription) family of transcription factors, where phosphorylation of STATs results in dimerization and nuclear import due to conformational changes and the exposure of a dimer-specific nuclear import signal [27]. It will be rewarding to investigate potential posttranslational modifications that modify the nucleocytoplasmic balance of NLRC5 as this may reveal alternative strategies to targeting the NTPase domain. Pharmacological intervention with NLRC5 activity may prove beneficial in various aspects of MHC class I biology, including vaccine development, transplant medicine, and cancer immunotherapy.
Highlights.
NLRC5 requires an intact NLS for its function as MHC class I transactivator.
Nuclear presence of NLRC5 is required for MHC class I induction.
Nucleotide-binding controls nuclear import and transactivation activity of NLRC5.
Acknowledgments
We thank Peter Cresswell, Peter van den Elsen, and Marja C.J.A. van Eggermond for providing reagents; Lisa Cameron for assistance with confocal microscopy. This work was supported by grants from the NIH (R01DK074738) and the Crohn’s and Colitis Foundation of America. K.S.K. is a recipient of the Investigator Award from the Cancer Research Institute and the Claudia Adams Barr Award and T.B.M is a recipient of the EMBO Long Term fellowship. The authors have no conflicting financial interests.
Abbreviations
- NBD
NLR proteins, nucleotide-binding domain
- LRR
leucine-rich repeat containing proteins
- CIITA
MHC class II transactivator
- β2M
β2-microglobulin
- NLS
nuclear localization signal
- RFX
regulatory factor X
- ATF1
activating transcription factor 1
- NF-Y
nuclear factor-Y
- LMB
leptomycin B
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Germain RN, Margulies DH. The biochemistry and cell biology of antigen processing and presentation. Annu Rev Immunol. 1993;11:403–50. doi: 10.1146/annurev.iy.11.040193.002155. [DOI] [PubMed] [Google Scholar]
- 2.Oldstone MB. Virus-lymphoid cell interactions. Proc Natl Acad Sci U S A. 1996;93:12756–8. doi: 10.1073/pnas.93.23.12756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steimle V, Otten LA, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome) Cell. 1993;75:135–46. [PubMed] [Google Scholar]
- 4.Beresford GW, Boss JM. CIITA coordinates multiple histone acetylation modifications at the HLA-DRA promoter. Nat Immunol. 2001;2:652–7. doi: 10.1038/89810. [DOI] [PubMed] [Google Scholar]
- 5.Masternak K, Muhlethaler-Mottet A, Villard J, Zufferey M, Steimle V, Reith W. CIITA is a transcriptional coactivator that is recruited to MHC class II promoters by multiple synergistic interactions with an enhanceosome complex. Genes Dev. 2000;14:1156–66. [PMC free article] [PubMed] [Google Scholar]
- 6.Reith W, Mach B. The bare lymphocyte syndrome and the regulation of MHC expression. Annu Rev Immunol. 2001;19:331–73. doi: 10.1146/annurev.immunol.19.1.331. [DOI] [PubMed] [Google Scholar]
- 7.Ting JP, Trowsdale J. Genetic control of MHC class II expression. Cell. 2002;109(Suppl):S21–33. doi: 10.1016/s0092-8674(02)00696-7. [DOI] [PubMed] [Google Scholar]
- 8.Boss JM, Jensen PE. Transcriptional regulation of the MHC class II antigen presentation pathway. Curr Opin Immunol. 2003;15:105–11. doi: 10.1016/s0952-7915(02)00015-8. [DOI] [PubMed] [Google Scholar]
- 9.Martin BK, Chin KC, Olsen JC, Skinner CA, Dey A, Ozato K, Ting JP. Induction of MHC class I expression by the MHC class II transactivator CIITA. Immunity. 1997;6:591–600. doi: 10.1016/s1074-7613(00)80347-7. [DOI] [PubMed] [Google Scholar]
- 10.van den Elsen PJ, Gobin SJ, van Eggermond MC, Peijnenburg A. Regulation of MHC class I and II gene transcription: differences and similarities. Immunogenetics. 1998;48:208–21. doi: 10.1007/s002510050425. [DOI] [PubMed] [Google Scholar]
- 11.van den Elsen PJ, Peijnenburg A, van Eggermond MC, Gobin SJ. Shared regulatory elements in the promoters of MHC class I and class II genes. Immunol Today. 1998;19:308–12. doi: 10.1016/s0167-5699(98)01287-0. [DOI] [PubMed] [Google Scholar]
- 12.Gobin SJ, Peijnenburg A, Keijsers V, van den Elsen PJ. Site alpha is crucial for two routes of IFN gamma-induced MHC class I transactivation: the ISRE-mediated route and a novel pathway involving CIITA. Immunity. 1997;6:601–11. doi: 10.1016/s1074-7613(00)80348-9. [DOI] [PubMed] [Google Scholar]
- 13.Benichou B, Strominger JL. Class II-antigen-negative patient and mutant B-cell lines represent at least three, and probably four, distinct genetic defects defined by complementation analysis. Proc Natl Acad Sci U S A. 1991;88:4285–8. doi: 10.1073/pnas.88.10.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang CH, Guerder S, Hong SC, van Ewijk W, Flavell RA. Mice lacking the MHC class II transactivator (CIITA) show tissue-specific impairment of MHC class II expression. Immunity. 1996;4:167–78. doi: 10.1016/s1074-7613(00)80681-0. [DOI] [PubMed] [Google Scholar]
- 15.Itoh-Lindstrom Y, Piskurich JF, Felix NJ, Wang Y, Brickey WJ, Platt JL, Koller BH, Ting JP. Reduced IL-4-, lipopolysaccharide-, and IFN-gamma-induced MHC class II expression in mice lacking class II transactivator due to targeted deletion of the GTP-binding domain. J Immunol. 1999;163:2425–31. [PubMed] [Google Scholar]
- 16.Williams GS, Malin M, Vremec D, Chang CH, Boyd R, Benoist C, Mathis D. Mice lacking the transcription factor CIITA--a second look. Int Immunol. 1998;10:1957–67. doi: 10.1093/intimm/10.12.1957. [DOI] [PubMed] [Google Scholar]
- 17.Meissner TB, Li A, Biswas A, Lee KH, Liu YJ, Bayir E, Iliopoulos D, van den Elsen PJ, Kobayashi KS. NLR family member NLRC5 is a transcriptional regulator of MHC class I genes. Proc Natl Acad Sci U S A. 2010;107:13794–9. doi: 10.1073/pnas.1008684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meissner TB, Li A, Kobayashi KS. NLRC5: a newly discovered MHC class I transactivator (CITA) Microbes Infect. 2011 doi: 10.1016/j.micinf.2011.12.007. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilmanski JM, Petnicki-Ocwieja T, Kobayashi KS. NLR proteins: integral members of innate immunity and mediators of inflammatory diseases. J Leukoc Biol. 2008;83:13–30. doi: 10.1189/jlb.0607402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ting JP, Lovering RC, Alnemri ES, Bertin J, Boss JM, Davis BK, Flavell RA, Girardin SE, Godzik A, Harton JA, Hoffman HM, Hugot JP, Inohara N, Mackenzie A, Maltais LJ, Nunez G, Ogura Y, Otten LA, Philpott D, Reed JC, Reith W, Schreiber S, Steimle V, Ward PA. The NLR gene family: a standard nomenclature. Immunity. 2008;28:285–7. doi: 10.1016/j.immuni.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bewry NN, Bolick SC, Wright KL, Harton JA. GTP-dependent recruitment of CIITA to the class II major histocompatibility complex promoter. J Biol Chem. 2007;282:26178–84. doi: 10.1074/jbc.M611747200. [DOI] [PubMed] [Google Scholar]
- 22.Harton JA, Cressman DE, Chin KC, Der CJ, Ting JP. GTP binding by class II transactivator: role in nuclear import. Science. 1999;285:1402–5. doi: 10.1126/science.285.5432.1402. [DOI] [PubMed] [Google Scholar]
- 23.Cressman DE, Chin KC, Taxman DJ, Ting JP. A defect in the nuclear translocation of CIITA causes a form of type II bare lymphocyte syndrome. Immunity. 1999;10:163–71. doi: 10.1016/s1074-7613(00)80017-5. [DOI] [PubMed] [Google Scholar]
- 24.Spilianakis C, Papamatheakis J, Kretsovali A. Acetylation by PCAF enhances CIITA nuclear accumulation and transactivation of major histocompatibility complex class II genes. Mol Cell Biol. 2000;20:8489–98. doi: 10.1128/mcb.20.22.8489-8498.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hake SB, Masternak K, Kammerbauer C, Janzen C, Reith W, Steimle V. CIITA leucine-rich repeats control nuclear localization, in vivo recruitment to the major histocompatibility complex (MHC) class II enhanceosome, and MHC class II gene transactivation. Mol Cell Biol. 2000;20:7716–25. doi: 10.1128/mcb.20.20.7716-7725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis BK, Roberts RA, Huang MT, Willingham SB, Conti BJ, Brickey WJ, Barker BR, Kwan M, Taxman DJ, Accavitti-Loper MA, Duncan JA, Ting JP. Cutting edge: NLRC5-dependent activation of the inflammasome. J Immunol. 2011;186:1333–7. doi: 10.4049/jimmunol.1003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vinkemeier U. Getting the message across, STAT! Design principles of a molecular signaling circuit. J Cell Biol. 2004;167:197–201. doi: 10.1083/jcb.200407163. [DOI] [PMC free article] [PubMed] [Google Scholar]