Some sugar residues of the pectinaceous polysaccharide backbone of primary cell walls are acetylated. This study identifies and characterizes a Populus pectin acetylesterase that liberates acetylesters from polysaccharides and unveils its biological roles in planta as a structural regulator of cell wall extensibility and digestibility that affects plant growth, development, and reproduction.
Abstract
Pectin is a major component of the primary cell wall of higher plants. Some galacturonyl residues in the backbone of pectinaceous polysaccharides are often O-acetylated at the C-2 or C-3 position, and the resulting acetylesters change dynamically during the growth and development of plants. The processes involve both enzymatic acetylation and deacetylation. Through genomic sequence analysis, we identified a pectin acetylesterase (PAE1) from black cottonwood (Populus trichocarpa). Recombinant Pt PAE1 exhibited preferential activity in releasing the acetate moiety from sugar beet (Beta vulgaris) and potato (Solanum tuberosum) pectin in vitro. Overexpressing Pt PAE1 in tobacco (Nicotiana tabacum) decreased the level of acetyl esters of pectin but not of xylan. Deacetylation engendered differential changes in the composition and/or structure of cell wall polysaccharides that subsequently impaired the cellular elongation of floral styles and filaments, the germination of pollen grains, and the growth of pollen tubes. Consequently, plants overexpressing PAE1 exhibited severe male sterility. Furthermore, in contrast to the conventional view, PAE1-mediated deacetylation substantially lowered the digestibility of pectin. Our data suggest that pectin acetylesterase functions as an important structural regulator in planta by modulating the precise status of pectin acetylation to affect the remodeling and physiochemical properties of the cell wall's polysaccharides, thereby affecting cell extensibility.
INTRODUCTION
The primary cell walls of plants are rigid yet dynamically organized networks comprising a mixture of polysaccharides and structural proteins, of which pectin is a major component (Carpita and McCann, 2000; Ridley et al., 2001; Mohnen, 2008). The pectinaceous polysaccharides consist of different structural domains, including the linear homogalacturonan (HG) regions of up to 200 residues of (1,4)-linked α-D-galacturonic acid (GalA) and the highly branched “hairy” regions, including rhamnogalacturonan I (RG-I), which encompasses the repeating disaccharide units of α-(1,2)-l-Rha-α-(1,4)-D-GalA, the highly substituted galacturonan RG-II domain, and the xylogalacturonan domain (Carpita and McCann, 2000; Mohnen, 2008). Some galacturonyl residues in the backbone of pectinaceous polysaccharides are often O-acetylated at the C-2 or C-3 hydroxyl, or methylesterified at the C-6 carboxyl, although the distribution of methyl- and acetylesters between HG and RG is still unclear (Kouwijzer et al., 1996; Perrone et al., 2002; Ralet et al., 2008). In the pectins of bamboo shoot (Phyllostachys edulis), flax (Linum usitatissimum), potato (Solanum tuberosum), cotton (Gossypium hirsutum), carrot (Daucus carota), tobacco (Nicotiana tabacum), and tomato (Solanum lycopersicum), acetyl groups reportedly are highly abundant in the RG-I domain (Komalavilas and Mort, 1989; Ishii, 1995, 1997; Schols and Voragen, 1996), whereas in the pectin of sugar beet (Beta vulgaris), ~75% of the acetyl groups belong to the HG backbone (Kouwijzer et al., 1996; Ralet et al., 2005).
Presumably, pectins are synthesized in the cis-Golgi, methylesterified in the medial- Golgi, substituted in the trans-Golgi, and then secreted into the cell wall in a highly esterified state (Micheli, 2001; Sterling et al., 2001; Mohnen, 2008). The function of pectin methylesterification has been explored extensively. Both its extent and distribution are found to be important for determining pectin’s physiochemical and biological properties (Peaucelle et al., 2008; Wolf et al., 2009). The enzymes responsible for regulating pectin methylesters are also well characterized (Tieman et al., 1992; Wen et al., 1999; Bosch and Hepler, 2005; Pelloux et al., 2007; Pelletier et al., 2010). By contrast, studies on the occurrence and functions of pectin acetylation are scarce (Pauly and Scheller, 2000; Manabe et al., 2011). Purportedly, pectin is acetylated between the Golgi and the cell wall during its exocytosis; then, the exported acetylated pectin is incorporated into cell walls (Liners et al., 1994; Pauly and Scheller, 2000; Scheller and Ulvskov, 2010). The extent and distribution of acetylesters vary according to the source of pectin (Schols and Voragen, 1994). Pectins from strawberry (Fragaria ananassa) and sugar beet contain a sizeable portion of acetyl groups, viz., 1.2 and 2.5% (w/w), respectively, equivalent to 5 and 10% acetylation of the available galacturonyl residues, while citrus fruit and apple (Malus domestica) pectins have fewer acetylesters, ~0.25 and 0.4%, respectively (Voragen et al., 1986). Moreover, the degree of O-acetylation of pectin changes during the growth and differentiation of plant tissues and in response to the environmental conditions (Liners et al., 1994; Vercauteren et al., 2002; Gou et al., 2008). The acetate content in the cell wall of juvenile leaves of black cottonwood (Populus trichocarpa) is much higher than that in mature and senescent leaves (Gou et al., 2008). The change in acetylation during development points to an in muro dynamic structural modification that might be important for the organization, metabolism, and function of polysaccharides in planta. However, there is limited information on the enzymatic processes of both the acetylation and deacetylation of polysaccharides and their biological effects on plant growth and development.
Earlier investigations suggested pectin acetylesters might be one crucial structural factor in regulating the polymer’s biophysical properties (Liners et al., 1994). For example, like methylester status, the degree of acetylation affected the gel formation of extracted pectins. Partial hydrolysis of the acetyl esters in beet pectins improved their gelation properties (Pippen et al., 1950; Williamson et al., 1990; Ralet et al., 2003). Deacetylation also increased pectin’s solubility in water by lowering the hydrophobicity of the polysaccharide backbone (Rombouts and Thibault, 1986). On the other hand, acetylesters demonstrably protect polysaccharides against enzymatic digestion (Biely et al., 1986; Schols and Voragen, 1994; Chen and Mart, 1996; Benen et al., 1999; Bonnin et al., 2003). Thus, removing those moieties either with digestive enzymes or by chemical saponification in vitro seems to be a prerequisite for degrading the polysaccharide core (Kauppinen et al., 1995).
The synthesis and dynamics of cell wall acetylesters involve at least two types of enzymes, acetyltransferases, which transacylate the sugar residues of polymers (Pauly and Scheller, 2000; Gille et al., 2011; Lee et al., 2011; Manabe et al., 2011), and acetylesterases, which cleave the ester bond between a glycosyl carbon and an acetyl group, thus releasing acetate from modified polysaccharides (Williamson, 1991; Bordenave et al., 1995). Pectin acetylesterase (EC 3.1.1.6; PAE) belongs to CAZy class 12 and 13 of the CE family (http://www.cazy.org/). The enzymes were isolated from bacteria (Shevchik and Hugouvieux-Cotte-Pattat, 1997, 2003), fungi (Kauppinen et al., 1995; Bonnin et al., 2008), and plants (Williamson, 1991; Bordenave et al., 1995; Christensen et al., 1996). Two different PAEs were purified from orange (Citrus sinensis) peel, one with a molecular mass (MW) of 29 to 30 kD and a pI of 5.1 (Williamson, 1991), the other with a molecular mass of 42 kD and a pI > 9 (Christensen et al., 1996). The PAE purified from the cell walls of mung bean (Vigna radiata) hypocotyls had physicochemical properties (molecular mass and pI) similar to those of the higher molecular mass acetylesterase from orange (Bordenave et al., 1995). The PAEs from both species exhibited the highest activity on synthetic substrates, such as triacetin and p-nitrophenyl acetate, and were active on the native pectin of sugar beet and flax (Williamson, 1991; Bordenave et al., 1995; Christensen et al., 1996). A cDNA encoding a putative PAE was isolated from mung bean seedlings, based on the peptide sequences of the purified protein (Breton et al., 1996). However, no further biochemical and biological analyses were made to ascertain its functions.
Approximately 10 putative PAE genes were deduced in the P. trichocarpa genome (Geisler-Lee et al., 2006); however, none are functionally characterized. In this study, we characterized one of those homologous genes. Expressing this gene in planta reduced the content of acetylester primarily in pectinaceous polysaccharides and, consequently, resulted in pleiotropic effects on the plant’s growth, development, and reproduction.
RESULTS
Gene Expression Analysis of Putative PAEs from P. trichocarpa
Previous analyses of genomic sequences in Populus recognized a set of putative carbohydrate esterase (CE) genes encoding pectin acetyltransferases (Geisler-Lee et al., 2006). To explore their functions, we retrieved the gene sequences from the P. trichocarpa genome. Sequence alignment and phylogenetic analysis showed that nine putative Populus CEs (excluding one partial sequence), together with 12 putative acetylesterase homologs from Arabidopsis thaliana (http://www.cazy.org/), clustered into three different clades. All plant esterase genes are distinct from those of bacteria and fungi. Among them, CE13_4, 5, and 7 are grouped with the PAE cDNA cloned from mung bean (Breton et al., 1996) and separate from the others (Figure 1; see Supplemental Data Set 1 online).
Figure 1.
Phylogenetic Analysis of Polysaccharide Acetylesterases.
The neighbor-joining tree was constructed from the aligned full-length amino acid sequences of putative carbohydrate acetylesterases from P. trichocarpa and Arabidopsis. The amino acid sequences of the following enzymes were also included: CAA67728 (Vr PAE, from V. radiata), AJ507215 (PaeX, from Erwinia chrysantemi), Y09828 (PaeY, from E. chrysantemi), BAA11692 (YxiM, from Bacillus subtilis), and CAA61858 (RAGA1, from A. aculeatus). Bar represents the output distance as number of substitutions per site (i.e., 0.2 substitutions per site).
These P. trichocarpa putative CE genes displayed distinct tissue-specific (see Supplemental Figures 1A to 1H online) and temporal (see Supplemental Figures 1I to 1P online) expression patterns, suggesting their potential distinct physiological functions. Among them, CE13_5 was highly expressed in young leaves and in the early developing nodes (see Supplemental Figures 1C and 1K online). Its homologous cDNA probe in Populus balsamifera was also highly expressed in female and male catkins, in addition to young leaves and seedlings, as found in a developmental tissue transcriptomic analysis (Wilkins et al., 2009; see Supplemental Figure 2 online). These expression patterns imply that CE13_5 may be involved in modifying primary cell walls in fast growing tissues. Accordingly, we cloned this gene for further functional analysis.
Recombinant Pt CE13_5 Exhibits Acetylesterase Activity in Vitro
Using RT-PCR, we isolated cDNA encoding CE13_5 from the total RNAs of the leaves and stems of P. trichocarpa. The cDNA clone (accession number HQ223420) contains 1185 nucleotides and encodes a polypeptide of 394 amino acid residues, with a deduced molecular mass of 42.8 kD and pI of 6.6.
The amino acid residues of CE13_5 share ~62% identity with the PAE from mung bean (Breton et al., 1996); both have a conserved GXSXG motif, characteristic of the Ser hydrolase superfamily. The encoded sequence of Pt CE13_5 differs from the bacterial PAEs, PaeY, PaeX, and YxiM (~2 to ~10% identities), and from the RG acetylesterase of Aspergillus aculeatus (~6% identity) (Figure 1), pointing to the distinct evolutionary origins of the polysaccharide acetylesterases.
To characterize the biochemical function of Pt CE13_5, we expressed the gene in Escherichia coli strain Rosetta (DE3) pLysS, a heterologous expression system with a tightly controlled level of protein expression. Although the recombinant protein was severely aggregated and sequestered into inclusion bodies, we recovered and purified a small portion of the soluble recombinant protein from the transgenic E. coli.
To examine its potential esterase activity on polysaccharides, we chemically derived the acetylated polysaccharides (Gou et al., 2008). The recombinant protein showed high activity in releasing acetyl moieties from acetylated pectate (polygalacturonan), xylan, and arabinogalactan, with the highest activity on the first one (see Supplemental Figure 3 online). Like the acetylesterases purified from plant species, the recombinant enzyme also displayed activity on the synthetic chemical 2-naphthol acetate (see Supplemental Figure 3 online).
Since chemical derivatization may entail the hyperacetylation of polysaccharides, and so might not represent the native acetylation pattern of cell wall polymers, we incubated Pt CE13_5 recombinant proteins with polysaccharides prepared from natural resources. These included the RG-I–enriched pectin from potato (Ishii, 1997), the HG-enriched pectin from sugar beet (Quéméner et al., 2003; Ralet et al., 2005), the birch wood (Betula spp) xylan, and a xylan/hemicellulose preparation from cell walls of tobacco leaves. We employed a mild alkaline treatment to calibrate the acetylester content in those polymers used for substrates (see Supplemental Figure 4 online). As depicted in Figure 2A, the recombinant protein showed high activity in releasing acetyl moieties from sugar beet and potato pectins but substantially lower activity with birch wood xylan and the tobacco xylan/hemicellulose preparation. Although the limited solubility of natural polymers (pectin and xylan) prevented a detailed kinetic analysis, determining the kinetic parameters using chemically acetylated polymers revealed that the recombinant Pt CE13_5 preferentially reacts with acetylated pectate over xylan (Table 1).
Figure 2.
Enzymatic Activity of Recombinant Pt PAE1.
(A) Acetylesterase activity of recombinant PAE1 with different natural carbohydrate polymers.
(B) Methylesters released by 2 μg pectin esterase (as positive control), Pt PAE1, and buffer (as negative control [Neg. Ctrl.]) within 15 min at 35°C.
(C) Ferulic acid released by alkaline treatment (2 n NaOH as positive control), 2 μg Pt PAE1, or buffer as negative control in 15 min at 35°C. Data represent mean of triplicate samples. The error bar represents se.
Table 1.
Kinetics of Recombinant Pt CE13-5
| Substrate | Vmax (μmole Acetate/mg/min) | Km (μg/mL)a |
| Acetylated pectate | 5.8 ± 0.5 | 6.3 ± 1.3 |
| Acetylated xylan | 5.1 ± 1.3 | 25.3 ± 8.6 |
Because the polymers lacked precise molecular mass, Km was not expressed in units of molar concentration.
To examine whether the recombinant enzyme has promiscuous methyl- or feruloyl- esterase activity, we incubated the enzyme with the highly methylesterified pectins from citrus fruits, potato, and sugar beet. We found that the Pt CE13_5 recombinant protein essentially showed no activity in releasing methylesters from all of those natural polymers compared with the pectin methylesterase (Figure 2B). Similarly, we did not detect any activity in removing the feruloyl moiety from the incubated sugar beet pectin, although a certain amount of ferulate is released from this polymer by mild alkaline treatment (Figure 2C). We thus designated Pt CE13_5 as P. trichocarpa pectin acetylesterase 1 (Pt PAE1).
Overexpression of PAE1 Reduces Acetyl Content in the Pectin of Transgenic Plants
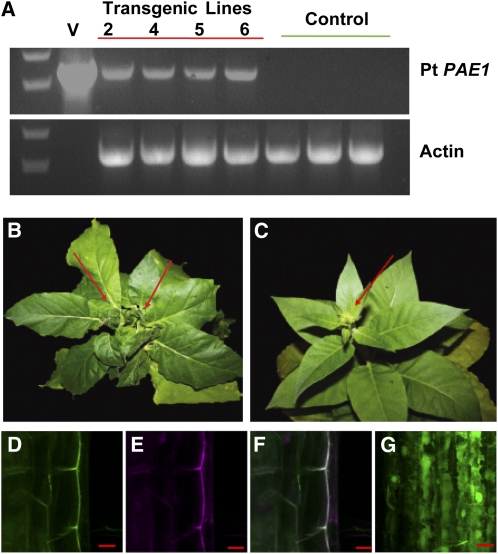
To investigate the biological function of Pt PAE1 in planta, we expressed its full-length cDNA in tobacco (Figure 3A). Overexpression of PAE altered the morphology of the tobacco plants in all of the independent transgenic lines. During vegetative growth, the primary apical buds of the Pt PAE1 transgenic plants frequently wilted, followed by the emergence of new primary and lateral buds (Figure 3B; see Supplemental Figure 5 online). The expanded leaves of the transgenic plants exhibited a wavy surface with curled edges, which differed from those of the wild type (Figure 3C; see Supplemental Figure 5 online).
Figure 3.
Overexpression of Pt PAE1 in Tobacco.
(A) RT-PCR examination of Pt PAE1 expression in transgenic tobacco lines. Full-length PAE1 cDNA was detected in the transgenic lines but not in the controls. V, transgenic vector (as positive control). The data represent the result from one of three replicates.
(B) and (C) Vegetative development of transgenic (B) and control (C) tobacco. Note that a couple of primary shoot apexes were apparent in the transgenic line, Pt PAE1_6 (arrows in [B]) but that only one was present in the control plant (arrow in [C]).
(D) to (F) Subcellular localization of Pt PAE1.
(D) Green fluorescence signal of GFP-PAE1 fusion monitored in the cells of 5-d-old dark-grown transgenic seedling.
(E) The cells stained with propidium iodide.
(F) The merged image of (D) and (E).
(G) Green fluorescence signal of the free GFP. Bars = 2 μm.
We fused Pt PAE1 at the C terminus of green fluorescent protein (GFP) and stably expressed this construct in tobacco, driven by the constitutive 35S cauliflower mosaic virus promoter. The fluorescence signals appeared primarily in the contours of the etiolated hypocotyl cells, in good agreement with propidium iodide staining, a convenient histological marker for monitoring the plant cell wall polysaccharides (McKenna et al., 2009) and in sharp contrast with that of free GFP (Figures 3D to 3G), indicating that PAE1 localized to cell walls.
Subsequently, we extracted individually the water- and acid-soluble pectins from the expanded young leaves of both transgenic and control tobacco plants (see Supplemental Figure 6 online) and isolated xylan from the xylem tissues via xylanase digestion. We readily detected acetyl esters in both the pectin and xylan fractions from the transgenic and control tobacco plants. However, their levels in both water- and acid-soluble pectins of most transgenic plants were significantly lower than in those of the control plants by about a 13 To ~42% and 8 to ~11%, respectively (Figures 4A and 4B). By contrast, the levels of acetyl moieties in xylanase-digested fractions showed either no change or even some increase in transgenic plants (Figure 4C). Meanwhile, the methylester content of pectin from transgenic tobacco leaves was notably increased, indicating potential metabolic compensation or carbon flux redirection (see Supplemental Figure 7 online).
Figure 4.
Cell Wall Acetylester Content of Pt PAE1 Overexpression Plants.
(A) and (B) Acetylester content of the water- and acid-soluble pectins from young leaves of the transgenic and control lines.
(C) Acetylester content of xylan from xylem cell walls. Data represent mean of triplicate samples. Error bar represents sd. Asterisks indicate a statistically significant change (P < 0.05) under Student’s t test.
These data suggest that Pt PAE1 encodes an active esterase that predominantly removes acetyl moieties from pectin in vivo.
Overexpression of Pt PAE1 Affects the Development of the Floral Tissues
As the transgenic tobacco plants approached reproductive growth, we found a number of developing or open flowers that had wilted or had fused petals (see Supplemental Figure 5 online). The lengths of the styles and filaments in the transgenic flowers were shorter than those in the flowers of the control plants (Figure 5A; see Supplemental Figure 8A online), indicating that the expression of Pt PAE1 retarded the elongation of both pistil and stamen tissues. Calculating the lengths of the epidermal cells at different positions of the stylar tract and filaments in flowers at flowering stage 12 (Drews et al., 1992) revealed that all epidermal cells of the filament and the cells of the basal and middle stylar tract of independent transgenic lines were significantly shorter than those of the control (Figure 5B), indicating that the expression of PAE1 depresses cell elongation. Consequently, the relative lengths of the stigma and anthers in transgenic flower were greatly altered. In control flowers, four of five anthers were at nearly the same height as the stigma or slightly above it (Figures 5C and 5D), whereas, in transgenic flowers, from flowering stage 11, the positions of all five anthers were lower than the stigma (Figure 5E), and this morphological change became more pronounced as flowering reached stage 12 (Figure 5F). The ratio of filament to pistil in transgenic flowers was markedly less than that of the controls (see Supplemental Figure 8B online), confirming the severe impairment of elongation of the filament cells in the transgenic plants.
Figure 5.
Development of Floral Tissues in Tobacco Plants Overexpressing Pt PAE1.
(A) Style and filaments in the developing flowers (at stage 12) of the control and transgenic (Pt PAE1) plants.
(B) The lengths of epidermal cells of the style and filament at the apical, middle, and basal parts of both tissues of independent transgenic lines. Data represent the average of 40 cells. Error bar denotes the sd. Asterisks indicate a statistically significant change (P < 0.05) under Student’s t test.
(C) to (F) Positioning of stigma and anthers in the flower of the wild-type control ([C] and [D]) and in the overexpression plants ([E] and [F]) at flowering stages 11 ([C] and [E]) and 12 ([D] and [F]). Bar = 5 mm.
[See online article for color version of this figure.]
To examine the potential changes of the cell wall composition, particularly of the acetylesters of the style and filaments, we collected these tissues from the transgenic and control tobacco plants at flowering stage 12 and imaged them via high-resolution Fourier transform infrared microspectroscopy (FTIR) (McCann et al., 1992). We calculated the ratio of the detected ester signal (integrated area from 1725 to 1775 cm−1; linear baseline from 900 to 1775 cm−1) to that of the polysaccharides (900 to 1180 cm−1; linear baseline from 900 to 1180 cm−1) to generate a false-color image on transverse sections of these organs. As depicted in Figures 6A and 6B, in a transverse section of the style of control plants, the epidermal cells of the transmitting tract exhibited a high ratio of esters to polysaccharides. By contrast, the ratio in the transgenic style was much decreased (Figures 6C and 6D). We observed similar patterns in the cell walls of filament cells of the control and transgenic plants (Figures 6E to 6G), denoting a diminution of total esters in the cell walls of those transgenic floral tissues.
Figure 6.
Cell Wall Esters of the Style and Filament.
(A) to (H) Optical and FTIR images of the basal tissues of the style and middle portion of the filament. The false-color images represent the ratio of ester to polysaccharide in the style (A) and filament (E) of control plants and the Pt PAE1 overexpression ([C] and [G]) plants. The corresponding optical images of the tissues are shown in (B), (D), (F), and (H).
(I) The representative FTIR spectra of the style, filament, and their corresponding control tissues. Each averaged spectrum was normalized with the area under the polysaccharide peaks so that the sizes of the ester peaks (at 1740 cm−1, arrow) were easily compared. Tg, transgenic.
(J) Content of pectin acetyl ester in the style and filament. Asterisks indicate a statistically significant change (P < 0.05) under Student's t test. Data represent mean of triplicate experiments. Error bar represents se.
To further ascertain the changes in esters in those transgenic tissues, we extracted pectins from styles and filaments and measured the contents of both acetyl- and methylester. We detected a significant reduction in acetylesters, calculated on the basis of equal amounts of pectin (measured for uronic acid), in the pectinaceous fractions of both the filaments and styles in transgenic plants compared with the controls (Figure 6J). However, there was no statistically significant difference in their methylester content, although the amount varied considerably in different lines (see Supplemental Figure 9 online). These data show that expressing PAE1 reduced pectin acetylation in fast-growing floral tissues.
To further analyze the effects of PAE1 expression on the composition of the cell walls of the style and filament, we depolymerized their cell wall polysaccharides and measured the content of neutral and acidic sugars, respectively, by gas chromatography–mass spectrometry and a colorimetric method. Uronic acid decreased significantly in the cell walls of both tissues in transgenic plants, indicating a change in the pectin matrix polymer or its extractability; meanwhile, we observed substantial increases in the hemicellulosic and cellulosic sugars, such as Gal, Xyl, and Glc in the filaments, and of Man and Gal in the style cell walls in three of four examined transgenic lines (Table 2). These data imply that PAE1-mediated deacetylation differentially affects the biogenesis and remodeling of the entire spectrum of cell wall polysaccharides in those fast-growing tissues.
Table 2.
Cell Wall Composition of Filament and Style
| Filament | Pt PAE1_2 | Pt PAE1_4 | Pt PAE1_5 | Pt PAE1_6 | Control |
| Rha | 1.79 ± 0.33 | 2.37 ± 0.16 | 2.88 ± 0.03 | 1.90 ± 0.82 | 1.93 ± 0.03 |
| Ara | 2.24 ± 0.39 | 2.93 ± 0.24 | 3.78 ± 0.05 | 3.85 ± 0.76 | 2.27 ± 0.08 |
| Xyl | 8.87 ± 1.97 | 14.05 ± 1.17** | 14.85 ± 0.67** | 19.33 ± 3.39** | 9.79 ± 0.19 |
| Man | 1.17 ± 0.06 | 1.36 ± 0.03 | 1.85 ± 0.05 | 1.55 ± 0.10 | 1.32 ± 0.13 |
| Glc | 94.93 ± 8.27** | 96.81 ± 3.65** | 105.34 ± 0.14** | 101.19 ± 6.30** | 78.78 ± 3.47 |
| Gal | 12.82 ± 1.19* | 13.45 ± 3.65** | 16.71 ± 0.14** | 17.42 ± 1.63** | 10.15 ± 3.47 |
| Uronic acid | 68.61 ± 20.06* | 119.60 ± 12.00* | 110.54 ± 8.91* | 122.91 ± 27.14 | 127.49 ± 14.20 |
| Style | Pt PAE1_2 | Pt PAE1_4 | Pt PAE1_5 | Pt PAE1_6 | Control |
| Rha | 1.05 ± 0.06 | 1.45 ± 0.09 | 1.97 ± 0.11 | 1. 56 ± 0.11 | 1.39 ± 0.12 |
| Fuc | 4.32 ± 0.35 | 5.84 ± 0.21 | 8.30 ± 1.18 | 6.94 ± 0.92 | 6.33 ± 0.06 |
| Xyl | 9.21 ± 1.17* | 10.15 ± 0.43** | 15.44 ± 2.18 | 14.06 ± 2.04 | 12.17 ± 0.10 |
| Man | 1.46 ± 0.06** | 1.19 ± 0.08* | 1.56 ± 0.08** | 1.22 ± 0.06* | 1.04 ± 0.03 |
| Glc | 90.61 ± 3.95** | 109.54 ± 3.01** | 130.04 ± 10.09 | 134.73 ± 15.23 | 129.34 ± 3.37 |
| Gal | 15.06 ± 0.79** | 21.27 ± 0.90** | 27.2 ± 2.71** | 24.84 ± 2.96** | 17.83 ± 0.45 |
| Uronic acid | 54.60 ± 7.63* | 59.89 ± 1.80* | 60.52 ± 2.66* | 52.36 ± 3.22* | 75.72 ± 2.27 |
The data represent mean (mg/g dry cell wall) of triplicate samples ± se. *t test, P value < 0.05; **t test, P value < 0.01. The t tests were the comparisons between individual transformants and the control line.
Overexpression of Pt PAE1 Affects the Development of Pollen Grains
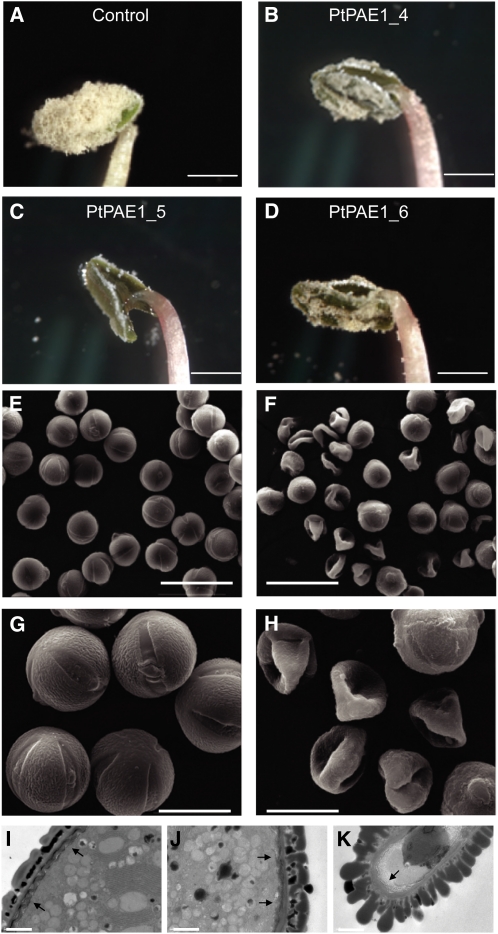
The expression of PAE1 also impaired the formation and development of pollen grains within the anthers. At flowering stage 12, the freshly dehiscent anthers of the control plants were fully filled with dehydrated mature pollen grains (Figure 7A); by contrast, there were fewer grains within the pollen sacs of transgenic plants (Figures 7B to 7D).
Figure 7.
Overexpression of Pt PAE1 Affects Pollen Grain Development in Tobacco.
(A) to (D) Mature pollen grains in the pollen sacs of the control (A) and transgenic ([B] to [D]) plants at flowering stage 12.
(E) to (H) Scanning electron microscopic images of the pollen grains of the control ([E] and [G]) and transgenic ([F] and [H]) plants.
(I) to (K) Transmission electron microscope images of the control (I), transgenic normal (J), and transgenic collapsed (K) pollen grains. Arrows point to the changes of the intine layer and the putative cellulose microfibrils.
Bars = 2.5 mm in (A) to (D) 50 μm in (E) and (F), 20 μm in (G) and (H), and 1 μm in (I) to (K).
[See online article for color version of this figure.]
We then chemically fixed grains from both control and transgenic plants and observed them under a scanning electron microscope. The pollen grains from the former had a uniform spherical shape and a regular exine pattern (Figures 7E and 7G), and the germination apertures were readily apparent (Figure 7G). By contrast, many pollen grains from transgenic plants had collapsed; the exine of cell walls displayed an irregular pattern, and the germination pores on the surface of the pollen grains had essentially disappeared (Figures 7F and 7H). We then observed these pollen grains by transmission electron microscopy. While the exine structure essentially remained in the normal shaped pollen grains of transgenic plants, the rod-like bacula (columella), tectum, and triphine of the exine had disappeared from the surface of the collapsed grains. Moreover, the intine and underlying electron-translucent layer, likely representing the cellulose microfibrils, of transgenic pollen grains appeared to be disorganized and looser than those of the controls (Figures 7I to 7K). These images suggested that expressing PAE1 altered structures of the pollen wall.
To examine the potential compositional changes in the cell walls of pollen grains, we analyzed transgenic and control mature pollen grains at flowering stage 12 via FTIR microspectroscopy after ethanol extraction. As Figure 8A depicts, the characteristic ester bond vibration at 1740 cm−1 of FTIR spectra (McCann et al., 2007) is less in the pollen from the transgenic lines. The ratio of the ester-bond signal to the total signals of polysaccharides ranged from 0.05 to 0.06 in transgenic plants, values significantly lower (t test, P < 0.001) than those in the wild type (0.14).
Figure 8.
Chemical Composition of Mature Pollen Grains.
(A) Average FTIR spectra of pollen grains from the transgenic (dotted lines) and control (solid line) plants. Each spectrum represents the average of 20 samples. The arrow indicates the peak of the acylester group.
(B) Acetic acid content of pollen grains isolated from transgenic and control plants. Asterisks indicate statistically significant changes under Student's t test (P < 0.01). Data represent the mean of triplicate samples. Error bar represents se.
We then released and quantified the wall-bound esters from the pollen grains. Acetylesters were readily detected in the cell walls of pollen grains of both the control and transgenic lines; we measured a significant reduction (up to 35%) of acetylester content in transgenic pollen grains compared with the control pollen (Figure 8B). However, the level of methylesters of both the control and transgenic pollen grains was below the detection limit, indicating that the cell walls of the mature pollen grains were less methylesterified than the leaves and floral tissues. In addition, we monitored the wall-bound ferulate ester in both groups of pollen grains and found no quantitative difference in ferulate ester contents between these groups. These data further confirm that the reduction in ester signal observed in the FTIR microspectroscopy analysis was mainly the result of an alteration in acetylester content of the pollen cell wall.
Overexpression of Pt PAE Hinders Pollen Germination and Pollen Tube Elongation
Subsequently, we examined the ability of pollen grains isolated from PAE1-overexpressing lines to germinate. We collected pollen from freshly dehisced flowers and incubated it for 4 h in growth medium. No pollen tubes emerged from the pollen grains of the transgenic lines (Figure 9A), but the majority of control grains germinated, and their emerging pollen tubes elongated (Figure 9B).
Figure 9.
Pollen Grain Germination and Pollen Tube Elongation.
(A) and (B) Germination of mature pollen from Pt PAE1 overexpression (A) and control (B) plants in solid germination medium.
(C) and (D) Pollen germination and tube growth of Pt PAE1 overexpression (C) and the control (D) plants in the transmitting tract of the wild-type style.
(E) to (H) In vitro pollen germination and pollen tube growth when wild-type pollen was treated with exogenous Pt PAE1 recombinant protein ([E] and [G]) or with BSA as the control ([F] and [H]).
Bars= 20 μm in (A) to (D) and 50 μm in (E) to (H).
To verify these in vitro observations, we used the grains from both the transgenic and the control lines to pollinate the wild-type stigma. Twenty-four hours later, few pollen tubes had emerged and grown from the transgenic pollen grains (Figure 9C, indicated with the blue fluorescence resulting from aniline blue staining) in contrast with the numerous tubes that emerged from the control lines (Figure 9D).
To confirm the effects of PAE1-mediated deacetylation on the germination and growth of pollen tubes, we incubated the pollen grains from the wild-type tobacco plants with purified recombinant PAE1 in growth medium. After 4 h, many pollen grains had germinated, and their pollen tubes grew in the medium supplied only with BSA as the control (Figures 9F and 9H). However, pollen grains incubated in medium with exogenous PAE swelled and most failed to produce tubes (Figures 9E and 9G). Moreover, a few emerged tubes were shorter and bulkier than those of the control (Figures 9G and 9H).
Overexpression of Pt PAE1 Depresses the Development of the Capsule
The pollen tube delivers sperm cells to the female gametophyte in the ovule. Failure of the pollen grain to germinate and of the tube to elongate in plants overexpressing Pt PAE1 severely inhibited fertilization and seed development.
Two weeks after anthesis, only a few seeds were set in the developing seedpods of transgenic plants (Figure 10A). Eventually, the transgenic lines yielded much smaller mature capsules (Figure 10C), with an average weight of >70% less than that of the wild type (Figure 10E), and the former contained few or no mature seeds.
Figure 10.
Development of the Seeds and Capsules of Pt PAE1 Transgenic Plants.
(A) to (D) Developing seeds in the capsules of the transgenic (A) and control (B) tobacco plants at 14 d postanthesis (the walls of capsules were peeled off for clearance) and the mature capsules of the transgenic (C) and the control (D) plants. Bar = 0.5 cm.
(E) Average weight of capsules and produced seeds (per capsule) of transgenic (Pt PAE1_2, 4, 5, and 6) and four independent control ([A] to [D]) plants. Data represent the mean of 10 capsules from each line. Error bar represents the sd. Asterisks indicate significant difference under pairwise comparison to the control line (A); P < 0.01, Student's t test.
[See online article for color version of this figure.]
The Overexpression of Pt PAE1 Lowers Pectin Digestibility
Reportedly, the acetylation of polysaccharides decreases their digestibility (Schols and Voragen, 1994; Chen and Mart, 1996; Benen et al., 1999; Bonnin et al., 2003). To clarify the effect of overexpression of Pt PAE1 on pectin digestibility, we isolated the acid-soluble pectins from the young leaves of the transgenic and the control plants and treated equal amounts of these pectins (based on quantification of uronic acids) with pectinase from Aspergillus that randomly hydrolyzes α-(1-4)-D-galactosiduronic linkages in pectin and other galacturonans (Pereira et al., 1992). We labeled the digested products with 2-anthranilic acid, a fluorophore that reacts specifically with the free reducing sugar of glycan to form a stable Schiff’s base, therefore enabling us to detect the labeled products via UV spectra (Sato et al., 1997, 2005). We then resolved the labeled hydrolysates by HPLC on a silica-based size exclusion column and monitored them with a UV diode array detector. Two predominant peaks representing the mono- and oligo-sugars were detected in all of the treated samples. In addition, a third peak characterized as the labeled citric acids from the digestive buffer was also resolved (Figure 11A). Quantification of the resulting hydrolysates revealed that the amount of released monomeric sugars in the pectin of the transgenic plants was ~60% less than that from the control plants (Figure 11B), indicating the lower digestibility of pectinaceous polysaccharides of the transgenic lines. To further clarify whether the reduced digestibility was caused directly by the deacetylation or was due to the indirect effect of the potential alteration of pectin structure or composition, following the expression of Pt PAE1, we incubated sugar beet pectin with or without recombinant Pt PAE1 and then treated the samples with pectinase. After resolving the samples by HPLC, monomeric sugars were readily observed in both samples (Figure 11C), but the level of this digestive product in the sample pretreated with Pt PAE1 was significantly lower than that of untreated sample (Figures 11C and 11D). The acetic acid released from sugar beet pectins by PAE1 was also labeled; its peak overlapped with that of citric acid from the buffer (Figures 11C and 11D).
Figure 11.
Effects of Pt PAE1 Overexpression on Pectin Digestibility.
(A) The digestion of leaf acid-soluble pectin from control (solid line) or transgenic (dotted lines) plants by pectinase.
(B) Quantification of monomeric and oligomeric sugars released in the reactions of (A).
(C) Pectinase digestion of sugar beet pectin with (dotted line) or without (solid line) recombinant PAE1 pretreatment.
(D) Quantification of the released monomeric and oligomeric sugars and acetic acid/citric acid in the reactions of (C). Data represent the mean of triplicate samples. Error bar represents sd. A/C, acetic acid and citric acid overlapping peak; C, citric acid; M, galacturonic acid; O, oligomeric sugar. *P < 0.05, Student’s t test; **P < 0.01, Student's t test.
These data demonstrate that PAE1-mediated deacetylation reduces the digestibility of pectin and hinders the release of small molecule sugars, a finding that sharply contrasts with previous studies on the acetylated hemicelluloses.
DISCUSSION
Poplar PAE1 Functions as a PAE
Continuous modification and remodeling of the composition and structure of cell walls is a prominent feature of the growth and development of plants (Ridley et al., 2001). Modulating the status of acetylesters is a ubiquitous process in the cell wall polysaccharides of higher plants, which necessitates the activity of polysaccharide acetylesterases (Williamson, 1991; Bordenave et al., 1995; Christensen et al., 1996). The distribution and preferential expression of a set of putative Populus carbohydrate acetylesterases in different tissues and at certain developmental stages in poplar stems (see Supplemental Figure 1 online) highlights their potential diverse and distinct functions in remodeling the complex configurations of the cell wall polysaccharides. Like the previously isolated PAE gene from mung bean (Breton et al., 1996), Pt PAE1 belongs to the Ser hydrolase superfamily, possessing a conserved lipase GXSXG motif, a characteristic of hydrolytic enzymes. Purified recombinant Pt PAE1 effectively releases acetate from different natural and synthetically acetylated carbohydrates, among which the protein showed a catalytic preference for acetylated pectin (Figure 2, Table 1). However, the enzyme did not remove methyl- or feruloylesters from polysaccharides that contain considerable amounts of such esterifications, suggesting it is specific in modulating polysaccharide acetylesters. Overexpressing this gene in tobacco significantly reduced the content of acetylesters of pectin in young leaves, the fast-growing floral tissues, and the pollen grains, in which pectin is the predominant cell wall component. Although the recombinant protein also showed low activity toward the acetylated nascent xylan in vitro, constitutive expression of Pt PAE1 in tobacco did not reduce the acetylation of xylan. In fact, in some examined transgenic plants, the acetylester content of xylan even increased slightly (Figure 4). This finding seems to implicate a complex yet largely unknown spatial and metabolic organization of polysaccharide modification. It also highlights the caution needed in interpreting the biological significance of in vitro enzyme assays. Overall, our in vitro and in vivo data suggest that Pt PAE1 primarily functions as a PAE.
PAE Modulates Extensibility of Cell Walls in Reproductive Tissues
Cell growth is intimately linked with the expansion of the cell walls (Parre and Geitmann, 2005). Previous studies demonstrated the dramatic consequences of the methylesterification status of pectin on cell wall’s texture and mechanical properties and, thereby, on cellular growth and cell shape (Bosch et al., 2005; Bosch and Hepler, 2005; Wolf et al., 2009). In this study, we found that Pt PAE1–mediated deacetylation greatly impeded pollen grain germination and tube elongation (Figure 9). In the pollen apertures and the apex of pollen tubes, the cell walls lack the callosic inner lining, and pectin is their major component (Geitmann and Steer, 2006). Although constitutive overexpression of PAE1 in planta caused complicated changes in cell wall composition and structure, the impairments on the germination of pollen grains and the extension of the pollen tube likely are dominated by the reduction in acetylester in the cell wall pectin. When exposing wild-type pollen grains to exogenous Pt PAE1 enzyme, the rehydrated pollen grains exhibited similar behavior as those transgenic pollen grains expressing Pt PAE1 (Figure 9). Although the inherent mechanisms underlying the inhibition of the pollen tube’s germination and elongation in transgenic plants may be complex, from a structural perspective, Pt PAE1–mediated pectin deacetylation might change the physiochemical properties of the cell wall’s polysaccharide networks, thereby lowering its extensibility. Kohn and Furda (1968) proposed that acetylation of the hydroxyl at C-2 and/or C-3 of galacturonic acid residues may impose steric hindrance, thus lowering the ability of pectic polysaccharides to associate intermolecularly through calcium ions. Conversely, deacetylation of pectin in transgenic plants might promote such associations, thus altering the mechanical properties and the configuration of the pectin network and consequently disturbing the programmed remodeling and structure of the primary cell walls, so affecting the cell walls’ asymmetric extension/elongation.
Expressing Pt PAE1 also disturbs the elongation of the style and filament tissues of flowers (Figure 5). The shorter epidermal cells of the staminal filaments and of the basal style in transgenic plants point to the limited expansion of their cell walls. The cells of these organs usually contain high levels of pectinaceous polysaccharides, although levels vary by species. By contrast, the rigid walls of the apical stylar cells have a lower pectin content and more cellulose and hemicelluloses (Marga et al., 2003). Expressing Pt PAE1 reduced the amount of acetylesters of the stylar and filament pectins (Figure 6). Along with deacetylation of the stylar and filament pectins, the extractable acidic polysaccharides were reduced; meanwhile, cellulosic and hemicellulosic components were increased in the cell walls of those reproductive tissues of transgenic plants (Table 2). These findings explain the retardation of cell elongation. Together, these results indicate that expansion of the plant cell wall requires a precise acetylation status of the polysaccharides; genetically perturbing such structural modification would affect the biogenesis, deposition, and remodeling of the entire polysaccharides in the cell wall.
Deacetylation of Pectin Reduces Its Digestibility
The digestibility of pectin is another factor that likely influences the cell wall's dynamic remodeling and, thereby, cellular growth and shape (Wolf et al., 2009). One of the proposed functions of pectin acetylation is to provide a steric barrier that prevents the pectinolytic enzymes from depolymerizing cell wall polysaccharides (Solms and Deuel, 1951; Rexova-Benkova et al., 1997; André-Leroux et al., 2009). Therefore, we anticipated that the deacetylation of pectin by Pt PAE1 would enhance its degradation. Surprisingly, we found that deacetylation considerably slowed the breakdown of pectin by microbial pectinase (Figure 11). The reduction in the digestibility of pectin by deacetylation was circumstantially confirmed in vitro using sugar beet pectin pretreated with recombinant Pt PAE1 (Figure 11). This experiment excluded the potential effects of the other composition and structure changes of pectin, caused by PAE1 overexpression in transgenic plants, on its digestibility. This finding implies that when hydrolyzing pectin polysaccharide, certain structural configurations of the polymer, including the precise pattern of acetylation, are a prerequisite for the action of pectolytic enzymes. The modulation of the digestibility of polysaccharides by PAE1 might be an additional or alternative factor that regulates cell wall extension in planta. Consistent with this assumption, global transcriptomic analyses of pollen germination and tubular growth in Arabidopsis revealed that the expression of putative PAEs is reciprocally coordinated with that of the cell wall–degrading enzymes (Wang et al., 2008; Qin et al., 2009). Compared with their expression levels in pollen grains, six putative PAE genes exhibited lower expression in the emerging tubes; this downregulation was accompanied by an increase in the gene expression of glycosyl hydrolase and pectin lyase (Wang et al., 2008).
Together, these data suggest that, like pectin methylesterases, PAE precisely regulates the acetylation status of polymers, which modulates the biophysical and physiological properties of the cell wall polysaccharides and thereby affects the plant’s growth, development, and reproduction.
METHODS
Plant Materials and Growth Conditions
Poplar ecotype Populus trichocarpa and tobacco (Nicotiana tabacum cv Xanthi) were used in this study. Poplar plants were grown in a greenhouse under conditions described previously (Yu et al., 2009). Tobacco plants were grown in a growth chamber at 22°C with a 16/8-h light/dark regime.
All chemicals were from Sigma-Aldrich unless otherwise stated.
Phylogenetic and Sequence Analysis
Phylogenetic analyses of putative polysaccharide acetylesterases were conducted on the full-length sequences of Populus and Arabidopsis thaliana putative homologs, together with those of the functionally characterized bacterial, fungal, and plant PAEs. Full-length amino acid sequences were first aligned by ClustalW version 1.83 (Thompson et al., 1994) with default parameters (http://www.ebi.ac.uk/Tools/clustalw/) and imported into the Molecular Evolutionary Genetics Analysis (MEGA) package, version 3.0 (Kumar et al., 2004). Phylogenetic analyses were conducted using the neighbor-joining method (Saitou and Nei, 1987) implemented in MEGA, with the option for pairwise deletion for handling alignment gaps and with the Poisson correction model for computing distance. For statistic reliability, bootstrap tests were undertaken with 1000 replicates. The final tree graphics were generated via the TreeView program (Page, 1996).
RNA Extraction and Quantitative RT-PCR
We collected tissues from poplar leaves, stems, roots, and apical buds as previously described (Yu et al., 2009). Total RNA was extracted by the hot borate method following Gou et al. (2008). Reverse transcription was performed with the SuperScript III first-strand synthesis system (Invitrogen) according to the manufacturer’s user manual. We performed real-time RT-PCR with iQ SYBR Green Supermix (Bio-Rad). The poplar ATPase1 gene served as the calibration internal standard (Yu et al., 2009). The normalized expression level in the leaf and first node was set at 100; the expression values of individual genes in different tissues were compared with those of the leaf and first node (set at 100) to give relative expression levels. The data are the mean of triplicate samples.
Protein and Plant Expression Vector Constructs, Tobacco Transformation, and Subcellular Localization Analysis
Full-length Pt CE13_5 cDNA was amplified from the stem’s reverse transcription product and subcloned into a Gateway cloning vector pDONR207 (Invitrogen). The gene was then transferred into the Gateway destination vector pHis9, derived from pET-28a, for protein expression, and both the Gateway binary vectors pMDC32 and pMDC43 (Curtis and Grossniklaus, 2003) for expressing this gene alone and for creating a CE13_5-GFP fusion in plant. The resulted pMDC32 construct constituted an expression cassette of Pt CE13_5, driven by a double 35S promoter. It was transferred first into the Agrobacterium tumefaciens GV3101strain that was then used for tobacco transformation through leaf disc infection (Horsch et al., 1985). Four independent transgenic lines, PtPAE1_2, PtPAE1_4, PtPAE1_5, and PtPAE1_6, were selected by means of hygromycin resistance (50 μg/mL) and planted in soil after rooting. The plants were maintained in a growth chamber under the conditions described. The resulted pMDC43 construct constituted a chimeric gene of Pt CE13_5 linked to the 3′ end of GFP under the 2× 35S promoter. The construct was transformed into tobacco via the same leaf disc infection procedure (Horsch et al., 1985) for subcellular localization analysis. The fluorescence signals of the expressed GFP-Pt CE13_5 fusion protein in dark-grown tobacco seedlings were observed under a Zeiss LSM 510 META NLO two-photon confocal laser scanning microscope (Carl Zeiss MicroImaging). The same materials were then stained with 10 μg/mL propidium iodide in water for 5 min and then observed by confocal microscopy. pCAMBIA1302 that harbors the free GFP gene, the original construct used to generate Gateway vector pMDC43 (Curtis and Grossniklaus, 2003), was expressed in tobacco and used as the control.
Enzymatic Assays
Recombinant protein was produced in Escherichia coli strain Rosetta (DE3) pLysS by induction with 0.5 mM isopropyl β-d-1-thiogalactopyranoside, overnight, at 18°C. The protein was purified with nickel-nitriloacetic acid resin from crude cell lysate. Sugar beet (Beta vulgaris) pectin was kindly provided by CP Kelco, birch wood (Betula spp) xylan was ordered from Sigma-Aldrich, and potato (Solanum tuberosum) pectin was prepared from the cell walls of potatoes as described (Melton and Smith, 2001b). The sugar beet pectin was further purified by precipitation with 70% ethanol. For substrate specificity assays, 2 μg recombinant proteins was incubated with 10 mg/mL pectin or 50 mg/mL xylan, in 100 mM Tris-HCl buffer, pH 7.0, at 35°C for 30 min. Acetate released from the reaction was immediately quantified using the Acetate Kinase Format Kit (K-ACETAK) from Megazyme (Megazyme International Ireland) according to the user’s manual.
To determine pectin methylesterase activity, the assay was performed using 1 mg/mL esterified pectin (>80% methyl esterification) from citrus fruit as the substrate and incubating with 2 μg recombinant protein, or the commercial pectin methylesterase (Sigma-Aldrich) as the control, in 100 mM Tris-HCl buffer, pH 7.0, at 35°C for 15 min. Methanol released was subsequently converted by alcohol oxidase (Sigma-Aldrich) and formaldehyde dehydrogenase (Sigma-Aldrich) coupling with the conversion of cofactor NAD to NADH. The produced NADH was measured at OD340 with methanol as standard (Grsic-Rausch and Rausch, 2004).
For assaying feruloylesterase activity, sugar beet pectin was incubated with 2 μg recombinant protein in 50 mM Tris-HCl buffer, pH 7, at 35°C, or treated with 2 n NaOH (as the control) for 15 min; then, the reaction mixtures were acidified to pH 4.0. The ferulic acid was extracted three times with 0.5 mL water-saturated ethyl acetate. The extracts were combined, dried under an N2 stream, dissolved in methanol, and quantified by HPLC according to the description by Gou et al. (2008).
Polysaccharide Acetylester Content Analyses
To quantify the content of acetylester bound on polysaccharides, we prepared cell wall materials from the young leaves, styles, filaments, and stems of transgenic and the wild-type control plants on the day of flowering. Water-soluble pectin was extracted with hot water (80°C), adjusted to pH 4, and then pelleted overnight in 70% ethanol at −20°C. To prepare acid-soluble pectin, after hot water extraction, the residuals were extracted with 0.04 M HCl (80°C), and the acid extracts were pelleted in 70% ethanol as mentioned before.
Two methods were adopted to prepare xylan/hemicellulose. The leaf’s cell wall was first treated with 1% ammonium oxalate at 37°C overnight to remove pectin. The residuals were then treated with 1% amylase overnight. Hemicellulose was extracted with DMSO at 70° overnight, and pelleted with a threefold volume of ethanol according to the description provided by Lee et al. (2011). Alternatively, xylan was isolated from xylem cell wall preparations by treating the samples with xylanase (Sigma-Aldrich) according to the methods described (Günl et al., 2010). The acetates bound on different polymers were released by incubation with 2 n NaOH at 37°C overnight. The hydrolysates were neutralized with HCl and analyzed with the Acetate Kinase Format Kit (Megazyme International Ireland).
Cell Wall Sugar Composition Analysis
Cell walls from the styles and filaments of transgenic and control plants on the day of flowering were prepared according to Günl et al. (2010). Pectin content was measured colorimetrically using galacturonic acid as the standard (Melton and Smith, 2001a). Neutral sugars were released from the prepared cell walls through sulfuric acid depolymerization and then converted to their alditol acetates (Melton and Smith, 2001b). These alditol acetates were separated on an Agilent gas chromatography–mass spectrometer (Agilent Technologies) equipped with a 30 mm × 0.25-mm (internal diameter) Agilent JandW HP-5MS capillary column; the initial oven temperature was maintained at 38°C for 30 s, increased to 170°C at 50°C/min, and then increased to 230°C at 2°C/min and held at 230°C for 5 min. The individual sugars were identified by comparison with authentic standard compounds; their quantification was based on the standard curves of each derivatized individual sugar made from the same gas chromatography–mass spectrometry run.
FTIR Imaging Analysis on Floral Tissues and Pollen Grains
The styles and filaments of the transgenic and wild-type flowers were collected on the day of flowering, frozen in liquid nitrogen, and kept at −80°C until use. The samples were embedded quickly in FSC22 Frozen Section Compound (Surgipath Medical Industries) and sectioned at 10-μm slices with a Leica CM1950 Cryo-microtome (Leica Microsystems). The sections were mounted on a barium-fluoride slide, washed with 70% ethanol thoroughly to remove the remaining embedding compound, and left to dry. We imaged the sections in the transmission mode with a Perkin-Elmer Spectrum Spotlight infrared microscope coupled to a Spectrum One FTIR Spectrometer (Perkin-Elmer). The images were collected with a 6.25-μm pixel resolution, a spectral resolution of 8 cm−1, and an integration time of 16 scans per pixel.
The wild-type and transgenic pollen grains, collected on the day of flowering, were kept in ethanol until use. For FTIR microspectroscopy analysis, we first stained the pollen grains with 1% ruthenium red for 5 min to monitor the methylester content and to facilitate their focus under the FTIR microscope. The grains were then flattened with a diamond compression cell (Spectra-Tech), washed with 70% ethanol, and scanned in transmission mode with a 12-μm square aperture, a spectral resolution of 4 cm−1, and integration time of 128 scans with a Perkin-Elmer Spectrum Spotlight IR microscope in “point” mode. Ten pollen grains were analyzed for each condition. The data sets from each sample were averaged in OMMIC software. To determine the relative acetylester levels, we determined the peak area of the carbonyl ester (1725 to 1775 cm−1) and the total polysaccharide (900 to 1180 cm−1) and then calculated the ratio of wall-bound esters to total polysaccharides.
The Pistil-to-Stamen Cell Length Ratio and Microscopy Analysis of the Pollen Grain
Petals were removed from flowers at stage 12. The length of the style and filament was measured from the receptacles of more than 20 flowers. After peeling off the epidermis from the basal, middle, and apical parts of the style and filament, we stained the material with 10 μg/mL propidium iodide for 10 min and observed it under an optical microscope (Leica). Cell length was measured for an average of 40 cells with an E-Ruler (http://www.mycnknow.com/). Dehiscent pollen grains at the same stage were observed under an optical microscope. More grains at the same developmental stage were fixed in 2.5% glutaraldehyde diluted with 0.1 M sodium phosphate buffer and then dehydrated in a graded ethanol series and dried with a Polaron critical-point drying apparatus (Polaron Instruments). They were mounted on aluminum stubs, sputter coated with gold, and imaged with a Quanta 200 environmental scanning electron microscope (FEI Company).
Pollen Tube Germination
Pollen grains collected on the day of flowering were germinated in growth medium containing 2% agarose according to Palanivelu and Preuss (2006). They were kept in a closed environment at room temperature for 4 h and then observed under a Leica DM 5500B microscope.
For in vivo germination, the anthers were removed at development stage 10 from wild-type flowers. Stage 12 pollen grains from these anthers and from overexpression plants were spread on wild-type stigmas on the day of flowering (i.e., stage 12). Twenty-four hours later, the stylar tissues were fixed in acetic acid, softened in 2 n NaOH for 24 h, stained with aniline blue for 24 h at 4°C, and then washed three times with water. The UV fluorescence of these tissues was then examined under a Zeiss LSM 510 META NLO two-photon confocal laser scanning microscope (Carl Zeiss MicroImaging).
Pectin Digestion and Anthranilamide Labeling
Eighty microliters of 0.5% (w/v) sugar beet pectin was incubated with 10 μg recombinant Pt PAE1 in 50 mM Tris-HCl buffer, pH 7.0, at 37°C overnight and then digested with 5 μL (25 units) pectinase (Sigma-Aldrich) in 200 mM citric acid buffer, pH 4.0, for 10 min at 37°C; alternatively, 50 μL of 0.5% acid-soluble pectin from the PAE1 transgenic and control plants was directly treated with 5 μL pectinase (Sigma-Aldrich). The products were pelleted by adding a sixfold volume of acetonitrile and sedimented at −20°C for 1 h. The recovered pellets were dissolved in 0.2 M 2-anthranilic acid (Sigma-Aldrich) in 1 M sodium cyanoborohydride (Sigma-Aldrich) to label the carbonyl carbon of the acyclic-reducing sugars. The labeled glycan was held at 65°C for 2 h. The products were then cooled to room temperature and pelleted again with sixfold acetonitrile, air-dried, and redissolved in 100 μL water. Twenty-microliter samples were injected into a BioSep-SEC-2000S column (Phenomenex) and separated in 0.2% acetic acid or formic acid buffer at a flow rate of 0.25 mL per minute with an Agilent 1100 HPLC system equipped with a UV diode-array detector (Agilent). UV absorbance was monitored at 254 nm and quantified using a standard curve of 2-anthranilic acid.
Accession Number
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession number HQ223420 for the identified PAE1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Expression of Populus Putative Polysaccharide Acetylesterase Genes.
Supplemental Figure 2. In Silico Expression of the Pt PAE1 Homologs in Populus balsamifera.
Supplemental Figure 3. Acetylesterase Activity of Recombinant Pt PAE1 with Different Chemically Acetylated Substrates.
Supplemental Figure 4. Acetylester Content of Natural Substrates Released by 2 n NaOH for 15 min at 35°C.
Supplemental Figure 5. Growth Phenotype of Independent Pt PAE1 Overexpression Lines.
Supplemental Figure 6. The Amount of Extracted Pectin from Leaf Cell Walls of Transgenic and Control Plants.
Supplemental Figure 7. Content of Methylester of Water-Soluble Pectin from Leaves of Transgenic and Control Plants.
Supplemental Figure 8. The Length of Style and Filaments.
Supplemental Figure 9. Content of Methylester in Water-Soluble Pectins Extracted from Style and Filament of Transgenic and Control Plants.
Supplemental Data Set 1. Multisequence Alignment of Polysaccharide Acetyltransferases.
Acknowledgments
We thank Simon Park, William Willis, and Randy Smith at the National Synchrotron Light Source for their help with FTIR microspectroscopy. Sugar beet pectin was kindly provided by CP Kelco U.S. This work was supported by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy through Grant DEAC0298CH10886 and by the National Science Foundation through Grant MCB-1051675 (to C.-J.L.), the Chinese Academy of Sciences/State Administration of Foreign Experts Affairs International Partnership Program for Creative Research Teams in Plant Metabolisms (to X.-Y.C), and the scholarship for distinguished overseas researcher from the National Science Foundation of China (31028003; to C.-J.L.). Use of the National Synchrotron light and confocal microscope at the Center of Functional Nanomaterials was supported by the Office of Basic Energy Sciences, U.S. Department of Energy, under Contract DEAC02-98CH10886.
AUTHOR CONTRIBUTIONS
C.-J.L. and J.-Y.G. designed experiments. J.-Y.G, L.M.M., G.H., and X.-H.Y. performed experiments. C.-J.L., J.-Y.G., and X.-Y.C. analyzed data. C.-J.L. wrote the article.
References
- André-Leroux G., Tessier D., Bonnin E. (2009). Endopolygalacturonases reveal molecular features for processivity pattern and tolerance towards acetylated pectin. Biochim. Biophys. Acta 1794: 5–13 [DOI] [PubMed] [Google Scholar]
- Benen J.A.E., Kester H.C.M., Visser J. (1999). Kinetic characterization of Aspergillus niger N400 endopolygalacturonases I, II and C. Eur. J. Biochem. 259: 577–585 [DOI] [PubMed] [Google Scholar]
- Biely P., MacKenzie C.R., Puls J., Schneider H. (1986). Cooperativity of esterases and xylanases in the enzymatic degradation of acetyl xylan. Biotechnol. 4: 731–733 [Google Scholar]
- Bonnin E., Clavurier K., Daniel S., Kauppinen S., Mikkelsen J.D.M., Thibault J.F. (2008). Pectin acetylesterases from Aspergillus are able to deacetyalte homogalacturonan as well as rhamnogalacturonan. Carbohydr. Polym. 74: 411–418 [Google Scholar]
- Bonnin E., Le Goff A., van Alebeek G.J.W.M., Voragen A.G.J., Thibault J.F. (2003). Mode of action of Fusarium moniliforme endopolygalacturonase towards acetylated pectin. Carbohydr. Polym. 52: 381–388 [Google Scholar]
- Bordenave M., Goldberg R., Huet J.C., Pernollet J.C. (1995). A novel protein from mung bean hypocotyl cell walls with acetyl esterase activity. Phytochemistry 38: 315–319 [DOI] [PubMed] [Google Scholar]
- Bosch M., Cheung A.Y., Hepler P.K. (2005). Pectin methylesterase, a regulator of pollen tube growth. Plant Physiol. 138: 1334–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M., Hepler P.K. (2005). Pectin methylesterases and pectin dynamics in pollen tubes. Plant Cell 17: 3219–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton C., Bordenave M., Richard L., Pernollet J.C., Huet J.C., Pérez S., Goldberg R. (1996). PCR cloning and expression analysis of a cDNA encoding a pectinacetylesterase from Vigna radiata L. FEBS Lett. 388: 139–142 [DOI] [PubMed] [Google Scholar]
- Carpita N.C., McCann M.C. (2000). The cell wall. In Biochemistry and Molecular Biology of Plants, B.B. Buchanan, W. Gruissem, and R.L. Jones, eds (Rockville, MD: America Society of Plant Physiology), pp. 52–108 [Google Scholar]
- Chen E.M.W., Mart A.J. (1996). Nature of sites hydrolyzable by endopolygalacturonase in partially-esterified homogalacturonans. Carbohydr. Polym. 29: 129–136 [Google Scholar]
- Christensen T.M.I.E., Nielsen J.E., Mikkelsen J.D. (1996). Isolation, characterization and immunolocallization of orange fruit acetyl esterase. In Pectins and Pectinases, J. Visser and A.G.J. Voragen, eds (Amsterdam: Elsevier Sciences), pp. 723–730 [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews G.N., Beals T.P., Bui A.Q., Goldberg R.B. (1992). Regional and cell-specific gene expression patterns during petal development. Plant Cell 4: 1383–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler-Lee J., et al. (2006). Poplar carbohydrate-active enzymes. Gene identification and expression analyses. Plant Physiol. 140: 946–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geitmann A., Steer M. (2006). The architecture and properties of the pollen tube cell wall. In Plant Cell Monogr. (3): The Pollen Tube, R. Malhó, ed (Berlin: Springer-Verlag), pp. 177–200 [Google Scholar]
- Gille S., de Souza A., Xiong G., Benz M., Cheng K., Schultink A., Reca I.-B., Pauly M. (2011). O-acetylation of Arabidopsis hemicellulose xyloglucan requires AXY4 or AXY4L, proteins with a TBL and DUF231 domain. Plant Cell 23: 4041–4053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou J.Y., Park S., Yu X.H., Miller L.M., Liu C.J. (2008). Compositional characterization and imaging of “wall-bound” acylesters of Populus trichocarpa reveal differential accumulation of acyl molecules in normal and reactive woods. Planta 229: 15–24 [DOI] [PubMed] [Google Scholar]
- Grsic-Rausch S., Rausch T. (2004). A coupled spectrophotometric enzyme assay for the determination of pectin methylesterase activity and its inhibition by proteinaceous inhibitors. Anal. Biochem. 333: 14–18 [DOI] [PubMed] [Google Scholar]
- Günl M., Gille S., Pauly M. (2010). OLIgo mass profiling (OLIMP) of extracellular polysaccharides. J. Vis. Exp. 40: 2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch R.B., Fry J.E., Hoffmann N.L., Eichholtz D., Rogers S.G., Fraley R.T. (1985). A simple and general method for transferring genes into plants. Science 227: 1229–1231 [DOI] [PubMed] [Google Scholar]
- Ishii T. (1995). Isolation and characterization of acetylated rhamnogalacturonan oligomersliberated from bamboo shoot cell-walls by Driselase. Mokuzai Gakkaishi 41: 561–572 [Google Scholar]
- Ishii T. (1997). O-acetylated oligosaccharides from pectins of potato tuber cell walls. Plant Physiol. 113: 1265–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppinen S., Christgau S., Kofod L.V., Halkier T., Dörreich K., Dalbøge H. (1995). Molecular cloning and characterization of a rhamnogalacturonan acetylesterase from Aspergillus aculeatus. Synergism between rhamnogalacturonan degrading enzymes. J. Biol. Chem. 270: 27172–27178 [DOI] [PubMed] [Google Scholar]
- Kohn R., Furda I. (1968). Binding of calcium ions to acetyl derivatives of pectin. Collect. Czech. Chem. Commun. 42: 731–744 [Google Scholar]
- Komalavilas P., Mort A.J. (1989). The acetylaton at O-3 of galacturonic acid in the rhamnose-rich portion of pectines. Carbohydr. Res. 189: 261–272 [Google Scholar]
- Kouwijzer M., Schols H.A., Pe'rez S. (1996). Acetylation of rhamnogalacturonan I and homogalacturonan: Theoretical calculations. In Pectins and Pectinases, J. Visser and A.G.J. Voragen, eds (Amsterdam: Elsevier Science), pp. 57–65 [Google Scholar]
- Kumar S., Tamura K., Nei M. (2004). MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5: 150–163 [DOI] [PubMed] [Google Scholar]
- Lee C., Teng Q., Zhong R., Ye Z.H. (2011). The four Arabidopsis reduced wall acetylation genes are expressed in secondary wall-containing cells and required for the acetylation of xylan. Plant Cell Physiol. 52: 1289–1301 [DOI] [PubMed] [Google Scholar]
- Liners F., Gaspar T., Van Cutsem P. (1994). Acetyl- and methyl-esterification of pectins of friable and compact sugar-beet calli: consequences for intercellular adhesion. Planta 192: 545–556 [Google Scholar]
- Manabe Y., et al. (2011). Loss-of-function mutation of REDUCED WALL ACETYLATION2 in Arabidopsis leads to reduced cell wall acetylation and increased resistance to Botrytis cinerea. Plant Physiol. 155: 1068–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marga F., Gallo A., Hasenstein K.H. (2003). Cell wall components affects mechanical properties: Studies with thistle flowers. Plant Physiol. Biochem. 41: 792–797 [Google Scholar]
- McCann M.C., Defernez M., Urbanowicz B.R., Tewari J.C., Langewisch T., Olek A., Wells B., Wilson R.H., Carpita N.C. (2007). Neural network analyses of infrared spectra for classifying cell wall architectures. Plant Physiol. 143: 1314–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann M.C., Hammouri M., Wilson R., Belton P., Roberts K. (1992). Fourier transform infrared microspectroscopy is a new way to look at plant cell walls. Plant Physiol. 100: 1940–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna S.T., Kunkel J.G., Bosch M., Rounds C.M., Vidali L., Winship L.J., Hepler P.K. (2009). Exocytosis precedes and predicts the increase in growth in oscillating pollen tubes. Plant Cell 21: 3026–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton L.D., Smith B.G. (2001a). Determination of the uronic acid content of plant cell walls using a colorimetric assay. In Current Protocols in Food Analytical Chemistry, A. Boyle and E. Harkins, eds (Hoboken, NJ: John Wiley & Sons), pp. E3.3.1–E3.3.4 [Google Scholar]
- Melton L.D., Smith B.G. (2001b). Determination of neutral sugars by gas chromatography of their alditol acetates. In Current Protocols in Food Analytical Chemistry, A. Boyle and E. Harkins, eds (Hoboken, NJ: John Wiley & Sons), pp. E3.2.1–E3.2.13 [Google Scholar]
- Micheli F. (2001). Pectin methylesterases: Cell wall enzymes with important roles in plant physiology. Trends Plant Sci. 6: 414–419 [DOI] [PubMed] [Google Scholar]
- Mohnen D. (2008). Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 11: 266–277 [DOI] [PubMed] [Google Scholar]
- Page R.D.M. (1996). TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Palanivelu R., Preuss D. (2006). Distinct short-range ovule signals attract or repel Arabidopsis thaliana pollen tubes in vitro. BMC Plant Biol. 6: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parre E., Geitmann A. (2005). Pectin and the role of the physical properties of the cell wall in pollen tube growth of Solanum chacoense. Planta 220: 582–592 [DOI] [PubMed] [Google Scholar]
- Pauly M., Scheller H.V. (2000). O-Acetylation of plant cell wall polysaccharides: identification and partial characterization of a rhamnogalacturonan O-acetyl-transferase from potato suspension-cultured cells. Planta 210: 659–667 [DOI] [PubMed] [Google Scholar]
- Peaucelle A., Louvet R., Johansen J.N., Höfte H., Laufs P., Pelloux J., Mouille G. (2008). Arabidopsis phyllotaxis is controlled by the methyl-esterification status of cell-wall pectins. Curr. Biol. 18: 1943–1948 [DOI] [PubMed] [Google Scholar]
- Pelletier S., et al. (2010). A role for pectin de-methylesterification in a developmentally regulated growth acceleration in dark-grown Arabidopsis hypocotyls. New Phytol. 188: 726–739 [DOI] [PubMed] [Google Scholar]
- Pelloux J., Rustérucci C., Mellerowicz E.J. (2007). New insights into pectin methylesterase structure and function. Trends Plant Sci. 12: 267–277 [DOI] [PubMed] [Google Scholar]
- Pereira S.S., Torres E.T., Gonzalez G.V., Rojas M.G. (1992). Effect of different carbon sources on the synthesis of pectinase by Aspergillus niger in submerged and solid state fermentation. Appl. Microb. Biotechnol. 39: 36–41 [Google Scholar]
- Perrone P., Hewage C.M., Thomson A.R., Bailey K., Sadler I.H., Fry S.C. (2002). Patterns of methyl and O-acetyl esterification in spinach pectins: New complexity. Phytochemistry 60: 67–77 [DOI] [PubMed] [Google Scholar]
- Pippen E.L., McCready R.M., Owens H.S. (1950). Gelation properties of partially acetylated pectins. J. Am. Chem. Soc. 72: 813–816 [Google Scholar]
- Qin Y., Leydon A.R., Manziello A., Pandey R., Mount D., Denic S., Vasic B., Johnson M.A., Palanivelu R. (2009). Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genet. 5: e1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quéméner B., Cabrera Pino J.C., Ralet M.C., Bonnin E., Thibault J.F. (2003). Assignment of acetyl groups to O-2 and/or O-3 of pectic oligogalacturonides using negative electrospray ionization ion trap mass spectrometry. J. Mass Spectrom. 38: 641–648 [DOI] [PubMed] [Google Scholar]
- Ralet M.C., Crépeau M.J., Bonnin E. (2008). Evidence for a blockwise distribution of acetyl groups onto homogalacturonans from a commercial sugar beet (Beta vulgaris) pectin. Phytochemistry 69: 1903–1909 [DOI] [PubMed] [Google Scholar]
- Ralet M.C., Crepeau M.J., Buchholt H.C., Thibault J.F. (2003). Polyelectrolyte behaviour and calcium binding properies of sugar beet pectins differing in their degrees of methylation and acetylation. Biochem. Eng. J. 16: 191–201 [Google Scholar]
- Ralet M.C., Cabrera J.C., Bonnin E., Quéméner B., Hellìn P., Thibault J.F. (2005). Mapping sugar beet pectin acetylation pattern. Phytochemistry 66: 1832–1843 [DOI] [PubMed] [Google Scholar]
- Rexova-Benkova L., Mrakova M., Luknar O., Kohn R. (1997). The role of sec-alcoholic groups of D-galacturonan in its degradation by endo-D-galacturonanase. Collect. Czech. Chem. Commun. 42: 3204–3213 [Google Scholar]
- Ridley B.L., O’Neill M.A., Mohnen D. (2001). Pectins: Structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 57: 929–967 [DOI] [PubMed] [Google Scholar]
- Rombouts F.M., Thibault J.-F. (1986). Enzymatic and chemical degradation and the fine structure of pectins from sugar-beet pulp. Carbohydr. Res. 154: 189–203 [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sato K., Sato K., Okubo A., Yamazaki S. (1997). Determination of monosaccharides derivatized with 2-aminobenzoic acid by capillary electrophoresis. Anal. Biochem. 251: 119–121 [DOI] [PubMed] [Google Scholar]
- Sato K., Sato K., Okubo A., Yamazaki S. (2005). Separation of 2-aminobenzoic acid-derivatized glycosaminoglycans and asparagine-linked glycans by capillary electrophoresis. Anal. Sci. 21: 21–24 [DOI] [PubMed] [Google Scholar]
- Scheller H.V., Ulvskov P. (2010). Hemicelluloses. Annu. Rev. Plant Biol. 61: 263–289 [DOI] [PubMed] [Google Scholar]
- Schols H.A., Voragen A.G. (1994). Occurrence of pectic hairy regions in various plant cell wall materials and their degradability by rhamnogalacturonase. Carbohydr. Res. 256: 83–95 [DOI] [PubMed] [Google Scholar]
- Schols H.A., Voragen A.G. (1996). Complex pectins: Structure elucidation using enzymes. In Pectins and Pectinases, J. Visser and A.G.J. Voragen, eds (Amsterdam: Elsevier Science), pp. 3–19 [Google Scholar]
- Shevchik V.E., Hugouvieux-Cotte-Pattat N. (1997). Identification of a bacterial pectin acetyl esterase in Erwinia chrysanthemi 3937. Mol. Microbiol. 24: 1285–1301 [DOI] [PubMed] [Google Scholar]
- Shevchik V.E., Hugouvieux-Cotte-Pattat N. (2003). PaeX, a second pectin acetylesterase of Erwinia chrysanthemi 3937. J. Bacteriol. 185: 3091–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solms J., Deuel H. (1951). Untersuchungen an acetylieter pektinsaure. Helv. Chim. Acta 34: 2242–2249 [Google Scholar]
- Sterling J.D., Quigley H.F., Orellana A., Mohnen D. (2001). The catalytic site of the pectin biosynthetic enzyme alpha-1,4-galacturonosyltransferase is located in the lumen of the Golgi. Plant Physiol. 127: 360–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman D.M., Harriman R.W., Ramamohan G., Handa A.K. (1992). An antisense pectin methylesterase gene alters pectin chemistry and soluble solids in tomato fruit. Plant Cell 4: 667–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercauteren I., de Almeida Engler J., De Groodt R., Gheysen G. (2002). An Arabidopsis thaliana pectin acetylesterase gene is upregulated in nematode feeding sites induced by root-knot and cyst nematodes. Mol. Plant Microbe Interact. 15: 404–407 [DOI] [PubMed] [Google Scholar]
- Voragen A.G.J., Schols H.A., Pilinik W. (1986). Determination of the degree of methylation and acetylation of pectins by h.p.l.c. Food Hydrocolloids 1: 65–70 [Google Scholar]
- Wang Y., Zhang W.Z., Song L.F., Zou J.J., Su Z., Wu W.H. (2008). Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol. 148: 1201–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen F., Zhu Y., Hawes M.C. (1999). Effect of pectin methylesterase gene expression on pea root development. Plant Cell 11: 1129–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins O., Nahal H., Foong J., Provart N.J., Campbell M.M. (2009). Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 149: 981–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson G. (1991). Purification and characterization of pectin acetylesterase from orange peel. Phytochemistry 30: 445–449 [Google Scholar]
- Williamson G., Faulds C.B., Matthew J.A., Archer D.B., Morris V.J., Brownsey G.J., Ridout M.J. (1990). Gelation of sugarbeet and citrus pectins using enzymes extracted from orange peel. Carbohydr. Polym. 13: 387–397 [Google Scholar]
- Wolf S., Mouille G., Pelloux J. (2009). Homogalacturonan methyl-esterification and plant development. Mol. Plant 2: 851–860 [DOI] [PubMed] [Google Scholar]
- Yu X.-H., Gou J.-Y., Liu C.J. (2009). BAHD superfamily of acyl-CoA dependent acyltransferases in Populus and Arabidopsis: Bioinformatics and gene expression. Plant Mol. Biol. 70: 421–442 [DOI] [PubMed] [Google Scholar]