This work shows that the rice blast fungus secretes a protein that can suppress plant defenses by affecting the way in which chitin, a component of fungal cell walls, is perceived by the rice plant.
Abstract
Plants use pattern recognition receptors to defend themselves from microbial pathogens. These receptors recognize pathogen-associated molecular patterns (PAMPs) and activate signaling pathways that lead to immunity. In rice (Oryza sativa), the chitin elicitor binding protein (CEBiP) recognizes chitin oligosaccharides released from the cell walls of fungal pathogens. Here, we show that the rice blast fungus Magnaporthe oryzae overcomes this first line of plant defense by secreting an effector protein, Secreted LysM Protein1 (Slp1), during invasion of new rice cells. We demonstrate that Slp1 accumulates at the interface between the fungal cell wall and the rice plasma membrane, can bind to chitin, and is able to suppress chitin-induced plant immune responses, including generation of reactive oxygen species and plant defense gene expression. Furthermore, we show that Slp1 competes with CEBiP for binding of chitin oligosaccharides. Slp1 is required by M. oryzae for full virulence and exerts a significant effect on tissue invasion and disease lesion expansion. By contrast, gene silencing of CEBiP in rice allows M. oryzae to cause rice blast disease in the absence of Slp1. We propose that Slp1 sequesters chitin oligosaccharides to prevent PAMP-triggered immunity in rice, thereby facilitating rapid spread of the fungus within host tissue.
INTRODUCTION
The filamentous fungus Magnaporthe oryzae is one of the most devastating plant pathogens, causing blast disease in a significant number of agronomically important crops, including rice (Oryza sativa), barley (Hordeum vulgare), and finger millet (Eleusine coracana) (Wilson and Talbot, 2009). To cause disease, infection structures called appressoria are required for penetration of the host plant (Talbot, 2003; Wilson and Talbot 2009). After penetration of the host surface, the fungal penetration peg differentiates to form a thin filamentous primary hypha and the fungus grows without causing disease symptoms. At this time, an intimate relationship between the host and pathogen is established, in which the host plasma membrane is not breached, but instead appears to become invaginated, thereby sealing the invading fungus in a host-derived plasma membrane, known as the extrainvasive hyphal membrane (EIHM) (Kankanala et al., 2007). Filamentous hyphae grow briefly within host cells before differentiating into bulbous secondary pseudohyphae, which propagate rapidly within the host cell (Kankanala et al., 2007). The fungus then moves into neighboring plant cells at pit field sites, potentially using plasmodesmata to traverse between rice cells (Kankanala et al., 2007). Rice blast disease symptoms only become visible following a prolonged biotrophic phase in which the fungus spreads extensively within rice tissue, suggesting that M. oryzae can evade host recognition and proliferate in living plant cells by active suppression of plant immunity.
During the early stages of infection, M. oryzae is believed to secrete effector proteins to suppress host defenses (Mosquera et al., 2009; Khang et al., 2010), although the precise function of rice blast effectors has not yet been determined. The best characterized M. oryzae effector, Avr-Pita, was first identified because it is recognized in rice cultivars carrying the Pi-ta resistance gene. The intracellular Pi-ta resistance gene product and Avr-Pita have been shown to interact directly (Jia et al., 2000), suggesting that Avr-Pita is secreted by the fungus and delivered across the host plasma membrane into rice cells. Avr-Pita is predicted to encode a metalloprotease, but its role in fungal virulence and the targets of its putative proteolytic activity have not yet been determined (Jia et al., 2000). Recent studies have confirmed that Avr-Pita is delivered into the cytoplasm of rice cells and have also led to the discovery of an infection structure, known as the biotrophic interfacial complex (BIC), which forms as a subapical bulbous structure at the periphery of invasive pseudohyphal cells (Mosquera et al., 2009; Khang et al., 2010). During biotrophic intracellular growth, this structure accumulates effector proteins by an unknown mechanism, and it has been proposed that BICs may be used to mediate the delivery of rice blast effectors into the host cytoplasm (Khang et al., 2010).
In this study, we set out to identify novel effectors secreted by the rice blast fungus. We were particularly interested in determining whether M. oryzae deploys effectors in the apoplast, the space between the fungal cell wall and the host plasma membrane. Secretion of apoplastic effectors is a common strategy of many extracellular fungal pathogens, but it is not clear whether intracellular colonizing fungi, such as M. oryzae, require extracellular effectors during tissue invasion (Mosquera et al., 2009; Jia et al., 2000).
The intercellular fungal pathogen Cladosporium fulvum, which causes leaf-mold disease of tomato (Solanum lycopersicum), colonizes the spaces between tomato spongy mesophyll cells and secretes several apoplastic effectors during colonization of tomato leaves (Thomma et al., 2005; van Esse et al., 2008). Many of these effectors also have Avr functions and are perceived by cognate Cf receptor gene products residing in the host plasma membrane (Wang et al., 2010). Effectors of C. fulvum are thought to be entirely apoplastic, which reflects the nature of pathogenic colonization. Interestingly, an effector known as Ecp6 was recently identified from C. fulvum that is secreted during infection (Bolton et al., 2008). Ecp6 contains LysM domains that have previously been implicated in carbohydrate binding and has been shown to bind chitin (de Jonge et al., 2010). Ecp6 may therefore suppress host recognition of chitin and pathogen-associated molecular pattern (PAMP)–triggered immunity through the scavenging of PAMP molecules (de Jonge et al., 2010). Although experiments have suggested a virulence function for this effector (Bolton et al., 2008), the cognate chitin elicitor receptor in tomato with which Ecp6 competes has yet to be identified.
In this report, we show that M. oryzae secretes a novel effector, which we have named Slp1, for Secreted LysM Protein 1. Intriguingly, although M. oryzae colonizes rice intracellularly, Slp1 shows strong similarity to C. fulvum Ecp6 and contains two LysM domains. Using live-cell imaging of rice tissue, we show that Slp1 specifically accumulates at the plant-fungal interface during the early stages of rice blast infections and that its delivery to this interface is vital for its biological function. We also demonstrate that Slp1 specifically binds chitin and is able to suppress chitin-triggered immunity in rice suspension cells, including the generation of reactive oxygen species (ROS). Slp1 competes for chitin binding with the rice pattern recognition receptor (PRR) chitin elicitor binding protein (CEBiP), which is required for chitin-triggered immunity in rice, acting in cooperation with the LysM receptor-like kinase Os-CERK1 (Shimizu et al., 2010). Finally, we show that Slp1 is important for rice blast disease and necessary for disease lesion expansion. When considered together, our results provide evidence that although the rice blast fungus invades and occupies plant cells, it must deploy an apoplastic effector to suppress PAMP-triggered immunity to facilitate its growth within rice tissue.
RESULTS
Slp1 Accumulates at the Plant-Fungal Interface during Biotrophic Growth
In this study, we set out to identify novel rice blast effector proteins secreted by invasive hyphae during plant infection. To visualize the host-pathogen interface directly, we first generated transgenic rice plants in which an LTi6B:green fluorescent protein (GFP) gene fusion (Kurup et al., 2005) was expressed, resulting in GFP becoming targeted to the rice plant plasma membrane. We found that the rice plasma membrane does invaginate around invasive hyphae within rice epidermal cells and becomes tightly apposed to the fungal cell wall, as shown in Figure 1A. It is clear, therefore, that the there is a close association between M. oryzae hyphal cell walls and the rice plasma membrane during plant infection. To identify potential effector-encoding genes involved in modulating the host–pathogen interaction, we identified genes encoding putatively secreted gene products that were upregulated during biotrophic growth compared with growth in axenic culture (Mosquera et al., 2009). One putative effector identified using these criteria was found to contain two putative LysM domains, which have previously been shown to bind carbohydrates in a number of proteins (Buist et al., 2008). We named this LysM domain–containing protein Slp1. Proteins containing LysM domains are ubiquitous in nature, and Slp1 shares significant homology with other predicted fungal LysM proteins (de Jonge and Thomma, 2009), including most notably the C. fulvum effector Ecp6, as shown in Supplemental Figures 1 and 2 online. Interestingly, the M. oryzae genome contains seven other LysM domain–containing proteins. One of these LysM proteins (gene ID MGG_03468), which we have called Slp2, was also found to show strong similarity to the C. fulvum Ecp6 protein, as shown in Supplemental Figure 2 online. However, we did not detect expression of SLP2 during plant infection (data not shown) and have not yet been able to find a clear phenotype for Δslp2 mutants. We have therefore focused our research effort on determining the biological role of Slp1 during biotrophic growth of M. oryzae.
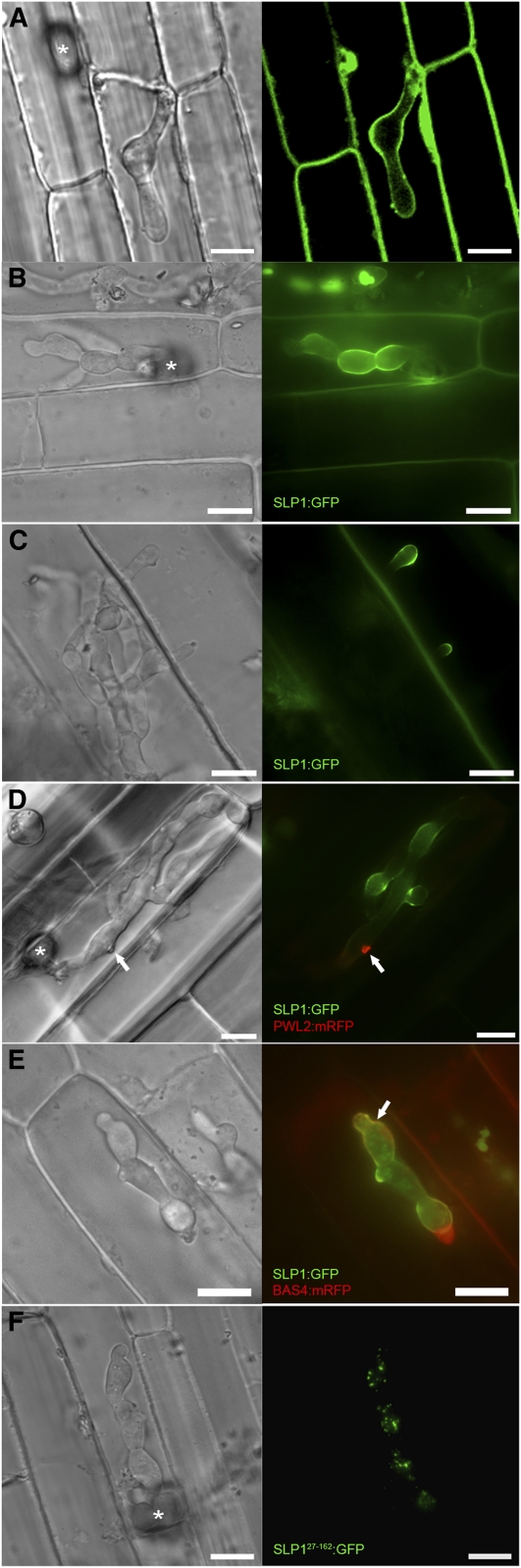
Figure 1.
Slp1 Accumulates at the Plant-Fungal Interface during Biotrophic Growth.
(A) Laser confocal micrograph of M. oryzae invasive hyphae-colonizing epidermal leaf cells of a transgenic line of rice expressing LTi6B:GFP. The rice cell plasma membrane becomes invaginated around the growing fungal hyphae.
(B) Cellular localization of Slp1:GFP in M. oryzae during biotrophic growth on epidermal rice cells at 24 HAI. Fluorescence was initially observed accumulating at the tips of invasive hyphae at the plant-fungal interface and was later found to surround invasive hyphae.
(C) At 36 HAI, Slp1:GFP fluorescence could be observed accumulating at the tips of filamentous hyphae-invading adjacent cells. At this time, fluorescence was no longer observed in initially infected host cells.
(D) Lack of colocalization between SLP1:GFP and PWL2:mRFP. A Guy11 M. oryzae transformant expressing both the SLP1:GFP and PWL2:mRFP constructs was used to visualize the cellular localization of Slp1 and the BIC-localized effector Pwl2 in planta. At 24 HAI, the Slp1-Gfp signal surrounded invasive hyphae, whereas Pwl2 accumulates at the BIC (white arrow).
(E) Partial colocalization of M. oryzae Slp1:GFP and Bas4:mRFP fusion proteins in the apoplastic space surrounding fungal invasive hyphae. White arrow indicates site of colocalization.
(F) Cellular localization of Slp27-162:GFP at 24 HAI on leaf sheath tissue. Slp27-162:GFP aggregates can be seen localizing within the cytoplasm of fungal invasive hyphae. White asterisk indicates the site of appressorium formation at the leaf surface.
Bars = 10 μm.
We hypothesized that Slp1 might act as an effector protein and decided to examine its localization during host tissue colonization (Mosquera et al., 2009; Khang et al., 2010). Putative apoplastic effectors secreted by intercellular fungal pathogens, such as C. fulvum, are invariably Cys rich (de Jonge et al., 2010), and as Slp1 contains six Cys residues, we further hypothesized that Slp1 might be secreted into the space between the fungal cell wall and the rice plasma membrane (see Supplemental Figure 2 online). To investigate the localization of Slp1, we engineered a strain of M. oryzae expressing a SLP1:GFP gene fusion under control of a native 2-kb promoter fragment. Live-cell imaging of infected rice leaf epidermis was performed to examine the cellular localization of Slp1 during fungal growth within rice cells. After the fungus penetrated the host cell, at ~24 to 28 h after inoculation (HAI), fluorescence could be observed to accumulate at the plant-fungal interface and was specifically observed to outline pseudohyphal fungal cells, as shown in Figure 1B. As the fungus moved into neighboring cells (at ~32 to 36 HAI), fluorescence was observed accumulating at the tips of invasive hyphae that were invading new host cells. At this time, fluorescence ceased to accumulate at the host-pathogen interface within the initially infected host cell (Figure 1C). At no stage was fluorescence observed within host cells nor could fluorescence be observed in other fungal structures, including conidia, germ tubes, or appressoria, as shown in Supplemental Figure 3 online. From these observations, we conclude that Slp1 is specifically expressed when the fungus is growing intracellularly within its host, a feature associated with putative rice blast effector proteins (Mosquera et al., 2009; Khang et al., 2010). Next, we wanted to examine whether the localization pattern of Slp1 differs from that of previously described rice blast effectors, which accumulate at BIC structures (Mosquera et al., 2009; Khang et al., 2010). We therefore engineered a strain of M. oryzae that simultaneously expressed a SLP1:GFP gene fusion and a Pathogenicity on Weeping Lovegrass2 (PWL2):monomeric red fluorescent protein (mRFP) gene fusion. Pwl2 is a previously characterized BIC-localized effector, known to be delivered into the cytoplasm of rice cells during plant infection by M. oryzae (Khang et al., 2010). We undertook live-cell imaging of infected rice epidermis, as shown in Figure 1D and Supplemental Movie 1 online. Interestingly, at >80% of infection sites observed between 24 and 32 HAI, colocalization between SLP1:GFP and PWL2:mRFP could not be observed (n > 100). Taken together, we conclude that Slp1 accumulates between the invaginated host plasma membrane and the fungal cell wall during initial invasion of rice cells and is therefore distinct from previously identified BIC-localized effectors.
Having established that Slp1 was not a BIC-localized effector protein, we were interested in trying to colocalize Slp1 with other rice blast effectors that appear to accumulate in the apoplast. One potential effector, presumed to be apoplastic in localization, is Bas4 (Mosquera et al., 2009; Khang et al., 2010). We therefore engineered an M. oryzae strain that simultaneously expresses SLP1:GFP and BAS4:mRFP. At 24 HAI, there did appear to be some colocalization between Slp1 and Bas4, as shown in Figure 1E. Although the two proteins appeared to colocalize, there were, however, significant areas where Slp1:GFP accumulated, but Bas4:mRFP did not. In view of our observation that Slp1 accumulates at the fungal-plant interface, we next investigated how the protein is delivered to the apoplast during biotrophic growth. SLP1 encodes a small secreted protein of 162 amino acids, with a predicted N-terminal signal peptide of 27 amino acids in length (based on SignalP 3.0 analysis). To test the significance of this secretion sequence, we engineered an M. oryzae strain in which the coding region of the first 27 amino acids of SLP1 was removed. A new start codon was introduced and the resulting coding region fused to GFP. Expression of the SLP127-162:GFP construct was driven by the native 2.0-kb SLP1 promoter fragment. Removal of the signal peptide prevented Slp1:GFP from reaching the tips of invasively growing hyphae, and Slp1 was no longer observed accumulating in the apoplastic space (Figure 1F). The resultant intracellular Slp127-162:GFP instead appeared to accumulate as aggregates in the fungal cytoplasm. Cellular mislocalization of SLP127-162:GFP is consistent with the hypothesis that Slp1 is an apoplastic effector, the secretion of which is dependent on a peptide sequence within the initial 27 amino acids.
Slp1 Is a Virulence Determinant in M. oryzae
To test the contribution of Slp1 to rice blast disease, a targeted gene deletion of SLP1 was performed in M. oryzae (see Supplemental Figure 4 online). Fungal spores of the resulting Δslp1 mutant and the isogenic wild-type Guy11 strain were harvested, adjusted to uniform concentrations, and applied to 21-d-old seedlings of the blast-susceptible rice cultivar CO-39 (Figure 2). Deletion of SLP1 significantly reduced the ability of M. oryzae to cause disease, and the symptoms of plants inoculated with Δslp1 spores were highly reduced when compared with plants infected with the wild-type Guy11 strain (Figure 2A). To quantify the reduction in virulence, both lesion density and lesion size were analyzed using image analysis software (ImageJ). The mean lesion size generated by the Δslp1 mutant was found to be significantly smaller than that of the Guy11 wild type (t test, P < 0.01) (Figure 2B). The mean lesion size for Guy11 was calculated to be 1.15 mm2 (±se 0.049, n > 100 lesions), while the mean lesion size of the Δslp1 mutant was calculated to be 0.31 mm2 (±se 0.025, n > 100). Additionally, the mean lesion density per unit area of the Δslp1 mutant (11.1 ± 5.7, n = 49) was found to be significantly lower than that of the wild type (40.7 ± 10.8, n = 28; t test, P < 0.01) (Figure 2C). Complementation analysis using the SLP1:GFP construct was performed, and reintroduction of the SLP1 gene was found to restore virulence to M. oryzae (see Supplemental Figure 5 online). Deletion of the Slp1 signal peptide prevented complementation of the Δslp1 mutant phenotype (see Supplemental Figure 6 online).
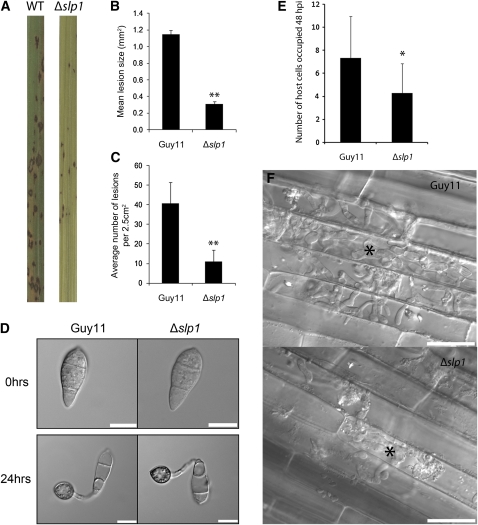
Figure 2.
SLP1 Is a Virulence Determinant in M. oryzae Required for Rice Tissue Invasion.
(A) Conidial suspensions of equal concentration (5 × 10−4 spores mL−1) from M. oryzae Guy11 (wild type [WT]) or Δslp1 mutants were used to inoculate 21-d-old seedlings of the blast susceptible rice cultivar CO-39. Disease symptoms were reduced on plants inoculated with Δslp1 mutants.
(B) Bar chart of mean lesion size of plants inoculated with Guy11 and the Δslp1 mutant. Mean lesion size was significantly reduced in plants inoculated with the Δslp1 mutant compared with the isogenic wild type (t test, P < 0.01). Error bars denote ± 1 se.
(C) Bar chart of mean lesion density of seedlings infected with Guy11 strain and the Δslp1 mutant per unit area. Mean lesion density was significantly reduced in Δslp1 mutant infections (t test, P < 0.01). Error bars denote 1 sd. Double asterisks in (B) and (C) denote P < 0.01 from two-tailed t test.
(D) Null Δslp1 mutants produce normal conidia and form appressoria in a time-dependent manner comparable to that of the wild-type Guy11 strain. Conidia of both Guy11 and Δslp1 mutant strains were harvested and set to a concentration of 5 × 10−4 spores mL−1. Spores were inoculated onto hydrophobic glass cover slips and incubated in a moist chamber at 26°C and examined by light microscopy. The morphology of conidia and appressoria was not altered in Δslp1 mutants. Bars = 10 μm.
(E) Bar chart showing the number of rice host cells occupied after 48 HAI with the Δslp1 mutant compared with Guy11. After 48 h, the number of host cells occupied by the fungus was recorded (n = 15 infection sites). At this time point, the number of host cells occupied by the Δslp1 mutant was found to be significantly lower than that of the wild-type Guy11 strain (two-tailed t test, P = 0.014). Asterisk denotes P < 0.05.
(F) Typical infection sites of rice leaf sheath inoculated with Δslp1 and Guy11, showing greater fungal proliferation and tissue invasion by the wild-type strain. Images were recorded 48 HAI. Asterisk marks the first infected host cell. Bar = 30 μm.
[See online article for color version of this figure.]
We also evaluated whether Δslp1 mutants were impaired in their ability to form functional infection structures or whether the virulence phenotype was simply a consequence of a reduction in fitness. We harvested spores of the Δslp1 mutant and wild-type strains and compared their ability to form appressoria on an inductive glass surface (Figure 2D). After 24 h, Δslp1 mutants were capable of forming mature appressoria in a manner identical to that of the wild-type M. oryzae strain. Vegetative growth rates and behavior in axenic culture were also identical to Guy11. From these observations, we conclude that the virulence phenotype of the Δslp1 mutant is associated with a reduced ability of the Δslp1 mutant to proliferate within host tissues, rather than a reduced capacity to make successful penetration structures. To test this idea, we examined and compared host tissues infected with a Δslp1 mutant compared with the isogenic Guy11. We initially counted the number of cells occupied by the fungus at 48 HAI and found that the number of host cells occupied by a Δslp1 mutant was significantly lower than the wild-type Guy11 strain (n = 15, two-tailed t test, P = 0.014) (Figure 2E). At 48 HAI, the mean number of host cells occupied by Δslp1 was found to be 4.29 cells (sd ± 2.5), while the mean number of cells occupied by Guy11 was 7.33 (sd ± 3.6). At 48 HAI, the Δslp1 mutant had only just started to colonize neighboring cells, while Guy11 had become well established at 48 HAI, with bulbous hyphae fully ramified in host tissues (Figure 2F). We conclude that Slp1 is necessary for efficient rice tissue invasion by M. oryzae to bring about rice blast disease.
M. oryzae Slp1 Is a Chitin Binding Protein
To define the biological function of Slp1, we cloned and overexpressed a SLP1 cDNA in Pichia pastoris (de Jonge et al., 2010). Recombinant Slp1 protein was isolated and purified. As Slp1 contains two putative LysM domains, we were initially interested to see whether the protein was capable of binding to specific polysaccharides. After incubating purified Slp1 protein with insoluble cell wall polysaccharides, we observed that Slp1 specifically coprecipitated with insoluble crab shell chitin and chitin beads and was detected in the insoluble pellet fraction following affinity precipitation (Figure 3). Slp1 did not, however, precipitate with any other tested cell wall polysaccharides, including chitosan (deacetylated chitin) and the plant cell wall polysaccharides cellulose and xylan, as evidenced by Slp1 remaining in the supernatant fraction after affinity precipitation (Figure 3A). Interestingly, not only did Slp1 appear to bind specifically to chitin and not to other polysaccharides, several bands were evident in both the pellet and supernatant fractions, suggesting that Slp1 is likely to be glycosylated or potentially forms oligomers. To ensure that these higher molecular weight protein bands were not contaminants from the protein overexpression system, mass spectrometry was performed on the gel fragments after in-gel trypsin digestion. Slp1 was detected in all of these experiments, confirming that the higher molecular weight protein bands were not due to expression artifacts and suggesting that Slp1 is likely to show abnormal electrophoretic mobility due to being a glycoprotein as reported for other LysM proteins (Kaku et al., 2006) or potentially to form multimers. We were interested in determining whether Slp1 had the capacity to form multimers based on a protein–protein interaction. We therefore performed high-stringency yeast two-hybrid analysis in which an SLP1 cDNA was simultaneously cloned into bait and prey vectors of the Matchmaker GAL4 two-hybrid system (Clontech). Using this system, we were able to detect a strong potential interaction between Slp1 monomers (see Supplemental Figure 7 online). This preliminary observation is consistent with the Slp1 effector having the capacity to form protein aggregates.
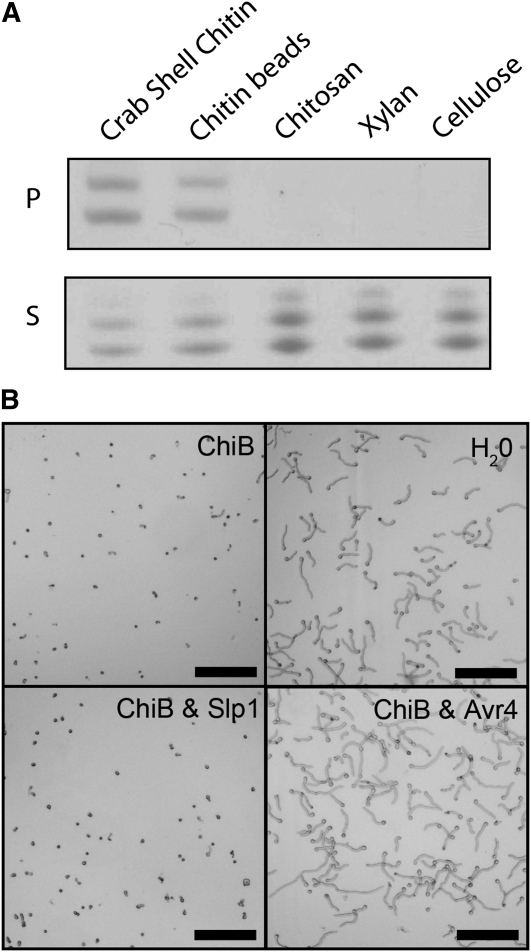
Figure 3.
Slp1 Binds Specifically to Chitin Oligosaccharides.
(A) Affinity precipitation experiments showing that Slp1 coprecipitates with insoluble crab shell chitin and chitin beads and was detected in the insoluble pellet fraction (P) following SDS-PAGE and Coomassie blue staining. Slp1 did not precipitate with other insoluble polysaccharides, including chitosan (deacetylated chitin) or the plant cell wall polysaccharides xylan and cellulose. Instead, Slp1 remained in the nonprecipitated supernatant fraction (S) after incubation with these polysaccharides.
(B) Slp1 does not provide protection from hyphal tip hydrolysis by chitinase enzymes. Micrographs of T. viride spores taken 24 h after addition of either water or crude extract of tomato leaves containing intracellular basic chitinases (ChiB). Pretreatment with 10 μM Avr4 prevented hydrolysis of T. viride hyphal tips by basic chitinase (Avr4 and ChiB), whereas pretreatment with 10 μM Slp1 (Slp1 and ChiB) did not. Bars = 10 μm.
As Slp1 appeared to be capable of binding chitin, we reasoned that Slp1 might bind to chitin in the fungal cell walls of invasive hyphae, thereby shielding hyphal tips from hydrolysis by plant-derived chitinases. Initial experiments demonstrated that when M. oryzae was grown in culture, it was not susceptible to disruption by crude extract of chitinase (data not shown). In many fungi, cell wall–incorporated proteins within a glucan matrix can reduce the accessibility of chitinase enzymes (Joosten et al., 1995; van den Burg et al., 2006). To address this, we used Trichoderma viride as a model species, which has been used widely to test this hypothesis (van den Burg et al., 2006; van Esse et al., 2007; de Jonge et al., 2010). We incubated T. viride spores with a crude extract of tomato leaves containing intracellular basic chitinases, in the presence or absence of the purified Slp1 protein, as shown in Figure 3B. Unlike the C. fulvum effector Avr4, which has previously been shown to protect hyphae from the hydrolysis of chitinases (van den Burg et al., 2006; van Esse et al., 2007), Slp1 was unable to protect T. viride spores from hydrolysis by chitinase enzymes. Previously, concentrations as low as 10 μM of Avr4 have been shown to provide hyphal tip protection from chitinase enzymes (Kaku et al., 2006; de Jonge and Thomma, 2009). In our experiments, even at concentrations of up to 100 μM of Slp1, protection from chitinases was not observed. Slp1 therefore shares characteristics with Ecp6, which also fails to protect fungal hyphae against hydrolysis by chitinases (de Jonge et al., 2010). Consequently, Slp1 is not likely to be involved in the protection of fungal hyphae from chitinases.
Slp1 Is a Competitive Inhibitor of the PRR Protein CEBiP and Suppresses Chitin-Induced Immune Responses in Rice Cells
During plant infection, the release of chitin oligosaccharides from hyphal tips can help to facilitate pathogen recognition by host plant cells (Kaku et al., 2006; van den Burg et al., 2006). Given the ability of Slp1 to bind chitin (Figure 3) and accumulate at the plant-fungal interface (Figure 1), we hypothesized that Slp1 might be involved in disrupting chitin-induced perception of the fungus in rice plants. To investigate whether Slp1 was capable of suppressing chitin-triggered immunity in rice cells, we tested whether Slp1 could suppress the chitin-induced oxidative burst (Yamaguchi et al., 2005). In the presence of nanomolar concentrations of an oligomer of N-acetyl glucosamine [(GlcNAc)8], rice suspension cells release ROS, which can be measured using luminol-dependent chemiluminescence. Upon incubating rice suspension cells with 1 nM (GlcNAc)8, chemiluminescence was detected after 20 min, as shown in Figure 4A. However, this oxidative burst was suppressed in the presence of a 10-fold molar excess of Slp1 (10 nM). Furthermore, we noticed that after 120 min, suppression of the oxidative burst was still observed in the presence of 10 nM Slp1, although suppression was much greater in the presence of a 100-fold molar excess of Slp1 (Figure 4A). This latter concentration of Slp1 was capable of suppressing the chitin-induced oxidative burst across all time points examined.
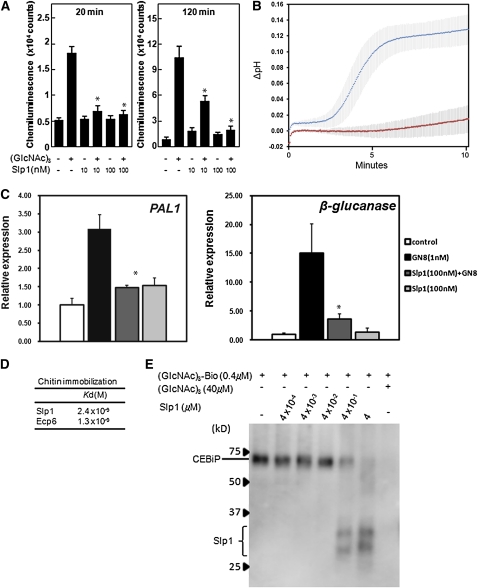
Figure 4.
Slp1 Is a Competitive Inhibitor of the PRR Protein CEBiP and Suppresses Chitin-Induced Immune Responses in Rice Cells.
(A) Slp1 inhibits the chitin-induced oxidative burst in rice suspension cells. Production of ROS 20 or 120 min after induction with 1 nM (GlcNAc)8 was determined in the absence or presence of Slp1 (10 and 100 nM). The experiment was performed twice with similar results. Mean with se of three replicate experiments is shown, and asterisks indicate significant differences (P < 0.01) when compared with the 1 nM (GlcNAc)8 treatment.
(B) Medium alkalinization of tomato cell suspensions is suppressed in the presence of Slp1. After treatment with 1 nM chitin oligosaccharides [(GlcNAc)6] (top line), the pH of the tomato cell suspensions increases after ~2 min. Upon incubation with 1 nM (GlcNAc)6 and a 10-fold molar excess of Slp1 (10 nM) (bottom line), medium alkalinization was inhibited. Error bars represent ± 1 sd of three independent replicate experiments.
(C) Expression of rice defense genes PAL1 and β-glucanase induced by GlcNAc is suppressed in the presence of Slp1. The bars display the relative transcript level of the chitin-responsive genes normalized to the constitutively expressed ubiquitin gene. The mean with se of two replicate experiments is shown, and asterisks indicate significant differences (P < 0.05) when compared with the 1 mM (GlcNAc)8 treatment.
(D) Affinities of fungal LysM effectors for (GlcNAc)8 determined by SPR analysis. Affinities between Ecp6 and Slp1 for (GlcNAc)8 were measured using the (GlcNAc)8-immobilized mode.
(E) Protein gel blot analysis using an antibiotin antibody showing affinity labeling of a microsomal membrane preparation (rice MF) from suspension-cultured rice cells containing the PRR CEBiP, with biotinylated (GlcNAc)8 [(GlcNAc)8-Bio], in the presence or absence of Slp1 and nonbiotinylated (GlcNAc)8. The experiment was performed twice with similar results.
[See online article for color version of this figure.]
To determine whether the ability of Slp1 to suppress chitin-triggered immunity was of wider significance, we also measured immunity responses in tomato suspension cells. In the presence of nanomolar concentrations of chitin oligosaccharides [(GlcNAc)6], plant cell suspensions have previously been shown to react by medium alkalinization (Felix et al., 1993). To test whether Slp1 might play a role in suppressing chitin-based responses in other plant species, we tested whether Slp1 could suppress a chitin-induced pH shift in tomato cell suspensions. We observed that in the presence of a 10-fold molar excess of Slp1 (10 nM), medium alkalinization of tomato suspensions cells was inhibited (Figure 4B). We therefore conclude that Slp1 is capable of suppressing chitin-induced immune responses in plant cells.
Chitin-triggered immunity is known to result in induction of pathogenesis-related genes, and we therefore sought to determine the effect of the Slp1 effector on induction of rice defense gene expression. We therefore performed quantitative RT-PCR (qRT-PCR) and examined changes in expression of the rice Phe ammonia lyase gene, PAL1, and the β-glucanase–encoding gene, rBG. In the presence of 1 nM (GlcNAc)8, expression of both PAL1 and β-glucanase increased significantly (Figure 4C). However, the increase in gene expression was suppressed when a 100-fold molar excess of Slp1 was also included, consistent with the role of Slp1 in preventing chitin-triggered immunity responses in rice.
In rice, the pattern recognition receptor LysM protein CEBiP resides at the rice plasma membrane and is able to bind to chitin oligosaccharides (Shibuya et al., 1996; Kaku et al., 2006). We hypothesized that Slp1 might therefore function to compete with the CEBiP recognition receptor residing at the invaginated rice cell membrane. CEBiP is a LysM domain–containing protein and interacts with the LysM receptor-like kinase protein CERK1 to bring about plant defense responses (Shimizu. et al., 2010). CeBiP has been shown to contribute to rice blast disease resistance (Kishimoto et al., 2010). We therefore performed a competition assay in which a microsomal membrane preparation containing the receptor protein CEBiP was isolated from rice suspension cells. When this membrane fraction was incubated with 0.4 μM biotinylated N-acetylchito-octaose (GlcNAc)8, labeling of CEBiP occurred (Figure 4E). When an equimolar amount of Slp1 (0.4 μM) was added, a significant portion of biotinylated (GlcNAc)8 bound to the effector, suggesting that Slp1 is capable of competing with CEBiP for chitin binding in this assay. When a 10-fold molar excess of Slp1 (4 μM) was added, binding of biotinylated (GlcNAc)8 to the membrane fraction containing CEBiP was almost entirely blocked and resulted in the almost exclusive labeling of Slp1 (Figure 4E).
We also determined the affinity kinetics of Slp1 for chitin oligosaccharides using surface plasmon resonance (SPR) technology. Using SPR, we tested for binding of Slp1 and Ecp6 to the ligand (chitin oligosaccharides [(GlcNAc)8]). Using the ligand-immobilized method, in which chitin oligosaccharides are immobilized to the sensor chip, we were able to calculate dissociation constants (Kd values) for both Slp1 and Ecp6 (Figure 4D). We estimated that the affinity for chitin oligosaccharides was similar for both Slp1 and Ecp6, with Kd values of 2.4 × 10−9 M and 1.3 × 10−9 M, respectively. Previously, the Kd value of CEBiP for chitin oligosaccharides was calculated as 2.9 × 10−8 M (Shibuya et al., 1996). Full rate constant values, including the Kd and Kon values, for the association of Slp1 for chitin oligosaccharides can be found in Supplemental Table 1 online. These results suggest that Slp1 and Ecp6 both show a high affinity for chitin oligosaccharides, which is consistent with the ability of Slp1 to act as competitive inhibitor of CEBiP. When all of these results are considered together, we conclude that the M. oryzae Slp1 protein competes directly with chitin receptor proteins in rice and is able to suppress chitin-induced immunity.
Targeted Gene Silencing of CEBiP in Rice Restores the Ability of Δslp1 Mutants of M. oryzae to Cause Rice Blast Disease
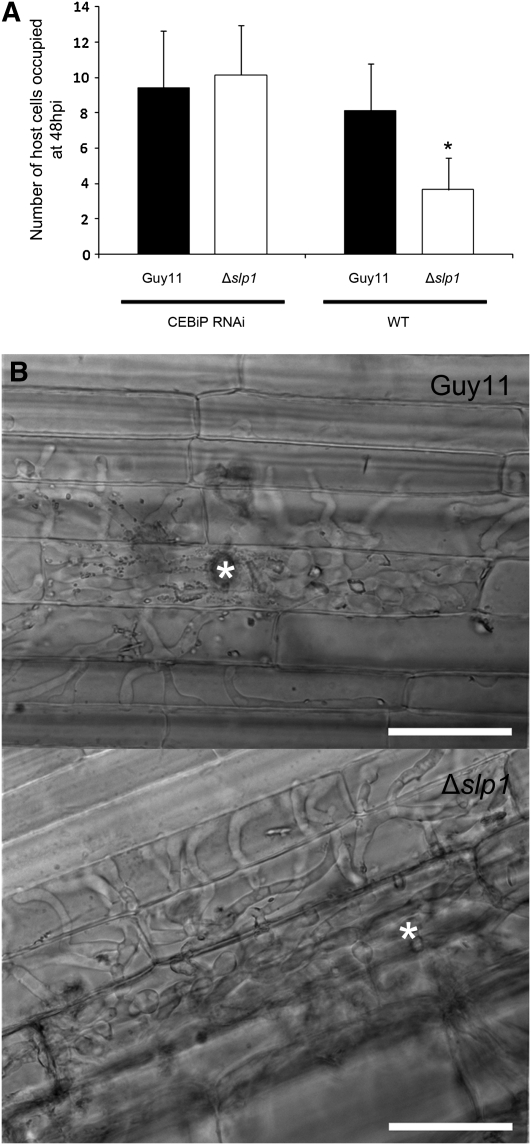
We were interested in establishing whether the ability of the Slp1 effector to act as a competitive inhibitor of CEBiP, thereby suppressing PAMP-triggered immunity, was the reason why M. oryzae Δslp1 mutants showed a significant reduction in their ability to cause rice blast disease. We therefore obtained transgenic rice lines of cultivar Nipponbare, in which the CEBiP-encoding gene had been silenced using RNA interference (RNAi; Kishimoto et al., 2010). These rice lines have previously been shown to lack chitin-triggered immune responses and to exhibit increased susceptibility to rice blast disease (Kishimoto et al., 2010). We inoculated the CEBiP RNAi plants, and corresponding wild-type Nipponbare rice lines, with the M. oryzae Δslp1 mutant and Guy11 strain. Strikingly, we observed that the Δslp1 mutant was as virulent as Guy11 when inoculated onto CEBiP RNAi plants (Figure 5). On CEBiP RNAi plants, the mean number of host cells occupied by the fungus at 48 HAI by the Guy11 and Δslp1 strain was 9.4 (sd ± 3.21) and 10.2 (sd ± 2.83), respectively. Furthermore, on CEBiP RNAi plants, no significant difference in host tissue colonization was observed between Guy11 and the Δslp1 mutant (two-tailed t test, n = 34 infection sites, P = 0.322). By contrast, when nonsilenced Nipponbare rice lines were inoculated, the mean number of host cells occupied by Guy11 and the Δslp1 mutant was 8.1 (sd ± 2.63) and 3.7 (sd ± 1.78), respectively. The mean number of host cells colonized by the Δslp1 mutant was significantly lower than the wild-type Guy11 (two-tailed t test, P < 0.01) (Figure 5). We also found that spray inoculation of CEBiP-RNAi seedlings with the Δslp1 mutant led to restoration of the number of disease lesions (data not shown). We conclude that it is the ability of Slp1 to act as a competitive inhibitor of CEBiP that is its principal function during rice blast disease and that this role is highly significant in determining the outcome of the host–pathogen interaction.
Figure 5.
The Ability of a Δslp1 Mutant to Cause Rice Blast Disease Is Restored When Inoculated onto a Rice Cultivar in Which CEBiP Has Been Silenced by RNAi.
(A) At 48 HAI, host cell colonization of the Δslp1 mutant was similar to that of Guy11 on a CEBiP RNAi line of cultivar Nipponbare (two-tailed t test, P = 0.323). On wild-type nontransformed Nipponbare, the Δslp1 mutant was significantly reduced in its ability to colonize host tissue (two-tailed t test, P < 0.01). Error bars represent 1 sd.
(B) Micrographs of typical infection sites of Guy11 and Δslp1 on leaf sheath tissue from the CEBiP RNAi line. White asterisks mark the initial site of host cell entry. Bars = 35 μm.
DISCUSSION
In this study, we set out to investigate the mechanisms used by the rice blast fungus to colonize living rice tissue. We focused on whether effector proteins secreted by M. oryzae during biotrophic growth could be involved in perturbing the way that rice plants initially detect the invading fungus by means of PAMP molecules, such as chitin oligosaccharides. Our results provide evidence that M. oryzae deploys an effector, Slp1, to suppress chitin-induced host defense responses in rice tissue and that this is significant in the development of rice blast disease. By contrast with previously described rice blast effectors (Jia et al., 2000; Khang et al., 2010), Slp1 accumulates in the apoplastic space at the plant-fungal interface and, in particular, is associated with colonization of new rice cells by the fungus during invasive growth. At this stage, however, we cannot exclude that Slp1 is secreted and subsequently becomes incorporated into the fungal cell wall matrix. Immunolocalization of Slp1 will help us to establish the precise localization of Slp1 at the plant-fungal interface. Having demonstrated a chitin binding role of Slp1, it is highly likely that Slp1 binds chitin oligosaccharides in the cell wall matrix in addition to free chitin oligosaccharides in the apoplastic space. We showed, by infecting a transgenic rice cultivar expressing a plant plasma membrane–targeted Lti6b-GFP, that M. oryzae invasive hyphae are encased by the invaginated plant cell membrane (or EIHM), suggesting an intimate interaction between the fungus and host, consistent with studies that have used the lipophilic styryl dye FM4-64 to investigate the nature of plant cell colonization by the rice blast fungus (Kankanala et al., 2007; Mosquera et al., 2009). Furthermore, the pattern of localization of Slp1 is strikingly different than that of BIC-localized (Khang et al., 2010) effector proteins that are subsequently delivered across the EIHM to the host cytoplasm (Khang et al., 2010). The pattern of localization of Slp1-GFP is similar to that of the putative rice blast effector protein BAS4 (Mosquera et al., 2009). Fusions of BAS4 to the fluorescent protein enhanced GFP have previously been shown to outline completely invasively growing biotrophic hyphae (Mosquera et al., 2009; Khang et al., 2010), although no function has yet been assigned to BAS4, and the significance of its apoplastic localization in rice blast disease has yet to be determined. The partial colocalization of Slp1-GFP and Bas4-mRFP reported here are consistent with Slp1 being apoplastically localized but highlight its secretion from actively growing invasive hyphal tips as they colonize new rice cells, which contrasts with the pattern of Bas4 secretion. These localization results support a distinct function for Slp1 compared with those of host cell–delivered effectors, which are likely to bind to intracellular targets within host plant cells (Jia et al., 2000). The molecular basis of effector translocation into host cells by fungal pathogens is not yet known, although a conserved mechanism involving phospholipid binding at the host cell membrane has been proposed in eukaryotic pathogens, such as oomycetes and fungi (Kale et al., 2010).
As a consequence of its ability to bind chitin, we set out determine whether Slp1 was capable of altering the chitin-triggered immune response of rice, the native host of M. oryzae. An increasing body of evidence has implicated the chitin elicitor receptor CEBiP in rice immunity (Kaku et al., 2006; Shimizu et al., 2010; Kishimoto et al., 2010). CEBiP is a plasma membrane glycoprotein that contains two LysM domains and shows high affinity for chito-oligosaccharides (Kaku et al., 2006). A reduction in CEBiP expression in cultured rice cells leads to a decrease in chitin elicitor-triggered defense responses, including ROS generation and expression of plant defense–associated genes (Kaku et al., 2006). CEBiP interacts with the chitin elicitor receptor kinase CERK1, which is also necessary for chitin-triggered immunity responses (Shimizu et al., 2010). In Arabidopsis thaliana, a similar LysM receptor-like kinase is implicated in chitin-triggered immunity and fungal resistance (Miya et al., 2007; Wan et al., 2008). CEBiP and CERK1 appear to form a plasma membrane heterooligomeric receptor complex in response to chitin oligosaccharides (Shimizu et al., 2010). Significantly, CEBiP is also directly implicated in resistance to rice blast disease; silencing of the CEBiP gene in rice allowed enhanced proliferation of M. oryzae in rice tissue, while expression of a novel CEBiP/Xa21 chimeric receptor led to more pronounced rice blast resistance (Kishimoto et al., 2010).
Evidence reported here indicates that Slp1 is capable of competing with the CEBiP receptor for chitin oligosaccharides [either (GlcNAc)6 or (GlcNAc)8] and suppressing chitin-triggered defense responses, such as ROS generation and induction of plant defense genes. We also showed that Slp1 is capable of suppressing chitin-triggered immune responses more generally because it is able to suppress medium alkalinization of tomato cell suspensions. SPR analysis also provided evidence that Slp1 has a similar affinity for chitin oligosaccharides as the C. fulvum effector Ecp6 (de Jonge et al., 2010) and predicted a higher affinity than previously reported for CEBiP (Shibuya et al., 1996). However, it is also worth noting that affinity measurements of chitin elicitor receptors vary significantly depending on the method used, with isothermal calorimetry and SPR (in either effector or ligand-immobilized modes) all showing significant variation (Shibuya et al., 1996; de Jonge et al., 2010). The Kd value of Ecp6 obtained by SPR using the effector-immobilized mode was, for instance, 3.8 × 10−7 M, while the value obtained previously by isothermal titration calorimetry was only 3.7 × 10−6 M (de Jonge et al., 2010). Generally the Kd values obtained by SPR have a tendency to give smaller values compared with other methods, probably because of the faster binding of the ligand to the immobilized protein on the sensor tip (Jecklin et al., 2009). Limitation of the accessible Kd range in the case of isothermal titration calorimetry may also contribute to the difficulty in comparing these values directly. At this stage, we cannot make any confident conclusions regarding the differential affinity of CEBiP and Slp1. It will be necessary in the future to measure the relative concentration of Slp1 and CEBiP directly at the rice–M. oryzae interface, although this is currently an extremely difficult technical challenge. However, we predict that Slp1 is likely to accumulate to a higher molar concentration at the host plasma membrane interface than the membrane-bound CEBiP receptor because the SLP1 gene is highly expressed during initial invasive growth of M. oryzae and live-cell imaging suggests that a significant amount of the effector is present at the host-pathogen interface (Figure 1). The significant reduction in virulence associated with the M. oryzae Δslp1 mutant and the reduced proliferation of the fungus in plant tissue were all consistent with a role for Slp1 in competitive inhibition of the CEBiP receptor, but we were keen to test this hypothesis directly. We therefore inoculated transgenic rice in which the CEBiP receptor gene had been silenced by RNAi (Kishimoto et al., 2010) with the Δslp1 mutant. The fact that the Δslp1 mutant caused rice blast normally in this CEBiP RNAi line provides strong evidence that in the absence of a chitin-triggered immune response to suppress, Slp1 does not serve any additional function during plant infection. Rather, the clear virulence phenotype associated with the Δslp1 mutant on normal wild-type rice cultivars must be associated with its ability to bind chitin and suppress CEBiP-mediated chitin-triggered immunity, consistent with the reduced ability of the mutant to colonize rice cells and the restoration of virulence by reintroduction and expression of SLP1. When considered together with the previously reported role of CEBiP in rice blast resistance (Kishimoto et al., 2010), it seems very likely that the interplay between Slp1 and CEBiP is pivotal in determining the progression of rice blast disease.
In addition to binding chitin, we suggest that Slp1 has the capacity to bind to itself, putatively forming homodimers. Recently, yeast two-hybrid analysis was used to demonstrate a positive interaction between the extracellular LysM receptor domains of CEBiP and CERK1 (Shimizu et al., 2010), although fungal LysM effector proteins that have recently been investigated have not appeared to share this property (de Jonge et al., 2010; Marshall et al., 2011). Although a biological reason for the multimerization of Slp1 in M. oryzae has not yet been demonstrated and our observations must be considered preliminary, it is possible that Slp1 forms multimers to provide an additional means of shielding bound chitin oligosaccharides or as a means of increasing its space-filling potential in the narrow apoplastic space around invasive hyphae, thereby enhancing its competitive inhibition of the host receptor CEBiP. Determining a crystal structure of Slp1 and studying its ability to form homodimers and multimeric complexes, in addition to its precise chitin oligomer binding characteristics, will enable a rigorous means of testing this hypothesis.
Previous identification of Ecp6 in the extracellular pathogen C. fulvum provided the first evidence that suppression of chitin-triggered immunity might be a means by which biotrophic fungal pathogens with this mode of tissue colonization overcome host defenses (de Jonge and Thomma, 2009; de Jonge et al., 2010). Although this and other studies have suggested a function for LysM effectors in suppressing chitin-based immune responses, there has not, until now, been evidence to link the presumed function of these proteins as suppressors of PAMP-triggered immunity to a role in plant disease. Our study has tested this idea and found evidence that the Slp1 effector of M. oryzae plays a role in rice blast disease due solely to its function in suppression of chitin-triggered defense responses. A large number of putatively secreted LysM domain–containing fungal proteins have been identified in fungi (de Jonge and Thomma, 2009), suggesting that overcoming this initial line of host defense may be fundamental to the successful infection of plants by pathogenic fungi.
METHODS
Fungal Strains, Growth Conditions, and DNA Analysis
Strains were grown on complete medium as described previously (Talbot et al., 1993). To carry out plant infection assays, spores were harvested from 10- to 14-d-old plate cultures in sterile distilled water and washed twice. Spores were counted using a hemocytometer (Corning) and confirmed using three independent cell counts. Rice plant infections were performed by spraying 21-d-old seedling of the rice blast susceptible rice cultivar CO-39 with spore suspensions at a concentration of 5 × 104 spores mL−1 in 0.2% gelatin, unless stated otherwise, and as described previously (Talbot et al., 1993). Disease symptoms were allowed to develop for 7 d, unless stated otherwise. Infected leaves were imaged using an Epson Workforce scanner at a resolution of 1200 dpi. Lesion size was determined using ImageJ, a freely available image analysis software package from the National Institutes of Health.
Generation and Infection of Transgenic Rice Cultivars
The LTi6B:GFP gene fusion targets GFP to the plant plasma membrane (Kurup et al., 2005) and was obtained from John Runions and Chris Hawes (Oxford Brookes University). The construct was transformed into rice callus into Oryza sativa cv Sasanishiki (Yoshida et al., 2009) using standard plant transformation protocols. Rice transformants were grown on 100 μg mL−1 hygromycin, confirmed by DNA gel blot, and expression checked by qRT-PCR, immunoblotting, and epifluorescence microscopy. T1 transformants were grown and backcrossed to generate stable T2 plants. For infection experiments, Magnaporthe oryzae conidia were harvested and inoculated onto rice leaf sheath at a concentration of 105 spores mL−1. Leaf tissue was incubated at 26°C in a moist chamber and fluorescence examined after 24 HAI by epifluorescence microscopy. Transgenic rice lines of cultivar Nipponbare, in which CEBiP had been silenced by RNAi, were as described previously (Kishimoto et al., 2010). CEBiP RNAi seeds were dehusked, surface sterilized using standard procedures, and grown on Murashige and Skoog media containing 25 mg L−1 hygromycin for 7 d before being transplanted to soil. For the inoculation of CEBiP RNAi plants, the Δslp1 mutant and Guy11 strains were inoculated at a density of 103 spores mL−1 onto 4-week-old leaf sheath tissue, as described previously (Mosquera et al., 2009). Leaf tissue was examined at 48 HAI, and the number of host cells occupied by biotrophically growing fungal hyphae in the upper epidermal leaf layer was counted. Experiments were repeated three times.
Phylogenetic Analysis
Phylogenetic tree construction used the phylogenetic analysis program PhyML (Dereeper et al., 2008). Phylogenies were constructed using the sequences shown in Supplemental Data Set 1 online, which were acquired based on a support value of 1 e−10 with Slp1. Sequence alignments were then generated using ClustalW (Chenna et al., 2003) and manually adjusted to optimize alignments.
Targeted Gene Replacement of SLP1
Targeted gene replacement of the M. oryzae SLP1 gene was performed using the split marker strategy as modified by Kershaw and Talbot (2009). Gene replacement was performed by replacing the 600-bp SLP1 locus with a hygromycin resistance selectable marker HPH, encoding a 1.4-kb hygromycin phosphotransferase resistance cassette. The two overlapping parts of the hph templates were PCR amplified using primers M13F with HY and M13R with YG (see Supplemental Table 2 online) as described previously (Kershaw and Talbot, 2009). A 1-kb DNA fragment upstream and downstream of the SLP1 open reading frame was additionally generated using the primers LF5′SLP1 and LF3′SLP1 and RF5′SLP1 and RF3′SLP1 amplified from genomic DNA of the Guy11 strain. A second-round PCR reaction was performed to fuse the overlapping split hph marker templates with the left and right flanking regions of the SLP1 locus. The wild-type M. oryzae Guy11 strain was then transformed with these deletion cassettes (2 μg of each flank). Putative transformants were selected in the presence of hygromycin B (200 μg mL−1) and checked by DNA gel blot analysis according to standard molecular techniques (Sambrook et al., 1989). Gene sequences and regions either side of SLP1 were retrieved from the M. oryzae genome database at the Broad Institute (http://www.broadinstitute.org/annotation/fungi/magnaporthe/). All primer sequences used in this study can be found in Supplemental Table 2 online.
Generation of SLP1:GFP and SLP127-162:GFP Gene Fusions
To generate an SLP1:GFP gene fusion, the SLP1 gene was amplified to include a 2-kb region upstream of the SLP1 start codon to additionally include the native promoter region using primers 5′SLP1:GFP with 3′SLP1:GFP (see Supplemental Table 2 online) amplified from genomic DNA. To generate an SLP127-162:GFP gene fusion, a 2-kb region upstream of the SLP1 start codon was amplified using the primers 5′SLP1:GFP and 3′SLP-Prom, and a 468-bp fragment was amplified using the primers 5′SLP1-nosp and 3′SLP1:GFP from genomic DNA (see Supplemental Table 2 online). These PCR fragments were then transformed with HindIII-digested pYSGFP-1 (Saunders et al., 2010) into Saccharomyces cerevisiae. In-frame gene fusions were created by gap-repair cloning based on homologous recombination in yeast (Oldenburg et al., 1997). Constructs were confirmed by sequencing through the gene fusion (MWG Operon) and then transformed into the M. oryzae Guy11 strain. At least three independent SLP1:GFP transformants were confirmed prior to experimental observations.
Light and Epifluorescence Microscopy
Epifluorescence microscopy was used to visualize GFP and RFP samples using a Zeiss Axioskop 2 microscope with differential interference contrast to image bright-field images. To visualize SLP1:GFP and PWL2:mRFP on leaf epidermis, conidia were harvested and inoculated onto rice leaf sheath tissue at a concentration of 105 spores mL−1 as described previously (Kankanala et al., 2007). Infected tissue was then excised and mounted onto a glass slide and observed using an IX81 inverted microscope (Olympus) and a UPlanSApo ×100/1.40 oil objective. Images were analyzed using the software package MetaMorph (Molecular Devices).
Production of Recombinant Slp1 Protein
RNA was extracted from infected leaf tissue 144 HAI. cDNA synthesis was performed on 500 ng of DNAase I (Invitrogen) treated RNA using the Affinityscript qPCR synthesis kit (Stratagene) according to the manufacturer’s guidelines. cDNA of SLP1 was cloned using the primers 5′ATG-SLP1 and 3′TAG-SLP1 and cloned into the vector pGEM-T (Promega) according to the manufacturer’s instructions. Affinity-tagged Slp1 was generated in the yeast Pichia pastoris by amplifying the SLP1 cDNA using primers 5′Slp-pic9 and 3′Slp1-pic9 to include a 5′ in-frame His6-FLAG-tag and subsequently cloned into vector pPIC9 (Invitrogen). Fermentation to produce recombinant Slp1 was performed as described in (Joosten et al., 1995; de Jonge et al., 2010). His6-FLAG–tagged Slp1 was purified using a Ni2+-NTA Superflow column (Qiagen) according to the manufacturer’s instructions.
Affinity Precipitation of Slp1 with Polysaccharides
The affinity of Slp1 for various polysaccharides was investigated by incubating 50 μg/mL of Slp1 with 5 mg of chitin beads (New England Biolabs), crab shell chitin, chitosan, xylan, or cellulose (Sigma-Aldrich) as described previously (de Jonge et al., 2010). Protein and the polysaccharide of interest were incubated at 4°C on a rocking platform in a final volume of 1 mL of water. After 16 h, the insoluble pellet fraction was centrifuged (5 min, 13,000g), and the supernatant was collected. The insoluble fraction was pelleted and rinsed a further three times in distilled sterile water to remove unbounded protein. Both the supernatant and the pelleted fractions were then boiled in 200 μL of 1% SDS solution before being examined by SDS-PAGE and Coomassie Brilliant Blue staining.
Cell Protection Assays Using Crude Extract of Chitinase from Tomato Leaves
Intracellular basis chitinases were extracted as described previously (Joosten et al., 1990, 1995). A 50-μL aliquot of Trichoderma viride spores was incubated overnight at room temperature at a concentration of 100 conidia mL−1. Recombinant Slp1 or Avr4 was then added to a final concentration of 10 or 100 μM (Joosten et al., 1995; de Jonge et al., 2010). After 2 h of incubation, 5 μL crude chitinase extract was added and spores were visualized microscopically after ~2 to 4 h.
Medium Alkalinization of Tomato Cell Suspensions
Suspension-cultured tomato (Solanum lycopersicum) cell line Msk8 was maintained as described previously (Felix et al., 1991). To examine medium alkalinization, 2.5 mL aliquots of Msk8 suspension cultured cells were placed into 12-well plates. This was placed on a rotary shaker at 200 rpm and left for 2 h to settle. On addition of either 1 nM (GlcNAc)6 or 1 nM (GlcNAc)6 and 10 nM Slp1, the pH of the cells, while shaking, was monitored continuously for 10 min using a glass electrode and recorded, as described by de Jonge et al. (2010). Prior to addition of the experimental to the cell medium, chitin oligosaccharides and recombinant protein were incubated at room temperature to equilibrate.
SPR Analysis
Affinities of LysM effectors to chitin oligosaccharides were analyzed by SPR measurements using a Biaore X100 instrument (GE Healthcare). In the effector-immobilized assay system, effectors were covalently immobilized by amine coupling to Sensor Chip CM5 according to the manufacturer’s protocol (GE Healthcare). Binding kinetics was measured by multicycle kinetics mode using Biacore X100 control software. In the (GlcNAc)8-immobilized assay system, biotinylated (GlcNAc)8 (Kaku et al., 2006) was coupled to a streptavidin preimmobilized sensor chip (Sensor Chip SA; GE Healthcare). Binding kinetics were measured by single-cycle kinetics mode using Biacore X100 control software. Either (GlcNAc)8 or effector solution was introduced onto the surface at a flow rate of 30 μL/min with HBS-EP+ buffer. The interaction was monitored at 25°C as the change in the SPR response. After monitoring for 2 min, the HBS-EP+ buffer was introduced onto the sensor chip to initiate dissociation.
Affinity Labeling of Rice Membranes with Biotinylated (GlcNAc)8
Affinity labeling with biotinylated (GlcNAc)8 was performed as described previously (Shinya et al., 2010). Suspension-cultured rice cells of O. sativa cv Nipponbare were maintained in a modified N-6 medium as described previously (Tsukada et al., 2002). A microsomal membrane preparation from suspension-cultured rice cells was mixed with biotinylated (GlcNAc)8 in the presence or absence of Slp1 and adjusted to 30 μL with binding buffer. After incubation for 1 h on ice, 3 μL of 3% ethylene glycol bis[succinimidylsuccinate] solution (Pierce) was added to the mixture and kept for 30 min. The reaction was stopped by the addition of 1 M Tris-HCl, mixed with SDS-PAGE sample buffer, boiled for 5 min, and used for SDS-PAGE. Immunoblotting was performed on an Immun-Blot polyvinylidene fluoride membrane (Bio-Rad Laboratories). Detection of biotinylated proteins was performed using a rabbit antibody against biotin (Bethyl Laboratories) as a primary antibody and horseradish peroxidase–conjugated goat anti-rabbit IgG (Chemicon International) as a secondary antibody. Biotinylated proteins were detected by the chemiluminescence with Immobilon Western Detection reagents (Millipore).
Measurement of ROS Generation and Gene Expression Analysis
ROS generation induced by elicitor treatment was analyzed by chemiluminescence due to the ferricyanide-catalyzed oxidation of luminol (5-amino-2,3-dihydro-1,4-phthalazinedione) (Desaki et al., 2006). Briefly, 40 mg of cultured cells was transferred into the 1 mL of fresh medium in a 2-mL centrifuge tube and preincubated for 30 min on a thermomixer shaker at 750 rpm. After the preincubation, (GlcNAc)8 was separately added to the culture medium in the absence or presence of Slp1. For gene expression studies using qRT-PCR, total RNA was prepared from each rice cultivar (40 mg) using an RNeasy plant mini kit (Qiagen) and subjected to cDNA synthesis using a QuantiTect reverse transcription kit (Qiagen). qRT-PCR was performed using TaqMan gene expression assay reagent using a model 7500 Fast Real-Time PCR system (Applied Biosystems). The 18S rRNA was used as an internal control to normalize the amount of mRNA. All primers used are shown in Supplemental Table 2 online.
Yeast Two-Hybrid Analysis
SLP1 cDNA was cloned into pGEMT using the primers 5′SLP1-BamHI and 3′SLP1-EcoRI. SLP1 cDNA was then digested and cloned as a BamHI-EcoRI fragment into the bait vectors pGBKT7 and pGADT7 (Clontech). Sequencing of both constructs was performed using T7 primer to ensure the constructs were in frame. Yeast two-hybrid analysis was then performed using the Matchmaker GAL4 Two-Hybrid System 3 (Clontech) according to the manufacturer’s instructions, as described previously (Wilson et al., 2010).
Accession Number
Sequence data from this article can be found in the GenBank/EMBL databases under accession number MGG10097.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogeny of Fungal LysM Proteins with a Bootstrap Support Value of 100.
Supplemental Figure 2. Multiple Sequence Alignment of the LysM Domains from a Number of Fungal Species.
Supplemental Figure 3. SLP1 Is Not Expressed in Conidia, Germ Tubes, or Incipient or Mature Appressoria.
Supplemental Figure 4. Targeted Gene Replacement of SLP1 and Confirmation by DNA Gel Blotting.
Supplemental Figure 5. Complementation Analysis of Δslp1 Mutants.
Supplemental Figure 6. SLP127-162:GFP Fails to Complement the Virulence Phenotype of the Δslp1 Mutant.
Supplemental Figure 7. High Stringency Yeast Two-Hybrid Analysis of a Putative Slp1-Slp1 Protein Interaction.
Supplemental Table 1. Rate Constants for Association and Dissociation of Slp1 with Chitin Oligomers Derived by Surface Plasmon Resonance Analysis.
Supplemental Table 2. Primers Used in This Work.
Supplemental Movie 1. Laser Confocal Micrograph of Invasive Hyphae of M. oryzae Showing Three-Dimensional Projection.
Supplemental Data Set 1. Text File of the Alignment of Fungal LysM Proteins Used for the Phylogenetic Analysis in Supplemental Figure 1.
Acknowledgments
T.A.M. was supported by a Sainsbury Studentship awarded to N.J.T. by the Gatsby Charitable Foundation. B.P.H.J.T. is supported by a Vidi grant of the Research Council for Earth and Life Sciences (ALW) of the Netherlands Organization for Scientific Research. N.S. is funded from the Program for Promotion of Basic Research Activities for Innovative Bioscience (PROBRAIN). We thank Barbara Valent (Kansas State University) for kindly donating the PWL2:mRFP construct.
AUTHOR CONTRIBUTIONS
T.A.M. and N.J.T. designed the project. T.A.M., A.K., T.S., L.S.R., I.O., H.S., R.T., Y.N., N.S., and B.P.H.J.T. performed experiments, and all authors contributed to analysis and interpretation of the results. T.A.M. and N.J.T. wrote the article with contributions, revisions, and edits from N.S. and B.P.H.J.T.
References
- Bolton M.D., et al. (2008). The novel Cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Mol. Microbiol. 69: 119–136 [DOI] [PubMed] [Google Scholar]
- Buist G., Steen A., Kok J., Kuipers O.P. (2008). LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 68: 838–847 [DOI] [PubMed] [Google Scholar]
- Chenna R., Sugawara H., Koike T., Lopez R., Gibson T.J., Higgins D.G., Thompson J.D. (2003). Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31: 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge R., Thomma B.P.H.J. (2009). Fungal LysM effectors: Extinguishers of host immunity? Trends Microbiol. 17: 151–157 [DOI] [PubMed] [Google Scholar]
- de Jonge R., van Esse H.P., Kombrink A., Shinya T., Desaki Y., Bours R., van der Krol S., Shibuya N., Joosten M.H., Thomma B.P. (2010). Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 329: 953–955 [DOI] [PubMed] [Google Scholar]
- Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.F., Guindon S., Lefort V., Lescot M., Claverie J.M., Gascuel O. (2008). Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36(Web Server issue): W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desaki Y., Miya A., Venkatesh B., Tsuyumu S., Yamane H., Kaku H., Minami E., Shibuya N. (2006). Bacterial lipopolysaccharides induce defense responses associated with programmed cell death in rice cells. Plant Cell Physiol. 47: 1530–1540 [DOI] [PubMed] [Google Scholar]
- Felix G., Grosskopf D.G., Regenass M., Basse C.W., Boller T. (1991). Elicitor-induced ethylene biosynthesis in tomato cells: characterization and use as a bioassay for elicitor action. Plant Physiol. 97: 19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G., Regenass M., Boller T. (1993). Specific perception of subnanomolar concentrations of chitin fragments by tomato cells: induction of extracellular alkalinisation, changes in protein phosphorylation, and establishment of a refractory state. Plant J. 4: 307–316 [Google Scholar]
- Jecklin M.C., Schauer S., Dumelin C.E., Zenobi R. (2009). Label-free determination of protein-ligand binding constants using mass spectrometry and validation using surface plasmon resonance and isothermal titration calorimetry. J. Mol. Recognit. 22: 319–329 [DOI] [PubMed] [Google Scholar]
- Jia Y., McAdams S.A., Bryan G.T., Hershey H.P., Valent B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19: 4004–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten M.H.A.J., Bergmans C.J.B., Meulenhoff E.J.S., Cornelissen B.J.C., De Wit P.J.G.M. (1990). Purification and serological characterization of three basic 15-kilodalton pathogenesis-related proteins from tomato. Plant Physiol. 94: 585–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten M.H.A.J., Verbakel H.M., Nettekoven M.E., Van Leeuwen J., van der Vossen R.T.M., de Wit P.J.G.M. (1995). The phytopathogenic fungus Cladosporium fulvum is not sensitive to the chitinase and β-1,3-glucanase defense proteins of its host, tomato. Physiol. Mol. Plant Pathol. 46: 45–59 [Google Scholar]
- Kaku H., Nishizawa Y., Ishii-Minami N., Akimoto-Tomiyama C., Dohmae N., Takio K., Minami E., Shibuya N. (2006). Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA 103: 11086–11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale S.D., et al. (2010). External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell 142: 284–295 [DOI] [PubMed] [Google Scholar]
- Kankanala P., Czymmek K., Valent B. (2007). Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19: 706–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershaw M.J., Talbot N.J. (2009). Genome-wide functional analysis reveals that infection-associated fungal autophagy is necessary for rice blast disease. Proc. Natl. Acad. Sci. USA 106: 15967–15972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khang C.H., Berruyer R., Giraldo M.C., Kankanala P., Park S.Y., Czymmek K., Kang S., Valent B. (2010). Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell 22: 1388–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto K., Kouzai Y., Kaku H., Shibuya N., Minami E., Nishizawa Y. (2010). Perception of the chitin oligosaccharides contributes to disease resistance to blast fungus Magnaporthe oryzae in rice. Plant J. 64: 343–354 [DOI] [PubMed] [Google Scholar]
- Kurup S., Runions J., Köhler U., Laplaze L., Hodge S., Haseloff J. (2005). Marking cell lineages in living tissues. Plant J. 42: 444–453 [DOI] [PubMed] [Google Scholar]
- Marshall R., Kombrink A., Motteram J., Loza-Reyes E., Lucas J., Hammond-Kosack K.E., Thomma B.P.H.J., Rudd J.J. (2011). Functional analysis of in planta expressed LysM effector homologues from the non-biotrophic fungus Mycosphaerella graminicola reveals varying contributions to virulence. Plant Physiol. 156: 756–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya A., Albert P., Shinya T., Desaki Y., Ichimura K., Shirasu K., Narusaka Y., Kawakami N., Kaku H., Shibuya N. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosquera G., Giraldo M.C., Khang C.H., Coughlan S., Valent B. (2009). Interaction transcriptome analysis identifies Magnaporthe oryzae BAS1-4 as biotrophy-associated secreted proteins in rice blast disease. Plant Cell 21: 1273–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg K.R., Vo K.T., Michaelis S., Paddon C. (1997). Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 25: 451–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press)
- Saunders D.G., Dagdas Y.F., Talbot N.J. (2010). Spatial uncoupling of mitosis and cytokinesis during appressorium-mediated plant infection by the rice blast fungus Magnaporthe oryzae. Plant Cell 22: 2417–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya N., Ebisu N., Kamada Y., Kaku H., Cohn J., Ito N. (1996). Localization and binding characteristics of a high-affinity binding site for N-acetylchitooligosaccharide elicitor in the plasma membrane from suspension-cultured rice cells suggest a role as a receptor for the elicitor signal at the cell surface. Plant Cell Physiol. 37: 894–898 [Google Scholar]
- Shimizu T., Nakano T., Takamizawa D., Desaki Y., Ishii-Minami N., Nishizawa Y., Minami E., Okada K., Yamane H., Kaku H., Shibuya N. (2010). Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64: 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinya T., Osada T., Desaki Y., Hatamoto M., Yamanaka Y., Hirano H., Takai R., Che F.S., Kaku H., Shibuya N. (2010). Characterization of receptor proteins using affinity cross-linking with biotinylated ligands. Plant Cell Physiol. 51: 262–270 [DOI] [PubMed] [Google Scholar]
- Talbot N.J. (2003). On the trail of a cereal killer: Exploring the biology of Magnaporthe grisea. Annu. Rev. Microbiol. 57: 177–202 [DOI] [PubMed] [Google Scholar]
- Talbot N.J., Ebbole D.J., Hamer J.E. (1993). Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 5: 1575–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma B.P.H.J., VAN Esse H.P., Crous P.W., DE Wit P.J.G.M. (2005). Cladosporium fulvum (syn. Passalora fulva), a highly specialized plant pathogen as a model for functional studies on plant pathogenic Mycosphaerellaceae. Mol. Plant Pathol. 6: 379–393 [DOI] [PubMed] [Google Scholar]
- Tsukada K., Ishizaka M., Fujisawa Y., Iwasaki Y., Yamaguchi T., Minami E., Shibuya N. (2002). Rice receptor for chitin oligosaccharides elicitor does not couple to heterotrimeric G-protein: elicitor responses of suspension cultured rice cells from Daikoku dwarf (d1) mutants lacking a functional G-protein α-subunit. Physiol. Plant. 116: 373–382 [Google Scholar]
- van den Burg H.A., Harrison S.J., Joosten M.H.A.J., Vervoort J., de Wit P.J.G.M. (2006). Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol. Plant Microbe Interact. 19: 1420–1430 [DOI] [PubMed] [Google Scholar]
- van Esse H.P., Bolton M.D., Stergiopoulos I., de Wit P.J.G.M., Thomma B.P.H.J. (2007). The chitin-binding Cladosporium fulvum effector protein Avr4 is a virulence factor. Mol. Plant Microbe Interact. 20: 1092–1101 [DOI] [PubMed] [Google Scholar]
- van Esse H.P., Van’t Klooster J.W., Bolton M.D., Yadeta K.A., van Baarlen P., Boeren S., Vervoort J., de Wit P.J.G.M., Thomma B.P.H.J. (2008). The Cladosporium fulvum virulence protein Avr2 inhibits host proteases required for basal defense. Plant Cell 20: 1948–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J., Zhang X.-C., Neece D., Ramonell K.M., Clough S., Kim S.Y., Stacey M.G., Stacey G. (2008). A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Fiers M., Ellendorff U., Wang Z., de Wit P.J.G.M., Angenent G.C., Thomma B.P.H.J. (2010). The diverse roles of extracellular leucine-rich repeat-containing receptor-like proteins in plants. Crit. Rev. Plant Sci. 29: 285–299 [Google Scholar]
- Wilson R.A., Gibson R.P., Quispe C.F., Littlechild J.A., Talbot N.J. (2010). An NADPH-dependent genetic switch regulates plant infection by the rice blast fungus. Proc. Natl. Acad. Sci. USA 107: 21902–21907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R.A., Talbot N.J. (2009). Under pressure: Investigating the biology of plant infection by Magnaporthe oryzae. Nat. Rev. Microbiol. 7: 185–195 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T., Minami E., Ueki J., Shibuya N. (2005). Elicitor-induced activation of phospholipases plays an important role for the induction of defense responses in suspension-cultured rice cells. Plant Cell Physiol. 46: 579–587 [DOI] [PubMed] [Google Scholar]
- Yoshida K., Saitoh H., Fujisawa S., Kanzaki H., Matsumura H., Yoshida K., Tosa Y., Chuma I., Takano Y., Win J., Kamoun S., Terauchi R. (2009). Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. Plant Cell 21: 1573–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]