This work identifies a nucleolar localization signal within the essential double-stranded RNA binding domain of the Cucumber mosaic virus silencing suppressor protein 2b. It also shows that direct 2b–ARGONAUTE interactions can redistribute both 2b and ARGONAUTE proteins in the nucleus but are not essential for 2b suppression of either posttranscriptional gene silencing or RNA-directed DNA methylation.
Abstract
Unique among the known plant and animal viral suppressors of RNA silencing, the 2b protein interacts directly with both small interfering RNA (siRNA) and ARGONAUTE1 (AGO1) and AGO4 proteins and is targeted to the nucleolus. However, it is largely unknown which regions of the 111-residue 2b protein determine these biochemical properties and how they contribute to its diverse silencing suppressor activities. Here, we identified a functional nucleolar localization signal encoded within the 61–amino acid N-terminal double-stranded RNA (dsRNA) binding domain (dsRBD) that exhibited high affinity for short and long dsRNA. However, physical interaction of 2b with AGOs required an essential 33-residue region C-terminal to the dsRBD and was sufficient to inhibit the in vitro AGO1 Slicer activity independently of its dsRNA binding activities. Furthermore, the direct 2b–AGO interaction was not essential for the 2b suppression of posttranscriptional gene silencing (PTGS) and RNA-directed DNA methylation (RdDM) in vivo. Lastly, we found that the 2b–AGO interactions in vivo also required the nucleolar targeting of 2b and had the potential to redistribute both the 2b and AGO proteins in nucleus. These findings together suggest that 2b may suppress PTGS and RdDM in vivo by binding and sequestering siRNA and the long dsRNA precursor in a process that is facilitated by its interactions with AGOs in the nucleolus.
INTRODUCTION
In RNA silencing, small RNAs (sRNAs) of 21 to 30 nucleotides in length are processed as duplexes from double-stranded RNA (dsRNA) by RNaseIII enzymes called Dicers to trigger RNA degradation, DNA methylation, heterochromatin formation, and protein translation inhibition in diverse eukaryotic organisms (Baulcombe, 2004, 2005; Matzke et al., 2004, 2007, 2009; Meister and Tuschl, 2004; Matzke and Birchler, 2005; Voinnet, 2005, 2008; Brodersen et al., 2008; Dunoyer and Voinnet, 2008; Chinnusamy and Zhu, 2009; Heo and Kim, 2009). In Arabidopsis thaliana, four Dicer-like proteins (DCLs) produce 21-nucleotide sRNAs, including siRNAs and microRNAs (miRNAs; via DCL4 or DCL1, respectively) and 22- and 24-nucleotide sRNAs (via DCL2 or DCL3, respectively) (Bartel, 2004; Dunoyer et al., 2005; Gasciolli et al., 2005; Xie et al., 2005; Brodersen and Voinnet, 2006). In the cytoplasm of plant cells, one selected sRNA strand is loaded onto one of 10 ARGONAUTE proteins (e.g., AGO1 or AGO7), forming an effector complex called an RNA-induced silencing complex (RISC) that targets transcripts for posttranscriptional gene silencing (PTGS), which is achieved specifically through RNA-RNA sequence recognition and base pairing (Vazquez et al., 2004; Allen et al., 2005; Yoshikawa et al., 2005; Vaucheret, 2008). AGO proteins are often named Slicer proteins because they cleave target transcripts at the duplex formed with the guide sRNA (Tolia and Joshua-Tor, 2007). In a plant nucleus, 24-nucleotide siRNAs, generated in a pathway involving a plant-specific DNA-directed RNA polymerase (Pol IV), RDR2 and DCL3 (Xie et al., 2004; Kanno et al., 2005; Matzke and Birchler, 2005; Zhang et al., 2007; Melnyk et al., 2011a) are loaded onto AGO4- or AGO6-containing effector complexes. These complexes are then recruited to target genomic regions, by RNA-RNA base pairing with Pol V–dependent transcripts from the target loci (Wierzbicki et al., 2009) to direct transcriptional silencing (TGS) and the de novo methylation of target genomic sequences, a process called RNA-directed DNA methylation (RdDM) (Zilberman et al., 2004; Li et al., 2006; Zheng et al., 2007; Matzke et al., 2009; Gao et al., 2010). RdDM induces the de novo methylation of cytosines in all sequence contexts (i.e., CpG, CpHpG, and CpHpH, where H is A, T, or G) at the region of siRNA-DNA sequence complementarity (Matzke et al., 2009).
In addition to regulating development, RNA silencing also functions as an antiviral mechanism in plants and invertebrates (Ding, 2010). Genetic studies have shown that multiple DCLs, RNA-dependent RNA polymerases (RDRs), and AGOs direct the antiviral defense in Arabidopsis. To invade their host effectively, viruses have evolved a vast array of proteins called viral suppressors of RNA silencing (VSR), which target different stages of the silencing process (Li and Ding, 2006; Ding and Voinnet, 2007; Diaz-Pendón and Ding, 2008; Burgyan and Havelda, 2011). Some VSRs, such as the tombusviral P19, act by binding and sequestering duplex siRNA to inhibit RISC assembly and cell-to-cell movement of RNA silencing (Chapman et al., 2004; Lakatos et al., 2006; Dunoyer et al., 2010), whereas others inhibit RNA silencing through interacting with the protein components of the RNA silencing machinery (Wu et al., 2010; Burgyan and Havelda, 2011).
The 2b protein encoded by Cucumber mosaic virus (CMV) was one of the first two VSRs reported in 1998 (Anandalakshmi et al., 1998; Béclin et al., 1998; Brigneti et al., 1998; Kasschau and Carrington, 1998; Xin et al., 1998). The 2b protein is unique among the known plant and animal VSRs because it directly interacts with both the RNA and protein components of the RNA silencing machinery (Zhang et al., 2006; Goto et al., 2007; González et al., 2010; Hamera et al., 2012). The interaction of CMV 2b with AGO1 and AGO4 from Arabidopsis has been demonstrated in vitro by glutathione S-transferase (GST) pull-down experiments and in vivo by coimmunoprecipitation and bimolecular fluorescence complementation assays, which is consistent with the observed activity of CMV 2b to suppress the in vitro slicer activity of AGO1 and AGO4 (Zhang et al., 2006; González et al., 2010; Hamera et al., 2012). CMV 2b and 2b of Tomato aspermy virus (TAV), which is from the same Cucumovirus genus as CMV, also bind to duplex siRNA in vitro (Goto et al., 2007; González et al., 2010; Hamera et al., 2012). The crystal structure for the N-terminal 69 amino acids of TAV 2b, which is highly conserved in sequence among cucumoviral 2b proteins (Ding et al., 1994), reveals a novel dsRNA binding domain folded into two long α-helices connected by a short linker (Goto et al., 2007; Chen et al., 2008). TAV 2b forms a dimer as indicated previously by immunoblot analysis of TAV 2b in infected plants (Shi et al., 1997) and has a mechanism similar to that of P19 to measure the length of the siRNA duplex by a pair of hook-like structures that depend on a Trp residue (Trp-50) from the C-terminal α-helix, which, however, is not conserved in other cucumoviral 2b proteins (Ding et al., 1994; Chen et al., 2008).
The 2b protein of CMV is active in vivo to suppress the RDR6-dependent RNA silencing that targets both infecting CMV and transgenes either in stable transgenic plants or delivered transiently by Agrobacterium tumefaciens coinfiltration in Nicotiana benthamiana (Diaz-Pendon et al., 2007; Zhang et al., 2008; Wang et al., 2011). Intriguingly, although the positive-strand RNA genome of CMV replicates exclusively in the cytoplasm, 2b is predominantly localized to the nucleus by single or double nuclear localization signals (NLSs) in subgroup II and I strains of CMV, respectively (Lucy et al., 2000; Wang et al., 2004). A recent study further demonstrated nucleolar localization of the 2b protein of Fny-CMV, a subgroup I isolate (González et al., 2010). It is also known that 2b suppression of transgene RNA silencing is associated with significantly reduced cytosine methylation of the transgene DNA and defective intercellular spread of transgene silencing (Lucy et al., 2000; Guo and Ding, 2002; Wang et al., 2004). However, it is largely unknown which regions of the 111-residue 2b protein of CMV determine these distinct biochemical properties and how each of them contributes to the diverse silencing suppressor activities of the 2b protein. We report in this study a detailed characterization of the 2b protein encoded by the severe Shan-Dong (SD) isolate from CMV subgroup I in terms of subcellular localization, RNA binding, AGO interactions, and suppression of PTGS and RdDM. Our analysis uncouples the domain requirements for dsRNA binding and nucleolar targeting from the physical interactions with AGO1, AGO4, and AGO6. Moreover, we report the identification of a nucleolar localization signal (NoLS) encoded within the dsRNA binding domain of SD2b. Interestingly, we found that the direct in vivo interactions of the SD2b protein with AGO1, AGO4, and AGO6 also require a functional NoLS and have the potential to redistribute both SD2b and AGO proteins in the nucleus.
RESULTS
Identification of a NoLS Encoded by the CMV 2b Protein
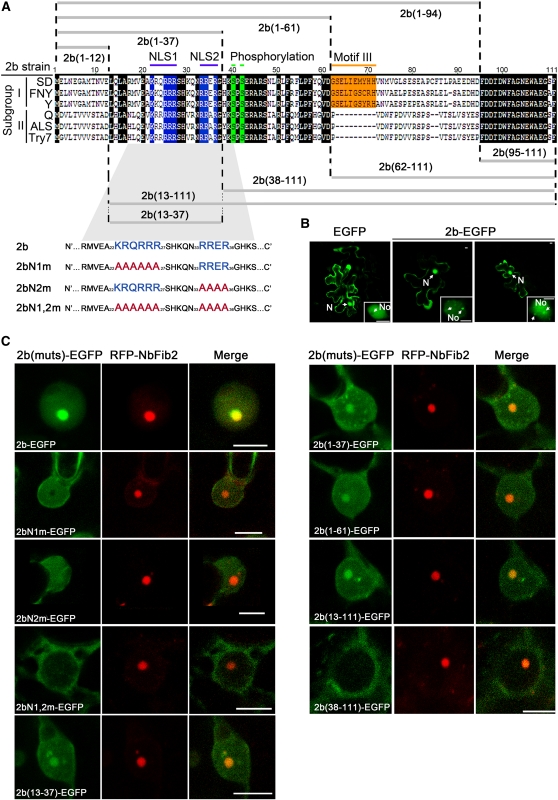
Previous studies have shown that the 2b protein encoded by CMV, which replicates in the cytoplasm, is localized to the nucleus and nucleolus (Lucy et al., 2000; Wang et al., 2004). The single and double NLSs contained in subgroup II and I CMV 2b proteins, respectively, have been characterized (Figure 1A). However, it is not clear if the CMV 2b protein contains a region that is necessary and sufficient for nucleolar localization. We examined the subcellular localization of SD2b fused at its C terminus with enhanced green fluorescent protein (EGFP) transiently expressed in Arabidopsis via bombardment. The green fluorescence of free EGFP from the control EGFP construct was observed in both the cytoplasm and nucleus but was excluded from the nucleolus, as could be observed in a close-up view (Figure 1B). We found that SD2b-EGFP was mainly detected in the nucleus with dense fluorescence in nucleoli, although some weak signal was found in nucleus-associated bodies (Figure 1B). Agrobacterium-mediated transient coexpression of SD2b-EGFP with fibrillarin, a marker of nucleolus (Kim et al., 2007), fused to red fluorescent protein (RFP-NbFib2) in N. benthamiana epidermal cells confirmed that SD2b localized in the nucleoli (Figure 1C). Thus, SD2b is also localized in the nucleolus, as found for the 2b protein of the Fny strain of CMV (González et al., 2010), which shares 89% identity with SD2b.
Figure 1.
Subcellular Localization of SD2b and Its Derivative Mutant Proteins.
(A) Alignment of 2b protein sequences from different subgroups of CMV and schematic of SD2b deletion mutants. The two NLS sequences, putative phosphorylation sites, and subgroup I–specific Motif III are highlighted in blue, green, and orange, respectively, and other conserved regions are highlighted in black. Two Arg-rich putative NLSs (blue letters) of the SD2b coding sequence were substituted with Ala residues (red letters) as previously reported (Lucy et al., 2000) and are also shown.
(B) Subcellular localization assays performed by bombarding 35S-SD2b-EGFP into 3-week-old Arabidopsis leaves. The empty EGFP vector was also bombarded as control. One of the typical cells from each bombardment assay for confocal microscopy analysis is presented. The nucleus (N), nucleolus (No), and possible nuclear bodies are labeled with arrows. Bars = 2.5 μm.
(C) Subcellular localization assays performed by coexpression of RFP-NbFib2 with 35S-SD2b-EGFP or mutant proteins fused with EGFP via Agrobacterium-mediated infiltration. One of the typical cells from each assay for confocal microscopy analysis is presented. Bars = 5 μm.
We generated Ala substitution mutants in either or both of the predicted NLSs of SD2b, 22KKQRRR27 and 33RRER36, yielding 2bN1m-EGFP, 2bN2m-EGFP, and 2bN1,2m-EGFP (Figure 1A). As expected from previous studies (Lucy et al., 2000; Wang et al., 2004), presence of either NLS allowed the accumulation of the fusion protein in the nucleus and removal of both NLSs in 2bN1,2m-EGFP or deleting the N-terminal 38 amino acids of SD2b that contains both NLSs in 2b(38-111)-EGFP led to the cytoplasmic localization of the resulting 2b fusion protein (Figure 1C). In contrast with SD2b-EGFP, however, the intense green fluorescence in the nucleolus was not observed in cells expressing either 2bN1m-EGFP or 2bN2m-EGFP (Figure 1C). Notably, 2bN1m-EGFP was specifically excluded from the nucleolus unlike 2bN2m-EGFP, which exhibited a uniform distribution throughout the nucleus (Figure 1C). Consistent with an earlier study (González et al., 2010), these observations indicate that the NLS-containing region of SD2b is necessary for the nucleolar targeting of the SD2b protein.
Several motifs composed mostly of Arg or Lys residues have been identified as both necessary and sufficient to target a protein to the nucleolus (Emmott and Hiscox, 2009). We noted that both NLSs of SD2b contain an (R/K)(R/K)X(R/K) motif that is conserved in CMV 2b proteins (Figure 1A) and appears twice in several characterized NoLSs (Horke et al., 2004). To determine if the N-terminal region of the 2b protein encodes an NoLS, we generated Agrobacterium infiltration constructs that coded for EGFP fused with amino acids 1 to 61, 1 to 37, 13 to 111, or 13 to 37 of SD2b (Figure 1A). We demonstrated strong nucleolar targeting for all of the 2b-GFP fusion proteins (Figure 1C), indicating that the region of SD2b from amino acid 13 to 37 is sufficient to direct the nucleolar targeting of the fusion proteins. Therefore, our findings together define the 13 to 37 region of SD2b as a functional NoLS.
The NoLS of the 2b Protein Is Part of the dsRNA Binding Domain
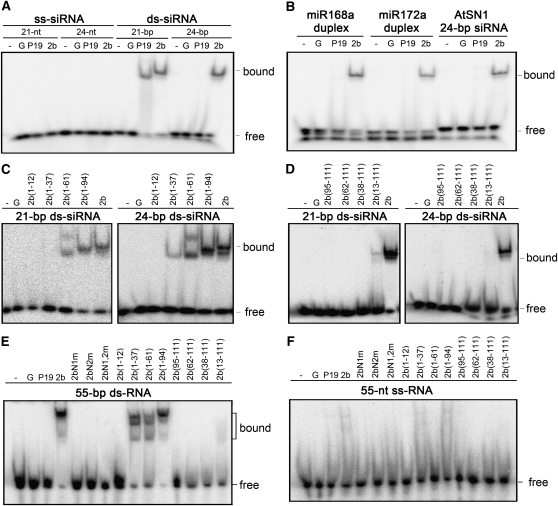
We next characterized the RNA binding activities of SD2b using electrophoretic mobility shift assays (EMSAs). To this end, SD2b, 2bN1m, 2bN2m, and 2bN1,2m as well as a panel of eight N- or C-terminal deletion mutants of SD2b (Figure 1A) were expressed and purified as GST fusion proteins (see Supplemental Figure 1A online). GST and GST-tagged P19, a tombusviral suppressor of RNA silencing (Silhavy et al., 2002), were used as controls. Similar to P19, SD2b did not bind to single-stranded RNA of 21, 24, or 55 nucleotides in length (Figures 2A and 2F; see Supplemental Figure 1D and Supplemental Table 1 online). GST-P19 efficiently bound the 21-bp but not 24-bp siRNA duplex, consistent with previous reports (Silhavy et al., 2002; Baulcombe and Molnár, 2004). We found that SD2b exhibited high affinity for 21- and 24-nucleotide siRNA duplexes (Figure 2A; see Supplemental Figure 1B online). Reciprocal-competing binding experiments further indicated that SD2b possesses a similar binding affinity for 21- and 24-nucleotide siRNA duplexes (see Supplemental Figure 1C online). By contrast, none of the NLS mutants was active in binding to either siRNA (see Supplemental Figure 1E online). These results are consistent with previous observations that 2b proteins encoded by two cucumoviruses, including a subgroup I strain of CMV, bind to duplexed siRNA in vitro (Goto et al., 2007; Chen et al., 2008).
Figure 2.
Gel Mobility Shift Assays for Detection of sRNA Binding Affinity of SD2b and Its Mutants.
sRNA binding affinity assays. GST-tagged SD2b ([A] and [B]), NLS mutants ([E] and [F]), and deletion mutants ([C] to [F]) were incubated with [γ-32P]-labeled synthetic 21- and 24-nucleotide siRNA ([A], [C], and [D]), miRNA duplexes (B), or with [γ-32P]-labeled synthetic 55-nucleotide-long ss or ds RNA ([E] and [F]) as indicated above the gels. GST-tagged P19 (P19), GST (G), and buffer (-) were used as controls. One nanomole of purified proteins was loaded in each assay. Bound and free probes are indicated. nt, nucleotides.
We found that SD2b also exhibited high affinities for 55-nucleotide-long dsRNA (Figure 2E) and for imperfect sRNA duplexes formed between miRNA and miRNA* (Figure 2B). We next determined the domain requirements for dsRNA binding by characterizing the three NLS mutants and the eight truncation mutants of SD2b in EMSA (Figures 2C and 2E). Alignment of the SD2b sequence to the crystal structure of the siRNA binding domain of TAV 2b (Chen et al., 2008) indicates that amino acids 8 to 36 and 41 to 58 of SD2b encode the α1 and α2 helices, respectively. We found that 2b(1-61) retained the ability to bind 21- and 24-nucleotide siRNA duplexes as well as 55-nucleotide dsRNA (Figures 2C and 2E), which is consistent with the prediction that 2b(1-61) contains the complete α1-linker-α2 structure involved in dsRNA binding. 2b(13-111) exhibited weak affinity for 21-nucleotide siRNA but showed no detectable affinity for either 24-nucleotide siRNA or 55-nucleotide dsRNA (Figures 2D and 2E). Moreover, deletion of 37 amino acids or longer from the N terminus abolished all of the dsRNA binding activities as did Ala substitutions in either or both of the NLS sequences between amino acids 13 and 37 (Figures 2C to 2E). These results indicate that deletions or mutations in the N-terminal α1 helix were deleterious to the dsRNA binding activity of the SD2b protein. Notably, although 2b(1-37) showed no detectable affinity for 21-nucleotide siRNA duplex (19-nucleotide dsRNA with 2-nucleotide 3′ overhangs), it was almost as active as 2b(1-61) in binding to 24-nucleotide siRNA duplex (22-nucleotide dsRNA with 2-nucleotide overhangs) and 55-nucleotide dsRNA (Figures 2C and 2E). Thus, it appeared that the region of SD2b encoding the short linker and the α2 helix was dispensable and the N-terminal α1 helix was sufficient for binding dsRNA of 24 and 55 nucleotides. As summarized in Figure 3, these results together indicate that the N-terminal half of SD2b contains overlapping domains required for nucleolar targeting and dsRNA binding. Since both 2b(1-37) and 2b(13-111) contained an intact NoLS, but exhibited defects in dsRNA binding, our results indicate that the NoLS is only part of a larger structural domain of SD2b required for binding to short and long dsRNA.
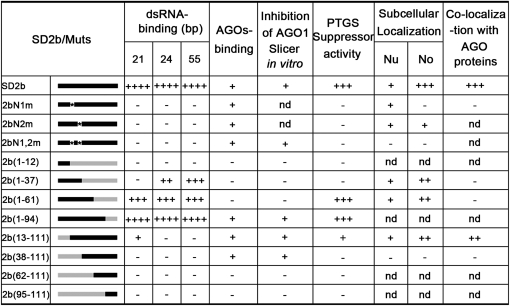
Figure 3.
Summary of Different Function Activities of SD2b and Its Derivative Mutants.
Muts, mutants; nd, no detection; No, nucleolus; Nu, nucleoplasm; +, positive activity; -, negative activity.
AGO Binding Requires a Domain of the 2b Protein Independent of Its dsRNA Binding
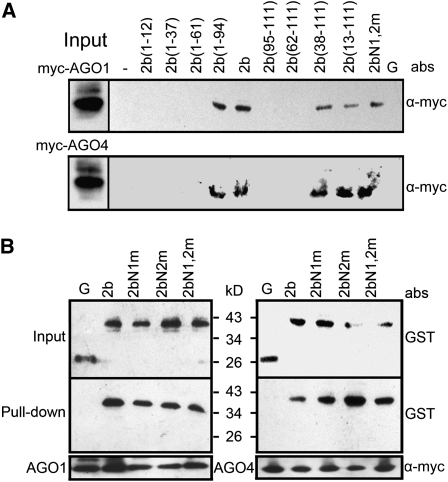
To detect the direct interaction between SD2b and AGOs, we generated epitope-tagged AGO1- and AGO4-expressing transgenic plants by transforming 35S-6myc-AGO1 and 35S-6myc-AGO4 plasmids into ago1-27 and ago4-2 mutants of Arabidopsis, respectively (Columbia-0 [Col-0] background; see Supplemental Figure 2 online). The expression of the AGO1 and AGO4 proteins was verified by immunoblot analysis (see Supplemental Figure 2 online). Complementation of ago1 and ago4 mutants by related myc-tagged AGO constructs was verified by examining the transgenic plants’ vegetal phenotypes and related siRNA hybridization (see Supplemental Figure 2 online). Purified, GST-tagged SD2b and its mutant fusion proteins were incubated with total protein extracts from 6myc-AGO1 or 6myc-AGO4 transgenic plants. Anti-α-myc antibody was then used to detect AGO proteins in the GST pulled-down products. We found that 2b(38-111), 2b(13-111), and 2b(1-94) displayed a binding affinity to both AGO1 and AGO4 similar to wild-type SD2b, whereas 2b(1-37), 2b(1-61), 2b(62-111), and other larger deletion mutants were all defective in direct interaction with AGOs (Figure 4A). Thus, the region of the SD2b encompassing residues 38 to 94 was essential for direct interaction of SD2b with AGOs, which, however, required neither the N-terminal α1 helix that was essential for dsRNA binding and included the NoLS nor the C-terminal 16 amino acids of SD2b that was relatively conserved among cucumoviral 2b proteins. As summarized in Figure 3, our findings together show that binding of the SD2b protein to AGOs requires a domain possibly between amino acids 38 and 94 and is independent of either dsRNA binding or nucleolar targeting.
Figure 4.
Detection of Domains of SD2b Required for Interaction with AGO Proteins.
Detection of AGO1 and AGO4 binding activities of deletion mutant (A) and SD2b NLS mutant proteins (B) by pull-down assay. Purified GST-tagged SD2b, NLS mutants, and deletion mutants (indicated on top of gels) incubated with total protein extracts from inflorescences of 6myc-AGO1 and 6myc-AGO4 transgenic Arabidopsis were pulled down using Glutathione Sepharose 4B beads (A) or anti-α -myc agarose beads (B). The pulled-down proteins were detected by immunoblot using anti-α-myc antibody (A) or anti-GST antibody (B). Incubation with GST protein (G) or buffer (-) serve as negative controls. abs, antibody. Input designates total protein extracts for immunoblot control.
In agreement with the above conclusion, mutating either or both of the NLSs of SD2b did not interfere with binding to either AGO1 or AGO4 in another assay using anti-GST antibody to pull down 6myc-AGO1 or 6myc-AGO4 (Figure 4B). Previous bimolecular fluorescence complementation experiments also suggested that NLS mutants of Fny-CMV 2b protein remain competent for interaction with AGO4 (González et al., 2010). Direct interaction of SD2b and AGO4 and AGO6 via the PAZ and PIWI domains conserved in all 10 Arabidopsis AGOs was confirmed by an in vitro assay (see Supplemental Figure 3 online), similar to the interaction with AGO1 reported previously (Zhang et al., 2006). This explains why SD2b physically interacted with each of the three AGOs examined.
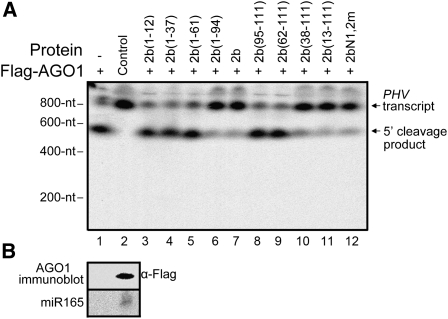
In Vitro Suppression of AGO1 Slicing Activity by the 2b Protein Requires Physical Interaction with AGOs but Is Independent of dsRNA Binding and Nucleolar Targeting
An in vitro reconstitution of the Arabidopsis AGO1-RISC described previously (Baumberger and Baulcombe, 2005; Zhang et al., 2006) was used to examine the slicing suppression activity of SD2b and its mutants. As expected, the cap-labeled PHAVOLUTA (PHV) RNA transcript, a target of miR165, was cleaved efficiently by the AGO1 complex immunoprecipitated from the total protein extracts of inflorescences from Flag-AGO1/ago1-36 plants by anti-α-FLAG M2 agarose beads but not by similar immunoprecipitated extracts from wild-type plants (Figure 5A, lanes 1 and 2). Figure 5B shows the presence of the endogenous miR165 and Flag-AGO1 in the complex coimmunoprecipitated from Flag-AGO1/ago1-36 plants. Prior incubation of GST-tagged SD2b with the AGO1 complex inhibited the miR165-guided cleavages of PHV RNA, leading to a markedly reduced production of the 5′-cleavage product (Figure 5A, lane 7). Four of the nine SD2b mutants examined, including 2b(13-111), 2b(38-111), 2b(1-94), and 2bN1,2m, were found to be potent suppressors of the in vitro slicing activity of AGO1 (Figure 5A). As summarized in Figure 3, all of the SD2b mutants capable of suppressing in vitro slicing retained the AGO1 binding activity. However, those suppressor-competent mutants included 2b(38-111) and 2bN1,2m, which were inactive in both dsRNA binding and nucleolar targeting (Figure 1C), as well as 2b(13-111), which retained only a weak binding affinity for 21-nucleotide siRNAs. These results indicate that the inhibition of AGO1 Slicer function by SD2b depends on the ability of SD2b to bind AGOs but is independent of its ability to bind to dsRNA or be targeted to the nucleolus.
Figure 5.
Detection of Inhibition Effects of SD2b and Its Mutations on AGO1-Mediated Cleavage Activity in Vitro.
(A) Effects of SD2b and its mutations (as indicated above the gel) on endogenous sRNA/Flag-AGO1–mediated cleavage activity in vitro. Flag-AGO1 obtained from inflorescences of Flag-AGO1/ago1-36 plants were incubated with buffer (lane 1) or GST-tagged SD2b or mutant proteins (lanes 3 to 12) and 32P-cap–labeled in vitro transcripts of PHV. Similar immunoprecipitated extracts from wild-type plants served as negative controls (lane 2). 32P-cap–labeled PHV RNA and 5′ cleavage products are indicated.
(B) Immunoprecipitates containing Flag-AGO1 protein were detected by immunoblot using a monoclonal antibody to Flag (top panel), and coimmunoprecipitated miR165 was detected by RNA gel blotting.
In Vivo Suppression of RNA Silencing by the 2b Protein Depends on dsRNA Binding but Is Independent of AGO Binding
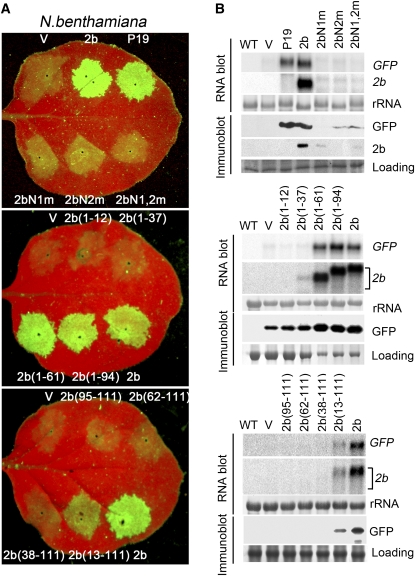
For the detection of the silencing suppression activity in vivo, we used an Agrobacterium coinfiltration assay in wild-type N. benthamiana leaves (Johansen and Carrington, 2001). RNA silencing against the infiltrated GFP transgene, which was RDR6 dependent, was observed at 4 d postagroinfiltration (DPA) in the infiltrated leaves when the 35S-GFP construct was coinfiltrated with the control (empty) binary plasmid (Figure 6). However, GFP silencing was suppressed when the binary plasmid encoding either SD2b or P19 was coinfiltrated (Figure 6A). We found that 2b(1-61) and 2b(1-94) were as active as SD2b in the coinfiltration assay and that 2b(13-111) retained partial suppressor activity (Figure 6A). However, the remaining nine mutants of SD2b failed to suppress GFP silencing as indicated by the lack of green fluorescence and accumulation of GFP protein and mRNAs in the coinfiltrated leaves (Figures 6A and 6B). We noted that all of the SD2b mutants defective in the suppression of GFP silencing were also expressed at low levels in the coinfiltrated leaves, as indicated by detection of SD2b mRNA and/or SD2b protein (Figure 6B). Thus, coinfiltration induced silencing against both the GFP and SD2b transgenes, which also inhibited the expression of those mutant SD2b transgenes that lost the silencing suppression activity. Consistent with this, coinfiltration with the tombusviral suppressor P19 restored the accumulation of most of the mutant SD2b mRNAs and of 2bN1m, 2bN2m, and 2bN1,2m proteins, which were among these mutant SD2b proteins detectable in immunoblots by the SD2b polyclonal antibodies (see Supplemental Figure 4 online). These results indicate that the SD2b mutant proteins were stable and lacked the silencing suppression activity to inhibit silencing targeting the coinfiltrated GFP and mutant SD2b transgenes.
Figure 6.
Analysis of Suppression of Transgene-Induced Silencing of SD2b Mutants in a Transient Expression System.
(A) GFP fluorescence in leaves of N. benthamiana plants coinfiltrated with 35S-GFP and 35S-SD2b-NLS mutants or 35S-SD2b deletion mutants (as indicated). Coinfiltrations of 35-GFP with vector control (V), 35S-SD2b, or 35S-P19 were used as controls. Photographs were taken under UV light at 4 DPA.
(B) RNA and protein gel blots analysis with samples extracted from infiltrated leaves at 4 DPA. 32P-labeled GFP and SD2b DNA probes were used. Anti-GFP monoclonal antibody and anti-SD2b polyclonal antibody were used to detect the accumulation of GFP and SD2b proteins in infiltrated leaves, respectively. Methylene blue–stained rRNA and Coomassie blue–stained protein are shown as mRNA and total protein loading controls.
As described above, 2b(1-61) retained the activity to bind long and short siRNAs and to be targeted to the nucleolus but lost the activity to bind AGOs or to suppress the in vitro slicer activity of AGO1. The N-terminal 69 amino acids of the TAV 2b protein that are folded into a dsRNA binding structure are also as effective as the wild-type TAV 2b in PTGS suppression (Chen et al., 2008). By contrast, 2b(13-111), which was partially active in the coinfiltration assay, exhibited no defect in binding to AGOs, suppressing the Slicer activity of AGO1 or localizing to the nucleolus compared with wild-type SD2b, but displayed weak 21-nucleotide siRNA binding activity and lost the ability to bind 24-nucleotide siRNA and long dsRNA (Figure 3). These results together strongly indicate that in vivo suppression of PTGS by the CMV 2b protein depends on its activity in dsRNA binding, in particular its ability to bind 21-nucleotide siRNA, and nucleolus targeting is insufficient. However, the activities of the SD2b protein either to bind AGO or to suppress the slicer activity of AGO1 appear dispensable for its silencing suppression in vivo.
Suppression of DNA Methylation by the 2b Protein Does Not Require Physical Interactions with AGOs
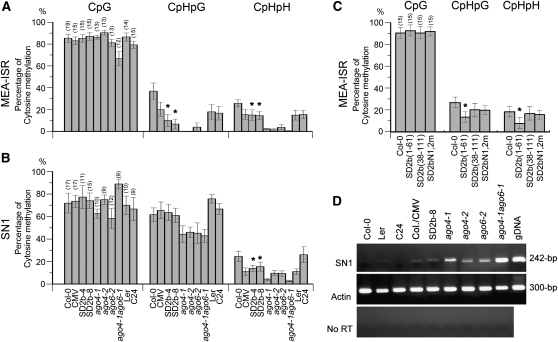
The in vivo nuclear localization and direct interactions with AGO of the SD2b protein suggest that SD2b may interfere with the RdDM in plants. To address this issue, coimmunoprecipitation experiments were performed in 6myc-tag transgenic Arabidopsis lines using anti-myc antibody. SD2b expressed from either the 6myc-SD2b transgene (see Supplemental Figure 5 online) or CMV infection was confirmed to interact with both AGO4 and AGO6 in vivo (see Supplemental Figure 6 online). DNA methylation of the well-characterized host endogenous genes at the MEA-ISR and SN1 loci (Zilberman et al., 2003) was examined in the 6myc-SD2b transgenic Arabidopsis plants by bisulfite genomic sequencing. As summarized in Figure 7A, the sequencing results revealed a similarly reduced non-CpG methylation at the MEA-ISR locus in two 6myc-SD2b lines (2b-4 and 2b-8) without affecting CpG symmetric methylation compared with that in control Col-0 (Figure 7A; see Supplemental Figure 7A online). At the SN1 locus, only CpHpH asymmetric methylation was affected in both lines (Figure 7B). Quantitative RT-PCR confirmed that reduction of SN1 DNA methylation increased its transcript expression in line 2b-8 (Figure 7D). DNA gel blots also showed hypomethylation only on CpHpH sites, but not CpG and CpHpG sites, of 5S rDNA in line 2b-8 (see Supplemental Figure 8 online). Taken together, reduction of CpHpH asymmetric methylation of endogenous genes was a common feature found in the two SD2b transgenic lines at the tested loci, which was in agreement with a recent report (Hamera et al., 2012).
Figure 7.
Analysis of Endogenous DNA Methylation in SD2b and SD2b Mutant Transgenic and SD-CMV–Infected Plants.
(A) to (C) Percentage of CG, CNG, and CNN methylation by bisulphite sequencing analysis of endogenous MEA-ISR and SN1 loci in the indicated genotypes. The number of clones analyzed is in parenthesis, and the original data are shown in Supplemental Figure 7 online. The statistical analysis was performed using the OriginPro 8 program. The bars denote the se of the means, and the corresponding values of SD2b-4, SD2b-8, and SD2b(1-61) (labeled with asterisks) are significantly different from those of Col-0 (one-way analysis of variance, P < 0.05).
(D) Quantitative RT-PCR showed increased SN1 transcript expression in line 2b-8 and SD-CMV–infected plants and ago mutant plants compared with their control background plants.
Similar reduction of methylation at the MEA-ISR locus was observed in transgenic plants expressing SD2b(1-61) but not in those expressing either SD2bN1,2m or SD2b(38-111) (Figure 7C; see Supplemental Figures 5A and 7B online). SD2b(1-61) plants also displayed mild developmental defects reminiscent of those visible in transgenic plants expressing SD2b (see Supplemental Figure 5A online). Since SD2b(1-61) lost the activity to directly interact with AGOs in vitro and in vivo, our results indicate that suppression of DNA methylation by the SD2b protein also requires the dsRNA binding domain but not the physical interaction with AGO proteins. In AGO-deficient mutant plants (ago4-1, ago4-2, and ago6-2), stronger reduction of CpHpH asymmetric methylation of the MEA-ISR, SN1, and 5S rDNA was observed in the parallel analysis (Figure 7; see Supplemental Figures 7A and 8 online), consistent with a previous report (Zilberman, 2008).
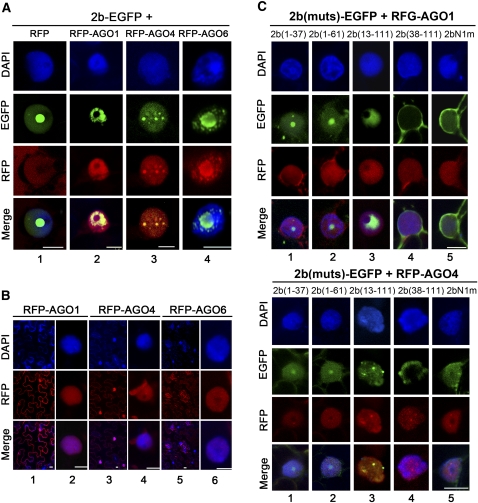
AGO-2b Interactions Have the Potential to Redistribute Both Proteins in the Nucleus
We next investigated if the direct interaction of the SD2b protein with AGOs had any biological implication in the host cell. Expression of RFP-AGO1 alone was distributed uniformly in the nucleus and in the cytoplasm (Figure 8B, columns 1 and 2) unlike SD2b-EGFP, which was mainly found in the nucleoli. However, coexpression of SD2b-EGFP and RFP-AGO1 resulted in their colocalization in the nucleus but absence from the nucleolus (Figure 8A, column 2). Redistribution of SD2b-EGFP in the nucleus out of the nucleolus was not observed when SD2b-EGFP was coexpressed with RFP (Figure 8A, column 1). Redistribution of SD2b-EGFP in the nucleus was also detected when SD2b-EGFP was coexpressed with RFP-AGO4 (Figure 8A, column 3) or with RFP-AGO6 (Figure 8A, column 4). We noted that the distribution patterns of AGO1, AGO4, and AGO6 were also altered when they were coexpressed with SD2b-EGFP (Figure 8).
Figure 8.
Colocalization Pattern of SD2b-AGO Complexes in N. benthamiana Leaf Epidermal Cells.
(A) Subcellular colocalization of SD2b-EGFP and RFP-AGO proteins. SD2b-EGFP coinfiltrated with empty RFP vector serves as control. One representative colocalization image is shown for each experiment.
(B) Subcellular location of RFP-AGO1, RFP-AGO4, and RFP-AGO6.
(C) Subcellular location of coexpression of SD2b derivative mutants with RFP-AGO1 (top panel) and RFP-AGO4 (bottom panel). GFP and RFP fluorescence were photographed at 3 DPA. DAPI staining was performed to represent the nuclei. Bars = 10 μm.
We observed similar nuclear colocalization and redistribution of SD2b and AGO proteins when RFP-AGO1 or RFP-AGO4 was coexpressed with 2b(13-111)-EGFP (Figure 8C, column 3), which physically interacted with AGOs and contained an intact NoLS but was defective in dsRNA binding. However, neither colocalization nor redistribution of SD2b and AGO proteins was detected when either RFP-AGO1 or RFP-AGO4 was coexpressed with 2b(1-37)-EGFP, 2b(1-61)-EGFP, or 2bN1m-EGFP (Figure 8C, columns 1, 2, and 5). None of these three 2b mutants physically interacted with AGOs, although 2bN1m-EGFP targeted to the nucleus (Figure 1C) and 2b(1-37)-EGFP and 2b(1-61)-EGFP contained an intact NoLS. We rarely observed nuclear colocalization of RFP-AGO1 or RFP-AGO4 with 2b(38-111)-EGFP, which physically interacted with AGOs but did not encode a NoLS. In the few cells where 2b(38-111)-EGFP and RFP-AGO proteins seemed to colocalize in the nuclear area, a close-up view demonstrated that the 2b(38-111)-EGFP was localized in the cytoplasm surrounding the nucleus but absent in the nucleus (Figure 8C, column 4). These findings indicate that in addition to nucleolar targeting, the activity of the SD2b protein to physically interact with AGOs is essential for the nuclear colocalization and redistribution of SD2b and AGO proteins.
DISCUSSION
In spite of being a small protein, 2b of CMV exhibits complex biochemical and subcellular targeting activities. These include binding to AGO1/AGO4 and short/long dsRNA and targeting to the nucleus and nucleolus. CMV 2b also has diverse silencing suppressor activities, including suppression of transgene-induced RNA silencing and DNA methylation, intercellular spread of RNA silencing, Slicer activity of AGO1 and AGO4, and host RDR6-dependent antiviral silencing. However, it is not clear what domains of 2b determine these activities and how each of these activities contributes to the in vivo silencing suppression. In this study, we defined the domain requirements for these biochemical activities of the SD-CMV 2b protein. Subsequent characterization of SD2b mutants defective in one or more of the biochemical activities in the suppression of AGO1 Slicer activity and transgene-induced RNA silencing and DNA methylation provides insights into the mechanism of the CMV 2b protein.
We found that the N-terminal half of the 2b protein encodes overlapping domains involved in binding to short and long dsRNA and in targeting to the nucleolus. We demonstrated high affinity binding of the SD2b protein to long dsRNA in addition to 21- to 24-nucleotide siRNA duplexes reported previously for the 2b proteins of CMV and TAV (Goto et al., 2007; Chen et al., 2008). The random selection of siRNA duplexes may explain why the 2b protein of Fny-CMV was active in siRNA binding in one study (Hamera et al., 2012) but inactive in another (Zhang et al., 2006). Characterization of 11 mutants of the SD2b protein in this work (Figure 3) revealed differential domain requirements for binding to 21-nucleotide siRNA duplex and to 24-nucleotide siRNA duplex/long dsRNA. Although the N-terminal 61 amino acids encoding the putative α1-short linker-α2 dsRNA binding domain were required for binding to 21-nucleotide siRNA duplex, only the N-terminal α1 helix of SD2b appeared to be essential for binding to 24-nucleotide siRNA duplex and long dsRNA. However, only the region encompassing amino acids 13 to 37 of the SD2b protein is both necessary and sufficient for nucleolar targeting. The NoLS of the SD2b protein first defined for a VSR shares a consensus sequence with other viral and cellular nucleolus-targeting proteins (Boyne and Whitehouse, 2006; Rajamäki and Valkonen, 2009) and includes the two closely spaced NLSs characterized in previous studies (Lucy et al., 2000; Wang et al., 2004). By contrast, deleting the entire N-terminal 37 amino acids or mutating either NLS or both within this region did not prevent the direct interaction of SD2b with either AGO1 or AGO4. These findings together demonstrate distinct domain requirements of the CMV 2b protein for dsRNA binding and physical interaction with AGO1/AGO4.
We detected active suppression of the in vitro slicing activity of AGO1 by all of the SD2b mutants that retained the ability to physically interact with AGO1/AGO4, including those mutants that are defective both in binding to dsRNA and targeting to the nucleolus: 2bN1,2m and 2b(38-111). These findings show that direct interaction with AGO1 is sufficient for 2b suppression of the in vitro AGO1 Slicer activity (Zhang et al., 2006). However, active suppression of cytoplasmic RDR6-dependent PTGS in coinfiltration assays was observed for 2b(1-61), which contained the complete dsRNA binding domain and retained the activities for binding long dsRNA and 21- and 24-nucleotide duplex siRNAs but lost the ability either to interact with AGOs or to suppress the AGO1 Slicer activity in vitro. Efficient suppression of RNA silencing in the same in vivo assay has also been reported for TAV 2b(1-69) (Chen et al., 2008). Notably, all except one of the SD2b mutants that remained active suppressors of the AGO1 Slicer activity in vitro were inactive in the coinfiltration assay (Figure 3). The exception was 2b(13-111), which exhibited a weak suppressor activity in the coinfiltration assay and was the only one among the AGO binding mutants that retained a weak activity to bind 21-nucleotide siRNA duplex. These results together indicate that the activity of the SD2b protein in dsRNA binding is essential, but the direct interaction with AGOs or its suppression of the AGO1 Slicer activity in vitro are dispensable, for the in vivo suppression of PTGS by the SD2b protein. The essential role of the NLS sequence as part of the dsRNA binding domain for in vivo silencing suppression explains why rescue of the nuclear targeting defect of a 2b NLS substitution mutant by adding a heterologous NLS at the C terminus failed to restore the silencing suppression activity (Lucy et al., 2000). Based on these findings, we propose that CMV 2b may act by binding and sequestering siRNA and its precursor long dsRNA during de novo dsRNA synthesis by the RDR6-dependent pathway. This model is consistent with the previous observations that CMV 2b inhibits both the RDR6-dependent amplification of viral siRNAs and the intercellular spread of transgene RNA silencing, which is now known to be mediated by the movement of siRNAs that are amplified by the RDR6 pathway (Dunoyer et al., 2005, 2010; Molnar et al., 2010; Melnyk et al., 2011b).
Similar to previous observations (Guo and Ding, 2002; Hamera et al., 2012), we found that stable expression of SD2b in transgenic Arabidopsis plants was correlated with reduction of DNA methylation at the MEA-ISR locus. Expression of 2b(1-61) was also effective, whereas no significant changes in DNA methylation at the MEA-ISR locus was observed in transgenic plants expressing either 2bN1,2m or 2b(38-111), both of which retained the ability to interact physically with AGOs and to suppress the AGO1 Slicer activity in vitro (Figures 3 and 7). These findings further indicate that suppression of DNA methylation by the CMV 2b protein requires the dsRNA binding activity but is independent of its ability to interact directly with AGOs. This suggests a similar role for the binding and sequestering siRNA and its precursor long dsRNA in the SD2b suppression of RdDM in the nucleus, which is supported in part by a recent observation that expression of CMV 2b was associated with the enrichment and retention of the labeled synthetic siRNA in the nucleus (Kanazawa et al., 2011). However, 2b expressed from a CMV-based vector carrying a host gene promoter sequence actually facilitated virus-induced TGS and DNA methylation (Kanazawa et al., 2011), similar in principle to PTGS of host genes targeted by potato virus X and geminiviral vectors, all of which express their own VSR to ensure a robust systemic viral infection (Lu et al., 2004; Carrillo-Tripp et al., 2006; Pandey et al., 2009; Zhang et al., 2011). Thus, VSRs inhibit antiviral silencing but become ineffective against silencing of a nuclear gene by the same pool of viral siRNAs, consistent with the idea that a replicating virus is much more difficult to silence and easier to suppress than a nuclear gene transcribed at a steady rate.
It is currently unknown how the 2b suppression of RdDM in the nucleus may contribute to the infection of a cytoplasmic virus whose replication involves only RNA. The physical interaction of SD2b with AGOs nonessential for its in vivo suppression activities requires the region encompassing residues 62 to 94. Although this region is highly variable in sequence, it is present among all of the cucumoviral 2b proteins (Ding et al., 1994), suggesting a possible in vivo function for CMV infection. We found in this study that the in vivo AGO–SD2b interaction depends also on the nucleolar targeting of the SD2b protein and has potential to redistribute both SD2b and AGOs in the nucleus by an unknown mechanism. In plants, production of heterochromatic siRNA, which is involved in TGS, is thought to occur in a region of the nucleolus or in nucleolus-associated body (D-body) due to the localization of protein components of the machinery and of siRNAs (Pontes and Pikaard, 2008). Moreover, cell-to-cell spread of RNA silencing and perception of the systemic silencing signal in plants requires the nuclear silencing pathway and the onset of systemic silencing was delayed in ago4-null mutant scions (Brosnan et al., 2007; Dunoyer et al., 2007; Smith et al., 2007). Thus, we propose that nucleolar targeting of SD2b and its physical interaction with AGO4 and AGO6 may enhance the activity of CMV SD2b in the suppression of the nuclear RdDM and the short- and long-distance spread of RNA silencing. However, we cannot rule out a possibility that physical interaction between the SD2b and AGO proteins and redistribution of the SD2b-AGO proteins may represent a plant defense response to counteract suppression of silencing induced by the nucleolar targeting of SD2b. Further investigation of the contribution to differences in CMV infectivity with different C-terminal deletion mutants of SD2b will be helpful to address this issue.
METHODS
Plant Material and Growth Conditions
The ago1-27, ago4-2, ago6-2, and Flag-AGO1/ago1-36 transgenic Arabidopsis thaliana was described previously (Morel et al., 2002; Baumberger and Baulcombe, 2005; Agorio and Vera, 2007). All transgenic plants and wild-type Col ecotype Arabidopsis were grown in glass house at 23°C and 16-h-light/8-h-dark cycles. Nicotiana benthamiana was grown in a glass house at 25°C and 16-h-light/8-h-dark cycles.
Construction of all plasmids, including: 35S-6myc-SD2b, 35S-6myc-AGO4, 35S-6myc-AGO6, 35S-6myc-AGO1, GST-P19, GST-tagged SD2b and its NLS and deletion mutant fusion protein constructs, SD2b and its NLS and deletion mutant EGFP fusion protein constructs, RFP-AGO1, RFP-AGO4, and His-tagged truncated peptide of AGO proteins constructs, as well as RFP-NbFib2, is described in Supplemental Methods 1.
Plant Transformation
All constructs for plant transformation were transformed into the Agrobacterium strain EHA105, and Arabidopsis transformations were performed using a standard floral dip method (Clough and Bent, 1998). 35S-6myc-SD2b, 35S-SD2b(1-61), 35S-SD2b(38-111), and 35S-SD2bN1,2m were transformed into Col-0 ecotype, and positive transformants were screened by Basta and hygromycin resistance. 35S-6myc-AGO1, 35S-6myc-AGO4, and 35S-6myc-AGO6 were transformed into ago1-27, ago4-2, and ago6-2 and positive transformants were screened on Murashige and Skoog medium containing 20 μg/L hygromycin and confirmed by immunoblots.
RNA Extraction and RNA Gel Blot Analysis
Plant total RNA used for RNA gel blot was extracted by the hot-phenol method as described (Fernández et al., 1997). For detection of GFP, SD2b, and TAS1C, DNA fragments were amplified by PCR with respective primers (see Supplemental Table 2 online) and labeled with [α-32P]dCTP using the Rediprime II system (GE Healthcare). For sRNA gel blots, total RNA was extracted using Trizol reagent (Invitrogen), and sRNA was obtained via lithium chloride and ethanol precipitation from total RNA. For detection of siRNAs, 30 μg sRNA was separated on 17% polyacrylamide-8 M urea gels. The probes (see Supplemental Table 2 online) were labeled with [γ-32P]ATP using polynucleotide kinase (NEB). Signal intensity was quantified using ImageQuant TL software (GE Healthcare).
EMSAs
Synthesized RNA oligos (see Supplemental Table 1 online) were radiolabeled in 50 pmol quantities with 0.3 μM [γ-32P]ATP and 20 units of T4 PNK (NEB). Annealing was performed by heating equimolar complementary single-stranded (ss) oligo mixtures in 10 mM Tris/HCl, pH 7.5, and 100 mM KCl at 99°C for 5 min followed by cooling to room temperature. Binding reactions were performed with 1 ng of radiolabeled ss/ds siRNA and 1 nmol of protein in binding buffer (20 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 300 mM NaCl, 0.1% Nonidet P-40, and cocktail). After 40 min at room temperature, 1 μL 50% glycerol and dye were added and protein-RNA complexes were resolved on 6% native polyacrylamide gel. The gels were then dried and exposed to a storage phosphor screen (GE Healthcare). For competition experiments, complexes of the protein-labeled ds siRNA were first formed. Synthesized ss oligos were 5′ phosphorylated and annealed as described above to form competitor ds siRNA. Then, competitors were added into the binding reaction at 10-, 100-, and 1000-fold excess. After an additional incubation for 30 min, the samples were subjected to 6% native polyacrylamide gel.
Expression and Purification of Recombinant Proteins
For Escherichia coli expression of fusion proteins, recombinant plasmids were transformed into BL21 cell and induced at 0.3 mM isopropyl ββ-d-1-thiogalactopyranoside (Sigma-Aldrich) in Luria-Bertani medium at 28°C for 3 to ~6 h. 6His- and GST-tagged fusion proteins were purified using Ni2+-nitriloacetic acid agarose beads (Qiagen) and Glutathione Sepharose 4B (GE Healthcare) by affinity columns, respectively, according to the manufacturer’s instructions.
Plant Total Protein Extraction, Antibody Preparation, and Immunoblots
Total plant protein extraction and immunoblotting were performed as described previously (Ying et al., 2010). SD2b polyclonal antiserum was prepared as previously described (Hou et al., 2011). AGO4 and AGO6 polyclonal antisera were generated by immunizing rabbits with purified His-tagged AGO4 and AGO6 PIWI peptides (see Supplemental Methods 1 online). Total protein extracts were resolved on 12% polyacrylamide gels and transferred onto polyvinylidene fluoride membranes (GE Healthcare). The membranes were blotted with the primary antibody α-Myc (9E10) (Santa Cruz Biotechnology) at a dilution of 1/1000; α-Flag (Sigma-Aldrich), His, GST, and GFP antibodies (Abmart) at 1/5000; and SD2b, AGO4, and AGO6 polyclonal antisera at 1/1000. After incubation with HRP-labeled secondary antibody (Abmart), the membrane was detected with Pierce ECL Western Blotting Substrate (Thermo Scientific).
In Vitro Pull-Down Assays
For SD2b and NLS mutant protein pull-down assays, 10 nmol of purified GST-tagged proteins was added into total protein extracts from inflorescences of 6myc-AGO1 and 6myc-AGO4 transgenic plants (0.5 g starting material for each sample). Forty microliters of anti-α-myc Agarose was then added, followed by overnight incubation at 4°C. After six vigorous washes, the immunoprecipitates were resuspended with loading buffer and denatured. The input and pulled-down proteins were detected by immunoblots using anti-GST antibody. The immunoprecipitated 6myc-AGO1 and 6myc-AGO4 was confirmed by anti-α-myc antibody immunoblots.
For SD2b deletion mutant pull-down assays, purified fusion proteins were incubated with 40 μL Glutathione Sepharose beads in PBS for 1 h rotating at room temperature. After three washes with PBS, the beads were then incubated with total protein extracts from inflorescences of 6myc-AGO1 and 6myc-AGO4 transgenic plants in immunoprecipitation buffer for 4 h at room temperature. Four washes followed and then the beads were resuspended with loading buffer and denatured protein complexes were detected as described above using anti-α-myc antibody.
In Vivo Coimmunoprecipitation
Coimmunoprecipitation was performed as previously reported (Azevedo et al., 2010) with minor modifications. Aerial tissues from wild-type and transgenic Arabidopsis (6myc-SD2b, 6myc-AGO4, and 6myc-AGO6) were homogenized in immunoprecipitation buffer. After centrifugation and filtration with a 0.45 μm filter, the clarified lysate was incubated with anti-myc agarose beads (Abmart) or anti-Flag M2 agarose beads (Sigma-Aldrich), and the complex was washed five times with immunoprecipitation buffer. The immunoprecipitates were denatured and subjected to immunoblot using corresponding antibodies.
AGO Slicer Activity
AGO1 Slicer assays were performed according to previous reports (Baumberger and Baulcombe, 2005; Qi et al., 2006; Zhang et al., 2006) with minor modifications. In brief, PHV mRNA was synthesized using an in vitro transcription kit (mMESSAGE mMACHINE T7; Ambion). After purification, mRNA was G-capped with [α-32P]GTP (3000 Ci/mmol; Perkin-Elmer) using Guanylyltransferase recombinant solution (Wako). After RNA purification with Quick Spin columns (Roche), the labeled mRNA was resuspended in 50 μL of RISC buffer. Immunoprecipitation of Flag-AGO1 was performed as described above with inflorescences of Flag-AGO1/ago1-36 and wild-type plants using α-FLAG M2 agarose beads. After two washes, Flag-AGO1 was eluted with 3×Flag peptide solution (Sigma-Aldrich; 250 μg/mL, 100 μL for 1 g of starting material). Twenty microliters of the eluate was incubated with 5 nmol of suppressor proteins for 3 h at 4°C. Two microliters of 32P-cap–labeled transcript substrate solution containing 0.25 μL of RNasin and 1 μL of 20 mM ATP was immediately added. After 3 h incubation at 25°C, RNA was recovered with Trizol reagent and separated on 8 M urea/5% polyacrylamide gels. The signals were detected via exposure to a storage phosphor screen (GE Healthcare).
Virus Inoculation and Transient Expression
Four-week-old Arabidopsis plants were inoculated with freshly prepared sap from wild-type SD-CMV–infected N. benthamiana leaves by rubbing mechanically. For suppressor activity assays, 5-week-old N. benthamiana leaves were infiltrated with culture of Agrobacterium tumefaciens EHA105 carrying 35S-GFP and SD2b or its mutant plasmids, and 35S-P19 and empty vector pBI121-GUS were coinfiltrated as control. The concentration of each culture was adjusted to OD600 0.5.
Subcellular Localization Assays
For SD2b subcellular localization assays, pBI221-SD2b-EGFP and pBI221-EGFP were bombarded into 3-week-old Arabidopsis leaf epidermal cells for transient expression. After 16 to 24 h culture on Murashige and Skoog medium in the dark, leaves were mounted on slides and visualized using a fluorescence microscope (Leica TCS SP5 II). Subcellular colocalization assays for SD2b and SD2b-derivative mutants were performed by infiltrating binary plasmids of pBI121 and pGDR vectors carrying related fragments into 5-week-old N. benthamiana leaves, which were maintained for 3 d at 25°C (16 h light/8 h dark). The nuclei were stained with 100 ng/mL 4′,6-diamidino-2-phenylindole (DAPI) for 10 min before confocal microscopy. Confocal fluorescence of GFP, RFP, and DAPI were captured with Leica TCS SP5 II confocal microscopes, and images were processed with Adobe Photoshop software (Adobe Systems).
DNA Bisulphite Sequencing Analysis
For bisulphite sequencing, 2 μg of Arabidopsis (Col-0) genomic DNA was digested with EcoRI and XhoI and used for bisulfate treatment using the EpiTect Bisulfite kit (Qiagen) according to handbook. The purified bisulphite-treated DNA was amplified by SN1- and MEA-ISR–specific primer pairs designed as previously described (Zilberman et al., 2003) (see Supplemental Table 2 online). PCR products were ligated into pGEM-Teasy, and nine to 19 individual clones were sequenced. The cytosine methylation analysis was performed via the CyMATE program (http://www.gmi.oeaw.ac.at/en/cymate-index/).
Quantitative RT-PCR
RT-PCR was performed as previously described (Gao et al., 2010) using M-MLV reverse transcriptase (Takara). cDNA was then subject to quantitative PCR using SN1-specific primers to amplify a 242-bp fragment with 25 thermal cycles. PCR reactions without reverse transcription were performed as controls to rule out DNA contamination. Three biological replicates were performed. SN1 expression was quantified at the logarithmic phase using the expression of the Actin housekeeping gene as an internal control. The primers are listed in Supplemental Table 2 online.
Accession Numbers
Sequence data from this article can be found in the GenBank database under the following accession numbers: At-AGO1 (NM 179453), At-AGO4 (NM 128262), At-AGO6 (128854), SD-CMV 2b (D86330), pBI221 (AF502128), and pCAMBIA1300 (AF234296).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Gel Mobility Shift Assay Detection of Small RNA Binding Affinity of SD2b.
Supplemental Figure 2. Phenotypes and Protein Expression of 6myc-AGO Transgenic Arabidopsis.
Supplemental Figure 3. Pull-Down Analysis of SD2b with Truncated AGO4 and AGO6 Proteins in Vitro.
Supplemental Figure 4. RNA Gel Blot and Immunoblot Analysis of Expression Levels of GFP, 2b, and 2b Mutant mRNA and the Corresponding Proteins in Coinfiltration Assays.
Supplemental Figure 5. Morphology Phenotypes and Expression Analysis of Transgenic Arabidopsis Plants Expressing SD2b and Mutants.
Supplemental Figure 6. Analysis of in Vivo Protein–Protein Interaction between SD2b and AGO Proteins.
Supplemental Figure 7. Bisulfite Sequencing Analysis of Methylated Cytosine Using the CyMATE Program.
Supplemental Figure 8. Analysis of 5S rDNA Methylation in SD2b Transgenic and SD-CMV–Infected Plants by DNA Gel Blots.
Supplemental Table 1. List and Structure of RNA Oligos Used for EMSA.
Supplemental Table 2. List of Primers and DNA Oligos Used in This Study.
Supplemental Methods 1. Plasmid Constructs, Genomic DNA Extraction and Blotting, and AGO Peptide Pull-Down Assay.
Acknowledgments
We thank David Baulcombe for Flag-AGO1/ago1-36 seeds, Yijun Qi for ago6-2 and ago4-1ago6-1 seeds, and Astrid Agorio for ago4-2 seeds. We thank Nam-Hai Chua for the pBA-6myc-AGO1 plasmid. This research was supported by grants from the National Science Foundation of China (Grant 31030009) and the Ministry of Science and Technology (Grant 2011CB100703).
AUTHOR CONTRIBUTIONS
H.-S.G. and C.-G.D. designed the research. C.-G.D., Y.-Y.F., B.-J.Z., W.-N.H., and H.Z. performed research. J.-H.Z. contributed new analytic/computational/etc. tools. H.-S.G., C.-G.D., and S.-W.D. analyzed the data and wrote the article.
References
- Agorio A., Vera P. (2007). ARGONAUTE4 is required for resistance to Pseudomonas syringae in Arabidopsis. Plant Cell 19: 3778–3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A.M., Carrington J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Anandalakshmi R., Pruss G.J., Ge X., Marathe R., Mallory A.C., Smith T.H., Vance V.B. (1998). A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 95: 13079–13084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo J., Garcia D., Pontier D., Ohnesorge S., Yu A., Garcia S., Braun L., Bergdoll M., Hakimi M.A., Lagrange T., Voinnet O. (2010). Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen-encoded GW repeat protein. Genes Dev. 24: 904–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Baulcombe D. (2004). RNA silencing in plants. Nature 431: 356–363 [DOI] [PubMed] [Google Scholar]
- Baulcombe D. (2005). RNA silencing. Trends Biochem. Sci. 30: 290–293 [DOI] [PubMed] [Google Scholar]
- Baulcombe D.C., Molnár A. (2004). Crystal structure of p19—A universal suppressor of RNA silencing. Trends Biochem. Sci. 29: 279–281 [DOI] [PubMed] [Google Scholar]
- Baumberger N., Baulcombe D.C. (2005). Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 102: 11928–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béclin C., Berthomé R., Palauqui J.C., Tepfer M., Vaucheret H. (1998). Infection of tobacco or Arabidopsis plants by CMV counteracts systemic post-transcriptional silencing of nonviral (trans)genes. Virology 252: 313–317 [DOI] [PubMed] [Google Scholar]
- Boyne J.R., Whitehouse A. (2006). Nucleolar trafficking is essential for nuclear export of intronless herpesvirus mRNA. Proc. Natl. Acad. Sci. USA 103: 15190–15195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigneti G., Voinnet O., Li W.X., Ji L.H., Ding S.W., Baulcombe D.C. (1998). Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17: 6739–6746 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Brodersen P., Sakvarelidze-Achard L., Bruun-Rasmussen M., Dunoyer P., Yamamoto Y.Y., Sieburth L., Voinnet O. (2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Brodersen P., Voinnet O. (2006). The diversity of RNA silencing pathways in plants. Trends Genet. 22: 268–280 [DOI] [PubMed] [Google Scholar]
- Brosnan C.A., Mitter N., Christie M., Smith N.A., Waterhouse P.M., Carroll B.J. (2007). Nuclear gene silencing directs reception of long-distance mRNA silencing in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 14741–14746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgyan J., Havelda Z. (2011). Viral suppressors of RNA silencing. Trends Plant Sci. 16: 265–272 [DOI] [PubMed] [Google Scholar]
- Carrillo-Tripp J., Shimada-Beltrán H., Rivera-Bustamante R. (2006). Use of geminiviral vectors for functional genomics. Curr. Opin. Plant Biol. 9: 209–215 [DOI] [PubMed] [Google Scholar]
- Chapman E.J., Prokhnevsky A.I., Gopinath K., Dolja V.V., Carrington J.C. (2004). Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 18: 1179–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.Y., Yang J., Lin C., Yuan Y.A. (2008). Structural basis for RNA-silencing suppression by Tomato aspermy virus protein 2b. EMBO Rep. 9: 754–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V., Zhu J.K. (2009). RNA-directed DNA methylation and demethylation in plants. Sci. China C Life Sci. 52: 331–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Díaz-Pendón J.A., Ding S.W. (2008). Direct and indirect roles of viral suppressors of RNA silencing in pathogenesis. Annu. Rev. Phytopathol. 46: 303–326 [DOI] [PubMed] [Google Scholar]
- Diaz-Pendon J.A., Li F., Li W.X., Ding S.W. (2007). Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell 19: 2053–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S.W. (2010). RNA-based antiviral immunity. Nat. Rev. Immunol. 10: 632–644 [DOI] [PubMed] [Google Scholar]
- Ding S.W., Anderson B.J., Haase H.R., Symons R.H. (1994). New overlapping gene encoded by the cucumber mosaic virus genome. Virology 198: 593–601 [DOI] [PubMed] [Google Scholar]
- Ding S.W., Voinnet O. (2007). Antiviral immunity directed by small RNAs. Cell 130: 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P., Himber C., Ruiz-Ferrer V., Alioua A., Voinnet O. (2007). Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nat. Genet. 39: 848–856 [DOI] [PubMed] [Google Scholar]
- Dunoyer P., Himber C., Voinnet O. (2005). DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat. Genet. 37: 1356–1360 [DOI] [PubMed] [Google Scholar]
- Dunoyer P., Schott G., Himber C., Meyer D., Takeda A., Carrington J.C., Voinnet O. (2010). Small RNA duplexes function as mobile silencing signals between plant cells. Science 328: 912–916 [DOI] [PubMed] [Google Scholar]
- Dunoyer P., Voinnet O. (2008). Mixing and matching: the essence of plant systemic silencing? Trends Genet. 24: 151–154 [DOI] [PubMed] [Google Scholar]
- Emmott E., Hiscox J.A. (2009). Nucleolar targeting: the hub of the matter. EMBO Rep. 10: 231–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández A., Guo H.S., Sáenz P., Simón-Buela L., Gómez de Cedrón M., García J.A. (1997). The motif V of plum pox potyvirus CI RNA helicase is involved in NTP hydrolysis and is essential for virus RNA replication. Nucleic Acids Res. 25: 4474–4480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., et al. (2010). An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature 465: 106–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasciolli V., Mallory A.C., Bartel D.P., Vaucheret H. (2005). Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 15: 1494–1500 [DOI] [PubMed] [Google Scholar]
- González I., Martínez L., Rakitina D.V., Lewsey M.G., Atencio F.A., Llave C., Kalinina N.O., Carr J.P., Palukaitis P., Canto T. (2010). Cucumber mosaic virus 2b protein subcellular targets and interactions: their significance to RNA silencing suppressor activity. Mol. Plant Microbe Interact. 23: 294–303 [DOI] [PubMed] [Google Scholar]
- Goto K., Kobori T., Kosaka Y., Natsuaki T., Masuta C. (2007). Characterization of silencing suppressor 2b of cucumber mosaic virus based on examination of its small RNA-binding abilities. Plant Cell Physiol. 48: 1050–1060 [DOI] [PubMed] [Google Scholar]
- Guo H.S., Ding S.W. (2002). A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 21: 398–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamera S., Song X., Su L., Chen X., Fang R. (2012). Cucumber mosaic virus suppressor 2b binds to AGO4-related small RNAs and impairs AGO4 activities. Plant J. 36: 104–115 [DOI] [PubMed] [Google Scholar]
- Heo I., Kim V.N. (2009). Regulating the regulators: posttranslational modifications of RNA silencing factors. Cell 139: 28–31 [DOI] [PubMed] [Google Scholar]
- Horke S., Reumann K., Schweizer M., Will H., Heise T. (2004). Nuclear trafficking of La protein depends on a newly identified nucleolar localization signal and the ability to bind RNA. J. Biol. Chem. 279: 26563–26570 [DOI] [PubMed] [Google Scholar]
- Hou W.N., Duan C.G., Fang R.X., Zhou X.Y., Guo H.S. (2011). Satellite RNA reduces expression of the 2b suppressor protein resulting in the attenuation of symptoms caused by Cucumber mosaic virus infection. Mol. Plant Pathol. 12: 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen L.K., Carrington J.C. (2001). Silencing on the spot. Induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol. 126: 930–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa A., Inaba J., Shimura H., Otagaki S., Tsukahara S., Matsuzawa A., Kim B.M., Goto K., Masuta C. (2011). Virus-mediated efficient induction of epigenetic modifications of endogenous genes with phenotypic changes in plants. Plant J. 65: 156–168 [DOI] [PubMed] [Google Scholar]
- Kanno T., Huettel B., Mette M.F., Aufsatz W., Jaligot E., Daxinger L., Kreil D.P., Matzke M., Matzke A.J. (2005). Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat. Genet. 37: 761–765 [DOI] [PubMed] [Google Scholar]
- Kasschau K.D., Carrington J.C. (1998). A counterdefensive strategy of plant viruses: Suppression of posttranscriptional gene silencing. Cell 95: 461–470 [DOI] [PubMed] [Google Scholar]
- Kim S.H., Macfarlane S., Kalinina N.O., Rakitina D.V., Ryabov E.V., Gillespie T., Haupt S., Brown J.W., Taliansky M. (2007). Interaction of a plant virus-encoded protein with the major nucleolar protein fibrillarin is required for systemic virus infection. Proc. Natl. Acad. Sci. USA 104: 11115–11120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakatos L., Csorba T., Pantaleo V., Chapman E.J., Carrington J.C., Liu Y.P., Dolja V.V., Calvino L.F., López-Moya J.J., Burgyán J. (2006). Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 25: 2768–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.F., Pontes O., El-Shami M., Henderson I.R., Bernatavichute Y.V., Chan S.W., Lagrange T., Pikaard C.S., Jacobsen S.E. (2006). An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell 126: 93–106 [DOI] [PubMed] [Google Scholar]
- Li F., Ding S.W. (2006). Virus counterdefense: Diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 60: 503–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Folimonov A., Shintaku M., Li W.X., Falk B.W., Dawson W.O., Ding S.W. (2004). Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc. Natl. Acad. Sci. USA 101: 15742–15747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucy A.P., Guo H.S., Li W.X., Ding S.W. (2000). Suppression of post-transcriptional gene silencing by a plant viral protein localized in the nucleus. EMBO J. 19: 1672–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M., Aufsatz W., Kanno T., Daxinger L., Papp I., Mette M.F., Matzke A.J. (2004). Genetic analysis of RNA-mediated transcriptional gene silencing. Biochim. Biophys. Acta 1677: 129–141 [DOI] [PubMed] [Google Scholar]
- Matzke M., Kanno T., Daxinger L., Huettel B., Matzke A.J. (2009). RNA-mediated chromatin-based silencing in plants. Curr. Opin. Cell Biol. 21: 367–376 [DOI] [PubMed] [Google Scholar]
- Matzke M., Kanno T., Huettel B., Daxinger L., Matzke A.J. (2007). Targets of RNA-directed DNA methylation. Curr. Opin. Plant Biol. 10: 512–519 [DOI] [PubMed] [Google Scholar]
- Matzke M.A., Birchler J.A. (2005). RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 6: 24–35 [DOI] [PubMed] [Google Scholar]
- Meister G., Tuschl T. (2004). Mechanisms of gene silencing by double-stranded RNA. Nature 431: 343–349 [DOI] [PubMed] [Google Scholar]
- Melnyk C.W., Molnar A., Bassett A., Baulcombe D.C. (2011a). Mobile 24 nt small RNAs direct transcriptional gene silencing in the root meristems of Arabidopsis thaliana. Curr. Biol. 21: 1678–1683 [DOI] [PubMed] [Google Scholar]
- Melnyk C.W., Molnar A., Baulcombe D.C. (2011b). Intercellular and systemic movement of RNA silencing signals. EMBO J 30: 3553–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A., Melnyk C.W., Bassett A., Hardcastle T.J., Dunn R., Baulcombe D.C. (2010). Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328: 872–875 [DOI] [PubMed] [Google Scholar]
- Morel J.B., Godon C., Mourrain P., Béclin C., Boutet S., Feuerbach F., Proux F., Vaucheret H. (2002). Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey P., Choudhury N.R., Mukherjee S.K. (2009). A geminiviral amplicon (VA) derived from Tomato leaf curl virus (ToLCV) can replicate in a wide variety of plant species and also acts as a VIGS vector. Virol. J. 6: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O., Pikaard C.S. (2008). siRNA and miRNA processing: New functions for Cajal bodies. Curr. Opin. Genet. Dev. 18: 197–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y.J., He X., Wang X.J., Kohany O., Jurka J., Hannon G.J. (2006). Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature 443: 1008–1012 [DOI] [PubMed] [Google Scholar]
- Rajamäki M.L., Valkonen J.P. (2009). Control of nuclear and nucleolar localization of nuclear inclusion protein a of picorna-like Potato virus A in Nicotiana species. Plant Cell 21: 2485–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi B.J., Ding S.W., Symons R.H. (1997). In vivo expression of an overlapping gene encoded by the cucumoviruses. J. Gen. Virol. 78: 237–241 [DOI] [PubMed] [Google Scholar]
- Silhavy D., Molnár A., Lucioli A., Szittya G., Hornyik C., Tavazza M., Burgyán J. (2002). A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 21: 3070–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L.M., Pontes O., Searle I., Yelina N., Yousafzai F.K., Herr A.J., Pikaard C.S., Baulcombe D.C. (2007). An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell 19: 1507–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolia N.H., Joshua-Tor L. (2007). Slicer and the argonautes. Nat. Chem. Biol. 3: 36–43 [DOI] [PubMed] [Google Scholar]
- Vaucheret H. (2008). Plant ARGONAUTES. Trends Plant Sci. 13: 350–358 [DOI] [PubMed] [Google Scholar]
- Vazquez F., Vaucheret H., Rajagopalan R., Lepers C., Gasciolli V., Mallory A.C., Hilbert J.L., Bartel D.P., Crété P. (2004). Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16: 69–79 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2005). Induction and suppression of RNA silencing: Insights from viral infections. Nat. Rev. Genet. 6: 206–220 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2008). Post-transcriptional RNA silencing in plant-microbe interactions: A touch of robustness and versatility. Curr. Opin. Plant Biol. 11: 464–470 [DOI] [PubMed] [Google Scholar]
- Wang X.B., Jovel J., Udomporn P., Wang Y., Wu Q., Li W.X., Gasciolli V., Vaucheret H., Ding S.W. (2011). The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana. Plant Cell 23: 1625–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Tzfira T., Gaba V., Citovsky V., Palukaitis P., Gal-On A. (2004). Functional analysis of the Cucumber mosaic virus 2b protein: Pathogenicity and nuclear localization. J. Gen. Virol. 85: 3135–3147 [DOI] [PubMed] [Google Scholar]
- Wierzbicki A.T., Ream T.S., Haag J.R., Pikaard C.S. (2009). RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat. Genet. 41: 630–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Wang X., Ding S.W. (2010). Viral suppressors of RNA-based viral immunity: Host targets. Cell Host Microbe 8: 12–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Allen E., Wilken A., Carrington J.C. (2005). DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102: 12984–12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H.W., Ji L.H., Scott S.W., Symons R.H., Ding S.W. (1998). Ilarviruses encode a Cucumovirus-like 2b gene that is absent in other genera within the Bromoviridae. J. Virol. 72: 6956–6959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying X.B., Dong L., Zhu H., Duan C.G., Du Q.S., Lv D.Q., Fang Y.Y., Garcia J.A., Fang R.X., Guo H.S. (2010). RNA-dependent RNA polymerase 1 from Nicotiana tabacum suppresses RNA silencing and enhances viral infection in Nicotiana benthamiana. Plant Cell 22: 1358–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M., Peragine A., Park M.Y., Poethig R.S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19: 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Du P., Lu L., Xiao Q., Wang W., Cao X., Ren B., Wei C., Li Y. (2008). Contrasting effects of HC-Pro and 2b viral suppressors from Sugarcane mosaic virus and Tomato aspermy cucumovirus on the accumulation of siRNAs. Virology 374: 351–360 [DOI] [PubMed] [Google Scholar]
- Zhang X., Henderson I.R., Lu C., Green P.J., Jacobsen S.E. (2007). Role of RNA polymerase IV in plant small RNA metabolism. Proc. Natl. Acad. Sci. USA 104: 4536–4541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yuan Y.R., Pei Y., Lin S.S., Tuschl T., Patel D.J., Chua N.H. (2006). Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 20: 3255–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Zhu J., Kapoor A., Zhu J.K. (2007). Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 26: 1691–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., et al. (2011). BSCTV C2 attenuates the degradation of SAMDC1 to suppress DNA methylation-mediated gene silencing in Arabidopsis. Plant Cell 23: 273–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D. (2008). The evolving functions of DNA methylation. Curr. Opin. Plant Biol. 11: 554–559 [DOI] [PubMed] [Google Scholar]
- Zilberman D., Cao X., Jacobsen S.E. (2003). ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716–719 [DOI] [PubMed] [Google Scholar]
- Zilberman D., Cao X., Johansen L.K., Xie Z., Carrington J.C., Jacobsen S.E. (2004). Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr. Biol. 14: 1214–1220 [DOI] [PubMed] [Google Scholar]