This article identifies the peroxisomal enzyme glycolate oxidase (GOX) as an essential element of nonhost resistance and provides evidence that apart from NADPH oxidase–generated H2O2, GOX is an alternate source for H2O2 production and plays a crucial role during both gene-for-gene and nonhost resistance by regulating genes from different defense signal transduction cascades.
Abstract
In contrast to gene-for-gene disease resistance, nonhost resistance governs defense responses to a broad range of potential pathogen species. To identify specific genes involved in the signal transduction cascade associated with nonhost disease resistance, we used a virus-induced gene-silencing screen in Nicotiana benthamiana, and identified the peroxisomal enzyme glycolate oxidase (GOX) as an essential component of nonhost resistance. GOX-silenced N. benthamiana and Arabidopsis thaliana GOX T-DNA insertion mutants are compromised for nonhost resistance. Moreover, Arabidopsis gox mutants have lower H2O2 accumulation, reduced callose deposition, and reduced electrolyte leakage upon inoculation with hypersensitive response–causing nonhost pathogens. Arabidopsis gox mutants were not affected in NADPH oxidase activity, and silencing of a gene encoding NADPH oxidase (Respiratory burst oxidase homolog) in the gox mutants did not further increase susceptibility to nonhost pathogens, suggesting that GOX functions independently from NADPH oxidase. In the two gox mutants examined (haox2 and gox3), the expression of several defense-related genes upon nonhost pathogen inoculation was decreased compared with wild-type plants. Here we show that GOX is an alternative source for the production of H2O2 during both gene-for-gene and nonhost resistance responses.
INTRODUCTION
All plants are generally resistant to a wide range of potential pathogens present in the environment, and the term nonhost resistance has been coined to define the resistance shown by an entire plant species to all isolates of a microbial species (Heath, 2000). A pathogen that cannot cause disease on a nonhost plant is referred to as a nonhost pathogen. The wide spectrum of nonhost resistance is in contrast with the more specific mechanism of gene-for-gene resistance, also known as effector-triggered immunity (ETI), which is mediated by the activity of pathogen effectors recognized by resistance (R) proteins (Jones and Dangl, 2006). The consequence of ETI is the elicitation of a localized programmed cell death (PCD) reaction known as the hypersensitive response (HR), which ultimately limits the spread of the pathogen (Hammond-Kosack and Jones, 1997). It has been proposed that nonhost resistance operates under two distinct mechanisms: type I and type II (Mysore and Ryu, 2004). In type I, the plant does not show any symptoms after inoculation with the nonhost pathogen, suggesting that pathogen growth is halted as a consequence of preformed or inducible defenses; in type II, a HR is triggered because the pathogen is able to disarm the first layers of defense, but is later recognized by the plant surveillance system (Mysore and Ryu, 2004).
Although nonhost resistance is not very well understood, it is known that the mechanism of nonhost resistance involves a first layer of defense that includes the plant cytoskeleton and constitutively produced peptides, proteins, and secondary metabolites with antimicrobial properties (Mysore and Ryu, 2004; Ellis, 2006; Lipka et al., 2008). The second layer of defense includes induction of plant defense-related genes upon perception of pathogen-associated molecular patterns (PAMPs) to prevent the spread of the pathogen and the initiation of disease. Among these are genes responsible for synthesis of phytoalexins (Thomma et al., 1999; Zhou et al., 1999; Loehrer et al., 2008), genes in defense-signaling transduction cascades involving the hormones salicylic acid (SA) (Mellersh and Heath, 2003; Loehrer et al., 2008), ethylene (ET) (Knoester et al., 1998; Geraats et al., 2003; Nasir et al., 2005), and jasmonic acid (JA) (Loehrer et al., 2008), as well as the MAP kinases: wound-induced protein kinase and SA-induced protein kinase (Sharma et al., 2003).
The HR associated with nonhost resistance is similar to the HR induced during gene-for-gene resistance and involves accumulation of reactive oxygen species (ROS), such as superoxide (O2−) and hydrogen peroxide (H2O2). Evidence for the role of ROS in triggering and/or executing the HR has been demonstrated by pharmacological studies showing that blocking ROS accumulation inhibited cell death (Levine et al., 1994). Furthermore, accumulation of H2O2 in catalase-deficient plants (Van Breusegem and Dat, 2006) or in transgenic plants expressing H2O2-generating enzymes activated cell death and increased protection against bacterial and oomycete pathogens (Wu et al., 1995). The effect of ROS in defense responses and the activation of the HR have been mainly associated with NADPH oxidase, which catalyzes the reduction of O2 into O2−. Further dismutation of O2− by the enzyme superoxide dismutase generates the most stable ROS, H2O2 (Lamb and Dixon, 1997). However, accumulated evidence for the role of NADPH oxidase in disease resistance is contradictory. Silencing of the genes encoding NADPH oxidases (RBOHA and RBOHB) in Nicotiana benthamiana eliminated H2O2 production, enhanced sporangia and disease lesion formation upon inoculation with Phytophthora infestans, and delayed or reduced HR cell death caused by the elicitor INF1 (Yoshioka et al., 2003). In Arabidopsis thaliana, the NADPH oxidase mutant respiratory burst oxidase homolog (rbohD) showed dramatic reduction in H2O2 accumulation but without significant effect on the HR (Torres et al., 2002). Furthermore, inoculation of rbohD mutants with both virulent and avirulent bacterial strains did not show a substantial difference in bacterial growth in comparison with wild-type plants (Torres et al., 2002; Chaouch et al., 2011). In contrast with the phenotypes observed in N. benthamiana, abolishment of H2O2 production in rbohD plants reduced, rather than increased, sporangiophore development and fungal biomass upon inoculation with Peronospora parasitica (Torres et al., 2002) and Alternaria brassicicola (Pogány et al., 2009), respectively. The latter work also showed that the rbohD mutant is not affected in stress-related responses (Pogány et al., 2009) and indicates that RBOHD actually suppresses cell death (Torres et al., 2002; Pogány et al., 2009).
In addition to the apoplastic H2O2 generated by the membrane-localized NADPH oxidase, H2O2 and other ROS are produced in various organelles and through different enzymatic reactions in plant cells (Van Breusegem and Dat, 2006). Among these are the peroxisomes, which provide a rich source of H2O2 through the glycolate oxidase (GOX) reaction (Foyer et al., 2009). GOX catalyzes the conversion of glycolate into glyoxylate during photorespiration with concomitant production of H2O2. The role of GOX in disease resistance has been tangentially proposed, but no mechanism was identified. For example, somatic hybrids between Brassica napus and Arabidopsis were produced to incorporate resistance in B. napus against Leptosphaeria maculans. Comparison of protein profiles between resistant and susceptible somatic hybrids revealed that GOX was abundantly present in resistant plants (Bohman et al., 2002). GOX was also shown to be induced in barley (Hordeum vulgare) upon inoculation with the pathogenic fungus Bipolaris sorokiniana (Schäfer et al., 2004), and melon cultivars resistant to the oomycete Pseudoperonospora cubensis had increased GOX enzyme activities (Taler et al., 2004).
In this article, we report the use of a virus-induced gene-silencing (VIGS)-based, fast-forward genetics approach (Baulcombe, 1999; Lu et al., 2003b; del Pozo et al., 2004) to identify plant genes that play a role in nonhost disease resistance. One of the genes identified through the screen encodes the photorespiratory enzyme, GOX. We demonstrated that the generation of H2O2 by GOX during nonhost resistance is independent of the oxidative burst mediated by NADPH oxidase.
RESULTS
VIGS-Based Screening Identifies Several N. benthamiana Genes Involved in Nonhost Disease Resistance
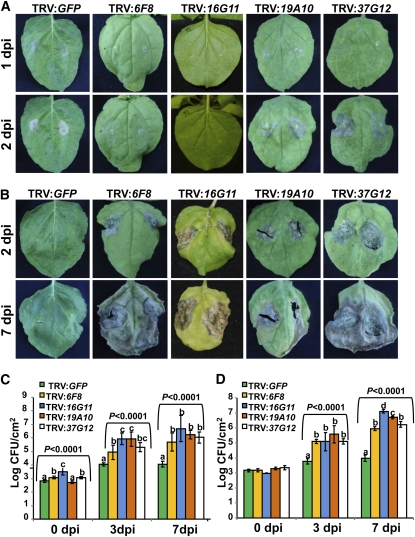
To identify genes that would enable us to dissect the complex phenomenon of nonhost disease resistance, we used a Tobacco rattle virus (TRV)-based VIGS system as a fast-forward genetics tool (Liu et al., 2002a; Liu et al., 2002b; Anand et al., 2007; Wangdi et al., 2010) to screen a normalized N. benthamiana Mixed Elicitor cDNA library (Anand et al., 2007). From the cDNA library, 3840 pTRV2 derivative clones (one gene per clone) were individually inoculated, in duplicate, along with pTRV1, into N. benthamiana plants. As a control, we used pTRV2 harboring the green fluorescent protein sequence (GFP) (GFP does not have any sequence similarity to plant DNA and therefore will not cause gene silencing). Three weeks after TRV inoculation, we infiltrated the upper gene-silenced leaves, using a needleless syringe, with a high inoculum (1 × 108 colony-forming units [cfu]/mL) of a type I nonhost pathogen, Pseudomonas syringae pv glycinea, or a type II nonhost pathogen, P. syringae pv tomato strain T1. After three rounds of screening, we identified 12 clones that, when silenced in N. benthamiana, showed alteration in the plant response when inoculated with nonhost bacterial pathogens. We selected four of those clones for further characterization. Upon syringe infiltration with P. syringae pv tomato strain T1 (a type II nonhost pathogen), the control plants (TRV:GFP) showed a typical HR characterized by necrosis limited around the inoculation site as early as 1 d after inoculation (DAI), whereas in some silenced lines, the HR started only after 2 or 3 DAI, or in some cases, inoculated leaves developed disease symptoms (Figure 1A). Upon inoculation with P. syringae pv glycinea (a type I nonhost pathogen), the control plants (TRV:GFP) did not show any visible symptoms, as expected; however, the silenced lines showed disease-associated necrosis around the site of inoculation as early as 2 DAI, and the necrosis extended throughout the leaf at 7 DAI (Figure 1B). At low levels of inoculum (1 × 104 cfu/mL), both type I and type II nonhost pathogens that were used showed increased bacterial accumulation in silenced plants starting at 3 DAI and reaching 10- to 100-fold more at 7 DAI in comparison with control plants (Figures 1C and 1D). These results demonstrated the use of VIGS-mediated fast-forward genetics to identify plant genes involved in nonhost disease resistance.
Figure 1.
A Fast-Forward Genetics Screen Using VIGS Allowed the Identification of Genes Involved in Nonhost Disease Resistance.
Silenced N. benthamiana plants and control plants (TRV:GFP) were challenged with nonhost pathogens P. syringae pv tomato strain T1 (A) and P. syringae pv glycinea (B) at 108 cfu/mL. HR or disease symptoms were evaluated at different times after inoculation. Bacterial growth in silenced lines was monitored by inoculating silenced and control plants with P. syringae pv tomato strain T1 (C) and P. syringae pv glycinea (D) at 104 cfu/mL. Four representative clones out of 12 are shown. Bars represent the mean and sd for four biological replicates in three independent experiments. Statistical significance for a particular time point was determined using one-way ANOVA, and P values from F test are indicated above bars. LSD test was used to test differences between treatments when statistical significance was found. Means with the same letter were not significantly different at P < 0.05. dpi, days postinoculation.
Silencing of GOX in N. benthamiana Compromises Nonhost Resistance and Also Affects Gene-for-Gene Resistance and PAMP-Mediated Immunity
We further characterized the cDNA clone TRV:16G11, which when silenced showed a very significant increase in the growth of nonhost bacteria compared with control plants. 16G11-silenced plants were delayed in the onset of the HR, which started at 48 h after inoculation and was not uniformly distributed around the site of inoculation. VIGS caused 80% downregulation of 16G11 mRNA as demonstrated by quantitative RT-PCR (qRT-PCR) (see Supplemental Figure 1 online).
The insert in TRV:16G11 was sequenced, and the sequence information was then analyzed to predict gene function. A BLASTX search against the J. Craig Venter Institute database revealed 90% identity to an Arabidopsis GOX gene, At3g14420. To facilitate a more comprehensive analysis of the N. benthamiana GOX homolog (NbGOX), we cloned the full-length NbGOX by Rapid Amplification of cDNA Ends (RACE). The cloned gene was then sequenced, and the translated amino acid sequence was then aligned with orthologous plant protein sequences using ClustalW (http://workbench.sdsc.edu). The alignment revealed a high degree of sequence conservation among GOX of various plant species (see Supplemental Figure 2 online). The full sequence of NbGOX was deposited in GenBank. In addition, we also sequenced the inserts in three other clones (6F8, 19A10, and 37G12).
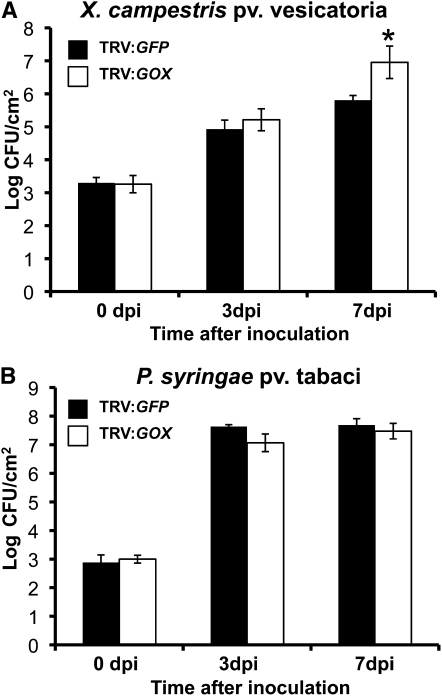
We challenged the GOX-silenced N. benthamiana plants with another unrelated nonhost pathogen, Xanthomonas campestris pv vesicatoria. Contrary to control plants inoculated with TRV:GFP, the growth of X. campestris pv vesicatoria in GOX-silenced plants was significantly greater at 7 DAI and was ~10-fold more than the control plants (Figure 2). We then checked whether GOX-silenced plants were hypersusceptible to a host pathogen, P. syringae pv tabaci. Interestingly, we observed that the population of the host pathogen at 3 and 7 DAI was similar in both control and GOX-silenced plants (Figure 2). In addition, populations of nonhost and host bacteria were monitored in GOX-silenced plants by inoculating GFPuv-labeled bacteria (Wang et al., 2007) at a lower concentration (3 × 104 cfu/mL) to prevent cell death associated with HR or disease. Under UV light, the growth of the nonhost pathogen P. syringae pv tomato strain T1 was visible as green fluorescent colonies in GOX-silenced lines (TRV:GOX) but not in the control plants (TRV:GFP) (see Supplemental Figure 3A online). GOX-silenced plants showed the same intensity of green fluorescence as that of control plants upon inoculation with the host pathogen P. syringae pv tabaci (see Supplemental Figure 3B online).
Figure 2.
Silencing of GOX in N. benthamiana Enhances Growth of Nonhost Pathogens, but Has No Effect on the Growth of a Host Pathogen.
Control plants (TRV:GFP) and GOX-silenced plants (TRV:GOX) were vacuum infiltrated with X. campestris pv vesicatoria (A) and P. syringae pv tabaci (B) at 104 cfu/mL, and bacterial populations were quantified at 0, 3, and 7 DAI. Data represent the mean and sd for four biological replicates in three independent experiments. Asterisk represents statistically significant value at P < 0.05 based on Student’s t test between TRV:GFP and TRV:GOX. dpi, days postinoculation.
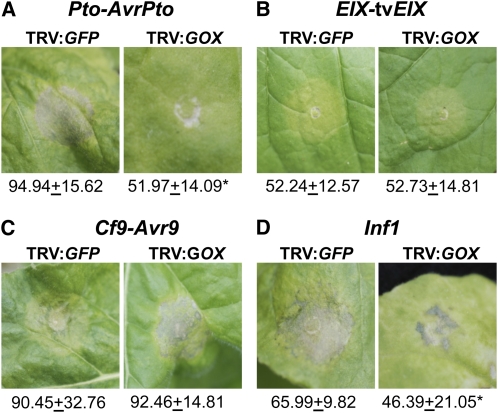
To determine whether silencing of GOX also delays HR associated with ETI and PAMP-triggered immunity, we induced an HR in fully expanded young leaves of GOX-silenced and control N. benthamiana plants by coexpressing several R genes and their cognate avirulence (Avr) genes using Agrobacterium (Figure 3). Coexpression of the R gene Pto (Tang et al., 1996) and avirulence gene AvrPto (Frederick et al., 1998) in control plants (TRV:GFP) produced the typical confluent tissue collapse that characterizes the HR, and consequently there was a considerable increase in autofluorescence caused by the release of phenolic compounds (Figure 3A; see Supplemental Figure 4A online) (Klement et al., 1990). By contrast, the HR in GOX-silenced plants (TRV:GOX) was considerably delayed, and the levels of autofluoresence were reduced by ~50% in comparison with control plants (Figure 3A; see Supplemental Figure 4A online). Other gene-for-gene combinations, such as EIX-tvEIX (Ron and Avni, 2004) and Cf9-Avr9 (Van der Hoorn et al., 2000), did not show any alterations in timing or intensity of HR in GOX-silenced plants when compared with control plants (Figures 3B and 3C; see Supplemental Figures 4B and 4C online). When the P. infestans gene-encoding elicitor protein INF1 (PAMP; Kamoun et al., 1998) was transiently expressed using Agrobacterium, HR development in GOX-silenced plants was slightly delayed, with a reduction in the levels of autofluorescence at 4 DAI when compared with control plants (Figure 3D; see Supplemental Figure 4D online). Taken together, these results suggest that GOX plays a role in nonhost disease resistance triggered by both type I and type II nonhost pathogens and also in the elicitation of the HR observed during Pto-AvrPto–mediated and INF1-mediated defense responses in N. benthamiana.
Figure 3.
Silencing of GOX in N. benthamiana Partially Compromises Gene-for-Gene Resistance and PAMP-Mediated Immunity.
GOX-silenced plants (TRV:GOX) and control plants (TRV:GFP) were coinfiltrated with A. tumefaciens strains carrying the R-Avr gene combinations Pto-AvrPto (A), EIX- tvEIX (B), Cf9-Avr9 (C), and with an Agrobacterium strain carrying the construct that expresses PAMP elicitor Inf1 (D). Symptoms of HR were evaluated at 4 DAI. For quantification of fluorescence intensity, 10 leaf disks (0.5 cm2) were observed under epifluorescence microscopy, and pictures were taken from 50 randomly chosen microscopic fields. Images were converted to gray scale, and mean gray value for the entire image was calculated using ImageJ (http://rsb.info.nih.gov/ij/). Numbers under the pictures represent means and sd of fluorescence intensity quantification (in arbitrary units). Asterisks indicate statistically significant difference between TRV:GFP and TRV:GOX using Student’s t test at P < 0.01.
Individual Null Mutations in Arabidopsis GOX Genes Compromise Nonhost Resistance
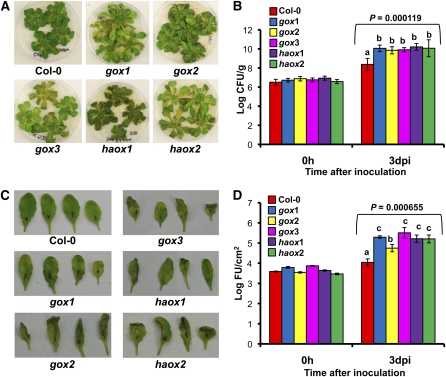
To genetically dissect the function of GOX in nonhost disease resistance, we decided to examine the role of its corresponding homologs in the tractable model plant, Arabidopsis. Computer-assisted predictions based on peroxisome targeting signals of the protein had previously identified five members of the GOX gene family in the Arabidopsis genome: GOX1 (At3g14420), GOX2 (At3g14415), GOX3 (At4g18360), HAOX1 (At3g14130), and HAOX2 (At3g14150) (Reumann et al., 2004). T-DNA insertion mutants for these genes were obtained and were confirmed to be null mutants (see Supplemental Figure 5 online).
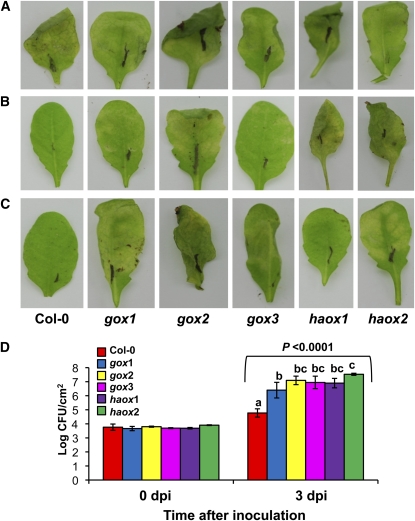
We conducted an initial screen to determine whether these mutant lines were susceptible to a nonhost pathogen by seedling flood-inoculation (Ishiga et al., 2011) of 4-week-old seedlings with the nonhost pathogen P. syringae pv syringae strain B728A. At 5 DAI, wild-type plants showed no disease symptoms, whereas all the gox mutants showed varying degrees of disease, from mild chlorosis in the upper leaves with necrosis in the lower leaves to total tissue collapse (Figure 4A). No symptoms were observed in mock-inoculated gox mutants (see Supplemental Figure 6 online). In addition, using this method of inoculation, the nonhost pathogen multiplied ~10-fold higher in all the gox mutants than in the wild-type Columbia ecotype (Col-0) (Figure 4B). All the gox mutants also showed high susceptibility to another nonhost pathogen, P. syringae pv tabaci, upon syringe-inoculation (Figure 4C) and supported more bacteria when compared with wild-type Col-0 (Figure 4D). Inoculation of gox mutant lines with the host pathogen P. syringae pv maculicola did not show any difference in bacterial growth in comparison with the wild type (see Supplemental Figure 7 online). These observations suggested that each of the individual gox mutants compromised nonhost resistance, and presumably the different members of the Arabidopsis GOX gene family do not have a redundant function with respect to nonhost resistance.
Figure 4.
Arabidopsis gox Mutants Show Increased Susceptibility to Nonhost Pathogens P. syringae pv syringae Strain B728A and P. syringae pv tabaci.
(A) Four-week old seedlings of wild-type Col-0 and T-DNA insertion mutants gox1, gox2, gox3, haox1, and haox2 were flood-inoculated with the nonhost pathogen P. syringae pv syringae strain B728A at 3 × 107 cfu/mL, and symptoms were evaluated at 5 DAI.
(B) Growth of P. syringae pv syringae strain B728A from flood inoculation was quantified in wild-type Col-0 and all the gox mutants at 0 and 3 DAI.
(C) Four-week old Col-0 and gox mutant plants were grown in soil and syringe-inoculated with P. syringae pv tabaci at 5 × 106 cfu/mL, and symptoms were evaluated after 3 d.
(D) Growth of P. syringae pv tabaci after syringe inoculation in wild-type Col-0 and all gox mutants was quantified at 0 and 3 DAI using a starting inoculum of 104 cfu/mL. Bars represent the mean and sd for four biological replicates in three independent experiments. Statistical significance for a particular time point was determined using one-way ANOVA, and P values from F test are indicated above bars. LSD test was used to test differences between treatments when statistical significance was found. Means with the same letter were not significantly different at P < 0.05. dpi, days postinoculation.
Gene-for-Gene Resistance Is Also Compromised in Arabidopsis gox Mutants
To determine whether GOX also plays a role in gene-for-gene resistance in Arabidopsis, we inoculated Col-0 and gox mutants using a needleless syringe at a low concentration (1 × 104 cfu/mL) with either the compatible strain P. syringae pv tomato strain DC3000 or the incompatible strains P. syringae pv tomato strain DC3000 (AvrB) and P. syringae pv tomato strain DC3000 (AvrRps4) carrying the avirulence genes AvrB and AvrRps4, respectively, and therefore rendering P. syringae pv tomato strain DC3000 avirulent in Arabidopsis (Innes et al., 1993; Hinsch and Staskawicz, 1996). As expected, the compatible strain P. syringae pv tomato strain DC3000 showed chlorosis and necrosis in wild-type Col-0 and in all gox mutant lines (Figure 5A). Inoculation with the avirulent strains P. syringae pv tomato strain DC3000 (AvrB) (Figure 5B) and P. syringae pv tomato strain DC3000 (AvrRps4) (Figure 5C) did not cause disease symptoms in the wild-type Col-0, whereas the gox mutant lines showed various degrees of disease symptoms ranging from mild chlorosis to severe necrosis, implicating Arabidopsis GOX in gene-for-gene resistance responses. P. syringae pv tomato strain DC3000 (AvrRps4) was chosen to further evaluate its growth in planta. The data revealed that gox mutant lines allowed at least 10-fold increase in bacterial titer at 3 DAI (Figure 5D). These results confirmed the observations in N. benthamiana that GOX plays a role in R gene–mediated resistance, suggesting a partial overlap between R gene and nonhost resistance mechanisms.
Figure 5.
Gene-for-Gene Resistance Is Compromised in Arabidopsis gox Mutants.
(A) to (C) Wild-type Col-0 and gox mutants were inoculated with the virulent pathogen P. syringae pv tomato strain DC3000 at 106 cfu/mL (A) and the avirulent pathogens P. syringae pv tomato strain DC3000 (AvrB) (B) and P. syringae pv tomato strain DC3000 (AvrRps4) (C). Photographs were taken after 5 d.
(D) Growth of P. syringae pv tomato strain DC3000 (AvrRps4) was monitored at 0 and 3 DAI using a starting inoculum of 104 cfu/mL. Data represents the mean and sd for four biological replicates in three independent experiments. Statistical significance for each time point was determined using one-way ANOVA, and P values from F test are indicated above bars. LSD test was used to determine differences between genotypes. Means with the same letter were not significantly different at P < 0.05. dpi, days postinoculation.
All the Members of the Arabidopsis GOX Gene Family Are Required for the Elicitation of Defense Responses upon Inoculation with a Nonhost Pathogen
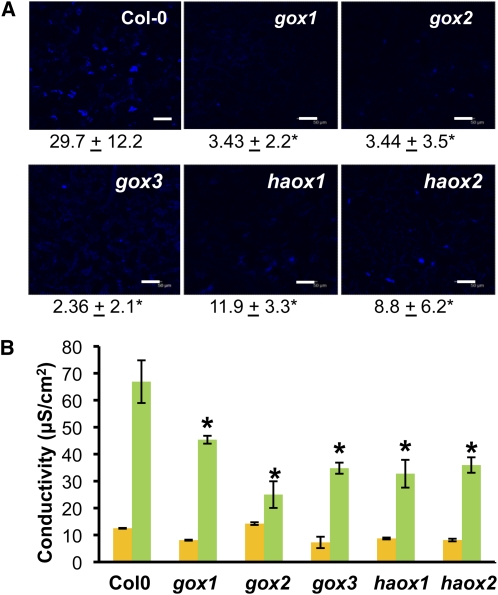
Callose deposition and HR are typical plant defense responses upon pathogen inoculation. We tested whether these responses were affected in the gox mutants upon inoculation with a nonhost pathogen. For callose deposition, wild-type Col-0 and gox mutants were infected with P. syringae pv tabaci at 106 cfu/mL and stained with aniline blue 48 h after inoculation. All the gox mutants showed a significant decrease in the numbers of callose deposits, with gox3 showing a 92% reduction followed by gox1 and gox2 with 88% reductions, whereas haox2 and haox1 were reduced by 70 and 60%, respectively (Figure 6A). Furthermore, the HR, quantified by the release of electrolytes, was reduced by 40 to 60% in gox mutants when compared with wild-type Col-0 (Figure 6B). These data clearly show that when inoculated with a nonhost pathogen, wild-type Col-0 is able to mount a defense response characterized by the HR and deposition of callose, whereas the intensity or timing of these responses in gox mutants are reduced or delayed as previously observed in N. benthamiana.
Figure 6.
Callose Deposition and HR Are Compromised in gox Mutants.
(A) Leaves inoculated with P. syringae pv tabaci at 106 cfu/mL were detached and stained with aniline blue 48 h after inoculation and observed under confocal microscopy. Numbers below the pictures represent the means and sd of callose deposits counted on images taken from 10 microscopic fields. Asterisks represent statistically significant differences between wild-type Col-0 and each one of the mutants based on Student’s t test at P < 0.05.
(B) Leaves were inoculated as described for (A) and used to monitor electrolyte leakage. Conductivity of mock-inoculated plants (yellow bars) and pathogen-inoculated plants (green bars) was measured 24 h after inoculation. Data represent the mean and sd for four biological replicates in three independent experiments. Asterisks represent statistically significant differences between wild-type Col-0 and each one of the mutants based on Student’s t test at P <0.05.
Bar = 50 μM.
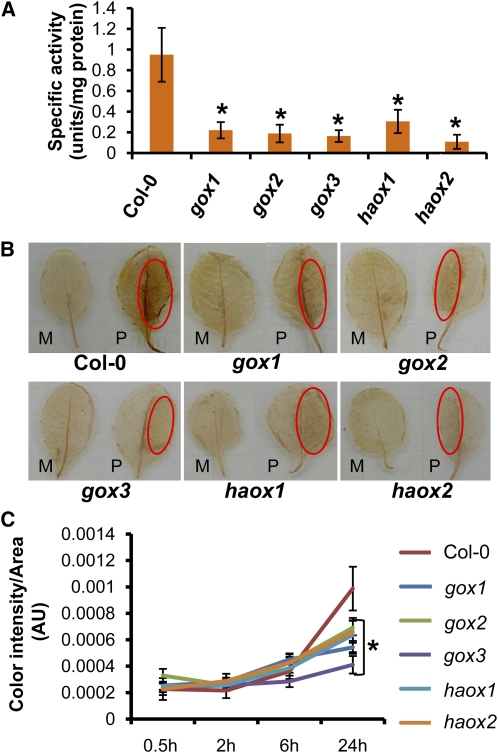
GOX Activity and Production of H2O2 Is Affected in gox Mutants
We confirmed that all the gox mutants indeed had significantly (~80%) less GOX activity, without pathogen inoculation, when compared with wild-type Col-0 (Figure 7A). Furthermore, the reaction catalyzed by GOX leads to production of H2O2, a key signaling molecule that triggers the process of localized cell death (HR) upon challenging with elicitors and avirulent pathogens (Levine et al., 1994). Because H2O2 is also available from different sources in plant cells (Neill et al., 2002), we hypothesized that GOX from peroxisomes might be the main source for H2O2 production during nonhost disease resistance responses.
Figure 7.
GOX Activity and Pathogen-Dependent H2O2 Production Are Affected in gox Mutants of Arabidopsis.
(A) Basal levels of GOX enzymatic activity in wild-type Col-0 and gox mutants. Protein extracts were added to a reaction mixture containing sodium glycolate, o-dianisidine cation, and horseradish peroxidase, and enzymatic activity was derived from a colorimetric reaction read at 440 nm. Bars represent the average and sd of measurements taken from four replicates. Asterisks represent a statistically significant difference between Col-0 and each one of the gox mutants based on Student’s t test (P < 0.05).
(B) Wild-type Col-0 and gox mutants were mock-inoculated with water (M) or with P. syringae pv tabaci (P) at 106 cfu/mL. Mock-inoculation was done on the entire leaf, but pathogen inoculation was done on one side of the leaf only (circled). After 24 h of inoculation, detached leaves were vacuum infiltrated with DAB (1 mg/mL) and incubated for 6 h as described. Leaves were cleared in 100% ethanol and preserved in 25% glycerol. Photographs were taken immediately afterward.
(C) H2O2 accumulation was quantified over a time-course experiment by collecting leaf samples for wild-type Col-0 and each of the gox mutants at 30 min, 2 h, 6 h, and 24 h after inoculation. Color intensity/area was measured as arbitrary units (AU) on photographs taken from all leaves after DAB staining. Mean and sd were calculated from measurements done on 20 leaves per time point for each of the genotypes. Asterisk represents statistically significant difference between Col-0 and each of the gox mutants after 24 h of inoculation based on Student’s t test (P < 0.05).
We monitored the accumulation of H2O2 upon syringe inoculation with the nonhost pathogen P. syringae pv tabaci at 106 cfu/mL, a concentration high enough to cause HR. At 24 h after inoculation, 3,3′-diaminobenzidine tetrahydrochloride (DAB) staining revealed a decreased accumulation of H2O2 in all the gox mutants when compared with the wild-type Col-0 (Figure 7B). No H2O2 accumulation was observed in Col-0 that was mock-inoculated, indicating that after 24 h, the H2O2 produced by wounding is no longer present (Figure 7B).
We conducted a time-course experiment to monitor the accumulation of H2O2 associated with GOX. Because H2O2 is unstable and direct quantification of H2O2 is cumbersome (Queval et al., 2008), we quantified H2O2 accumulation indirectly as the color intensity associated with DAB staining as done previously (Torres et al., 2005) (Figure 7C). Col-0 and all the gox mutants showed almost equivalent levels of H2O2 accumulation at 30 min, 2 h, and 6 h after inoculation. However, at 24 h after inoculation, there was a significantly higher H2O2 accumulation in wild-type Col-0 when compared with gox mutants, indicating that the timing of H2O2 generated by GOX follows the second phase of the oxidative burst associated with NADPH oxidase activity (Lamb and Dixon, 1997).
GOX Function in Nonhost Resistance Is Independent of NADPH Oxidase Activity
We used three independent approaches to confirm that the Arabidopsis gox phenotypes are not caused by a defect in NADPH oxidase: i) we used VIGS to silence the NADPH oxidase-encoding gene RBOHD in wild-type Col-0 and in gox mutants to evaluate disease development and bacterial growth; ii) we measured basal levels of NADPH oxidase enzymatic activity in wild-type Col-0 and all the gox mutants; and iii) we measured the basal levels of gene expression of the NADPH oxidase-encoding gene RBOHD in wild-type Col-0 and all the gox mutants.
TRV-based VIGS was used to silence RBOHD as described (Lu et al., 2003). Transcripts of RBOHD were quantified by qRT-PCR in the silenced and control plants. Although we designed constructs to specifically target RBOHD, we also observed silencing of RBOHF to some extent (because of high similarity) that was variable among different lines when inoculated with TRV:RBOHD. After TRV:RBOHD inoculation, we obtained ~25 to 60% downregulation of RBOHD in wild-type Col-0 and all gox mutants as quantified by qRT-PCR (see Supplemental Figure 8A online).
Previous results have shown that the rbohD mutant is compromised in the accumulation of H2O2 after 6 h of inoculation with an incompatible (HR-causing) pathogen, P. syringae pv tomato strain DC3000 (AvrRpm1) (Torres et al., 2002). We tested whether the levels of downregulation of RBOHD caused by VIGS in wild-type Col-0 were enough to affect the accumulation of H2O2 after 6 h of inoculation with P. syringae pv tomato strain DC3000 (AvrRpm1). Silenced and control lines were inoculated with P. syringae pv tomato strain DC3000 (AvrRpm1) at 2 × 107 cfu/mL, and after 6 h of inoculation, inoculated leaves were harvested for DAB staining as described earlier. We observed that RBOHD-silenced Col-0 plants had a statistically significant minor reduction in H2O2 accumulation (see Supplemental Figure 8B online). However, silencing of RBOHD in wild-type Col-0 did not significantly affect the accumulation of H2O2 associated with GOX after 24 h of inoculation with a nonhost pathogen, P. syringae pv tabaci (see Supplemental Figure 8C online).
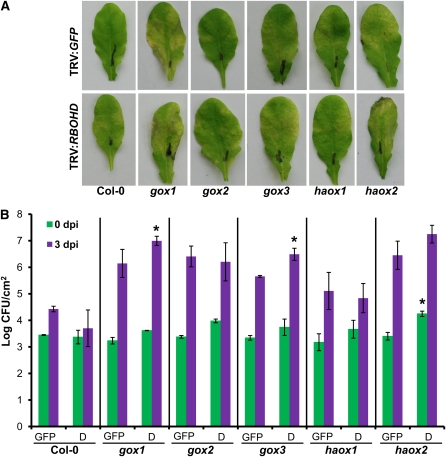
Two weeks after inoculation with TRV:GFP or TRV:RBOHD, Arabidopsis (Col-0) plants were challenged with a nonhost pathogen, P. syringae pv tabaci, to observe symptom development and to quantify bacterial growth in planta. Wild-type Col-0 plants inoculated with TRV:RBOHD showed mild disease symptoms. Silencing of RBOHD in the gox mutants did not have any significant additive effect regarding disease symptoms in gox mutants (Figure 8A). However, given the high variability and the nonquantitative nature of the symptoms, we decided to examine the multiplication of the nonhost pathogen P. syringae pv tabaci in planta. Silencing of RBOHD did not have any effect on bacterial growth in wild-type Col-0, gox2, haox1, and haox2 mutant plants when compared with nonsilenced control (TRV:GFP) (Figure 8B). The observed very slight increase in bacterial growth after silencing of RBOHD in gox1 and gox3 backgrounds warrants further investigation.
Figure 8.
Effect of Silencing RBOHD on Disease Symptom Development in Wild-Type Col-0 and gox Mutants.
(A) Two-week-old Arabidopsis plants were infiltrated with TRV:RBOHD. TRV:GFP-infiltrated plant used as control. Two weeks after silencing, plants were inoculated with the nonhost pathogen P. syringae pv tabaci at 105 cfu/mL, and disease symptoms were photographed at 5 DAI.
(B) Silenced plants were inoculated with P. syringae pv tabaci at a concentration of 104 cfu/mL, and bacterial growth was examined at 0 DAI (green bars) and 3 DAI (purple). Bars represent the mean and sd of bacterial growth from three independent experiments in control plants (GFP) compared with RBOHD-silenced plants (D). Asterisks represent statistically significant values between control plants (GFP) and RBOHD-silenced plants (D) for equivalent time points within a genotype at P < 0.05 based on Student’s t test. dpi, days postinoculation.
To rule out that GOX mutation affected the activity of NADPH oxidase, we isolated membrane fractions from wild-type Col-0 and all the gox mutants to measure NADPH oxidase activity; the enzymatic assay was based on the reduction of sodium,3′-[1-[phenylamino-carbonyl]-3,4-tetrazolium]-bis(4-methoxy-6-nitro)benzenesulfonic acid hydrate (XTT) upon the generation of O2− by NADPH oxidase. gox1 didn’t show any difference in NADPH oxidase activity when compared with Col-0, but gox3, haox1, and haox2 mutants showed a slight increase (~1.3-fold to ~1.5-fold), although none of them were compromised in NADPH oxidase activity in comparison with wild-type Col-0 (see Supplemental Figure 9A online). It is unlikely that a slight increase in NADPH oxidase activity would be the cause for compromised disease resistance in gox mutants. In addition, when we measured the basal levels of gene expression of RBOHD in wild-type Col-0 and all the gox mutant backgrounds, we found that only in gox3 was there a small (~1.6-fold) but significant increase in expression (see Supplemental Figure 9B online). We therefore concluded that the compromised disease resistance in gox mutants is caused by the reduced H2O2 or GOX activity and is independent of the activity of NADPH oxidase.
Genes Encoding GOX Exhibit Differential Patterns of Gene Expression in Response to a Challenge with Nonhost Pathogen
As mentioned earlier, Arabidopsis contains five members of the GOX gene family; GOX1 and GOX2 are contiguous on one arm of chromosome 3, and their nucleotide and amino acid sequences are highly similar (see Supplemental Figure 10 online); HAOX1 and HAOX2 are separated from each other by one gene in the other arm of the same chromosome 3, and their sequences are also highly similar (see Supplemental Figure 10 online). By contrast, GOX3 is not duplicated, and is located on chromosome 4 (www.Arabidopsis.org). To elucidate the functions of the different copies of GOX, we used three different criteria to mine the publicly available expression data (GENEVESTIGATOR; http://www.genevestigator.ethz.ch) (Zimmermann et al., 2004): 1) levels of expression in leaves; 2) gene expression associated with pathogens and elicitors; and 3) gene expression associated with PCD for its relationship to the HR. These criteria revealed distinct patterns of gene expression among the GOX genes. GOX1 and GOX2 were shown to be highly expressed in leaves but repressed after treatment with nonhost, virulent, or avirulent pathogens as well as elicitors and are highly induced by PCD (Zimmermann et al., 2004). GOX3 was shown to be expressed at lower levels in leaves but significantly induced after inoculation with nonhost pathogens (Blumeria graminis, Botrytis cinerea, P. infestans), virulent pathogens (P. syringae, Erysiphae cichoracearum), or avirulent pathogens (P. syringae AvrRpm1), and repressed by elicitors, such as harpin (HrpZ), flagellin (flg22), lipopolysaccharide, and PCD (Zimmermann et al., 2004). HAOX1 and HAOX2 were also expressed at low levels in leaves and induced after inoculation with the avirulent pathogen P. syringae (AvrRps4) and by the elicitors HrpZ, flg22, lipopolysaccharide, and after PCD (Zimmermann et al., 2004). We used qRT-PCR to monitor the expression of GOX genes in wild-type Col-0 plants over a time-course (0 to 24 h) experiment after inoculation with the nonhost pathogen, P. syringae pv tabaci (Table 1). GOX1 and GOX2 were downregulated after inoculation, whereas GOX3, HAOX1, and HAOX2 showed significant increases in gene expression ranging from approximately sevenfold to ninefold induction after 24 h. Therefore, GOX3, HAOX1, and HAOX2 were induced by nonhost pathogens, and we speculate that these genes play a major role in regulating nonhost defense responses. However, because we observed striking phenotypes with all the gox mutants, we suspect that all the genes are important to different degrees during nonhost disease resistance.
Table 1.
Expression of GOX Genes in Wild-Type Col-0 upon Inoculation with a Nonhost Pathogen
| Gene | Relative Gene Expressiona (0 h after inoculation) | Relative Gene Expression (1 h after inoculation) | Relative Gene Expression (4 h after inoculation) | Relative Gene Expression (24 h after inoculation) |
| GOX1 | 0.67 ± 0.07 | 0.92 ± 0.08 | 0.69 ± 0.06 | 0.28 ± 0.01 |
| GOX2 | 0.60 ± 0.09 | 0.51 ± 0.1 | 0.43 ± 0.04 | 0.44 ± 0.02 |
| GOX3 | 0.77 ± 0.12 | 0.86 ± 0.09 | 0.95 ± 0.12 | 5.58 ± 1.23 |
| HAOX1 | 0.80 ± 0.06 | 0.83 ± 0.13 | 0.81 ± 0.09 | 7.43 ± 0.85 |
| HAOX2 | 0.51 ± 0.08 | 1.36 ± 0.22 | 1.85 ± 0.12 | 8.99 ± 0.41 |
Gene-specific expression values were normalized using the expression of Elongation factor 1α (EF1α) and Ubiquitin 5 (UBQ5).
Genes Encoding GOX Act Together to Confer Nonhost Disease Resistance
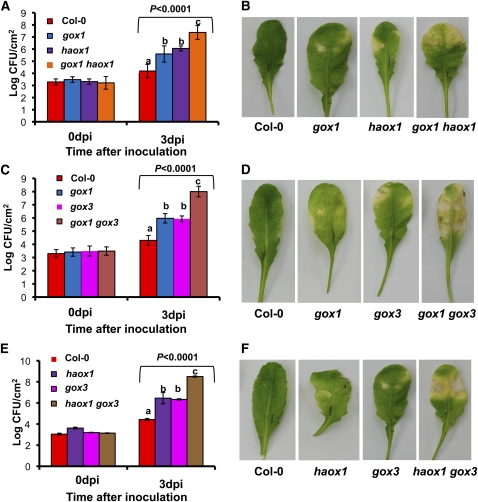
To determine whether GOX genes function additively, we developed the following double mutant lines: gox1 haox1, gox1 gox3, gox3 haox1, and selected double homozygous mutants for further analysis. Because GOX1 and GOX2 are adjacent on the same chromosome, we could not make double mutants for these genes. The same applies to HAOX1 and HAOX2. Wild-type Col-0, single mutants, and double mutants were inoculated with the nonhost pathogen P. syringae pv tabaci at 104 cfu/mL, and bacterial multiplication was monitored at 0 and 3 DAI. The gox1 haox1 double mutant showed a ~10-fold increase in bacterial numbers at 3 DAI in comparison with the single mutant parents gox1 and haox1 (Figure 9A), with moderate increase in symptoms (Figure 9B). Interestingly, both gox1 gox3 (Figure 9C) and haox1 gox3 (Figure 9E) supported ~100-fold more bacteria at 3 DAI, and the symptoms were dramatically and consistently greater in comparison with their single-mutant parents (Figures 9D and 9F). Thus, it seems that GOX1, GOX3, and HAOX1 quantitatively contribute to confer nonhost resistance. Furthermore, these data confirm the lack of redundancy among GOX gene copies.
Figure 9.
Arabidopsis gox Double Mutants Have Additive Effects.
Four-week-old wild-type (Col-0), single mutants gox1, haox1, and gox3, and three different double homozygous mutants (gox1 haox1, gox1 gox3, and haox1 gox3) were syringe-inoculated with the nonhost pathogen P. syringae pv tabaci at 104 cfu/mL to quantify bacterial growth at 0 and 3 DAI (A), (C), and (E). Bars represent the mean and sd of three independent experiments. Statistical significance for each time point was determined using one-way ANOVA, and P values from F test are indicated above bars. LSD test was used to determine differences between genotypes. Means with the same letter within a time point were not significantly different at P < 0.05. P. syringae pv tabaci was also inoculated at 106 cfu/mL to observe symptom development, and the photographs were taken at 5 DAI (B), (D), and (F). dpi, days postinoculation.
HAOX2 and GOX3 Play a Role in Activating Defense Signal Transduction Cascades
Earlier, we speculated that there might be a significant role for GOX3 and HAOX2 in nonhost resistance, because of their high levels of expression during inoculation with a nonhost pathogen (Table 1). Therefore, we wanted to investigate whether gox3 and haox2 mutants were affected in the expression of defense-related genes from known signal transduction cascades. Using qRT-PCR, the expression of defense-related genes in wild-type Col-0, haox2, and gox3 mutants upon inoculation with the nonhost pathogen P. syringae pv tabaci were compared (Table 2). We chose NHO1 because it has been shown to be required for nonhost disease resistance (Lu et al., 2001). Additional genes representing other plant defense–mediated signal transduction cascades—COI1 (JA pathway [Xie et al., 1998]), EIN3 (ET pathway [Roman et al., 1995; Chao et al., 1997]), EDS1 (Glazebrook et al., 1996), RAR1 (Muskett et al., 2002), PAD4 (Zhou et al., 1998), and SAG101 (SA pathway [Feys et al., 2005])—and genes associated with downstream responses, such as NPR1 (Dong, 2004) and PR-1 (Glazebrook et al., 1997), were chosen for transcript profiling. We also included both splice variants of WRKY4, a gene that has a role in defense responses against pathogens (Vandenabeele et al., 2003; Lai et al., 2008).
Table 2.
Expression of Various Defense-Related Genes in gox3 and haox2 Mutants after 24 h of Inoculation with the Nonhost Pathogen P. syringae pv tabaci
| Gene | Col-0 24 h Gene Expression Relative to Col-0 at 0 h (Pathogen/Mock Ratio) | gox3 24 h Gene Expression Relative to Col-0 at 0 h (Pathogen/Mock Ratio) | haox2 24 h Gene Expression Relative to Col-0 at 0 h (Pathogen/Mock Ratio) | F Test P Value |
| NHO1 | 1.81 ± 0.2a | 1.34 ± 0.08b | 0.71 ± 0.05c | <0.0001 |
| COI1 | 1.05 ± 0.09a | 0.71 ± 0.06b | 0.17 ± 0.13c | <0.0001 |
| EDS1 | 1.62 ± 0.41 | 0.84 ± 0.06 | 1.27 ± 0.18 | 0.15 |
| PAD4 | 13.2 ± 2.0a | 2.07 ± 0.7b | 1.13 ± 0.33b | <0.0001 |
| RAR1 | 1.15 ± 0.06a | 1.09 ± 0.11a | 0.17 ± 0.03b | <0.0001 |
| NPR1 | 2.8 ± 1.1a | 1.24 ± 0.13b | 0.08 ± 0.01c | 0.0002 |
| SAG101 | 2 ± 0.33a | 1.12 ± 0.07b | 1.4 ± 0.1a,b | 0.0249 |
| WRKY4A | 0.49 ± 0.05 | 0.39 ± 0.01 | 0.41 ± 0.09 | 0.7248 |
| WRKY4B | 2.17 ± 0.41a | 1.73 ± 0.33a,b | 0.77 ± 0.06b | 0.03 |
| EIN3 | 1.17 ± 0.06a | 0.98 ± 0.23a | 0.34 ± 0.08b | 0.0026 |
| PR1 | 260.2 ± 49.5a | 69.06 ± 26.45b | 5.65 ± 0.87c | <0.0001 |
| UBQ5 | 1.49 ± 0.21 | 1.56 ± 0.11 | 1.54 ± 0.3 | 0.9689 |
Statistical significance was determined using one-way ANOVA followed by Duncan test to establish differences between genotypes. Means with the same letter are not significantly different. Col-0 at 0 h was arbitrarily set to 1.
In wild-type Col-0, out of 11 defense-related genes tested, COI1, RAR1, and EIN3 were not induced. WRKY4A was repressed, whereas the remaining genes were induced after 24 h of inoculation with the nonhost pathogen P. syringae pv tabaci (Table 2). By contrast, all the genes that were induced in wild-type Col-0 were either not induced or induced at lesser levels or in some cases suppressed in gox3 and haox2 mutants (Table 2). Interestingly, COI1, RAR1, EIN3, which were not induced in the wild type, were instead significantly suppressed in haox2 mutants. WRKY4A was suppressed in Col-0 and both mutants without any significant differences among them, whereas WRKY4B was only significantly downregulated in the haox2 mutant. These results indicate that both mutants affect defense responses associated with different pathways, but the haox2 mutant seemed to be more severely compromised in inducing such responses.
DISCUSSION
We used VIGS in a fast-forward genetics screen (Baulcombe, 1999; Lu et al., 2003b; del Pozo et al., 2004) in N. benthamiana to identify plant genes that play a role in nonhost disease resistance (Figure 1). Silencing of GOX delayed the onset of the HR (Figure 1A; see Supplemental Figure 1 online), allowed the growth of the nonhost pathogens P. syringae pv tomato strain T1 (Figure 1C; see Supplemental Figure 3 online), P. syringae pv glycinea (Figure 1D) and X. campestris pv vesicatoria (Figure 2), and also affected the gene-for-gene resistance triggered by Pto-AvrPto and the elicitation of HR mediated by INF1 (Figure 3; see Supplemental Figure 4 online). All these data suggest that GOX represents a convergence point for signaling pathways originating from biological interactions with nonhost pathogens, incompatible pathogens, and elicitors. We do not understand, however, the basis of the specificity of response regarding Pto-AvrPto interaction and not the other interactions tested. Perhaps, as in Arabidopsis, there are different N. benthamiana genes encoding GOX that remain to be identified that account for that specificity, or maybe the other gene-for-gene interactions transduce the signal independently of GOX.
Similar to N. benthamiana, we observed that Arabidopsis mutants in GOX genes are compromised in nonhost disease resistance toward P. syringae pv tabaci and P. syringae pv syringae strain B728A (Figure 4) and in gene-for-gene resistance against the P. syringae pv tomato strain DC3000 strains harboring the avirulence genes AvrB and AvrRps4 (Figure 5). Upon inoculation with the nonhost pathogen P. syringae pv tabaci, all the gox mutants had dramatic reductions in callose deposition and cell death (quantified by electrolyte leakage; Figure 6) as well as in the levels of H2O2 (Figure 7B). These responses in the gox mutants are different from those observed in the NADPH oxidase mutant (rbohD) inoculated with the avirulent pathogen P. syringae pv tomato strain DC3000 (AvrRpm1). At concentrations high enough to elicit the HR, the rbohD mutant showed decreased H2O2 levels but only an 8% reduction in electrolyte leakage when compared with the wild-type Col-0 (Torres et al., 2002). Interestingly, the decrease in H2O2 accumulation caused by reduction of GOX appeared at 24 h after inoculation, after the oxidative burst attributed to NADPH oxidase (Figure 7C). Using different assays, we demonstrated that gox-dependent phenotypes are largely independent of NADPH oxidase function. Silencing of RBOHD (see Supplemental Figure 8 online) increased symptom development in wild-type Col-0 without significant difference in the bacterial growth in planta (Figures 8A and 8B). This indicates that the symptoms observed are not associated with the pathogen per se but with general susceptibility to environmental factors and is in agreement with the role of NADPH oxidase in wounding responses (Miller et al., 2009). Increased symptom development was also observed in gox3 and haox2, but only in the gox3 background was there a significant increase in bacterial titers (Figures 8A and 8B). We found that the extent of downregulation of RBOHD was enough to partially compromise H2O2 accumulation associated with the incompatible pathogen P. syringae pv tomato strain DC3000 (AvrRpm1) as reported before for the rbohD mutant (Torres, et al., 2002) (see Supplemental Figure 8B online). Furthermore, in contrast with the nonspecific response (wounding) of NADPH oxidase, the responses mediated by GOX are specific to certain plant–microbe interactions mediated by nonhost pathogens and avirulent pathogens, but not to host pathogens, and their effects are manifested 24 h after inoculation.
We showed that although all the GOX gene family members in Arabidopsis are essential for nonhost disease resistance, only GOX3, HAOX1, and HAOX2 are induced after inoculation with a nonhost pathogen (Table 1). This finding might indicate that these genes are part of a hierarchy wherein HAOX2 and perhaps HAOX1 have a main role during plant–pathogen interactions and activate diverse downstream signaling cascades, whereas GOX3 plays a secondary role activating specific defense-related genes. When the Arabidopsis plant is challenged with pathogens or elicitors, GOX3, HAOX1, and HAOX2 are induced (Table 1). GOX3 seems to partially and quantitatively affect NHO1 and SA-related defense responses (Table 2) and HAOX2 seems to affect all defense pathways tested, including SA, JA, and ET. We propose that the H2O2 generated by HAOX2 activates the SA pathway, mediated by PAD4, NPR1, and PR-1. PAD4 and SAG101 act together to amplify the SA defenses that eventually lead to PCD (Feys et al., 2005), and PAD4 has been shown to have pleiotropic effects on the regulation of NPR1 and PR-1 during defense responses (Zhou et al., 1998). Furthermore, these genes are known to be regulated by ROS (Rustérucci et al., 2001), and their importance in nonhost resistance to trigger the HR has been reported (Lipka et al., 2005). Additional support for the involvement of SA in the cascade initiated by GOX has been provided by recent work with an Arabidopsis cat2 mutant that is unable to metabolize the H2O2 produced by GOX (Chaouch et al., 2010). Consequently, the cat2 mutant accumulates high levels of H2O2 under photorespiratory conditions (long days), which trigger lesion formation reminiscent of the HR and show increased resistance to P. syringae pv tomato strain DC3000 in comparison with wild-type Col-0 (Chaouch et al., 2010). Interestingly, the cat2 mutant also showed significant accumulation of SA and increased expression of NHO1, a gene specifically associated with nonhost disease resistance (Lu et al., 2001; Kang et al., 2003) against Pseudomonas and presumably responsible for activating genes specific for nonhost resistance either directly or through PR-1. Our data showing that NHO1 is significantly downregulated in haox2 are in agreement with this finding. HAOX2 mutation also affected the expression of the transcription factor EIN3 (Table 2), which is involved in the ET signal transduction cascade (Chao et al., 1997). ET and SA coordinately induce several defense-related genes (Schenk et al., 2000), and ET can potentiate the SA-mediated induction of PR-1 (Lawton et al., 1994), suggesting that indeed ET and SA can act together to activate defense responses, such as PR-1 induction.
The nonhost pathogen used in this study, P. syringae pv tabaci, is a biotrophic pathogen that usually suppresses or fails to elicit the JA signal transduction pathway (Zimmerli et al., 2004). As expected, COI1 and EIN3 were not induced in wild-type Col-0 after 24 h of inoculation (Table 2). COI1 was downregulated in the haox2 mutant (Table 2), suggesting that HAOX2 is involved in its induction perhaps when SA and JA act synergistically (Nomura et al., 2006). We also showed that only one of the splice forms of the transcription factor WRKY4 (WRKY4B) is induced upon pathogen inoculation in wild-type Col-0 but is downregulated in the Athaox2 mutant (Table 2), indicating that HAOX2 contributes to WRKY4B induction. This finding is in contrast with the proposed negative role for WRKY4A in defense responses against P. syringae pv tomato strain DC3000 (Lai et al., 2008). It would be interesting to test whether WRKY4, as a transcription factor, is specifically and directly responsive to the H2O2 generated by HAOX2 and to identify its target genes associated with plant immunity.
Although GOX1 and GOX2 were shown to be downregulated by pathogen (Table 1), their role of nonhost resistance is undeniable as evidenced by the phenotypes of single mutants (Figure 4) and their quantitative effect in double mutants (Figure 9). More investigation is needed to understand how they are involved in the process. GOX1 and GOX2 are proposed to be involved in basic metabolism (photorespiration; Foyer et al., 2009), and hence it is likely that their mechanism of action involves remodeling of metabolism, as observed for RBOHF (Chaouch et al., 2011). Thus, their role in nonhost disease resistance may not be directly related to the production of H2O2 but to the interplay between soluble sugars and ROS (Couée et al., 2006). Alternatively, it is possible that these genes operate at different times during the interaction, and their downregulation upon pathogen infection might be a mechanism to energetically favor defense responses. A similar observation was made when microarray analysis was used to study Arabidopsis gene expression upon treatment with host and nonhost powdery mildews (Zimmerli et al., 2004). In addition, because GOX1 and GOX2 were shown to be highly induced by PCD (Zimmermann et al., 2004), it is likely that these genes might also be involved in the control of the HR to restore homeostasis and return to normal metabolic conditions, as has been proposed for RBOHD (Torres et al., 2005; Pogány et al., 2009) and RBOHF (Chaouch et al., 2011). The suppression of cell death by RBOHD and RBOHF (Chaouch et al., 2011) and the negative regulation of general stress-related genes and PR-1 associated with RBOHD after fungal infection (Pogány et al., 2009) or elicitor treatment (Galletti et al., 2008) is in sharp contrast with the positive regulation of defense responses leading to HR associated with HAOX2 and GOX3. Intriguingly, both RBOHF and HAOX2 are essential for SA accumulation, supporting the widely accepted view of crosstalk among ROS produced from different sources (Torres et al., 2006).
In conclusion, we have shown that the peroxisomal enzyme GOX plays a paramount role in nonhost resistance and some cases of gene-for-gene–mediated resistance in two different plant species. The mechanism of resistance is associated with the production of H2O2, which occurs during photorespiration through the conversion of glycolate to glyoxylate by GOX. We propose that the H2O2 generated specifically by HAOX2 and GOX3 activates components of the SA signal transduction cascade and also seems to regulate JA and ET pathways. GOX1 and GOX2 are known to play an essential role during basic metabolism (Foyer et al., 2009), and they also seem to play a secondary or indirect role during defense responses, although they are essential in the transition between defense responses and basic metabolism to restore homeostasis.
METHODS
Bacterial Strains and Arabidopsis thaliana Mutants
Agrobacterium tumefaciens strains were grown at 28°C in Luria-Bertani medium supplemented with rifampicin (25 μg/mL) and kanamycin (50 μg/mL). Pseudomonas syringae strains were grown in King’s B medium at 30°C supplemented with rifampicin (25 μg/mL), kanamycin (50 μg/mL), or streptomycin (50 μg/mL) when needed; Xanthomonas campestris pv vesicatoria was grown in Luria-Bertani medium. Bacterial strains used in this study are listed in Supplemental Table 1 online.
We obtained T-DNA insertions in GOX1 (SAIL177_G11), GOX2 (SALK_044052), HAOX1 (SAIL84_A04), and HAOX2 (SALK102409) from the ABRC (Alonso and Stepanova, 2003), and the T-DNA insertion in GOX3 (Gabi-Kat_523D09) was obtained from the European Arabidopsis Stock Centre (Rosso et al., 2003).
Fast-Forward Genetic Screening Using VIGS to Identify Genes Required for Nonhost Resistance
We used a normalized pTRV2-cDNA library in A. tumefaciens strain GV2260 containing clones from RNA isolated from mixed-elicitor–treated leaves of Nicotiana benthamiana (Anand et al., 2007). Agroinoculation for VIGS was performed using the toothpick method described previously (Anand et al., 2007; Wangdi et al., 2010). Two to 3 weeks after inoculation, fully expanded leaves of both silenced plants and vector control plants were infiltrated (using a needleless syringe) with a type I nonhost pathogen, P. syringae pv glycinea, and a type II nonhost pathogen, P. syringae pv tomato strain T1, at 3 × 108 cfu/mL. HR and/or disease symptoms were observed between 2 and 7 DAI.
HR Assays in N. benthamiana
To study the role of GOX in gene-for-gene resistance and PAMP-mediated immunity, silenced and control plants were coinfiltrated with a mixture of Agrobacterium strains containing various combinations of Avr-R genes (35S:AvrPto-35S:Pto; 35S:Avr9- 35S:Cf9; and 35S:tvEIX-35S:Eix2; Tang et al., 1996; Frederick et al., 1998; Van der Hoorn et al., 2000; Ron and Avni, 2004). In addition, an Agrobacterium strain carrying the PAMP-inducing Inf1 (35S:Inf1; Kamoun et al., 1997) was also infiltrated. HR symptoms were visually scored at 4 DAI, and samples were taken at 5 DAI for quantification. Quantification of the HR was done microscopically by monitoring the autofluorescence associated with the ensuing cell death (Klement et al., 1990). Ten disks (0.5 cm2) per treatment were collected and observed under an epifluorescence microscope using a GFP filter (excitation, 480 nm; emission, 535 nm). A total of 50 images per sample were collected from random microscopic fields and were analyzed by ImageJ software (http://rsb.info.nih.gov/ij/) by converting the images to gray scale and measuring the mean gray value for the entire image.
Cloning of Full-Length GOX from N. benthamiana
The clone TRV:16G11 was sequenced, and that partial sequence was used to design primers to clone the full-length gene using FirstChoice RLM-RACE Kit (Ambion) by following the manufacturer’s instructions. Briefly, total RNA was extracted from N. benthamiana leaves using the RNeasy Kit (Qiagen). To clone the 5′ end, total RNA was first treated with calf intestine alkaline phosphatase to remove free 5′-phosphates from fragmented mRNA, rRNA, and tRNA. After phenol extraction, the RNA was then treated with tobacco (Nicotiana tabacum) acid pyrophosphatase to remove the cap structure from the full-length mRNA. A 45-base RNA adapter oligonucleotide was then ligated to the RNA using T4 RNA ligase at 37°C for 1 h. The ligated RNA was used for reverse transcription at 42°C for 1 h using random decamers provided by the manufacturer. The cDNA was then used to amplify the 5′ end of the GOX gene by nested PCR using the following primer sets: 5′RACE (inner) and 5′RACE (outer). The PCR products were purified and cloned into pGEM-T vector (Promega). To clone the 3′ end of GOX, first-strand cDNA was directly synthesized from total RNA using the supplied 3′ RACE adapter. The cDNA was then subjected to nested PCR using the following primers: 3′RACE (outer) and 3′ RACE (inner) in addition to the supplied primers. The cloned ends of GOX were sequenced and used for full gene cloning. All primer sequences used for cloning are shown in Supplemental Table 2 online.
Quantification of Bacterial Growth in N. benthamiana
To follow the kinetics of bacterial growth, GOX-silenced and control N. benthamiana plants were vacuum infiltrated with host and nonhost pathogens at 3 × 104 cfu/mL to achieve uniform infection. At 0, 3, and 7 DAI, two leaf samples from four biological replicates were collected using a 0.5-cm2 core borer; leaf samples were ground, subjected to serial dilution, plated on King’s B agar medium supplemented with appropriate antibiotics, and incubated at 28°C for 2 d for bacterial colony counting. For visualization of bacterial growth using GFPuv-labeled strains, plants were syringe-inoculated at 104 cfu/mL, and observations were made at 7 DAI as described previously (Wang et al., 2007).
In Planta Inoculation to Evaluate Symptom Development and Bacterial Growth in Arabidopsis
Two methods and two nonhost pathogens were used to study symptom development in Arabidopsis: flood inoculation that mimics natural infection and syringe inoculation to accomplish uniform infection. For flood inoculation, 4-week-old plants grown in Murashige and Skoog plates were incubated for 5 min with 40 mL of a bacterial suspension containing P. syringae pv syringae strain B728A at a concentration of 3 × 107 cfu/mL (Ishiga et al., 2011). Symptoms were observed after 5 d. To examine bacterial growth, the entire rosette was harvested, ground, and serially diluted as described (Uppalapati et al., 2008; Ishiga et al., 2011).
For syringe inoculation, 6-week-old plants were infiltrated with a needleless syringe on the abaxial side of the leaves with the nonhost pathogen P. syringae pv tabaci at a concentration of 5 × 106 cfu/mL, and symptoms were evaluated after 3 d. This method was also used to examine bacterial growth in planta by nonhost pathogens as well as by host pathogens. For that purpose, bacteria were infiltrated at a concentration of 104 cfu/mL. At 0 and 3 DAI, two leaf samples from four biological replicates were collected, and the bacteria were quantified in a similar fashion as described above for N. benthamiana.
Enzymatic Assays
A total of 1 g of tissue harvested from 6-week-old Arabidopsis plants was resuspended in 6 mL of protein extraction buffer (0.25 M Suc, 50 mM Hepes-KOH, pH 7.2, 3 mM EDTA, 1 mM DTT, 0.6% polyvinylpyrrolidone, 3.6 mM l-Cys, 0. mM MgCl2, and complete-EDTA–free proteinase inhibitor cocktail [Roche Applied Science]), vortexed, and filtered through cheesecloth. The filtrate was centrifuged at 4°C at 10,000g for 45 min. Supernatants were transferred to new tubes and used directly to measure GOX activity or to extract membranes for NADPH oxidase activity assay.
Spectrophotometric assay to measure GOX activity was performed using sodium glycolate as substrate and by detecting the formation of the o-dianisidine radical cation at 440 nm as described previously (Macheroux et al., 1991). To measure NADPH oxidase activity, membrane fractions were separated from supernatants by centrifugation at 203,000g for 60 min, and pellets were then resuspended in 10 mM Tris-HCl (pH 7.4) (Sagi and Fluhr 2001). The NADPH-dependent O2− generating activity in the membrane fraction was determined after the reduction of XTT by O2−. The assay mixture contained 50 mM Tris-HCl buffer (pH 7.5), 0.5 mM XTT, 100 mM NADPH, and 15 to 30 μg of membrane proteins. XTT reduction was determined at 492 nm, and rates of O2− generation were calculated using an extinction coefficient of 2.16 × 104 M−1cm−1 (Jiang et al., 2002).
Detection of H2O2
Six-week-old Arabidopsis plants were inoculated with either P. syringae pv tabaci or P. syringae pv tomato strain DC3000 (AvrRpm1) at 106 or 107 cfu/mL or mock-inoculated. Only one-half of each leaf was inoculated. Leaves were detached and stained with a solution of DAB (1 mg/mL) for 6 h as described (Thordal-Christensen et al., 1997). To quantify the accumulation of H2O2 over time, 20 leaves were collected at 30 min, 2 h, 6 h, and 24 h after inoculation. After DAB staining, leaves were cleared in 100% ethanol and preserved in 25% glycerol. Stained leaves were scanned, and images analyzed by ImageJ software (http://rsb.info.nih.gov/ij/) by converting the images to gray scale and measuring the color intensity in the inoculated area as reported previously (Torres et al., 2005).
Callose Deposition
Wild-type Col-0 and Arabidopsis gox mutants were infected with the nonhost pathogen P. syringae pv tabaci at 106 cfu/mL. Leaves were detached, cleared, and stained with 0.1% aniline blue as described (Kvitko et al., 2009). After destaining, leaves were observed under a Leica TCS SP2 AOBS Confocal Laser Scanning Microscope with 4′,6-diamidino-2-phenylindole filter (Leica Microsystems). Callose deposits were counted on images taken from 10 random microscopic fields at 20× magnification using ImageJ software.
Electrolyte Leakage
Inoculated leaf samples were collected at 24 h after inoculation. Two leaf disks (0.5 cm2 each) were collected in triplicate for each sample, vacuum infiltrated with 25 mL water, and shaken for 1 h (López-Solanilla et al., 2004). Conductivity was measured with the Orion pHuture MMS555A conductivity meter (Thermo Electron).
qRT-PCR
Whole leaf samples from 4-week-old N. benthamiana or 6-week-old Arabidopsis were harvested and ground in liquid nitrogen. RNA was extracted using the RNeasy Plant Mini Kit (Qiagen) and treated with DNaseI (Ambion) before cDNA synthesis. Two micrograms of RNA were used for cDNA synthesis using Omniscript (Qiagen). qRT-PCR primers were designed using Primer Express Software v 3.0 (Applied Biosystems) and are listed in Supplemental Table 2 online. Primers for GOX1, GOX2, GOX3, HAOX1, and HAOX2 were designed to anneal in regions of low sequence similarity, among the GOX family members, toward the 3′ untranslated region. For each sample, three biological replicates with three technical replicates were used.
VIGS of RBOHD in Arabidopsis
We used TRV-based VIGS to downregulate the expression of the NADPH oxidase-encoding gene, RBOHD, in wild-type Col-0 and all five gox mutants described in this article. Primers AtrbohD3′F and AtrbohD3′R (see Supplemental Table 2 online) were used to clone ~350 bp fragment of RBOHD in TRV2. TRV:RBOHD was infiltrated initially in N. benthamiana as described for VIGS above. Sap from inoculated leaves was used as a source of virions to inoculate 3-week old Arabidopsis plants as previously described (Lu et al., 2003). Silencing of RBOHD gene was confirmed by qRT-PCR 2 weeks after inoculation. Silenced plants were syringe-inoculated with the nonhost pathogen P. syringae pv tabaci for symptom evaluation and for quantification of bacterial growth at the concentrations previously described.
Statistical Analyses
When indicated, Student’s t test was used for pairwise comparison between treatments. For multiple comparisons, one-way analysis of variance (ANOVA) was used to determine statistical significance among treatments at P < 0.05. If statistical significance based on the P value of the F test was found, the least significant difference (LSD) or Duncan tests at a P value < 0.05 were used to test differences between treatments. SAS Enterprise (SAS Institute) was used for statistical analyses.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: GOX1 (At3g14420), GOX2 (At3g14415), GOX3 (At4g1836), HAOX1 (At3g14130), HAOX2 (At3g14150), NHOI (At1g80460), COI1 (At2g39940), EDS1 (At3g48090), PAD4 (At3g52430), RAR1 (At5g51700), NPR1 (At1g64280), WRKY4A (At1g13960), EIN3 (At3g20770), PR1 (At2g14610), UBQ5 (At3g62250). N. benthamiana sequences: GOX (HQ110098); clone 6F8 (JN688263), clone 19A10 (JN688262), clone 37G12 (JN688264).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Silencing of GOX in N. benthamiana Affects the Elicitation of the HR.
Supplemental Figure 2. Deduced Amino Acid Alignment of Several Orthologous Protein Sequences of GOX.
Supplemental Figure 3. Silencing of GOX Compromises Nonhost Resistance in N. benthamiana.
Supplemental Figure 4. Silencing of GOX in N. benthamiana Compromises HR-Dependent Responses during Pto-AvrPto Interaction and after Transient Expression of Inf1.
Supplemental Figure 5. Confirmation of Gene Knockout in Glycolate Oxidase T-DNA Insertion Mutants (gox).
Supplemental Figure 6. Phenotypes of Wild-Type Col-0 and gox Mutants Grown on Murashige and Skoog Plates and Mock-Inoculated.
Supplemental Figure 7. gox Mutants Do Not Enhance Susceptibility to the Host Pathogen P. syringae pv maculicola in Arabidopsis.
Supplemental Figure 8. RBOHD Was Downregulated in Wild-Type Col-0 and All gox Mutants after VIGS.
Supplemental Figure 9. NADPH Oxidase Activity and Expression Are Not Reduced in gox Mutants.
Supplemental Figure 10. Similarity among Arabidopsis Genes Encoding GOX.
Supplemental Table 1. Bacterial Strains Used in This Study.
Supplemental Table 2. Primers Used in This Study.
Acknowledgments
We thank Jeff Dangl for providing P. syringae pv tomato strain DC3000 (AvrRpm1), Bethany Bishop for double mutant screening, Dong-Man Khu for assistance with statistical analyses, Vagner Benedito for assistance with qRT-PCR analysis, Hee-Kyung Lee for technical assistance, and Colleen Elles and Janie Gallaway for plant care. We also thank Richard Dixon, Kelly Craven, Million Tadege, and Seonghee Lee for critical reading of the article and Gregory Martin for providing the N. benthamiana VIGS-cDNA library. This work was supported by the Samuel Roberts Noble Foundation and in part by a grant from the Oklahoma Center for the Advancement of Science and Technology (PSB09-020). C.-M.R. acknowledges funding from the Next-Generation BioGreen 21 Program (grant no. PJ008170), Rural Development Administration, South Korea. The Leica AOBS confocal system used in this study was purchased using a National Science Foundation Equipment Grant (DBI 0400580).
AUTHOR CONTRIBUTIONS
C.M.R. designed experiments, performed research, and wrote the article; M.S.-K., K.W., C.-M.R., and A.K. performed research; and K.S.M. designed experiments and wrote the article.
References
- Alonso J.M., Stepanova A.N. (2003). T-DNA mutagenesis in Arabidopsis. Methods Mol. Biol. 236: 177–188 [DOI] [PubMed] [Google Scholar]
- Anand A., Vaghchhipawala Z., Ryu C.M., Kang L., Wang K., del-Pozo O., Martin G.B., Mysore K.S. (2007). Identification and characterization of plant genes involved in Agrobacterium-mediated plant transformation by virus-induced gene silencing. Mol. Plant Microbe Interact. 20: 41–52 [DOI] [PubMed] [Google Scholar]
- Baulcombe D.C. (1999). Fast forward genetics based on virus-induced gene silencing. Curr. Opin. Plant Biol. 2: 109–113 [DOI] [PubMed] [Google Scholar]
- Bohman S., Wang M., Dixelius C. (2002). Arabidopsis thaliana-derived resistance against Leptosphaeria maculans in a Brassica napus genomic background. Theor. Appl. Genet. 105: 498–504 [DOI] [PubMed] [Google Scholar]
- Chao Q., Rothenberg M., Solano R., Roman G., Terzaghi W., Ecker J.R. (1997). Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144 [DOI] [PubMed] [Google Scholar]
- Chaouch S., Queval G., Noctor G. (Nov 25, 2011). AtRbohF is a crucial modulator of defence-associated metabolism and a key actor in the interplay between intracellular oxidative stress and pathogenesis responses in Arabidopsis. Plant J http://dx.doi.org/10.1111/j.1365-313X.2011.04816.x [DOI] [PubMed] [Google Scholar]
- Chaouch S., Queval G., Vanderauwera S., Mhamdi A., Vandorpe M., Langlois-Meurinne M., Van Breusegem F., Saindrenan P., Noctor G. (2010). Peroxisomal hydrogen peroxide is coupled to biotic defense responses by ISOCHORISMATE SYNTHASE1 in a daylength-related manner. Plant Physiol. 153: 1692–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couée I., Sulmon C., Gouesbet G., El Amrani A. (2006). Involvement of soluble sugars in reactive oxygen species balance and responses to oxidative stress in plants. J. Exp. Bot. 57: 449–459 [DOI] [PubMed] [Google Scholar]
- del Pozo O., Pedley K.F., Martin G.B. (2004). MAPKKKalpha is a positive regulator of cell death associated with both plant immunity and disease. EMBO J. 23: 3072–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X. (2004). NPR1, all things considered. Curr. Opin. Plant Biol. 7: 547–552 [DOI] [PubMed] [Google Scholar]
- Ellis J. (2006). Insights into nonhost disease resistance: Can they assist disease control in agriculture? Plant Cell 18: 523–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B.J., Wiermer M., Bhat R.A., Moisan L.J., Medina-Escobar N., Neu C., Cabral A., Parker J.E. (2005). Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17: 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer C.H., Bloom A.J., Queval G., Noctor G. (2009). Photorespiratory metabolism: Genes, mutants, energetics, and redox signaling. Annu. Rev. Plant Biol. 60: 455–484 [DOI] [PubMed] [Google Scholar]
- Frederick R.D., Thilmony R.L., Sessa G., Martin G.B. (1998). Recognition specificity for the bacterial avirulence protein AvrPto is determined by Thr-204 in the activation loop of the tomato Pto kinase. Mol. Cell 2: 241–245 [DOI] [PubMed] [Google Scholar]
- Galletti R., Denoux C., Gambetta S., Dewdney J., Ausubel F.M., De Lorenzo G., Ferrari S. (2008). The AtrbohD-mediated oxidative burst elicited by oligogalacturonides in Arabidopsis is dispensable for the activation of defense responses effective against Botrytis cinerea. Plant Physiol. 148: 1695–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraats B.P., Bakker P.A., Lawrence C.B., Achuo E.A., Höfte M., van Loon L.C. (2003). Ethylene-insensitive tobacco shows differentially altered susceptibility to different pathogens. Phytopathology 93: 813–821 [DOI] [PubMed] [Google Scholar]
- Glazebrook J., Rogers E.E., Ausubel F.M. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143: 973–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J., Rogers E.E., Ausubel F.M. (1997). Use of Arabidopsis for genetic dissection of plant defense responses. Annu. Rev. Genet. 31: 547–569 [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack K.E., Jones J.D. (1997). Plant disease resistance genes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 575–607 [DOI] [PubMed] [Google Scholar]
- Heath M.C. (2000). Nonhost resistance and nonspecific plant defenses. Curr. Opin. Plant Biol. 3: 315–319 [DOI] [PubMed] [Google Scholar]
- Hinsch M., Staskawicz B. (1996). Identification of a new Arabidopsis disease resistance locus, RPs4, and cloning of the corresponding avirulence gene, avrRps4, from Pseudomonas syringae pv. pisi. Mol. Plant Microbe Interact. 9: 55–61 [DOI] [PubMed] [Google Scholar]
- Innes R.W., Bisgrove S.R., Smith N.M., Bent A.F., Staskawicz B.J., Liu Y.C. (1993). Identification of a disease resistance locus in Arabidopsis that is functionally homologous to the RPG1 locus of soybean. Plant J. 4: 813–820 [DOI] [PubMed] [Google Scholar]
- Ishiga Y., Ishiga T., Uppalapati S.R., Mysore K.S. (2011). Arabidopsis seedling flood-inoculation technique: A rapid and reliable assay for studying plant-bacterial interactions. Plant Methods 7: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Zhang J. (2002). Involvement of plasma membrane NADPH oxidase in abscisic acid and water stress-induced antioxidant defense in leaves of maize seedlings. Planta 215: 1022–1030 [DOI] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kamoun S., Lindqvist H., Govers F. (1997). A novel class of elicitin-like genes from Phytophthora infestans. Mol. Plant Microbe Interact. 10: 1028–1030 [DOI] [PubMed] [Google Scholar]
- Kamoun S., van West P., Vleeshouwers V.G., de Groot K.E., Govers F. (1998). Resistance of Nicotiana benthamiana to Phytophthora infestans is mediated by the recognition of the elicitor protein INF1. Plant Cell 10: 1413–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang L., Li J., Zhao T., Xiao F., Tang X., Thilmony R., He S., Zhou J.M. (2003). Interplay of the Arabidopsis nonhost resistance gene NHO1 with bacterial virulence. Proc. Natl. Acad. Sci. USA 100: 3519–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klement Z., Stall R.E., Novacky A.J., Ersek T., Fett W.F., Huang J.S., Beckman C.H. (1990). Mechanism of resistance. In Methods in Phytobacteriology, Z. Klement, K. Rudolph, and D.C. Sands, eds (Budapest, Hungary: Akademiai Kiado), pp. 469–473 [Google Scholar]
- Knoester M., van Loon L.C., van den Heuvel J., Hennig J., Bol J.F., Linthorst H.J. (1998). Ethylene-insensitive tobacco lacks nonhost resistance against soil-borne fungi. Proc. Natl. Acad. Sci. USA 95: 1933–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvitko B.H., Park D.H., Velásquez A.C., Wei C.F., Russell A.B., Martin G.B., Schneider D.J., Collmer A. (2009). Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 5: e1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z., Vinod K., Zheng Z., Fan B., Chen Z. (2008). Roles of Arabidopsis WRKY3 and WRKY4 transcription factors in plant responses to pathogens. BMC Plant Biol. 8: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C., Dixon R.A. (1997). The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 251–275 [DOI] [PubMed] [Google Scholar]
- Lawton K.A., Potter S.L., Uknes S., Ryals J. (1994). Acquired resistance signal transduction in Arabidopsis is ethylene independent. Plant Cell 6: 581–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A., Tenhaken R., Dixon R., Lamb C. (1994). H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79: 583–593 [DOI] [PubMed] [Google Scholar]
- Lipka U., Fuchs R., Lipka V. (2008). Arabidopsis non-host resistance to powdery mildews. Curr. Opin. Plant Biol. 11: 404–411 [DOI] [PubMed] [Google Scholar]
- Lipka V., et al. (2005). Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310: 1180–1183 [DOI] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Dinesh-Kumar S.P. (2002a). Virus-induced gene silencing in tomato. Plant J. 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Marathe R., Dinesh-Kumar S.P. (2002b). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30: 415–429 [DOI] [PubMed] [Google Scholar]
- Loehrer M., Langenbach C., Goellner K., Conrath U., Schaffrath U. (2008). Characterization of nonhost resistance of Arabidopsis to the Asian soybean rust. Mol. Plant Microbe Interact. 21: 1421–1430 [DOI] [PubMed] [Google Scholar]
- López-Solanilla E., Bronstein P.A., Schneider A.R., Collmer A. (2004). HopPtoN is a Pseudomonas syringae Hrp (type III secretion system) cysteine protease effector that suppresses pathogen-induced necrosis associated with both compatible and incompatible plant interactions. Mol. Microbiol. 54: 353–365 [DOI] [PubMed] [Google Scholar]
- Lu M., Tang X., Zhou J.M. (2001). Arabidopsis NHO1 is required for general resistance against Pseudomonas bacteria. Plant Cell 13: 437–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Martin-Hernandez A.M., Peart J.R., Malcuit I., Baulcombe D.C. (2003). Virus-induced gene silencing in plants. Methods 30: 296–303 [DOI] [PubMed] [Google Scholar]
- Macheroux P., Massey V., Thiele D.J., Volokita M. (1991). Expression of spinach glycolate oxidase in Saccharomyces cerevisiae: Purification and characterization. Biochemistry 30: 4612–4619 [DOI] [PubMed] [Google Scholar]
- Mellersh D.G., Heath M.C. (2003). An investigation into the involvement of defense signaling pathways in components of the nonhost resistance of Arabidopsis thaliana to rust fungi also reveals a model system for studying rust fungal compatibility. Mol. Plant Microbe Interact. 16: 398–404 [DOI] [PubMed] [Google Scholar]
- Miller G., Schlauch K., Tam R., Cortes D., Torres M.A., Shulaev V., Dangl J.L., Mittler R. (2009). The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci. Signal. 2: ra45. [DOI] [PubMed] [Google Scholar]
- Muskett P.R., Kahn K., Austin M.J., Moisan L.J., Sadanandom A., Shirasu K., Jones J.D., Parker J.E. (2002). Arabidopsis RAR1 exerts rate-limiting control of R gene-mediated defenses against multiple pathogens. Plant Cell 14: 979–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysore K.S., Ryu C.M. (2004). Nonhost resistance: How much do we know? Trends Plant Sci. 9: 97–104 [DOI] [PubMed] [Google Scholar]
- Nasir K.H., et al. (2005). High-throughput in planta expression screening identifies a class II ethylene-responsive element binding factor-like protein that regulates plant cell death and non-host resistance. Plant J. 43: 491–505 [DOI] [PubMed] [Google Scholar]
- Neill S.J., Desikan R., Clarke A., Hurst R.D., Hancock J.T. (2002). Hydrogen peroxide and nitric oxide as signalling molecules in plants. J. Exp. Bot. 53: 1237–1247 [PubMed] [Google Scholar]
- Nomura K., Debroy S., Lee Y.H., Pumplin N., Jones J., He S.Y. (2006). A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313: 220–223 [DOI] [PubMed] [Google Scholar]
- Pogány M., von Rad U., Grün S., Dongó A., Pintye A., Simoneau P., Bahnweg G., Kiss L., Barna B., Durner J. (2009). Dual roles of reactive oxygen species and NADPH oxidase RBOHD in an Arabidopsis-Alternaria pathosystem. Plant Physiol. 151: 1459–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queval G., Hager J., Gakière B., Noctor G. (2008). Why are literature data for H2O2 contents so variable? A discussion of potential difficulties in the quantitative assay of leaf extracts. J. Exp. Bot. 59: 135–146 [DOI] [PubMed] [Google Scholar]
- Reumann S., Ma C., Lemke S., Babujee L. (2004). AraPerox. A database of putative Arabidopsis proteins from plant peroxisomes. Plant Physiol. 136: 2587–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman G., Lubarsky B., Kieber J.J., Rothenberg M., Ecker J.R. (1995). Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: Five novel mutant loci integrated into a stress response pathway. Genetics 139: 1393–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron M., Avni A. (2004). The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16: 1604–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso M.G., Li Y., Strizhov N., Reiss B., Dekker K., Weisshaar B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Rustérucci C., Aviv D.H., Holt B.F., III, Dangl J.L., Parker J.E. (2001). The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell 13: 2211–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi M., Fluhr R. (2001). Superoxide production by plant homologues of the gp91 (phox) NADPH oxidase. Modulation of activity by calcium and by tobacco mosaic virus. Plant Physiol. 126: 1281–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer P., Hückelhoven R., Kogel K.H. (2004). The white barley mutant albostrians shows a supersusceptible but symptomless interaction phenotype with the hemibiotrophic fungus Bipolaris sorokiniana. Mol. Plant Microbe Interact. 17: 366–373 [DOI] [PubMed] [Google Scholar]
- Schenk P.M., Kazan K., Wilson I., Anderson J.P., Richmond T., Somerville S.C., Manners J.M. (2000). Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 97: 11655–11660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P.C., Ito A., Shimizu T., Terauchi R., Kamoun S., Saitoh H. (2003). Virus-induced silencing of WIPK and SIPK genes reduces resistance to a bacterial pathogen, but has no effect on the INF1-induced hypersensitive response (HR) in Nicotiana benthamiana. Mol. Genet. Genomics 269: 583–591 [DOI] [PubMed] [Google Scholar]
- Taler D., Galperin M., Benjamin I., Cohen Y., Kenigsbuch D. (2004). Plant eR genes that encode photorespiratory enzymes confer resistance against disease. Plant Cell 16: 172–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Frederick R.D., Zhou J., Halterman D.A., Jia Y., Martin G.B. (1996). Initiation of plant disease resistance by physical interaction of AvrPto and Pto Kinase. Science 274: 2060–2063 [DOI] [PubMed] [Google Scholar]
- Thomma B.P., Nelissen I., Eggermont K., Broekaert W.F. (1999). Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. Plant J. 19: 163–171 [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen H., Zang Z., Wei Y., Collinge D.B. (1997). Subcellular localization of H2O2 in plants, H2O2 accumulation in papillae and hypersensitive response during barley-powdery mildew interaction. Plant J. 11: 1187–1194 [Google Scholar]
- Torres M.A., Dangl J.L., Jones J.D. (2002). Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 99: 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M.A., Jones J.D., Dangl J.L. (2005). Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat. Genet. 37: 1130–1134 [DOI] [PubMed] [Google Scholar]
- Torres M.A., Jones J.D., Dangl J.L. (2006). Reactive oxygen species signaling in response to pathogens. Plant Physiol. 141: 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppalapati S.R., Ishiga Y., Wangdi T., Urbanczyk-Wochniak E., Ishiga T., Mysore K.S., Bender C.L. (2008). Pathogenicity of Pseudomonas syringae pv. tomato on tomato seedlings: Phenotypic and gene expression analyses of the virulence function of coronatine. Mol. Plant Microbe Interact. 21: 383–395 [DOI] [PubMed] [Google Scholar]
- Van Breusegem F., Dat J.F. (2006). Reactive oxygen species in plant cell death. Plant Physiol. 141: 384–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Hoorn R.A., Laurent F., Roth R., De Wit P.J. (2000). Agroinfiltration is a versatile tool that facilitates comparative analyses of Avr9/Cf-9-induced and Avr4/Cf-4-induced necrosis. Mol. Plant Microbe Interact. 13: 439–446 [DOI] [PubMed] [Google Scholar]
- Vandenabeele S., Van Der Kelen K., Dat J., Gadjev I., Boonefaes T., Morsa S., Rottiers P., Slooten L., Van Montagu M., Zabeau M., Inze D., Van Breusegem F. (2003). A comprehensive analysis of hydrogen peroxide-induced gene expression in tobacco. Proc. Natl. Acad. Sci. USA 100: 16113–16118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Kang L., Anand A., Lazarovits G., Mysore K.S. (2007). Monitoring in planta bacterial infection at both cellular and whole-plant levels using the green fluorescent protein variant GFPuv. New Phytol. 174: 212–223 [DOI] [PubMed] [Google Scholar]
- Wangdi T., Uppalapati S.R., Nagaraj S., Ryu C.M., Bender C.L., Mysore K.S. (2010). A virus-induced gene silencing screen identifies a role for Thylakoid Formation1 in Pseudomonas syringae pv tomato symptom development in tomato and Arabidopsis. Plant Physiol. 152: 281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Shortt B.J., Lawrence E.B., Levine E.B., Fitzsimmons K.C., Shah D.M. (1995). Disease resistance conferred by expression of a gene encoding H2O2-generating glucose oxidase in transgenic potato plants. Plant Cell 7: 1357–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D.X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Yoshioka H., Numata N., Nakajima K., Katou S., Kawakita K., Rowland O., Jones J.D., Doke N. (2003). Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 15: 706–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N., Tootle T.L., Glazebrook J. (1999). Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell 11: 2419–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N., Tootle T.L., Tsui F., Klessig D.F., Glazebrook J. (1998). PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10: 1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerli L., Stein M., Lipka V., Schulze-Lefert P., Somerville S. (2004). Host and non-host pathogens elicit different jasmonate/ethylene responses in Arabidopsis. Plant J. 40: 633–646 [DOI] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]