A novel differential RNA-seq approach was used to obtain a genome-wide map of transcription start sites in the chloroplast genome of barley (Hordeum vulgare). Noncoding RNAs undergo extensive synthesis in plastids, and promoters used by different types of RNA polymerases in green and white ribosome-free plastids of the barley mutant line albostrians were identified.
Abstract
Gene expression in plastids of higher plants is dependent on two different transcription machineries, a plastid-encoded bacterial-type RNA polymerase (PEP) and a nuclear-encoded phage-type RNA polymerase (NEP), which recognize distinct types of promoters. The division of labor between PEP and NEP during plastid development and in mature chloroplasts is unclear due to a lack of comprehensive information on promoter usage. Here, we present a thorough investigation into the distribution of PEP and NEP promoters within the plastid genome of barley (Hordeum vulgare). Using a novel differential RNA sequencing approach, which discriminates between primary and processed transcripts, we obtained a genome-wide map of transcription start sites in plastids of mature first leaves. PEP-lacking plastids of the albostrians mutant allowed for the unambiguous identification of NEP promoters. We observed that the chloroplast genome contains many more promoters than genes. According to our data, most genes (including genes coding for photosynthesis proteins) have both PEP and NEP promoters. We also detected numerous transcription start sites within operons, indicating transcriptional uncoupling of genes in polycistronic gene clusters. Moreover, we mapped many transcription start sites in intergenic regions and opposite to annotated genes, demonstrating the existence of numerous noncoding RNA candidates.
INTRODUCTION
Chloroplasts are the characteristic organelles of plants and, with their photosynthetic activity, form the basis of the autotrophic lifestyle. They have evolved from a cyanobacterial endosymbiont into an organelle firmly integrated in the metabolism of the plant cell. During evolution, the majority of the cyanobacterial genes were lost or transferred to the nucleus of the host cell, while several genes mainly required for photosynthesis and gene expression were retained in the organellar genome (Kleine et al., 2009). The plastid genome (plastome) contains functional rpo genes coding for homologs of the cyanobacterial RNA polymerase subunits α, β, β’, and β’’, which form the core of the plastid-encoded plastid RNA polymerase (PEP; Ohyama et al., 1986; Shinozaki et al., 1986; Sijben-Müller et al., 1986; Hajdukiewicz et al., 1997; Liere et al., 2011). Nuclear-encoded sigma factors interact with PEP to confer promoter recognition (Liu and Troxler, 1996; Tanaka et al., 1996, 1997; Schweer et al., 2010; Lerbs-Mache, 2011). The promoters used by PEP are similar to Escherichia coli σ70 promoters, consisting of −35 and −10 consensus elements (Gatenby et al., 1981; Gruissem and Zurawski, 1985; Strittmatter et al., 1985; Liere and Börner, 2007). In stark contrast with the eubacterial RNA polymerase, PEP is not sufficient to transcribe all genes in plastids of higher plants. A second polymerase, denoted nuclear-encoded plastid RNA polymerase (NEP), was found to participate in plastid transcription (Siemenroth et al., 1981; Hess et al., 1993; Allison et al., 1996). NEP is represented by one or more phage-type RNA polymerase (Hedtke et al., 1997, 2000). It recognizes distinct types of promoters with sequence similarity to plant mitochondrial promoters (Allison et al., 1996; Vera et al., 1996; Liere and Börner, 2007). NEP promoters were found to be more active in early leaf development, while transcription activity of PEP is reported to increase during chloroplast maturation (Courtois et al., 2007; Swiatecka-Hagenbruch et al., 2008). According to the current view, based on studies of only a small subset of plastid genes, most genes coding for housekeeping proteins are transcribed by both PEP and NEP; genes of photosystems I and II are completely dependent on PEP transcription; and a few housekeeping genes, including the rpoB operon, are transcribed exclusively from NEP promoters (Hajdukiewicz et al., 1997; Swiatecka-Hagenbruch et al., 2007). However, transcripts initiated by NEP cover the entire plastome in leaves of transplastomic tobacco (Nicotiana tabacum) plants manipulated to lack PEP activity, suggesting a more general function of NEP in chloroplast transcription (Legen et al., 2002).
Noncoding RNAs (ncRNAs), including antisense RNAs (asRNAs) as well as RNAs transcribed from intergenic regions, play important roles in posttranscriptional regulation in both prokaryotes and eukaryotes. Cyanobacteria, the chloroplast progenitors, have a plethora of noncoding RNAs at their disposal to regulate gene expression (Georg and Hess, 2011). However, only a few potentially regulatory RNAs have been reported to be transcribed from plastid genes (Nishimura et al., 2004; Georg et al., 2010; Hotto et al., 2010; Zghidi-Abouzid et al., 2011). Therefore, chloroplasts may contain additional hitherto undetected genes for regulatory ncRNAs.
Differential RNA sequencing (dRNA-seq) is a recently established method designed to selectively identify primary transcripts (Sharma et al., 2010) and already proven to be a powerful tool for mapping transcription start sites (TSSs) and to identify ncRNAs in several bacterial and archaeal species (Jäger et al., 2009; Sharma et al., 2010; Mitschke et al., 2011). The method is based on the comparison of terminator exonuclease (TEX)–treated (TEX+) and nontreated (TEX−) RNA samples. TEX degrades RNAs with a 5′ monophosphate (i.e., processed transcripts) but not with a 5′ triphosphate or 5′ CAP structure (i.e., primary, unprocessed transcripts). The comparison of cDNA libraries generated from TEX− and TEX+ samples can therefore be exploited to identify the protected primary transcripts and their TSSs. The phosphorylation status of processed 5′ ends of plastid transcripts has not been investigated, but enzymes homologous to bacterial processing RNases are suggested to be involved in the processing of chloroplast transcripts (Stern et al., 2010; Walter et al., 2010), and the T4 ligase requiring 5′ monophosphates ligates oligonucleotides to processed plastid transcripts (Swiatecka-Hagenbruch et al., 2007). Hence, 5′ ends of processed chloroplast transcripts most likely also have a monophosphate while the primary transcripts carry a triphosphate, as in bacteria. Therefore, it should be possible to distinguish and map both primary and processed RNA 5′ termini in chloroplasts using TEX-based dRNA-seq. So far, only a few plastid TSSs were mapped and their promoters studied. This is mainly due to the tedious techniques previously available, such as primer extension analysis, RNase protection assay, in vitro capping by guanyltransferase, and 5′-rapid amplification of cDNA ends (RACE), which can be applied to the study of individual targets (Kapoor et al., 1997; Hübschmann and Börner, 1998; Liere and Maliga, 1999; Swiatecka-Hagenbruch et al., 2007). The major advantage of dRNA-seq over conventional TSS mapping techniques is that it allows for high-throughput, genome-wide, and, thus, cost-effective TSS mapping.
Here, we address the questions of the existence of ncRNAs and the division of labor between PEP and NEP in plastids of the barley (Hordeum vulgare) line albostrians. This mutant line is characterized by green, white, and striped leaves with identical genotypes. Plastids in white leaves and white parts of striped leaves are ribosome deficient and lack all plastid-encoded proteins, including the core subunits of PEP (Hess et al., 1993). Therefore, transcription in these mutant plastids is performed exclusively by NEP. Using dRNA-seq (Sharma et al., 2010), we mapped TSSs in green (transcription by both PEP and NEP) and white (transcription only by NEP) albostrians plastids. We obtained a genome-wide map of the PEP and NEP initiation sites and detected numerous TSSs within operons, in intergenic spaces, and opposite to annotated genes, indicating transcriptional uncoupling within polycistronic transcription units and suggesting the generation of many ncRNA candidates.
RESULTS
dRNA-seq Analysis of the Chloroplast Primary Transcriptome
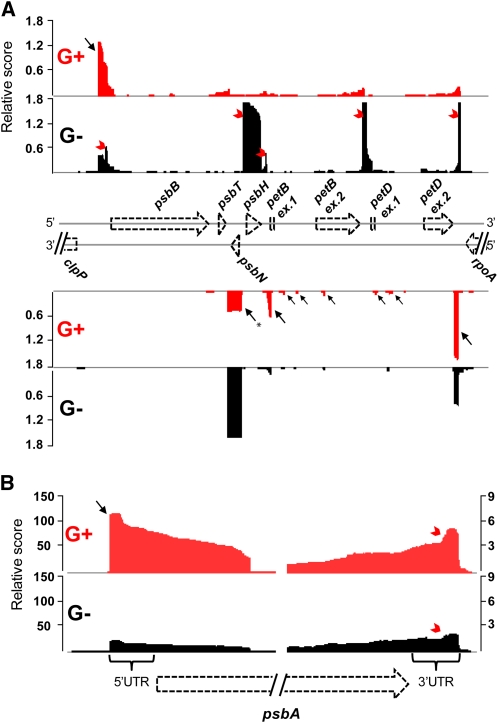
We analyzed the primary transcriptome of green (G) and white (W) albostrians plastids of mature first leaves by dRNA-seq (Sharma et al., 2010). To discriminate between primary and processed transcripts, we constructed and compared two differential cDNA libraries per plastid type. G and W libraries were generated from untreated total plastid RNA and thus contained both primary and processed transcripts. The G+ and W+ libraries were generated from TEX-treated plastid RNA and were thus enriched for primary transcripts (see Supplemental Figure 1 online). We analyzed a similar number of sequence reads (~70,000) with ≥18 nucleotides for each library. Using the WU-BLAST algorithm (http://blast.wustl.edu/), we mapped 94.5% (G+) and 79.1% (G−) of the sequence reads from green plastids and 14.3% (W+) and 7.1% (W−) of the reads from white plastids to the chloroplast genome of barley (NC_008590). Due to the small amount of available white mutant leaves, we had to use plastid samples more contaminated by nuclei and mitochondria than in the case of green leaves; consequently, a lower percentage of reads arose from plastids in the white libraries. Nevertheless, the obtained reads from both W libraries were sufficient to provide a picture of the transcriptome of PEP-lacking plastids. For each library, graphs representing the number of mapped sequence reads per nucleotide were calculated and visualized using the Integrated Genome Browser (see Supplemental Figure 2 online). The TEX treatment eliminates processed transcripts and thereby enriches for primary transcripts in relative terms. The resulting characteristic difference in the cDNA distribution in (+) compared with (–) libraries revealed TSSs on a global scale. As a typical example, the dRNA-seq data for the psbB operon in green plastids is shown in Figure 1A. This operon comprises five genes transcribed into a large precursor mRNA, which is extensively processed. In the G− library (black), several cDNA peaks (indicated by red arrowheads) are visible, which are completely eliminated by the exonuclease treatment and missing in the G+ library (red). They correspond to the previously described major processing sites of this operon (Westhoff, 1985; Chen and Stern, 1991; Barkan et al., 1994; Felder et al., 2001; Pfalz et al., 2009). Many of the other 5′ ends susceptible to degradation by TEX in our dRNA-seq analysis are also in agreement with previously published data on chloroplast processing. Additionally, we verified nine of them as 5′-monophosphates using 5′-RACE or circularization RT-PCR (cRT-PCR) (see Supplemental Data Set 1 online). Moreover, we detected short sequences associated with most of the processed 5′ ends to accumulate in the untreated samples, suggesting protection from degradation by nucleases. This is related to the proposed involvement of pentatrico peptide repeat (and related) proteins in defining the ends of chloroplast RNAs (Pfalz et al., 2009; Prikryl et al., 2011). A detailed analysis of these small RNAs is presented in a separate publication (Zhelyazkova et al., 2011). In the G+ library, the cDNAs are clustered toward the nuclease-protected 5′ end of the primary transcript, indicating the TSS of the psbB operon (black arrow). The TSS mapped by dRNA-seq (see Supplemental Data Set 2A) is in agreement with the one previously found in spinach (Spinacia oleracea; Westhoff, 1985) and was verified here by 5′-RACE (see Supplemental Data Set 3 online). In total, 11 out of the 12 TSSs, which were previously determined by alternative methods, could be detected and defined with high accuracy by dRNA-seq (up to ±2-nucleotide difference; see Supplemental Table 1 and Supplemental References 1 online), demonstrating the sensitivity and accuracy of this approach for mapping chloroplast TSSs.
Figure 1.
The Barley Chloroplast Transcriptome Revealed by dRNA-seq.
cDNA reads from libraries enriched [red, (+) libraries] and nonenriched [black, (−) libraries] for primary transcripts by TEX treatment were mapped to the barley chloroplast genome. Graphs were normalized to the number of mapped reads per library and visualized using the Integrated Genome Browser. Genes are depicted by dashed, unfilled arrows/rectangles and triangles below the graph files. The y axis indicates per mill (a 10th of a percent) mapped reads per genome position.
(A) cDNA reads of green (G+/−) libraries mapped onto the psbB operon region. Both strands are represented. The psbB operon is encoded on the plus strand. The cDNA peaks in G−, which are no longer detectable after the TEX treatment, are indicated by red arrowheads. They represent major processing sites of the psbB operon. In the G+ library, the cDNAs are clustered toward the nuclease-protected 5′ end of the primary transcript, indicating the TSS of the psbB operon (black arrow). The transcription profile of psbN, encoded within the psbB operon but on the opposite strand, is also depicted. TpsbN-46 (black arrow with an asterisk) is one of the examples that was not detected as enriched transcripts in the TEX-treated G+ library. Multiple TSSs upstream of psbN and antisense to the psbB operon were mapped.
(B) cDNA reads of green (G+/−) libraries mapped onto psbA. The TSS of psbA, indicated by a black arrow, is visible after TEX treatment. The 3′ terminal hairpin RNAs, marked by red arrowheads, are resistant to TEX and reveal the 3′ end of psbA mRNA.
As another example, the dRNA-seq profile for psbA in green plastids is shown in Figure 1B. After TEX treatment, we observed a more than fourfold enrichment of cDNAs clustered at the nuclease-protected 5′ end of the primary transcript. The TSS mapped by dRNA-seq is in agreement with already published data (Boyer and Mullet, 1988). Unexpectedly, we observed a distinctive stepwise accumulation of cDNAs in proximity to the 3′ end of the psbA open reading frame (ORF) in both the (−) and (+) libraries but more pronounced in the latter (Figure 1B, red arrowheads). The majority of the 3′ ends of these cDNA reads matched precisely the last base pair of the stem-loop structure formed at the 3′ end of the psbA mRNA (Memon et al., 1996; see Supplemental Figure 3 online). Stem-loop structures participate in mRNA 3′ end formation by blocking 3′ to 5′ exonuclease activity (Stern et al., 2010). We mapped comparable TEX-resistant cDNA accumulations near the 3′ ends of 14 genes. These may mark the 3′ ends of the corresponding mRNAs. A high probability of stem-loop structure formation was predicted in all cases. Two of them were experimentally verified as 3′ ends and another three correspond to previously mapped mRNA 3′ termini (see Supplemental Table 2 online).
Annotation and Classification of Potential TSSs
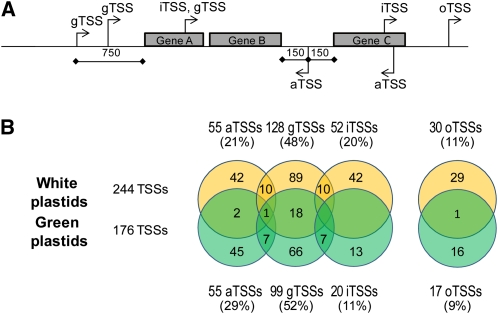
In total, we manually assigned 176 and 244 potential TSSs in green and white albostrians plastids, respectively (see Supplemental Data Set 2 online). Of these, 76 (green) and 91% (white) were at least twofold enriched in our (+) versus (−) libraries. In both green and white plastids, we identified more TSSs than one might expect for a genome comprising 113 genes (Saski et al., 2007; see Methods), many of which clustered in polycistronic transcription units (Kanno and Hirai, 1993). The initiating nucleotide was a purine in 91 (green) and 84% (white) of the TSSs. In both green and white plastids, A was preferred over G as an initiating nucleotide, and this preference was more pronounced in white plastids. The TSSs were further grouped into four categories based on their genomic location: gTSSs (gene TSSs), found within the 750-nucleotide region upstream of annotated genes; iTSSs (internal TSSs) found within annotated genes and giving rise to sense transcripts; aTSSs (antisense TSSs) located on the opposite strand within or up to 150 bp up/downstream of annotated genes and giving rise to antisense transcripts; and oTSSs (orphan TSSs), TSSs that cannot be assigned to any of the above categories. Several TSSs could be assigned to more than one category (Figure 2).
Figure 2.
Classification and Category Assignment of TSSs Based on dRNA-seq.
(A) Schematic representation of the annotation and category assignment of TSSs based on their genomic location in the barley plastome. In certain cases, a TSS can be assigned to more than one category (e.g., iTSS and gTSS). An aTSS that is mapped more than 150 nucleotides downstream of an annotation but is represented by cDNAs that overlap with the 3′ UTR of the gene is still considered as an aTSS.
(B) Distribution and overlap among TSS categories in green and white plastids. A total of 244 and 176 TSSs were mapped in white and green plastids, respectively. TSSs were further grouped into four categories. The number and percentage (in parentheses) of TSSs assigned to each category is given. Twenty-one TSSs in white and 15 TSSs in green could be assigned to more than one category. Twenty-two TSSs were found in libraries of both green and white plastids.
[See online article for color version of this figure.]
Theoretically, a processed 5′ end might be protected from TEX digestion by stable structures resulting from intramolecular base pairing. In such a case, a 5′ end could be misannotated as a putative TSS instead of a processing site. Therefore, we bioinformatically analyzed the first 50 and 100 nucleotides of all potential primary transcripts mapped in green and white plastids for the probability of stable structure formation. On average, the number of nucleotides potentially participating in base pair interactions was low in the region immediately downstream of the mapped TSSs. Thus, stable stem loops are unlikely to be formed near the 5′ end of most analyzed transcripts and hence could not act as a barrier to digestion by TEX (see Supplemental Figure 4 online). Moreover, 52 TSSs, among them examples with stable stem-loops at their 5′ end, were selected and 40 TSSs (77%) were successfully verified by 5′-RACE analysis as major 5′-PPP ends in independent RNA preparations from green and white plastids (see Supplemental Data Set 3 online). Thus, we conclude that our TEX-based RNA-seq approach reliably identifies TSSs and discriminates them from processing sites in RNA preparations from green and white leaves. Strikingly, only 22 TSSs were found to be identical in green (both PEP and NEP present) and white (only NEP present) plastids (see Supplemental Table 3 online). Since NEP activity was reported to be higher in white compared with green leaves (Emanuel et al., 2004), chloroplasts of green leaves are not expected to exhibit the activities of NEP promoters that are not found in white plastids. Thus, at least 154 of the 176 TSSs (88%) in green barley chloroplasts should have originated from PEP activity. That is, PEP is by far the dominating RNA polymerase in chloroplasts of mature leaves.
Promoter Sequences
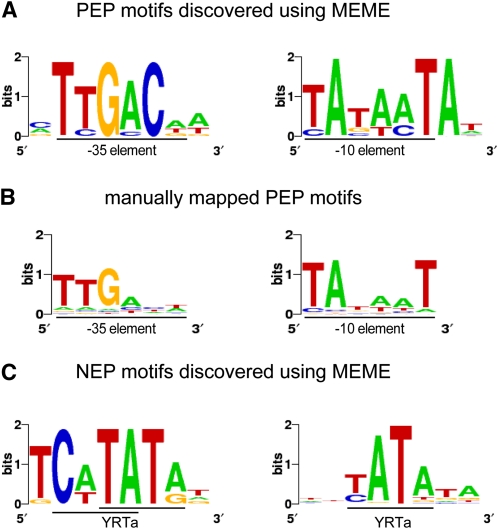
The −1- to −25-nucleotide and −26- to −50-nucleotide upstream regions of all TSSs mapped in green and white plastids were screened separately for potential promoter motifs with lengths of 3 to 9 nucleotides using the Multiple Expectation-Maximization for Motif Elicitation (MEME) tool (Bailey et al., 2009). Two 8-nucleotide-long motifs were discovered in the −1- to −25-nucleotide and −26- to −50-nucleotide upstream regions of only 44 (25%) and 20 (11%) of the TSSs, respectively, in green plastids (Figure 3A). These two motifs were found to be significantly overrepresented (P value = 2.434e-05 and 0.008552, respectively) in pre-TSS stretches of green plastids compared with those of white plastids and show high similarity to the previously described −10 and −35 PEP promoter consensus hexamers (Liere and Börner, 2007). Since it is possible that some of the −10 and −35 elements are more variable and therefore hard to be identified using MEME default cutoff values, we performed a manual search for the occurrence of these motifs. Indeed, we were able to map a −10 element (TAtaaT; uppercase letters depict overrepresented nucleotide >1 bit) three to nine nucleotides upstream of the transcription start point in 156 TSSs (89% of the TSSs) in green leaves. The −35 region was again found to be less conserved than the −10 box. A ttGact motif 15 to 21 nucleotides upstream of the −10 element was mapped in 70% (109/156) of the TSSs (Figure 3B).
Figure 3.
Sequence Logos of Promoter Motifs Detected in Green and White Plastids.
Logos were visualized using WebLogo (http://weblogo.berkeley.edu/).
(A) In green plastids, MEME analysis discovered a −10 (right) and −35 (left) PEP consensus element upstream of 44 and 20 TSSs, respectively. The motifs were found to be significantly enriched in green pre-TSS sequences.
(B) A manual search for the PEP promoter elements detected the −10 box (right) in 156 TSSs and the −35 box (left) in 109 of the TSSs with a mapped −10 element.
(C) Two versions of the YRTa motif were discovered by MEME in white plastids. A TCaTATat motif (left) was found upstream of 22 of the white TSSs and YATata (right) upstream of 151 (62%) TSSs.
[See online article for color version of this figure.]
A MEME search of sequences from white plastids revealed an 8-nucleotide-long motif in the −1 to −25 upstream region of 22 TSSs (Figure 3C, left). The motif was found to be significantly overrepresented (P value = 0.009847) in white compared with green pre-TSS stretches and resembled an extended version of the YRTa motif, the most frequently observed NEP promoter motif (Liere and Börner, 2007). An additional search limited only to the first 10 nucleotides upstream of the TSSs revealed a second motif that was significantly more predominant (P value = 2.058e-05) in white pre-TSS regions. The motif represents another version of the YRTa motif and was found upstream of 151 (62%) of the white TSSs (Figure 3C, right). MEME did not detect any additional motifs in the upstream regions or even when the analysis was extended to the −50 to +25 regions around the TSSs in white plastids. Moreover, a manual search for the YRTa motif increased its detection to only 73%.
The Primary Transcriptome of Annotated Genes
Eighty-nine of the 113 chloroplast genes are part of 20 operons (polycistronic gene clusters), while 24 genes are transcribed monocistronically in barley plastids (see Supplemental Figure 5 online and Methods; gene count and operon annotation of the barley chloroplast genome). We found TSSs upstream of 15 and 14 operons and of 19 and 20 separate genes in green and white plastids, respectively (see Supplemental Figure 5 and Supplemental Data Set 4 online). We were able to detect the 5′-P ends for the majority of genes/operons without visible TSSs, suggesting either fast mRNA maturation or low abundance of their primary transcripts. Only in the cases of rpl32, ndhH, and ndhF was neither a TSS nor a 5′-P detectable among the ~70,000 evaluated sequences per green sample. In samples of white plastids, we did not detect any transcripts of trnL-UAA, ndhH, ccsA, and psbE-F-L-J, perhaps owing to their low synthesis or fast degradation. The genes of transcripts not found in white plastids may have no NEP promoter. Moreover, 44 gTSSs in green and 58 in white leaves were found within operons and thus may function in uncoupling those gene clusters into smaller polycistronic and monocistronic transcription units. Between two and six TSSs were found for 25 and 35 genes in green and white plastids, respectively (see Supplemental Data Set 4 online), with two being the most common case. The average length of the 5′ untranslated region (UTR; the region between an ORF and its corresponding TSS) was 235 nucleotides in green and 232 nucleotides in white plastids.
A major goal of this study was to elucidate the division of labor between PEP and NEP. Eighty gTSSs were found in RNA libraries of green but not white leaf material, and these TSSs therefore represented potential primary PEP transcripts. Nineteen gTSSs were found in both green and white plastids (see Supplemental Table 3 online). Among them were TrpoB-147, Trpl23-71, and Trps15-228 (TSSs are named after the downstream-located gene and the number of nucleotides between the TSS and the start codon of the ORF), which were previously reported to be NEP-dependent TSSs in green plastids (Hübschmann and Börner, 1998; Liere and Maliga, 1999; Swiatecka-Hagenbruch et al., 2007). These TSSs were significantly overrepresented in the W+ compared with the G+ library. Identical gTSSs were found in both green and white libraries in the case of the photosynthesis genes psbA, psaA, and psbN, which were shown to be transcribed from PEP promoters in chloroplasts (Zghidi et al., 2007; Swiatecka-Hagenbruch et al., 2007). TpsbA-80 and TpsbN-46 were additionally verified by 5′-RACE (see Supplemental Data Set 3 online). The identification of these TSSs in white samples cannot be explained by a contamination with green chloroplasts, since the PEP-dependent TSSs of other abundant RNAs (e.g., trnG-UCC, psbE, and petN) were not detected in samples from white tissue. It seems more likely that the promoter regions of TpsbA-80, TpsaA-209, and TpsbN-43 represent overlapping motifs recognized by both PEP and NEP. Indeed, we found promoter motifs for both polymerases upstream of TpsbA-80 and TpsbN-43. Consequently, we cannot conclusively say if PEP, NEP, or both polymerases transcribe several genes in green barley chloroplasts (see Supplemental Table 3 online; marked as unclear). The TSSs enriched in white but not green plastids could indicate potential NEP promoters used in green plastids (e.g., Trps2-152, TndhB-275, and TndhI-99; see Supplemental Table 3 online).
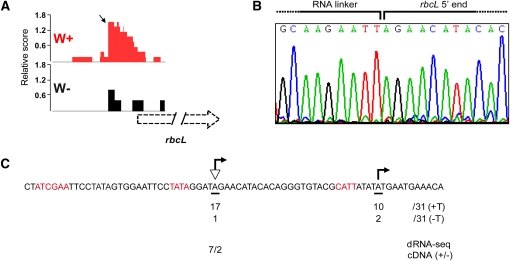
Previous data suggested that several genes, such as atpB, rrn16, and rps4, have both PEP and NEP promoters (Swiatecka-Hagenbruch et al., 2007). According to our dRNA-seq data, it is a common feature of plastid genes to have promoters for both polymerases (see Supplemental Figure 5 and Supplemental Data Set 4 online). Only for 11 genes (trnL-UAA, trnM-CAU, trnN-GUU, psbE-F-L-J, trnT, petN, trnS-UGA, and trnQ-UGG) with primary transcripts in green plastids were no TSSs identified in the RNA of white plastids. Those genes may indeed have no NEP promoter, though we cannot rule out that NEP-transcribed primary transcripts exist from these genes, but below the level of detection in our experiments. Genes coding for subunits of photosynthetic enzymes, such as photosystem I, photosystem II, and ribulose-1,5-bis-phosphate carboxylase/oxygenase, were previously suggested to be exclusively transcribed by PEP (Hajdukiewicz et al., 1997). However, we found TSSs for numerous photosynthesis genes in white tissue. For example, rbcL is transcribed by a typical NEP promoter with an YRTa motif in white plastids (Figure 4). We also found TSSs for psbB, psbD, psbM, and other photosynthesis genes in white libraries. The psaJ NEP promoter was even among the most active promoters in white plastids (see Supplemental Data Set 2B online). Some of the potential NEP promoters for annotated genes were found as internal promoters within upstream genes (see below).
Figure 4.
The rbcL Gene Is Transcribed from a NEP Promoter in White Plastids.
(A) cDNA reads of white (W+/−) dRNA-seq libraries mapped onto rbcL. The TSS of rbcL is indicated by a black arrow.
(B) A chromatogram displaying the sequence at the ligation site of a cloned 5′-RACE product. The 3′ end of the linker and the 5′ end of the rbcL primary transcript are shown. The primary 5′ end determined by 5′-RACE supports the TSS revealed by dRNA-seq analysis.
(C) Promoter region of rbcL used in white plastids. The TSS identified by dRNA-seq and 5′-RACE are marked by a triangle and arrows, respectively. The sequence motifs shown in red represent putative NEP promoters. An additional TSS immediately downstream of the TSS detected by dRNA-seq was detected by 5′-RACE analysis. The number of clones supporting each TSS in T+ (TAP+) and T− (TAP−) reactions, as well as the total number of sequenced clones (number after the slash), are given. The numbers of cDNAs in (+/−) dRNA-seq are also listed.
We detected a more than twofold enrichment of potential primary transcripts starting exactly at position +1 (relative to the gene start, the mature 5′ end normally generated by RNaseP) for six and 10 trn genes in green and white libraries, respectively. Five of them were identical between green and white libraries (see Supplemental Data Set 2 and Supplemental Table 3 online). These TSSs could either result from upstream promoters or indicate transcription from internal promoters as proposed for several chloroplast tRNA genes in spinach (Gruissem et al., 1986; Cheng et al., 1997) and mustard (Sinapis alba; Neuhaus and Link, 1990; Nickelsen and Link, 1990; Liere and Link, 1994). They might be false-positive TSS candidates due to the particular structure of tRNAs in which the amino acid acceptor stem might protect some mature 5′ ends of tRNAs from the attack of TEX.
Internal TSSs
We mapped numerous TSSs within annotated genes (iTSSs) in RNA samples of both green and white plastids (see Supplemental Data Set 2 online). Twenty iTSSs were found in green plastids, seven of which also qualified as TSSs of downstream genes in operons, suggesting a potential role of these TSSs in uncoupling transcription units. Indeed, among them is the previously identified TpsbC-194 (Sexton et al., 1990). Ten out of the detected 52 iTSSs in white plastids could also be considered as potential TSSs of downstream genes. For example, we observed an iTSS (TrpoC1-599) in the rpoB gene in white leaves, which might serve as a gTSS for the transcription of the downstream rpoC1. 5′-RACE analysis detected TrpoC1-599 in samples from both green and white plastids (see Supplemental Data Set 3 online).
Potential Regulatory ncRNAs
We mapped numerous TSSs within or up to 150 bp up or downstream of annotated genes on the opposite strand (aTSSs), as well as in intergenic regions (oTSSs) in green and white plastids (see Supplemental Data Set 2 online), indicating the synthesis of many ncRNAs. Two examples are presented in Figure 5. We found 55 aTSSs in both green and white plastids leading to antisense transcripts to 40 and 45 genes, respectively. A total of 9 and 11% of the TSSs in green and white plastids, respectively, were mapped to intergenic regions and represent oTSSs. The set of aTSSs and oTSSs mapped in green libraries let us predict 60 ncRNA candidates (denoted Hv_nc) in mature barley chloroplasts (see Supplemental Data Set 5 online). 5′-RACE analysis revealed two potential ncRNAs, the TSSs of which were solely visible in white dRNA-seq libraries, to be also present in green plastids with the same initiating nucleotide (see Supplemental Data Set 3 online). Therefore, it is possible that more of the aTSSs or oTSSs exclusively found in white libraries are actually TSSs of ncRNAs also expressed in green plastids. In addition, some of the more distant gTSSs could be used for ncRNA synthesis rather than expression of annotated genes. We also cannot rule out the possibility that the TSSs of part of the predicted ncRNAs could be driving transcription of further downstream located genes. However, the 3′ ends of seven ncRNA candidates were mapped precisely by 3′-RACE analysis, thus verifying the synthesis of ncRNAs from these TSSs (see Supplemental Data Set 5 online).
Figure 5.
ncRNA Candidates in Mature Plastids.
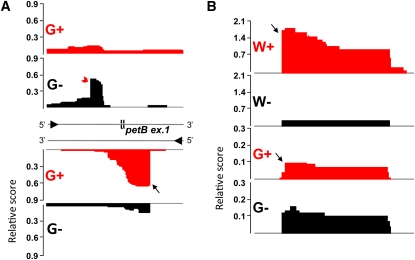
(A) cDNA reads of green (G+/−) libraries mapped onto the petB exon 1. The detected processed 5′ end of petB is indicated by a red arrowhead and the TSS of the petB antisense transcript, as_petB (Hv_nc40), is marked by a black arrow. The direction of transcription is indicated by black arrowheads on the DNA strands. The as_petB RNA is complementary to the intron binding sites 1 and 2 of the petB intron (group II intron).
(B) dRNA-seq reveals an ncRNA (Hv_nc50) in the intergenic region between trnL-CAA and trnI-CAU in both green and white plastids. The detected oTSSs are indicated by black arrows.
DISCUSSION
We used the TEX-based dRNA-seq approach (Sharma et al., 2010) to investigate the primary transcriptome of chloroplasts. We could detect with high accuracy 11 out of the 12 barley chloroplast TSSs previously determined by alternative methods. Moreover, we verified, by 5′-RACE analysis, 40 novel TSSs identified by dRNA-seq. Hence, dRNA-seq can serve as a valuable method for high-throughput and accurate mapping of TSSs in the chloroplast genome. Our data also indicate that the majority, if not all, processed chloroplast transcripts have a monophosphorylated 5′ end, since they were efficiently eliminated by TEX. Only a small number of TSSs, mapped during this study by 5′-RACE or described previously, were not detectable as enriched transcripts in the TEX-treated G+ library (see Supplemental Data Set 2A online; Not enriched). A possible reason for the missing enrichment could be identical primary and processed 5′ ends, which would hinder their unequivocal identification by dRNA-seq. Identical initiation and processing sites were described for several transcripts in Arabidopsis thaliana mitochondria (Kühn et al., 2005). Recently it was demonstrated that the RNA polymerase of the Gram-negative bacterium Pseudomonas aeruginosa may use nanoRNAs as primers for transcription initiation, thus generating primary transcripts with 5′ ends similar to processed ones (Goldman et al., 2011). Even though there is no indication that such a process takes place in plastids, a potential similar function of PEP, the bacteria-type RNA polymerase, would explain why no enrichment after TEX treatment of some primary transcripts was observed only in green libraries. Furthermore, RppH, a member of the Nudix (for nucleoside diphosphates linked to some moiety X) hydrolase enzyme family, removes the β and γ phosphates of the initiating nucleotide in primary transcripts and thereby initiates rapid RNA decay in E. coli (Deana et al., 2008). An analogous activity is found in Bacillus subtilis (Richards et al., 2011). In Arabidopsis there are 27 genes encoding Nudix hydrolase homologs, with seven and eight of them being targeted to mitochondria and chloroplasts, respectively. Organellar-type Nudix hydrolases showed pyrophosphohydrolase activity toward various substrates, such as ADP-Rib, ADP-Glc, CoA, and NADH (Ogawa et al., 2008). However, it still remains to be elucidated if one or more of these enzymes also exhibit RppH-like activity and generate monophosphorylated 5′ ends of organellar transcripts.
The nuclear and mitochondrial genomes of higher plants contain sequences of chloroplast origin that still may show a high degree of similarity or even identity to the original sequence (Noutsos et al., 2005; Hao and Palmer, 2009). Though usually not active, some might be transcribed and contained in our samples, in particular in those from the less pure plastids isolated from white leaves. We would expect such chloroplast-like transcripts from the nucleus to carry nonchloroplast sequences at the 5′ end, permitting their detection. In the case of chloroplast-like transcripts of mitochondrial origin, the situation is more complicated, since the mitochondrial RNA polymerases recognize NEP promoters (i.e., complete chloroplast sequences could be transcribed in mitochondria). The lack of a complete mitochondrial genome sequence of barley, however, precludes the analysis of such mitochondria-borne sequences. Arguably, one should be able to detect such sequences in our cDNA libraries because they too might be chimeric (i.e., are fused to nonchloroplast sequences) So far, we have not found chimeric sequences of this type in white or green libraries. We tested in particular tRNAs for slightly deviating sequences, since plant mitochondrial genomes have active tRNA genes of chloroplast origin that are nearly identical in both genomes. As a rule, however, mitochondrial transcripts are of much lower abundance than chloroplast transcripts, which could be the reason why we have not detected indications for transcripts of potential mitochondrial origin.
According to our data, several features of chloroplast transcription that have previously been demonstrated only for a few genes or operons can be regarded as characteristic for most genes/operons. (1) Multiple promoters drive transcription of many individual genes and operons. (2) Internal promoters (gTSSs) were mapped in most operons. While bacteria commonly use polycistronic mRNAs in translation and only rarely cleave them to generate less complex transcripts, plastid polycistronic mRNAs may undergo processing, resulting in smaller polycistronic or monocistronic RNAs. If the intercistronic processing has evolved to facilitate translation on plastid ribosomes, is still a matter of debate (Barkan, 2011). The presence of numerous operon-internal TSSs is another way to generate less complex transcripts and to adjust the abundance of individual mRNAs transcribed from the same operon. (3) The fact that 5′ UTRs play a role in posttranscriptional regulation of gene expression from processing to translation (Stern et al., 2010) is supported by our observation that most chloroplast mRNAs have long 5′ UTRs. Thus, binding of more than one protein to the UTR might be a common phenomenon. Alternatively, such long 5′ UTRs could indicate the necessity for complex folding of this region, which might be a requirement for riboswitches or the binding of certain proteins. Usage of multiple promoters allows for the transcription of mRNAs with different lengths of 5′ UTRs from the same gene/operon and might be a way to control stability and function of the RNAs. (4) PEP is the dominating RNA polymerase in green leaves. We included RNA of white leaves of the albostrians mutant in our analyses to learn more about the division of labor between PEP and NEP in plastid gene expression. Our data clearly demonstrate that only a minority of transcripts in mature chloroplasts originates from NEP activity.
Promoter Motifs in Green and White Plastids
In green plastids, both PEP and NEP primary transcripts exist, with the former being the predominant type (at least 88% of all primary transcripts). Consequently, MEME analysis detected the typical motifs of PEP promoters as the only conserved sequences in the upstream region of the TSSs mapped in green plastids. However, the −10 and −35 boxes were discovered upstream of only 25and 11%, respectively, of all TSSs. We identified as a main reason for the poor detection by MEME the high variability of the nucleotides in the PEP promoter elements, since a manual search with relaxed parameters (see Methods) increased their detection: the −10 box was mapped in 89% of the TSSs, while the −35 box in 70% of the TSSs with a −10 element. Still, the lower conservation of the −35 box suggests that PEP transcription can occur independently of the −35 element and relies on motifs with extremely low similarity to the E. coli σ70 −35 element or other variable cis-elements.
All TSSs in white plastids are generated by NEP activity. However, we detected the YRTa motif typical for NEP type Ia and Ib promoters (Liere et al., 2011) upstream of only 73% of the TSSs in white leaves. Similarly, upstream of many TSSs in higher plant mitochondria, where transcription is performed by NEP-related phage-type RNA polymerases, no conserved promoter motifs could be detected. (Liere et al., 2011). We also found no evidence for a conserved GAA-box in a regular distance to the YRTa motif in the barley NEP promoters as described for so-called type Ib NEP promoters in dicots (Liere and Börner, 2007). Transcription from tobacco PclpP-53, the only example of a NEP class II promoter, also was shown to be dependent on promoter elements located downstream of the TSSs (−5 to +25; Sriraman et al., 1998). MEME did not detect any downstream motif when the analyzed region around the TSSs in white leaves was extended to −50 to +25 (i.e., a consensus motif for type II promoters could not be observed), leaving type Ia the only NEP promoter type clearly identifiable in barley plastids. If this is related to the fact that barley and other grasses have only one NEP polymerase (RPOTp) in contrast with eudicots, which have two NEP polymerases (RPOTp and RPOTmp; Liere et al., 2011), remains to be further analyzed.
Function of PEP and NEP
A surprising finding was the extremely high number of promoters used in white leaves in the absence of PEP. We performed a computer-aided analysis of the 5′ sequences of all identified primary transcripts in green and white samples for their potential to form stable stem-loop structures. Stem-loop structures might protect the RNA molecules from degradation by TEX and could therefore result in misidentification of processed transcripts as primary ones. According to this analysis, such structures are unlikely to be formed near the 5′ end of most analyzed transcripts in both plastid types (see Supplemental Figure 4 online). Additionally, we selected 30 TSSs of white plastids for further analysis by 5′-RACE analysis and could verify 25 of them. We also found no evidence for the occurrence of double-stranded RNA, often protected from exonucleolytic attack, which would be represented in our libraries by cDNA reads precisely complementary to each other. Thus, the TEX-resistant 5′ ends should indeed be TSSs, and we can exclude the possibility that the identified primary transcripts in white plastids would represent RNAs generated by processing or degradation of larger read-through transcripts initiated at only a few NEP promoters.
We mapped altogether 244 TSSs in white plastids, among them 128 gTTSs upstream of 70 plastid genes. Many of the NEP-transcribed genes encode proteins of the photosynthetic apparatus and were previously thought to be expressed solely by PEP (Hajdukiewicz et al., 1997; Swiatecka-Hagenbruch et al., 2007). Transcription of all genes was previously observed in ΔrpoB (PEP-deficient) tobacco plastids and mainly attributed to spurious transcription initiation by NEP throughout the plastome (Legen et al., 2002). However, our high-resolution TSS mapping analysis revealed that RNAs in white plastids are not transcribed from randomly distributed AT-rich sequences or YRTa motifs but rather result from distinct initiation sites localized directly upstream of the genes. Moreover, we observed that NEP is able to generate virtually the same mRNAs as PEP, with similar 5′ and 3′ UTR lengths and, in the case of psaA, psbA, and psbN, even the same 5′ primary end. Therefore, it is still unclear why NEP cannot rescue the albino phenotype in ΔrpoB tobacco plastids (i.e., PEP-deficient mutants with functional translation). The amount of transcripts generated by NEP instead of PEP might be simply insufficient to meet the demands for the extensive translation required for greening (Siemenroth et al., 1981; Allison et al., 1996). Alternatively or additionally, NEP may not transcribe a small number of genes for which we found TSSs in green but not in white plastids. Furthermore, mRNA stability and even translational efficiency may be correlated with differential RNAP usage (Legen et al., 2002; Cahoon et al., 2004).
The activation of a NEP promoter was described in Arabidopsis to compensate for abolished transcription from the atpB PEP promoter (Schweer et al., 2006). We observed that, in the absence of PEP, at least 222 NEP promoters that were undetected by our analysis in green plastids are used in white tissue. Therefore, activation of transcription by NEP in the absence of PEP might serve as a general rescue mechanism in higher plants. The observed stimulation of NEP-dependent transcription might be connected to the higher accumulation of RPOTp transcripts in white albostrians leaves (Emanuel et al., 2004). Additionally or alternatively, PEP, being the more abundant and active RNA polymerase in mature chloroplasts, might hinder the access of NEP to its promoters if they are located in close proximity to PEP promoters (Lyubetsky et al., 2011). We indeed observed many examples of NEP promoters in the neighborhood of PEP promoters. Therefore, it would be interesting to experimentally compare the strength of binding of NEP and PEP to their promoters and to see to what degree NEP transcription is stimulated if PEP activity is reduced but not fully lacking.
The question remains as to whether the numerous NEP promoters found in white mutant plastids are also active in normal wild-type plastids. We could show by 5′-RACE analysis that several TSSs solely detected in W libraries are also used in wild-type chloroplasts, suggesting that there might exist quite a large number of NEP promoters with weak activities in green leaves that were below the level of detection by our dRNA-seq analysis. Moreover, the Arabidopsis RPOTmp gene coding for a mitochondrial and chloroplast (NEP) RNA polymerase is particularly active in nongreen tissues and in very young leaves (Emanuel et al., 2006). Similarly, the expression of the previously described NEP-dependent genes was distinctly higher in developing compared with mature chloroplasts (Courtois et al., 2007; Zoschke et al., 2007). Therefore, it should be worth investigating developing chloroplasts for the usage of the NEP promoters exclusively detected in white albostrians plastids.
Plastid ncRNAs
Only a few reports support the existence of regulatory ncRNAs in chloroplasts. They focus on single RNAs and do not point to ncRNAs as a more general regulatory principle (Nishimura et al., 2004; Georg et al., 2010; Hotto et al., 2010; Zghidi-Abouzid et al., 2011). One study identified 12 ncRNAs in tobacco chloroplasts (Lung et al., 2006). However, none of them was proven to originate from independent RNA genes, rather than being products of processing or degradation of other transcripts. Another study demonstrated the interference of an artificial asRNA with the editing of the corresponding sense RNA in chloroplasts (Hegeman et al., 2005). Moreover, asRNAs generated as by-products of read-through transcription and subsequent processing can interfere with the translational efficiency of the target mRNAs. Such asRNAs are normally eliminated by RNase J (Sharwood et al., 2011).
In contrast with the situation in chloroplasts, numerous bacterial ncRNAs with regulatory functions have been described. Recent studies revealed a high degree of antisense and noncoding transcription in a cyanobacterium (Georg et al., 2009; Mitschke et al., 2011). Here, we provide evidence for extensive ncRNA synthesis in plastids. We detected numerous ncRNAs from intergenic regions as well as antisense transcripts to ~35% of all genes in green plastids. We verified the existence of several selected ncRNAs by an alternative approach. We speculate that some of the asRNAs detected in this study may have a function in regulating chloroplast gene expression, since they are complementary to known important regulatory regions of chloroplast mRNAs, like the 5′ UTR (e.g., as_atpI, as_rps15, and as_psaA), and may function to repress transcription and translation as in bacteria. Moreover, other asRNA candidates were found to be complementary to essential regions involved in base-pairing interactions required for splicing of group II introns in plastids [e.g., as_petB(1) as_trnV, and as_rpl2]. Like most of the annotated plastid genes, various ncRNA candidates have both PEP and NEP promoters. Differences in the composition and abundance of ncRNAs in green and white samples might indicate functions in different tissues or at different stages of chloroplast development. Elucidation of the function and relevance of ncRNAs in plastids may turn out to be a new and exciting field of chloroplast biology.
METHODS
Plant Material
The barley mutant line albostrians (Hordeum vulgare cv ‘Haisa’) was grown for 11 d in soil at 23°C in a growth chamber with a photoperiod of 16 h (light intensity: 150 μE s−1 m−2). The progeny of homozygous albostrians plants consists of green, white, and striped seedlings in a ratio of ~1:1:8. The recessive nuclear albostrians allele is proposed to cause the loss of ribosomes in about half of the plastids early in development. Consequently, green tissue contains normally developed green chloroplasts, while white tissue contains ribosome-deficient plastids, and both types of tissue have identical genotypes (Hagemann and Scholz, 1962; Börner et al., 1976; Hess et al., 1993). The first leaves from completely green and white seedlings were harvested and used directly for plastid isolation.
Plastid Isolation
Plastids were isolated as previously described (Zubo et al., 2008). Briefly, 10 μg of leaf material were homogenized in 90 mL of homogenization buffer (0.33 M sorbitol, 50 mM Tricine, pH 8.0, 2 mM EDTA, and 5 mM β-mercaptoethanol). The homogenate was filtered through two layers of Miracloth (Calbiochem-Behring) and centrifuged at 4000 rpm (green) or 10,000 rpm (white) for 10 min. The pellet was resuspended in 1.5 mL of homogenization buffer and fractionated in a 30%/70% (green) or 10%/20%/70% (white) discontinuous Percoll gradient (GE Healthcare) by centrifugation at 6000 rpm (green) or 12,000 rpm (white) for 30 min. Intact chloroplasts from green leaves were collected at the interphase between 30 and 70% of Percoll. Plastids from white leaves were collected at the interphase between 20 and 70% (intact plastids) and 10 and 20% (semi-intact), and the mixture of intact and semi-intact plastids was used in the subsequent steps. All procedures were performed at 4°C. Plastids were washed two times in homogenization buffer, pelleted, and stored at −80°C.
RNA Extraction
Total RNA was extracted from isolated plastids using TRIzol (Invitrogen) following the manufacturer’s protocol. The RNA concentration was determined by optical density with a spectrophotometer at 260 nm (Nanodrop), and the integrity of rRNA bands was verified by electrophoresis on 1% denaturating agarose gels containing 1.7 M formaldehyde. The RNA was stored at −80°C.
dRNA-seq, Read Mapping, and Data Visualization
Depletion of processed RNAs was performed as previously described (Sharma et al., 2010). Briefly, genomic DNA contamination was removed from total RNA from green and white plastids using gDNA Wipeout buffer (Qiagen). For depletion of processed transcripts, 7 μg of RNA from each sample was treated with TEX (Epicentre; TER51020) or in buffer alone for 60 min at 30°C. One unit of TEX was used per 1 μg total chloroplast RNA. Following organic extraction (25:24:1 [v/v] phenol/chloroform/isoamyalcohol), RNA was recovered by overnight precipitation with 2.5 volumes of ethanol/0.1 M sodium acetate, pH 6.5. RNA was further treated with 1 unit tobacco acid pyrophosphatase (TAP; Epicentre; T19100) for 1 h at 37°C to generate 5′-monophosphates for linker ligation and again purified by organic extraction and precipitation as described above. cDNA library preparation and 454 pyrosequencing were performed as previously described (Berezikov et al., 2006) but omitting size fractionation. Sequencing was performed on Roche 454 FLX machines at the Max Planck Institute for Molecular Genetics (Berlin, Germany).
For mapping of cDNAs, 5′-linker and poly(A)-tail clipped reads of at least 18 nucleotides were aligned to the barley chloroplast genome (NC_008590) using WU-BLAST 2.0 (http://blast.wustl.edu/) with the following parameters: −B = 1; −V = 1; −m = 1; −n = −3; −Q = 3; −R = 3; −gspmax = 1; −hspmax = 1; −mformat = 2; and −e = 0.0001. For each library, graphs representing the number of mapped reads per nucleotide were calculated and visualized using the Integrated Genome Browser version 6.1 software from Affymetrix as previously described (Sittka et al., 2008). The graphs were normalized to the total number of mapped reads in each library, and the y axis indicates per mill mapped reads at a given position.
Annotation and Classification of TSSs
A 5′ end was considered a TSS when it was both represented by at least two cDNA reads starting at the same genomic position (nucleotide) and found enriched after the TEX treatment. A 5′ end represented by one cDNA was considered only as a gTSS when it extended into an annotated gene and a consensus promoter was predicted upstream. A 5′ end represented by less cDNAs in (+) than (−) green libraries (see Supplemental Data Set 3A online; column: Not enriched) was still accepted as a TSS if (1) it agreed with already published data, (2) was verified as a TSS by 5′-RACE (see Supplemental Data Set 2 online), or (3) it was enriched after TEX treatment in white libraries. We accepted TSS of trn genes mapped to +1 relative to the gene start only if they were found to be at least twofold enriched in (+) versus (−) libraries. Stepwise accumulation of cDNAs enriched in our (+) libraries with similar 3′ ends mapped shortly downstream of annotated ORFs were regarded as signals corresponding to 3′ UTRs. Several enriched 5′ ends mapped within five consecutive nucleotides were considered a single TSS and denoted by the genomic position of the most abundant of the 5′ ends in (+) libraries. The less abundant 5′ ends are listed in the Comments section of Supplemental Data Set 3 online.
TSSs were grouped into four categories based on their location with respect to annotated genes in the barley plastome (Figure 2). The categories were assigned as previously described (Sharma et al., 2010) with the following modifications. We increased the distance of gTSSs to the annotated genes from 500 to 750 nucleotides since chloroplast genes can be transcribed by both PEP and NEP from promoters located far upstream (Liere and Börner, 2007). In barley, the most distant experimentally verified gene initiation site is PpsbD-711 (Berends Sexton et al., 1990; Sexton et al., 1990), which was also mapped here as a TSS by dRNA-seq (see Supplemental Table 1 online). gTSSs with cDNA reads that did not reach into a downstream gene were referred to as “disconnected” (see Comments section of Supplemental Data Set 3 online). Since we detected a big average length of UTRs in green and white plastids, the distance of an aTSS to the upstream/downstream annotated gene on the opposite strand was increased from 100 to 150 nucleotides. An aTSS mapped more than 150 nucleotides downstream of an annotated gene was still considered an aTSS if it was represented by cDNAs overlapping with the 3′ UTR of the gene.
Secondary Structure Prediction of the 5′ Regions of Primary Transcripts
Minimum free energy secondary structures of the 5′ regions of the 176 and 244 primary transcripts mapped in green and white plastids, respectively, were predicted using RNAfold (part of the Vienna RNA Package version 1.8.5; Zuker and Stiegler, 1981; McCaskill, 1990; Hofacker et al., 1994; Hofacker and Stadler, 2006; Bompfünewerer et al., 2008) using the default parameters and the “-p” flag to retrieve the partition function and base pairing probability matrix. The analysis was performed within the first 50 and 100 nucleotides of the 5′ region. Based on the minimum free energy structures, mountain plot values representing the number of enclosing nucleotides per nucleotide position were calculated using a small Python script (http://www.python.org/). The mountain plot distributions were visualized as box plots using R (http://www.r-project.org/).
Promoter Analysis
The −1- to −25-, −26- to −50-, −1- to −10-, and −50- to +25-nucleotide regions of 176 and 244 TSSs mapped by dRNA-seq (see Supplemental Data Sets 2A and 2B online) in green and white plastids, respectively, were screened for common motifs with lengths of 3 to 9 nucleotides using MEME (Bailey et al., 2009) version 4.7.0 with default cutoff values. MEME outputs were corrected for the expected distances of the detected motifs with respect to +1 (Liere and Börner, 2007). The occurrence of detected MEME motifs was further examined using the Motif Alignment and Search Tool (MAST; part of the MEME suite). A Pearson's χ2 test with Yates' continuity correction of MAST scores was used to determine if the motifs were significantly overrepresented in the analyzed sequences of green or white plastids (P value < 0.05 = significant). The manual search for PEP promoter motifs was performed by enforcing 50% similarity (three out of six) to all of the nucleotides and 60% similarity (two out of three) to the overrepresented ones in the published consensus −10 (TAtaaT) and −35 (TTGaca; Liere and Börner, 2007). In case of the −10 box, only hexamers with at least one adenine residue were considered. The distance of the −10 element to the TSSs was restricted from 3 to 9 nucleotides, since these were the extremes found in already published data on plastid promoters (Sun et al., 1989; Swiatecka-Hagenbruch et al., 2007). A −35 element was expected 15 to 21 nucleotides upstream of the −10 box (Harley and Reynolds, 1987). The upstream sequences of white TSSs in which no motif was detected by MEME were manually screened for the presence of a YRTa motif 2 to 5 nucleotides upstream of the TSSs. All sequence logos were generated using WebLogo (http://weblogo.berkeley.edu/; Crooks et al., 2004).
Gene Count and Operon Annotation of the Barley Chloroplast Genome
There are 113 unique genes on the barley chloroplast genome, 78 of which are protein coding and 37 of which code for tRNAs or rRNAs (Saski et al., 2007). Maturation of rps12 mRNA involves trans-splicing of 5′-rps12 and 3′-rps12 (Hildebrand et al., 1988), and the respective genes were therefore considered as two separate genes. Genes from inverted repeat regions were counted only once. Two trn genes are annotated in the region from 15209 to 15338 on the barley chloroplast genome (NC_008590): trnM-CAU (15209-15267) and trnT-GGU (15275-15338) (NC_008590). However, we detected cDNAs corresponding only to trnT-GGU, mapping to position 15203 to 15274. Moreover, trnK-UUU was also reannotated: exon 1, 4460 to 4425; exon 2, 1945 to 1910. The absence of trnM-CAU and the new annotation of trnT-GGU and trnK-UUU were supported by the tRNAscan-SE program (http://lowelab.ucsc.edu/tRNAscan-SE/; Lowe and Eddy, 1997). Gene content and gene order of the barley chloroplast genome were found to be identical to those of rice (Oryza sativa; Saski et al., 2007). According to experimental data, 17 polycistronic and 22 monocistronic transcripts were detected in rice (Kanno and Hirai, 1993). Here, we accept the same organization for barley chloroplast genes with the following modifications: (1) trnE-Y was regarded as an operon, since the corresponding polycistronic transcript was detected in our analysis; (2) rpoC1-C2 bicistronic transcript was expanded to rpoB-rpoC1-rpoC2 (Hudson et al., 1988); (3) clpP- rps12 5′ operon was corrected to clpP- rps12 5′- rpl20 (Hübschmann et al., 1996); (3) trnK exon 1-matK-trnK exon 2 was regarded as a polycistronic transcript; (4) psbM, psbN, and ndhF were considered to be monocistronic transcripts as previously shown (Kawaguchi et al., 1992; Casano et al., 2001); (5) trnG-trnfM was considered to be a polycistronic transcript (Oliver and Poulsen, 1984); (6) psaI, rpl23 (HvsvCp031), trnV-GAC, and trnS-UGA failed to be detected in rice as parts of any of the nearby transcription units and are therefore assumed to be transcribed monocistronically. In summary, we assume that 89 barley plastid genes are grouped into 20 operons, while 24 genes are transcribed as monocistronic RNAs (see Supplemental Data Set 4 online).
5′-RACE, 3′-RACE, and cRT-PCR
The 5′ ends were identified by 5′-RACE analysis as previously described (Kühn et al., 2005). Briefly, 1 μg of total RNA from green and white plastids was treated with TAP (Epicentre) or in buffer alone for 60 min at 37°C. Following organic extraction (25:24:1 [v/v/v] phenol/chloroform/isoamyalcohol), RNA was recovered by overnight precipitation with 3 volumes of ethanol/3 M sodium acetate, pH 5.2. RNA was then ligated to the 5′ RNA linker (5R_L; see Supplemental Data Set 6 online) and again purified by organic extraction and precipitation as described above. RT was performed with up to five target specific reverse primers and followed by another step of organic extraction and precipitation. Target- and linker-specific primers were used to amplify the products of the RT reaction. Nested PCRs were performed. PCR products were analyzed on gels, and products of interest were excised, purified, and ligated into the pGEM-T vector (Promega). Ligation products were transformed into Escherichia coli TOP10 (Invitrogen). Bacterial clones containing the plasmid inserts were subjected to DNA sequencing with primer M13_rev (vector-specific primer). The templates for the sequencing reactions were prepared using the TempliPhi500 amplification kit (GE Healthcare) following the manufacturer’s protocol.
The 3′ ends were verified by 3′-RACE using the abundant chloroplast transcript of rrn16 (monophosphorylated) as 3′ linker. One microgram of total RNA from green and white plastids was self-ligated for 60 min at 37°C using 40 units of T4 RNA ligase (Epicentre) in the presence of 1 mM ATP and 40 units of RNase inhibitor (Fermentas) in the appropriate buffer. Control reactions were set up without adding ligase. Following organic extraction (25:24:1 [v/v/v] phenol/chloroform/isoamyalcohol), RNA was recovered by overnight precipitation with 3 volumes of ethanol/3 M sodium acetate, pH 5.2. RNA was then reverse-transcribed using rrn16-specific primer (3R_1; see Supplemental Data Set 6 online) and SuperScript III (Invitrogen) according to the manufacturer’s protocol. Following organic extraction, cDNA was recovered as described above. Target (forward) and rrn16 (reverse) specific primers were used for amplification of the products of the RT reaction. Annealing steps were performed at 55°C in the first PCR and 58°C in the subsequent nested PCR. PCR products were further handled as described above.
cRT-PCR was performed as previously described (Pfalz et al., 2009).
Oligonucleotides
A list of DNA and RNA oligonucleotides used for 5′-RACE, 3′-RACE, and cRT-PCR is provided in Supplemental Data Set 6 online.
Data Files
Graph files of blasted sequence reads equal to/bigger than 18 nucleotides and TSS and processing sites annotations in G and W libraries (.gff files) are available at http://www2.hu-berlin.de/biologie/genetics/Graph%20files.zip.
Accession Number
The dRNA-seq data from this article have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (Edgar et al., 2002) under accession number GSE33773 (http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSE33773).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Experimental Setup and Overview of Sequenced and Mapped Reads.
Supplemental Figure 2. Mapped Reads of Green (G) and White (W) dRNA-seq Libraries.
Supplemental Figure 3. Detection of 3′ Terminal Hairpin RNAs in TEX-Treated Samples.
Supplemental Figure 4. Prediction of Stable Structure Formation at the 5′ Ends of Primary Transcripts.
Supplemental Figure 5. Operon and TSS Map of the Barley Chloroplast Genome.
Supplemental Table 1. Comparison of TSSs Determined by dRNA-seq with Previously Mapped Primary Ends.
Supplemental Table 2. Potential mRNA 3′ Termini Revealed by Hairpin RNAs Resistant to TEX Treatment.
Supplemental Table 3. Identical TSSs in G and W dRNA-seq Libraries.
Supplemental Data Set 1. Processed 5′ Ends Revealed by dRNA-seq.
Supplemental Data Set 2. List of TSSs Revealed by dRNA-seq of Barley Plastids.
Supplemental Data Set 3. Verification of TSSs by 5′-RACE Analysis.
Supplemental Data Set 4. Operon Map of the Barley Chloroplast Genome.
Supplemental Data Set 5. ncRNA Candidates in Mature Barley Chloroplasts.
Supplemental Data Set 6. Oligonucleotides Used in 5′-RACE, 3′-RACE, and cRT-PCR.
Supplemental References 1. References for Supplemental Figures 3 and 5, Supplemental Tables 1 and 2, and Supplemental Data Set 1.
Acknowledgments
We thank Alice Barkan (University of Oregon, Eugene) for critical reading of a previous version of this manuscript. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (SFB 429) to K.L. and T.B. P.Z. is a fellow of the Helmholtz Graduate School “Molecular Cell Biology” at the Max Delbrück Center for Molecular Medicine and Humboldt University, Berlin, Germany.
AUTHOR CONTRIBUTIONS
T.B. designed the research. P.Z., C.M.S. and K.U.F. performed research. P.Z., C.M.S., K.U.F., K.L., J.V., and T.B. analyzed the data. P.Z., C.M.S., K.U.F., K.L., and T.B. wrote the article.
References
- Allison L.A., Simon L.D., Maliga P. (1996). Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J. 15: 2802–2809 [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. (2009). MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 37(Web Server issue): W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (2011). Expression of plastid genes: Organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 155: 1520–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A., Walker M., Nolasco M., Johnson D. (1994). A nuclear mutation in maize blocks the processing and translation of several chloroplast mRNAs and provides evidence for the differential translation of alternative mRNA forms. EMBO J. 13: 3170–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berends Sexton T., Jones J.T., Mullet J.E. (1990). Sequence and transcriptional analysis of the barley ctDNA region upstream of psbD-psbC encoding trnK(UUU), rps16, trnQ(UUG), psbK, psbI, and trnS(GCU). Curr. Genet. 17: 445–454 [DOI] [PubMed] [Google Scholar]
- Berezikov E., Thuemmler F., van Laake L.W., Kondova I., Bontrop R., Cuppen E., Plasterk R.H. (2006). Diversity of microRNAs in human and chimpanzee brain. Nat. Genet. 38: 1375–1377 [DOI] [PubMed] [Google Scholar]
- Bompfünewerer A.F., Backofen R., Bernhart S.H., Hertel J., Hofacker I.L., Stadler P.F., Will S. (2008). Variations on RNA folding and alignment: Lessons from Benasque. J. Math. Biol. 56: 129–144 [DOI] [PubMed] [Google Scholar]
- Börner T., Schumann B., Hagemann R. (1976). Biochemical studies on a plastid ribosome deficient mutant of Hordeum vulgare. Genetics and Biogenesis of Chloroplasts and Mitochondria, Bücher T., Neupert W., Sebald W., Werner S., (Amsterdam: Elsevier/North Holland; ), pp. 41–48 [Google Scholar]
- Boyer S.K., Mullet J.E. (1988). Sequence and transcript map of barley chloroplast psbA gene. Nucleic Acids Res. 16: 8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon A.B., Harris F.M., Stern D.B. (2004). Analysis of developing maize plastids reveals two mRNA stability classes correlating with RNA polymerase type. EMBO Rep. 5: 801–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casano L.M., Martín M., Sabater B. (2001). Hydrogen peroxide mediates the induction of chloroplastic Ndh complex under photooxidative stress in barley. Plant Physiol. 125: 1450–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.C., Stern D.B. (1991). Specific binding of chloroplast proteins in vitro to the 3′ untranslated region of spinach chloroplast petD mRNA. Mol. Cell. Biol. 11: 4380–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.S., Lin C.H., Chen L.J. (1997). Transcription and processing of the gene for spinach chloroplast threonine tRNA in a homologous in vitro system. Biochem. Biophys. Res. Commun. 233: 380–385 [DOI] [PubMed] [Google Scholar]
- Courtois F., Merendino L., Demarsy E., Mache R., Lerbs-Mache S. (2007). Phage-type RNA polymerase RPOTmp transcribes the rrn operon from the PC promoter at early developmental stages in Arabidopsis. Plant Physiol. 145: 712–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. (2004). WebLogo: A sequence logo generator. Genome Res. 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deana A., Celesnik H., Belasco J.G. (2008). The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature 451: 355–358 [DOI] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A.E. (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuel C., von Groll U., Müller M., Börner T., Weihe A. (2006). Development- and tissue-specific expression of the RpoT gene family of Arabidopsis encoding mitochondrial and plastid RNA polymerases. Planta 223: 998–1009 [DOI] [PubMed] [Google Scholar]
- Emanuel C., Weihe A., Graner A., Hess W.R., Börner T. (2004). Chloroplast development affects expression of phage-type RNA polymerases in barley leaves. Plant J. 38: 460–47215086795 [Google Scholar]
- Felder S., Meierhoff K., Sane A.P., Meurer J., Driemel C., Plücken H., Klaff P., Stein B., Bechtold N., Westhoff P. (2001). The nucleus-encoded HCF107 gene of Arabidopsis provides a link between intercistronic RNA processing and the accumulation of translation-competent psbH transcripts in chloroplasts. Plant Cell 13: 2127–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatenby A.A., Castleton J.A., Saul M.W. (1981). Expression in E. coli of maize and wheat chloroplast genes for large subunit of ribulose bisphosphate carboxylase. Nature 291: 117–121 [DOI] [PubMed] [Google Scholar]
- Georg J., Hess W.R. (2011). Regulatory RNAs in cyanobacteria: Developmental decisions, stress responses and a plethora of chromosomally encoded cis-antisense RNAs. Biol. Chem. 392: 291–297 [DOI] [PubMed] [Google Scholar]
- Georg J., Honsel A., Voss B., Rennenberg H., Hess W.R. (2010). A long antisense RNA in plant chloroplasts. New Phytol. 186: 615–622 [DOI] [PubMed] [Google Scholar]
- Georg J., Voss B., Scholz I., Mitschke J., Wilde A., Hess W.R. (2009). Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol. Syst. Biol. 5: 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman S.R., Sharp J.S., Vvedenskaya I.O., Livny J., Dove S.L., Nickels B.E. (2011). NanoRNAs prime transcription initiation in vivo. Mol. Cell 42: 817–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruissem W., Elsner-Menzel C., Latshaw S., Narita J.O., Schaffer M.A., Zurawski G. (1986). A subpopulation of spinach chloroplast tRNA genes does not require upstream promoter elements for transcription. Nucleic Acids Res. 14: 7541–7556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruissem W., Zurawski G. (1985). Identification and mutational analysis of the promoter for a spinach chloroplast transfer RNA gene. EMBO J. 4: 1637–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann R., Scholz F. (1962). A case of gene induced mutations of the plasmotype in barley. Theor. Appl. Genet. 32: 50–59 [Google Scholar]
- Hajdukiewicz P.T., Allison L.A., Maliga P. (1997). The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 16: 4041–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao W., Palmer J.D. (2009). Fine-scale mergers of chloroplast and mitochondrial genes create functional, transcompartmentally chimeric mitochondrial genes. Proc. Natl. Acad. Sci. USA 106: 16728–16733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley C.B., Reynolds R.P. (1987). Analysis of E. coli promoter sequences. Nucleic Acids Res. 15: 2343–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedtke B., Börner T., Weihe A. (1997). Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science 277: 809–811 [DOI] [PubMed] [Google Scholar]
- Hedtke B., Börner T., Weihe A. (2000). One RNA polymerase serving two genomes. EMBO Rep. 1: 435–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegeman C.E., Halter C.P., Owens T.G., Hanson M.R. (2005). Expression of complementary RNA from chloroplast transgenes affects editing efficiency of transgene and endogenous chloroplast transcripts. Nucleic Acids Res. 33: 1454–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess W.R., Prombona A., Fieder B., Subramanian A.R., Börner T. (1993). Chloroplast rps15 and the rpoB/C1/C2 gene cluster are strongly transcribed in ribosome-deficient plastids: evidence for a functioning non-chloroplast-encoded RNA polymerase. EMBO J. 12: 563–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand M., Hallick R.B., Passavant C.W., Bourque D.P. (1988). Trans-splicing in chloroplasts: The rps 12 loci of Nicotiana tabacum. Proc. Natl. Acad. Sci. USA 85: 372–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker I.L., Fontana W., Stadler P.F., Bonhoeffer L.S., Tacker M., Schuster P. (1994). Fast folding and comparison of RNA secondary structures. Monatshefte f. Chemie 125: 167–188 [Google Scholar]
- Hofacker I.L., Stadler P.F. (2006). Memory efficient folding algorithms for circular RNA secondary structures. Bioinformatics 22: 1172–1176 [DOI] [PubMed] [Google Scholar]
- Hotto A.M., Huston Z.E., Stern D.B. (2010). Overexpression of a natural chloroplast-encoded antisense RNA in tobacco destabilizes 5S rRNA and retards plant growth. BMC Plant Biol. 10: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübschmann T., Börner T. (1998). Characterisation of transcript initiation sites in ribosome-deficient barley plastids. Plant Mol. Biol. 36: 493–496 [DOI] [PubMed] [Google Scholar]
- Hübschmann T., Hess W.R., Börner T. (1996). Impaired splicing of the rps12 transcript in ribosome-deficient plastids. Plant Mol. Biol. 30: 109–123 [DOI] [PubMed] [Google Scholar]
- Hudson G.S., Holton T.A., Whitfield P.R., Bottomley W. (1988). Spinach chloroplast rpoBC genes encode three subunits of the chloroplast RNA polymerase. J. Mol. Biol. 200: 639–654 [DOI] [PubMed] [Google Scholar]
- Jäger D., Sharma C.M., Thomsen J., Ehlers C., Vogel J., Schmitz R.A. (2009). Deep sequencing analysis of the Methanosarcina mazei Gö1 transcriptome in response to nitrogen availability. Proc. Natl. Acad. Sci. USA 106: 21878–21882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno A., Hirai A. (1993). A transcription map of the chloroplast genome from rice (Oryza sativa). Curr. Genet. 23: 166–174 [DOI] [PubMed] [Google Scholar]
- Kapoor S., Suzuki J.Y., Sugiura M. (1997). Identification and functional significance of a new class of non-consensus-type plastid promoters. Plant J. 11: 327–337 [DOI] [PubMed] [Google Scholar]
- Kawaguchi H., Fukuda I., Shiina T., Toyoshima Y. (1992). Dynamical behavior of psb gene transcripts in greening wheat seedlings. I. Time course of accumulation of the psbA through psbN gene transcripts during light-induced greening. Plant Mol. Biol. 20: 695–704 [DOI] [PubMed] [Google Scholar]
- Kleine T., Maier U.G., Leister D. (2009). DNA transfer from organelles to the nucleus: the idiosyncratic genetics of endosymbiosis. Annu. Rev. Plant Biol. 60: 115–138 [DOI] [PubMed] [Google Scholar]
- Kühn K., Weihe A., Börner T. (2005). Multiple promoters are a common feature of mitochondrial genes in Arabidopsis. Nucleic Acids Res. 33: 337–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legen J., Kemp S., Krause K., Profanter B., Herrmann R.G., Maier R.M. (2002). Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J. 31: 171–188 [DOI] [PubMed] [Google Scholar]
- Lerbs-Mache S. (2011). Function of plastid sigma factors in higher plants: Regulation of gene expression or just preservation of constitutive transcription? Plant Mol. Biol. 76: 235–249 [DOI] [PubMed] [Google Scholar]
- Liere K., Börner T. (2007). Transcription and transcriptional regulation in plastids. Cell and Molecular Biology of Plastids, Bock R., (Berlin/Heidelberg: Springer; ), pp. 121–174 [Google Scholar]
- Liere K., Link G. (1994). Structure and expression characteristics of the chloroplast DNA region containing the split gene for tRNA(Gly) (UCC) from mustard (Sinapis alba L.). Curr. Genet. 26: 557–563 [DOI] [PubMed] [Google Scholar]
- Liere K., Maliga P. (1999). In vitro characterization of the tobacco rpoB promoter reveals a core sequence motif conserved between phage-type plastid and plant mitochondrial promoters. EMBO J. 18: 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liere K., Weihe A., Börner T. (2011). The transcription machineries of plant mitochondria and chloroplasts: Composition, function, and regulation. J. Plant Physiol. 168: 1345–1360 [DOI] [PubMed] [Google Scholar]
- Liu B., Troxler R.F. (1996). Molecular characterization of a positively photoregulated nuclear gene for a chloroplast RNA polymerase sigma factor in Cyanidium caldarium. Proc. Natl. Acad. Sci. USA 93: 3313–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T.M., Eddy S.R. (1997). tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung B., Zemann A., Madej M.J., Schuelke M., Techritz S., Ruf S., Bock R., Hüttenhofer A. (2006). Identification of small non-coding RNAs from mitochondria and chloroplasts. Nucleic Acids Res. 34: 3842–3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubetsky V.A., Zverkov O.A., Rubanov L.I., Seliverstov A.V. (2011). Modeling RNA polymerase competition: The effect of σ-subunit knockout and heat shock on gene transcription level. Biol. Direct 6: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaskill J.S. (1990). The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers 29: 1105–1119 [DOI] [PubMed] [Google Scholar]
- Memon A.R., Meng B., Mullet J.E. (1996). RNA-binding proteins of 37/38 kDa bind specifically to the barley chloroplast psbA 3′-end untranslated RNA. Plant Mol. Biol. 30: 1195–1205 [DOI] [PubMed] [Google Scholar]
- Mitschke J., Georg J., Scholz I., Sharma C.M., Dienst D., Bantscheff J., Voss B., Steglich C., Wilde A., Vogel J., Hess W.R. (2011). An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc. Natl. Acad. Sci. USA 108: 2124–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus H., Link G. (1990). The chloroplast psbK operon from mustard (Sinapis alba L.): Multiple transcripts during seedling development and evidence for divergent overlapping transcription. Curr. Genet. 18: 377–383 [DOI] [PubMed] [Google Scholar]
- Nickelsen J., Link G. (1990). Nucleotide sequence of the mustard chloroplast genes trnH and rps19′. Nucleic Acids Res. 18: 1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y., Kikis E.A., Zimmer S.L., Komine Y., Stern D.B. (2004). Antisense transcript and RNA processing alterations suppress instability of polyadenylated mRNA in chlamydomonas chloroplasts. Plant Cell 16: 2849–2869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noutsos C., Richly E., Leister D. (2005). Generation and evolutionary fate of insertions of organelle DNA in the nuclear genomes of flowering plants. Genome Res. 15: 616–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T., Yoshimura K., Miyake H., Ishikawa K., Ito D., Tanabe N., Shigeoka S. (2008). Molecular characterization of organelle-type Nudix hydrolases in Arabidopsis. Plant Physiol. 148: 1412–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama K., et al. (1986). Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature 322: 572–574 [Google Scholar]
- Oliver R., Poulsen C. (1984). Structure of a heavily transcribed region of barley chloroplast DNA. Transfer RNA genes for serine (UGA), glycine (GCC, UCC), formyl-methionine and threonine (GGU). Carlsberg Res. Commun. 49: 647–673 [Google Scholar]
- Pfalz J., Bayraktar O.A., Prikryl J., Barkan A. (2009). Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 28: 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prikryl J., Rojas M., Schuster G., Barkan A. (2011). Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl. Acad. Sci. USA 108: 415–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards J., Liu Q., Pellegrini O., Celesnik H., Yao S., Bechhofer D.H., Condon C., Belasco J.G. (2011). An RNA pyrophosphohydrolase triggers 5′-exonucleolytic degradation of mRNA in Bacillus subtilis. Mol. Cell 43: 940–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saski C., Lee S.B., Fjellheim S., Guda C., Jansen R.K., Luo H., Tomkins J., Rognli O.A., Daniell H., Clarke J.L. (2007). Complete chloroplast genome sequences of Hordeum vulgare, Sorghum bicolor and Agrostis stolonifera, and comparative analyses with other grass genomes. Theor. Appl. Genet. 115: 571–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweer J., Loschelder H., Link G. (2006). A promoter switch that can rescue a plant sigma factor mutant. FEBS Lett. 580: 6617–6622 [DOI] [PubMed] [Google Scholar]
- Schweer J., Türkeri H., Kolpack A., Link G. (2010). Role and regulation of plastid sigma factors and their functional interactors during chloroplast transcription - Recent lessons from Arabidopsis thaliana. Eur. J. Cell Biol. 89: 940–946 [DOI] [PubMed] [Google Scholar]
- Sexton T.B., Christopher D.A., Mullet J.E. (1990). Light-induced switch in barley psbD-psbC promoter utilization: A novel mechanism regulating chloroplast gene expression. EMBO J. 9: 4485–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma C.M., Hoffmann S., Darfeuille F., Reignier J., Findeiss S., Sittka A., Chabas S., Reiche K., Hackermüller J., Reinhardt R., Stadler P.F., Vogel J. (2010). The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464: 250–255 [DOI] [PubMed] [Google Scholar]
- Sharwood R.E., Halpert M., Luro S., Schuster G., Stern D.B. (2011). Chloroplast RNase J compensates for inefficient transcription termination by removal of antisense RNA. RNA 17: 2165–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K., et al. (1986). The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO J. 5: 2043–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemenroth A., Wollgiehn R., Neumann D., Börner T. (1981). Synthesis of ribosomal RNA in ribosome-deficient plastids of the mutant “albostrians” of Hordeum vulgare L. Planta 153: 547–555 [DOI] [PubMed] [Google Scholar]
- Sijben-Müller G., Hallick R.B., Alt J., Westhoff P., Herrmann R.G. (1986). Spinach plastid genes coding for initiation factor IF-1, ribosomal protein S11 and RNA polymerase alpha-subunit. Nucleic Acids Res. 14: 1029–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittka A., Lucchini S., Papenfort K., Sharma C.M., Rolle K., Binnewies T.T., Hinton J.C., Vogel J. (2008). Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 4: e1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriraman P., Silhavy D., Maliga P. (1998). The phage-type PclpP-53 plastid promoter comprises sequences downstream of the transcription initiation site. Nucleic Acids Res. 26: 4874–4879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D.B., Goldschmidt-Clermont M., Hanson M.R. (2010). Chloroplast RNA metabolism. Annu. Rev. Plant Biol. 61: 125–155 [DOI] [PubMed] [Google Scholar]
- Strittmatter G., Gozdzicka-Jozefiak A., Kössel H. (1985). Identification of an rRNA operon promoter from Zea mays chloroplasts which excludes the proximal tRNAValGAC from the primary transcript. EMBO J. 4: 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun E., Wu B.W., Tewari K.K. (1989). In vitro analysis of the pea chloroplast 16S rRNA gene promoter. Mol. Cell. Biol. 9: 5650–5659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiatecka-Hagenbruch M., Emanuel C., Hedtke B., Liere K., Börner T. (2008). Impaired function of the phage-type RNA polymerase RpoTp in transcription of chloroplast genes is compensated by a second phage-type RNA polymerase. Nucleic Acids Res. 36: 785–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiatecka-Hagenbruch M., Liere K., Börner T. (2007). High diversity of plastidial promoters in Arabidopsis thaliana. Mol. Genet. Genomics 277: 725–734 [DOI] [PubMed] [Google Scholar]
- Tanaka K., Oikawa K., Ohta N., Kuroiwa H., Kuroiwa T., Takahashi H. (1996). Nuclear encoding of a chloroplast RNA polymerase sigma subunit in a red alga. Science 272: 1932–1935 [DOI] [PubMed] [Google Scholar]
- Tanaka K., Tozawa Y., Mochizuki N., Shinozaki K., Nagatani A., Wakasa K., Takahashi H. (1997). Characterization of three cDNA species encoding plastid RNA polymerase sigma factors in Arabidopsis thaliana: Evidence for the sigma factor heterogeneity in higher plant plastids. FEBS Lett. 413: 309–313 [DOI] [PubMed] [Google Scholar]
- Vera A., Hirose T., Sugiura M. (1996). A ribosomal protein gene (rpl32) from tobacco chloroplast DNA is transcribed from alternative promoters: similarities in promoter region organization in plastid housekeeping genes. Mol. Gen. Genet. 251: 518–525 [DOI] [PubMed] [Google Scholar]
- Walter M., Piepenburg K., Schöttler M.A., Petersen K., Kahlau S., Tiller N., Drechsel O., Weingartner M., Kudla J., Bock R. (2010). Knockout of the plastid RNase E leads to defective RNA processing and chloroplast ribosome deficiency. Plant J. 64: 851–863 [DOI] [PubMed] [Google Scholar]
- Westhoff P. (1985). Transcription of the gene encoding the 51 kd chlorophyll a-apoprotein of the photosystem II reaction centre from spinach. Mol. Gen. Genet. 201: 115–123 [Google Scholar]
- Zghidi W., Merendino L., Cottet A., Mache R., Lerbs-Mache S. (2007). Nucleus-encoded plastid sigma factor SIG3 transcribes specifically the psbN gene in plastids. Nucleic Acids Res. 35: 455–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zghidi-Abouzid O., Merendino L., Buhr F., Malik Ghulam M., Lerbs-Mache S. (2011). Characterization of plastid psbT sense and antisense RNAs. Nucleic Acids Res. 39: 5379–5387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelyazkova P., Hammani K., Rojas M., Voelker R., Vargas-Suárez M., Börner T., Barkan A. (December 8, 2011). Protein-mediated protection as the predominant mechanism for defining processed mRNA termini in land plant chloroplasts. Nucleic Acids Res. http://dx.doi.org/10.1093/nar/gkr1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R., Liere K., Börner T. (2007). From seedling to mature plant: Arabidopsis plastidial genome copy number, RNA accumulation and transcription are differentially regulated during leaf development. Plant J. 50: 710–722 [DOI] [PubMed] [Google Scholar]
- Zubo Y.O., Yamburenko M.V., Selivankina S.Y., Shakirova F.M., Avalbaev A.M., Kudryakova N.V., Zubkova N.K., Liere K., Kulaeva O.N., Kusnetsov V.V., Börner T. (2008). Cytokinin stimulates chloroplast transcription in detached barley leaves. Plant Physiol. 148: 1082–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. (1981). Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 9: 133–148 [DOI] [PMC free article] [PubMed] [Google Scholar]