This work describes two new plant-specific proteins, termed ATI1 and ATI2, which bind Atg8f, a central protein of the autophagy machinery. Upon carbon starvation, these proteins are associated with previously undefined spherical bodies that move dynamically on the endoplasmic reticulum network.
Abstract
Atg8 is a central protein in bulk starvation–induced autophagy, but it is also specifically associated with multiple protein targets under various physiological conditions to regulate their selective turnover by the autophagy machinery. Here, we describe two new closely related Arabidopsis thaliana Atg8-interacting proteins (ATI1 and ATI2) that are unique to plants. We show that under favorable growth conditions, ATI1 and ATI2 are partially associated with the endoplasmic reticulum (ER) membrane network, whereas upon exposure to carbon starvation, they become mainly associated with newly identified spherical compartments that dynamically move along the ER network. These compartments are morphologically distinct from previously reported spindle-shaped ER bodies and, in contrast to them, do not contain ER-lumenal markers possessing a C-terminal HDEL sequence. Organelle and autophagosome-specific markers show that the bodies containing ATI1 are distinct from Golgi, mitochondria, peroxisomes, and classical autophagosomes. The final destination of the ATI1 bodies is the central vacuole, indicating that they may operate in selective turnover of specific proteins. ATI1 and ATI2 gene expression is elevated during late seed maturation and desiccation. We further demonstrate that ATI1 overexpression or suppression of both ATI1 and ATI2, respectively, stimulate or inhibit seed germination in the presence of the germination-inhibiting hormone abscisic acid.
INTRODUCTION
Plant growth depends on multiple factors, among which are stresses such as limiting light periods, limiting levels of nitrogen, and exposure to salt and drought (Masclaux-Daubresse et al., 2010; Mittler and Blumwald, 2010). Exposure to stress has multiple physiological and metabolic impacts and also leads to deprivation of energy (Baena-González and Sheen, 2008; Masclaux-Daubresse et al., 2010). Hence, plants have evolved convergent stress-associated processes that protect them from these stresses (Baena-González and Sheen, 2008; Masclaux-Daubresse et al., 2010). One of the central cellular machineries that regulates plant growth under energy-depleting stresses is macroautophagy, hereafter referred to as autophagy. Plant autophagy is classically associated with the bulk turnover of macromolecules and organelles in the vacuole upon energy deprivation (Yoshimoto, 2010). Yet, plant autophagy was also shown to be involved in multiple other physiological and developmental processes, such as metabolism, senescence, stress tolerance, and innate immune response (Thompson and Vierstra, 2005; Bassham, 2009; Hayward and Dinesh-Kumar, 2010). Autophagy can be either nonselective, causing massive degradation of cellular components, or selective, that is, regulating specific cellular remodeling events during development and upon exposure to various stresses (Meijer et al., 2007; Bassham, 2009). One of the core proteins in the autophagy machinery is Atg8, serving as a central component in the formation of autophagosomes in yeast, mammals, and plants (Nakatogawa et al., 2007; Bassham, 2009; Weidberg et al., 2010). Atg8 proteins have also been shown to be involved in the selective turnover of protein aggregates and unwanted or malfunctioning organelles (Pankiv et al., 2007; Kirkin et al., 2009; Ichimura and Komatsu, 2010). Arabidopsis thaliana possesses nine Atg8 isoforms (Atg8a to Atg8i). Expression of a green fluorescent protein (GFP)-Atg8f fusion construct in transgenic Arabidopsis plants was shown to alter the response of the plants to hormones and abiotic stresses (Slavikova et al., 2008). A recent report also showed that the controlled turnover of an Arabidopsis multistress regulator protein, termed TSPO, occurs through its binding to Atg8 (Vanhee et al., 2011).
One of the critical factors for the autophagy process is the selection of cargo to be turned over. In mammals, two autophagic cargo receptors, p62 and NBR1, are known to recognize specific ubiquitinated substrates for degradation (Noda et al., 2010; Johansen and Lamark, 2011). Recently, a functional hybrid protein of p62 and NBR1 was also identified in Arabidopsis (At-NBR1; Svenning et al., 2011). These autophagic cargo receptors, which naturally possess almost no sequence similarity to each other, were shown to contain a common Atg8-interacting motif (AIM), generally referred to as a W/YXXL/I/V-like motif, for direct interaction with Atg8 family proteins (Noda et al., 2008; Kirkin et al., 2009; Okamoto et al., 2009; Novak et al., 2010; Svenning et al., 2011). Additional AIM-containing selective autophagy receptors, involved in mitochondrial clearance, have also been identified in mammals and yeast (Okamoto et al., 2009; Novak et al., 2010). It is thus plausible to assume the existence of additional Atg8-interacting proteins in plants, serving multiple functions associated with selective autophagy.
In this report, we describe the identification of two closely related previously uncharacterized Arabidopsis Atg8-interacting proteins (ATI1 and ATI2), each of which contains two putative AIMs located on either side of a predicted transmembrane domain. Both ATIs interact with the Arabidopsis Atg8f isoform (as a representative) as elucidated using both the yeast two-hybrid approach and the in vivo bimolecular fluorescence complementation (BiFC) approach (Bracha-Drori et al., 2004). We also show that under favorable (nonstress) growth conditions, the ATI1 and ATI2 proteins are partially associated with the endoplasmic reticulum (ER) network, while exposure to carbon starvation triggers their association with the surface of newly identified spherical ER-associated compartments that are morphologically distinct from another previously reported spindle-shaped ER-derived compartment (Matsushima et al., 2002, 2003). In addition, we demonstrate the functional role of ATI1 and ATI2 in seed germination in response to exogenous abscisic acid (ABA) treatment.
RESULTS
ATI1 and ATI2, Two Closely Related Plant-Specific Proteins with No Functional Annotation, Possess Two Putative AIMs and Bind the Autophagy-Associated Atg8f Isoform in Two Independent in Vivo Systems
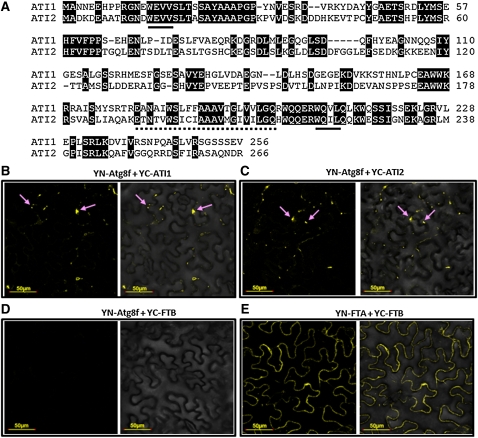
To discover previously unidentified plant proteins that interact with the autophagy-associated Atg8 protein family, we used the Arabidopsis Atg8f isoform (a representative of nine Arabidopsis Atg8 isoforms) as bait in a yeast two-hybrid approach. The prey cDNA library was prepared from a dark-grown Arabidopsis cell suspension (Németh et al., 1998). Several positive cDNA clones were detected in the yeast two-hybrid screen, and the interaction of their encoded proteins with Atg8f was reconfirmed several times with one on one yeast two-hybrid analysis (data not shown). Sequencing of the cDNA in the positive clones revealed that two of them match the Arabidopsis loci At2g45980 and At4g00355, which encode two closely related proteins with no functional annotation. Due to their detectable interaction with Atg8f in the yeast two-hybrid assay, we named these proteins ATI1 and ATI2, respectively. Protein sequence analysis showed that ATI1 and ATI2 share 44% identity and 67% similarity in their amino acid sequences (Figure 1A; the two proteins) and that homologous proteins to ATI1 and ATI2 are present only in plants, but not in the animal kingdom (see Supplemental Figure 1 online). In addition, a distant ATI1/ATI2 homolog is present in the moss Physcomitrella patens (see Supplemental Figure 1 online). The ATI1 and ATI2 proteins each contain a predicted single transmembrane domain and two putative AIMs (Noda et al., 2010), the more canonical one located upstream, and a second one located downstream, of the predicted transmembrane domain (Figure 1A; see Supplemental Figure 2 online). The presence of a pair of putative AIMs in the sequences of both ATIs further supports their possible interaction with Atg8 family proteins.
Figure 1.
ATI1 and ATI2 Are Newly Identified Plant-Specific Proteins That Bind Atg8f.
(A) Amino acid sequence alignment between ATI1 and ATI2. Conserved residues are shaded. Consensus AIMs are underlined with continues lines. The putative single transmembrane domain is underlined with a dashed line.
(B) and (C) BiFC analysis, including transient coexpression in N. benthamiana leaves of YN-Atg8f with either YC-ATI1 (B) or YC-ATI2 (C). YFP fluorescence, detected as yellow bodies in the cytosol (pink arrows), demonstrates the interaction of ATI1 and ATI2 with Atg8f.
(D) A negative control for BiFC analysis, including transient coexpression of YN-Atg8f with a YFP fusion with nonrelated protein, YC-FTB, shows no fluorescence.
(E) A positive control for BiFC analysis, including transient coexpression of YN-FTA with YC-FTB, reconstructing the cytoplasmic PFT protein (Bracha-Drori et al., 2004), shows homogenous cytosolic fluorescence. (B) to (E) are fluorescence images (left side of each panel) and combined fluorescence and transmittance image (right side of each panel).
To confirm further the interaction of ATI1 and ATI2 with Atg8f, we used the in vivo BiFC approach (Bracha-Drori et al., 2004) in which fusion proteins were generated linking ATI1 or ATI2 with one half of the marker yellow fluorescent protein (YFP; YC-ATI1 and YC-ATI2) and Atg8f with the second half of the YFP protein (YN-Atg8f). Assuming that ATI1 and ATI2 are localized to a membrane, the direction of the fusions was based on the hypothesis that the more canonical AIM, which is located in the N-terminal part of the protein, would face the cytoplasm, where Atg8f is localized. Each of these two sets of combined constructs (YN-Atg8f plus YC-ATI1 and YN-Atg8f plus YC-ATI2) was then transiently coexpressed in Nicotiana benthamiana leaves. Interaction between Atg8f and either ATI1 or ATI2 was expected to bring the two halves of the YFP polypeptide in close proximity and generate yellow fluorescence. As shown in Figures 1B and 1C, cotransformation of either YN-Atg8f plus YC-ATI1 or YN-Atg8f plus YC-ATI2 constructs generated scattered YFP fluorescence in the epidermis cells of N. benthamiana leaves, implying that Atg8f interacts with either ATI1 or ATI2 in vivo, confirming the yeast two-hybrid results. As a negative control, YC-FTB (β-subunit of the Arabidopsis protein farnesyl transferase [PFT]; see Methods) and YN-Atg8f were cotransformed into N. benthamiana leaves and did not generate any YFP fluorescence (Figure 1D). To demonstrate that YC-FTB is a functional fusion, it was cotransformed with YN-FTA (α-subunit of the Arabidopsis PFT) into N. benthamiana leaves, resulting in uniform cytoplasmic YFP fluorescence (Figure 1E), as expected from the PFT-soluble cytoplasmic protein (Bracha-Drori et al., 2004). The BiFC results also supported our supposition that the more canonical AIM located at the N-terminal part of ATI1 or ATI2 is apparently located at the cytoplasmic side of the putative compartment to which ATI1 or ATI2 might be localized.
We next examined the expression pattern of the ATI1 and ATI2 genes using real expression data from the publicly available, microarray-derived expression databases GENEVESTIGATOR and BAR. Expression of these two genes appears to be induced by various stresses, such as carbon and nitrogen starvation as well as salt and drought stresses (see Supplemental Figures 3 and 4 online), all of which are also commonly known to cause energy deprivation and associate with autophagy in plants (Bassham, 2007; Baena-González and Sheen, 2008; Masclaux-Daubresse et al., 2010). The developmental and stress-induced gene expression patterns of ATI1 and ATI2 are comparable to the gene expression pattern of Atg8f (see Supplemental Figures 3 and 4 online). In addition, combined analysis of ATTED-II and PAGEMAN overrepresentation analysis showed that ATI1 and ATI2 are coexpressed mainly with genes associated with the ubiquitin degradation system, autophagy, and posttranslational modifications (see Supplemental Figure 5 online).
Under Favorable Growth Conditions, ATI1 and ATI2 Are Partially Colocalized with the ER
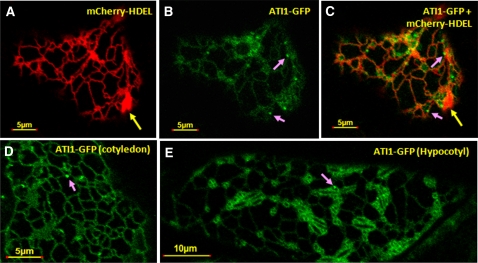
To test the intracellular localization of ATI1 and ATI2, we produced ATI1-GFP, ATI2-GFP, and ATI2-monomeric red fluorescent protein (mRFP) fusion constructs transcribed by the 35S promoter (Pro35S). Transient coexpression of ATI1-GFP and ATI2-mRFP in cotyledons of Arabidopsis seedlings (Li et al., 2009) showed that these two proteins were largely colocalized with each other (see Supplemental Figures 6A to 6C online). Next, each of the ATI1-GFP or ATI2-GFP constructs was transiently coexpressed together with a mCherry-HDEL protein marker of the ER lumen (Nelson et al., 2007) in cotyledons of Arabidopsis seedlings, grown under favorable growth conditions. In cells of these cotyledons, mCherry-HDEL (red color) was visualized in the confocal microscope as a typical network of ER cisterna (Figure 2A, red network). In addition, the mCherry-HDEL marker stained some spindle-shaped bodies (Figures 2A and 2C, yellow arrows; see also Figures 3B and 3C), which apparently represent the ER-located spindle-shaped bodies that have been previously described (Hawes et al., 2001; Matsushima et al., 2003). The ATI1-GFP fusion protein was partially colocalized with the mCherry-HDEL protein on the ER network (cf. Figures 2A and 2B; also see Figure 2C depicting the merged image of 2A and 2B). Both the ATI1-GFP and ATI2-GFP fusion proteins showed a partial colocalization pattern with the mCherry-HDEL marker (cf. Figures 2A to 2C to Supplemental Figures 6D to 6F online).
Figure 2.
ATI1 Is Partially Colocalized to the ER Network.
(A) The mCherry-HDEL marker (Nelson et al., 2007) labels an ER network as well as the previously described spindle-shape bodies (yellow arrow) (Matsushima et al., 2003).
(B) and (C) The mCherry-HDEL ER marker was transiently coexpressed with ATI1-GFP (B) showing partial colocalization (C) with the ER network and in different body structures on the ER network as indicated by pink arrows.
(D) and (E) ATI1 is localized to the ER in cotyledon (D) and hypocotyl (E) cells of transgenic Arabidopsis plants stably overexpressing ATI1-GFP. Body structures on the ER network are also detected in ATI1-GFP transgenic plants (pink arrows).
Figure 3.
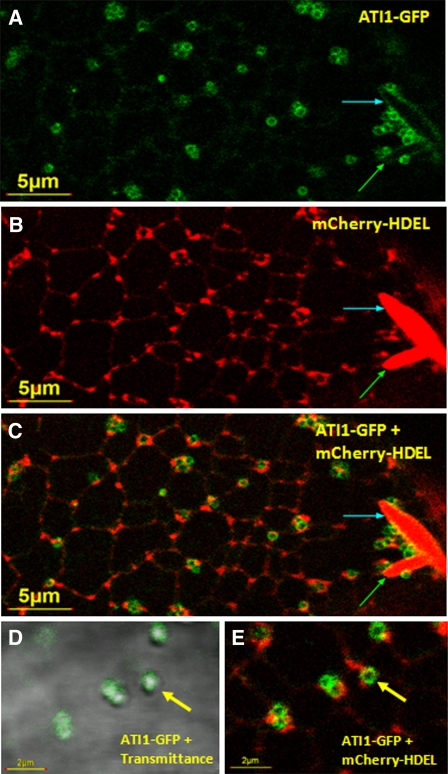
Carbon Starvation–Induced Spherical Bodies Containing ATI1-GFP Are Localized in Close Proximity to the ER Network.
Confocal microscopy analysis of hypocotyl epidermis cells of 7-d-old transgenic Arabidopsis plants stably expressing ATI1-GFP (A), mCherry-HDEL (B), and combined ATI1-GFP plus mCherry-HDEL (C). Combined transmission and GFP fluorescence image at a larger magnification shows the localization of ATI1-GFP on the surface of the spherical bodies (D) and the localization of these spherical bodies in close proximity to the ER (E). Previously described spindle-shaped ER bodies (Matsushima et al., 2003) are seen (A) to (C) (red bodies marked by blue and green arrows). The newly identified bodies (yellow arrows in [D] and [E]) marked with ATI1-GFP on their surface (green bodies) are localized in close proximity to the ER network.
To corroborate the partial colocalization of ATI1 and ATI2 with the ER, we generated transgenic plants stably expressing each of the Pro35S:ATI1-GFP or Pro35S:ATI2-GFP proteins alone and also individually transformed the Pro35S:ATI1-GFP or Pro35S:ATI2-GFP construct into plants stably expressing the Pro35S:mCherry-HDEL (Nelson et al., 2007). In all subsequent analyses of the plants expressing ATI1 and ATI2, these two proteins showed very similar expression and in situ localization patterns; hence, in the following results, we focus on ATI1 as a representative. We first analyzed the intracellular localization of ATI1-GFP in cotyledons and hypocotyls of transgenic plants stably expressing the Pro35S:ATI1-GFP construct and grown under favorable growth conditions (Figures 2D and 2E). In both tissues, the stably expressed ATI1-GFP appeared to be partially associated with the ER network, resembling the images obtained with the mCherry-HDEL ER marker in the transient expression analysis in cotyledons. Notably, in both tissues, ATI1-GFP also appeared to be associated with sporadic small round bodies that were located on the ER network (Figures 2B to 2E, bodies marked by pink arrows).
Exposure of Seedlings to Carbon Starvation Stimulates the Formation of Newly Identified Spherical Bodies Containing the ATI1-GFP Polypeptide
Since expression of ATI1 is induced by various stresses that cause energy deprivation and promote autophagy (see Supplemental Figure 3 online), and also because this protein interacts with the Atg8f isoform, we tested the effect of carbon starvation (causing energy deprivation) on the intracellular localization of this protein. To address this issue, we germinated seeds of the wild type and the double transgenic plants expressing ATI1-GFP and mCherry-HDEL on agar plates lacking sugars, grew them for 6 d in a controlled growth room, and then exposed them to darkness for additional 24 h (see Methods). Hypocotyls and cotyledons of these plants were then subjected to confocal microscopy analysis. Exposure to carbon starvation stimulated the formation of newly identified small spherical bodies (~0.5 to 1 μM in diameter) labeled with ATI1-GFP on their surface (Figure 3A) that were located in close proximity to the ER (Figures 3B, 3C, and 3E) and were also detected in a merge of a transmission image with a ATI-GFP fluorescence image (Figure 3D; for a larger magnification image, see Supplemental Figures 7E and 7F online). These bodies were also detected in a sole transmission image of carbon starved hypocotyl and cotyledon epidermis cells of wild-type plants (see Supplemental Figure 8 online), indicating that they appear naturally and are not a result of our transgenic approach. A certain number of these spherical bodies were also detected in ATI1-GFP plants grown under favorable growth conditions (Figures 2B to 2E, bodies marked by pink arrows). However, a statistical quantification showed a significantly higher number of spherical bodies detected in ATI1-GFP plants grown under carbon starvation conditions compared with nonstressed plants (see Supplemental Figure 9 online).
No mCherry-HDEL fluorescence was detected inside the ATI1-containing bodies (hereafter referred to as ATI1 bodies), suggesting they do not contain ER lumen (Figures 3C and 3E). This was also corroborated by analyzing hypocotyl epidermis cells of transgenic plants stably expressing GFP-HDEL (Nelson et al., 2007) exposed to carbon starvation using confocal microscopy. As shown within the yellow ovules in Supplemental Figures 10A to 10C online, the ATI1 bodies in the transmission image (panel A) were not labeled with GFP-HDEL (panels B and C). The ATI1 bodies are also clearly distinct from the previously identified spindle-shaped bodies (Matsushima et al., 2003) by their differential morphology (Figure 3C; see Supplemental Figure 10D online) and also by the fact that the ATI1 bodies are not labeled with the mCherry-HDEL or GFP-HDEL markers (Figures 3C and 3E; see Supplemental Figures 10E and 10F online) and are more dynamic than the ER bodies. The dynamic movement of ATI1 bodies in comparison to the movement of ER bodies is seen in Supplemental Movie 1 online.
ATI1 Bodies Dynamically Move along the ER Network
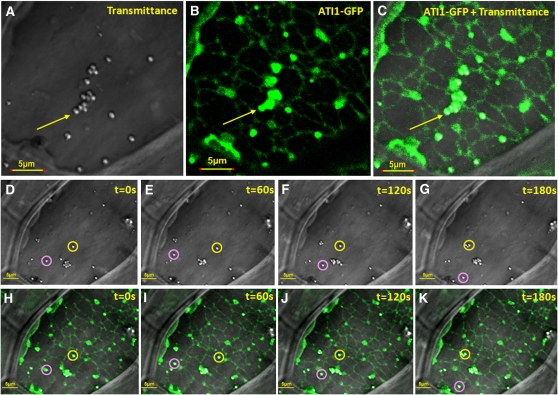
ATI1-GFP fluorescence showed that the ATI1 bodies are located in close proximity to the ER network but are not entirely colocalized with this network (Figures 3A to 3C). This is also clearly shown in larger magnification (Figure 3E). To confirm these observations further, we generated lower-resolution images with relatively higher laser intensity (for better visualization of the ER network) of hypocotyl epidermis cells of the same lines of seedlings expressing only ATI1-GFP examined before and after exposure to carbon starvation. Under this condition of higher laser intensity, it was shown that ATI1 bodies are located on the ER network (Figures 4A to 4C). Figures 4D to 4K show the transmission images (panels D to G) and ATI1-GFP florescence images (panels H to K) derived from a time-lapse image series. The two depicted bodies marked by pink and yellow circles clearly move on the ER network as determined by the changing distances between the pink and yellow circles along panels D to G and H to K. The dynamic movement of the ATI1 bodies on the ER network is also clearly visualized in Supplemental Movies 2 and 3 online. Under the higher laser intensity conditions, ATI1-GFP fluorescence was detected on the entire volume of the ATI1 bodies (Figure 4). The reason for this is not clear but may indicate that a small amount of GFP may also be localized inside these vesicles due to proteolysis of the GFP moiety from ATI1-GFP.
Figure 4.
ATI1 Bodies, Induced upon Exposure to Carbon Starvation, Dynamically Move along the ER Membrane Network.
(A) to (C) Depicted steady state light transmittance (A), ATI1-GFP fluorescence (B), and a merge of these images (C) taken from confocal microscopy analysis that was also used for a time-lapse experiment shown in (D) to (K). The images were taken from 7-d-old transgenic Arabidopsis seedling hypocotyls stably overexpressing ATI1-GFP and exposed to carbon starvation. The yellow arrows indicate a typical group of spherical bodies containing ATI1-GFP, which associate with the ER network that is also labeled by the same ATI1-GFP fluorescence.
(D) to (K) Movement of ATI1-GFP–labeled bodies on the ER network. The time-lapse movement (pictures taken every 60 s) is illustrated by the observation that the depicted two bodies, localized in yellow and pink circles, exist in different locations from each other in (D) to (K). The fluorescence images ([B], [C], and [H] to [K]) were taken in relatively high laser intensity to visualize the ER membrane network, rendering the resolution of GFP fluorescence lower compared with Figure 3. As a result, the ATI1-GFP fluorescence appears to cover both the surface and the interior of the spherical bodies. A movie depicting the dynamic movement of the bodies in the cell is available in Supplemental Movie 2 online.
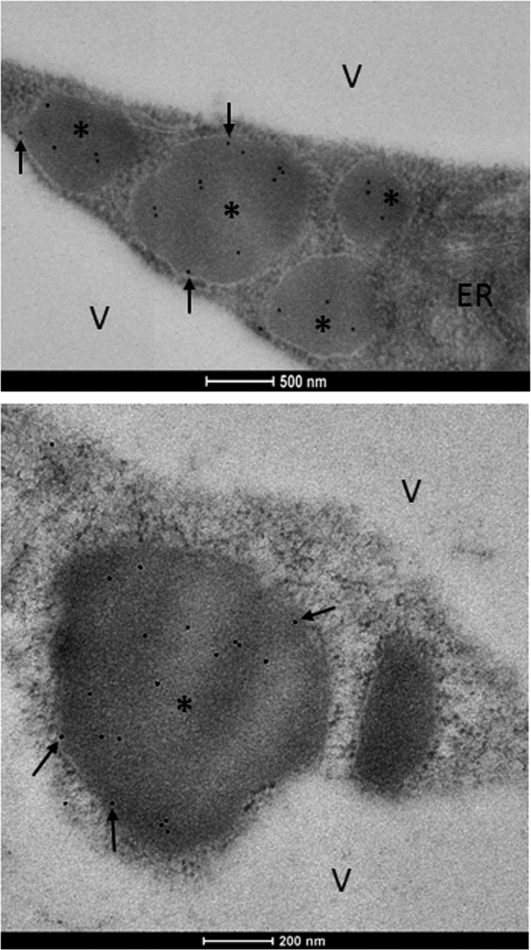
ATI1 Bodies Identification by Electron Microscopy
To determine the ultrastructure of the ATI1 bodies in ATI1-GFP transgenic Arabidopsis cells, proximal regions of hypocotyls taken from carbon-starved seedlings were subjected to a relatively mild chemical fixation and then ultrathin sections were immunogold labeled with anti-GFP antibodies (see Methods). The labeled sections were then subjected to electron microscopy analysis. Anti-GFP antibodies labeled almost exclusively a significant number of spherical compartments located in the cytosol, in close proximity to the ER (Figure 5), and essentially no labeling was observed in any other regions, indicating the specific enriched localization of ATI1 and ATI2 inside the spherical compartments. Moreover, in many of the micrographs, more than one of the spherical labeled compartments could be observed (data not shown). The average estimated diameter of the spherical bodies was generally 0.5 to 1 μm, in agreement with the size measured using confocal microscopy (see Supplemental Figure 7 online). The black arrows in Figure 5 mark the gold labeling of ATI1-GFP molecules located on the surface of the bodies. We hypothesize that the nonsurface molecule labeling may result either from ATI1-GFP proteolysis events or internalization of ATI1-GFP inside the bodies (see Discussion).
Figure 5.
Spherical-Shaped ATI1 Bodies in Epidermal Cells of Carbon-Starved Transgenic Arabidopsis Hypocotyls Observed by Electron Microscopy.
Electron micrographs of ATI1-GFP hypocotyls showing the characteristic spherical structures of ATI1 bodies located close to ER structure. Black dots are immunogold labeling of GFP antibodies used to identify the ATI1-GFP protein. ATI1 bodies are marked with asterisks. Black arrows mark the ATI1-GFP molecules localized to the surface of the bodies. V, vacuole. Bars = 0.5 μm in the top panel and 0.2 μm in the bottom panel.
ATI1 Bodies Are Distinct from Golgi, Mitochondria, and Peroxisomes
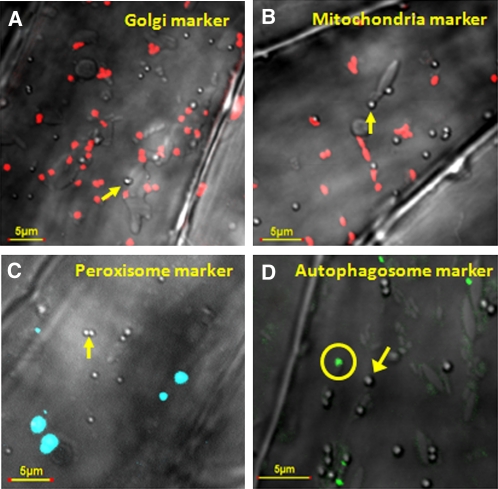
To test whether the ATI1 bodies are related to any other dynamic cellular organelles, we used transgenic Arabidopsis plants stably expressing one of the following constructs: (1) the Golgi marker GmMan1-mCherry, (2) the mitochondrial marker ScCOX4-mCherry (Nelson et al., 2007), and (3) the peroxisome marker AtPEX5-CFP (Tian et al., 2004). Seeds of these transgenic plants were germinated and exposed to carbon starvation and then subjected to confocal microscopy analysis. The merged transmission images of the ATI1 bodies and the fluorescence of the different organelles (red for Golgi or mitochondria and blue for the peroxisomes) are shown in Figures 6A to 6C. These images illustrate that ATI1 bodies are clearly not colocalized with any of these organelles, implying that they represent a distinct compartment unrelated to the Golgi, mitochondria, or peroxisomes.
Figure 6.
ATI1 Bodies Are Not Colocalized with Golgi-, Mitochondria-, or Peroxisome-Specific Markers nor with an Autophagosome Marker.
Combined transmission images of a confocal microscope showing spherical ATI1 bodies (indicated by yellow arrows) and images with fluorescent markers of Golgi (A), mitochondria (B), peroxisomes (C), and autophagosomes (D). Images were taken from hypocotyls of 7-d-old transgenic Arabidopsis seedlings stably overexpressing the Golgi marker GmMan1-mCherry (red bodies), the mitochondrial marker ScCOX4-mCherry (red bodies), the peroxisome marker AtPEX5-CFP (blue bodies), and the autophagosome marker GFP-Atg8f (green bodies) following exposure to carbon starvation. The spherical bodies seen by the transmission images are not colocalized with any of these marker proteins. The autophagosome is indicated by a yellow circle.
ATI1 Bodies Are Distinct from Classical Autophagosomes, and Only a Small Proportion of Them Physically Associate with Atg8f at Any Given Time Point
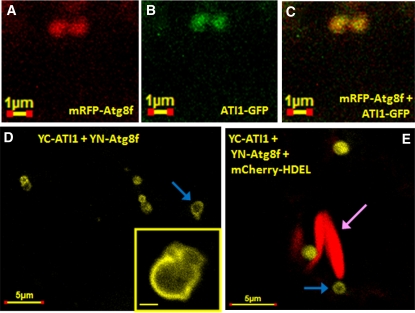
To elucidate the relationship between the ATI1 bodies and the classical Atg8-containing autophagosomes, we first generated Pro35S:GFP-Atg8f and Pro35S:mRFP-Atg8f autophagosome marker constructs, based on previous works demonstrating that Atg8 isoforms are efficient autophagosome markers (Yoshimoto et al., 2004; Contento et al., 2005; Sláviková et al., 2005; Thompson et al., 2005; Xiong et al., 2007). Seedlings of transgenic plants stably expressing GFP-Atg8f were exposed to carbon starvation, and their hypocotyl epidermis cells were then subjected to confocal microscopy analysis. As shown in Figure 6D, the ATI1 bodies detected in the transmission images (a depicted ATI1 body is marked by a yellow arrow) were clearly distinct in structure and generally not colocalized with classical autophagosomes marked by GFP-Atg8f (autophagosomes are marked by yellow circle). Yet, when transiently coexpressed in cotyledons of Arabidopsis seedlings exposed to carbon starvation (see Methods), a relatively rare number of images showed colocalization of the autophagosome marker mRFP-Atg8f and ATI1-GFP (Figures 7A to 7C). To elucidate further the temporary interaction of Atg8f with the ATI1 bodies, we used the BiFC method (Bracha-Drori et al., 2004) in which we transiently coexpressed the YN-Atg8f together with YC-ATI1 in cotyledons of transgenic Arabidopsis seedlings stably expressing mCherry-HDEL. The seedlings were exposed to carbon starvation and their cotyledons analyzed by confocal microscopy. YFP fluorescence was detected on the surface of spherical bodies containing ATI1 (Figures 7D and 7E, ATI1 bodies marked by blue arrows) revealing that ATI1 interacts with Atg8f in vivo. The YFP-labeled bodies were also distinct from the mCherry-labeled spindle-shaped ER bodies (Matsushima et al., 2003) (Figure 7E, ER body marked by pink arrow).
Figure 7.
Atg8f, a Protein Marker for Autophagosomes, Is Infrequently Colocalized with the ATI1 Bodies Following Exposure to Carbon Starvation.
(A) to (C) A rare case depicts the colocalization of mRFP-Atg8f (red in [A]) and ATI1-GFP (green in [B]) in the same bodies (yellow in [C]) using a transient expression assay that includes exposure to darkness and mild carbon starvation (see Methods).
(D) and (E) BiFC analysis of YC-ATI1 and YN-Atg8f split YFP interaction in mCherry-HDEL transgenic Arabidopsis cotyledons using the same transient expression assay described for (A) to (C). The YFP fluorescence on the surface of the spherical bodies indicates the interaction of ATI1 with Atg8f. A magnification of the body is provided in a small square on the bottom right part of (D). The yellow spherical bodies (indicated by blue arrows) are generally found in the vicinity of two spindle-shaped bodies (indicated by red mCherry color and a pink arrow) emphasizing their size and shape differences.
ATI1 Bodies Are Transported into the Central Vacuole
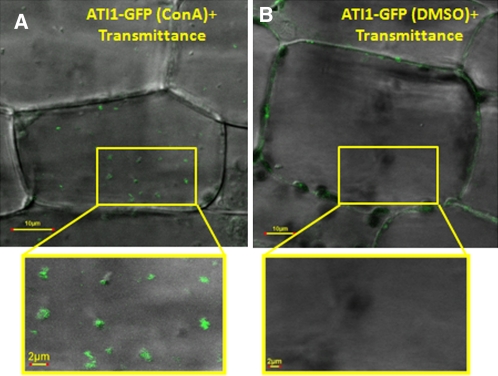
Starvation-induced plant autophagosomes are initially formed in the cytosol where they engulf cytoplasm and various organelles and transport them to the vacuole for degradation (Bassham, 2009). It was thus interesting to test whether the ATI1 bodies, which initially localize to the ER network, are transported into the vacuole. To answer this question, seedlings of the transgenic plants expressing ATI1-GFP were exposed to carbon starvation and then treated with Concanamycin A (ConA), which inhibits the activity of vacuolar proteases (Dröse et al., 2001) and enables visualization of GFP inside the plant central vacuole (Tamura et al., 2003; Yoshimoto et al., 2004; Sláviková et al., 2005; Ishida et al., 2008). The plants were then subjected to confocal microscopy analysis. Small bodies, exhibiting the green fluorescence of ATI1-GFP, were visualized inside vacuoles after treatment with ConA (Figure 8; see Supplemental Movie 4 online), indicating that the ATI1 bodies that are located on the ER network upon exposure to carbon starvation are further transported into the vacuole.
Figure 8.
Bodies Containing ATI1-GFP Are Found inside the Vacuole of Hypocotyl Cells of Plants Exposed to Carbon Starvation.
Combined transmission and fluorescence images of hypocotyls of transgenic Arabidopsis cell stably expressing ATI1-GFP exposed to carbon starvation followed by ConA treatment or DMSO treatment as control (see Methods). ConA-treated cells accumulated small GFP-labeled bodies in the central vacuole (A), while DMSO-treated cell vacuoles remained clear (B). The magnified area inside the yellow boxes depicts a section of the central vacuole in each image.
ATI Expression Levels Affect Seed Germination in the Presence of Exogenous ABA
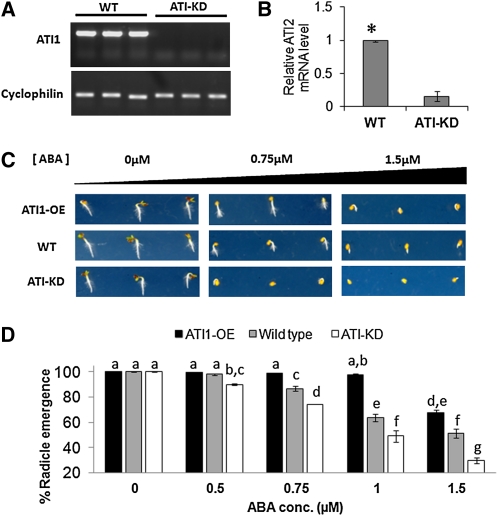
ABA is a plant hormone that affects several important processes throughout the plant life cycle (Giraudat et al., 1994). One of its key functions is to maintain seed dormancy and to prevent precocious germination (Bewley, 1997). This is done through the moderate accumulation of ABA during seed development, with the highest ABA levels in the dry seed. During imbibition and germination, there is a drastic reduction in the ABA content of the seedling (Braybrook and Harada, 2008). The expression levels of both ATI genes follow the accumulation of ABA in the developing seed, reaching the highest expression level in dry seeds (see Supplemental Figure 11 online). Moreover, a concomitant decrease in ATI1/2 gene expression during imbibition and germination also follows the ABA decrease in the same developmental stages (see Supplemental Figure 11 online). Those observations led us to speculate that ATIs are involved in ABA degradation during early seed germination. To test this hypothesis, we first used Pro35S:ATI1-GFP plants (ATI1-OE, described earlier). Because we hypothesized that both ATI1 and ATI2 proteins have the same function, a double knockout line was needed to elucidate any possible phenotype. Whereas a T-DNA knockout line for ATI1 was available (SAIL_404_D09) (Alonso et al., 2003), no T-DNA knockout lines for ATI2 were found. Thus, we screened and isolated an RNA interference (RNAi) line with significant reduction in ATI2 expression (see Methods). Both ATI1-KO and ATI2-RNAi plants were characterized using RT-PCR and real-time PCR, respectively (data not shown) and the lowest-expressing lines were manually crossed. F3 progeny were screened again using the same methods, and a stable line with no expression of ATI1 (Figure 9A) and a significant reduction of ~80% in the expression of ATI2 (Figure 9B) was identified. This plant line was designated ATI-KD and was used for further studies. Next, seeds of wild-type, ATI1-OE, and ATI-KD plants were sown on agar plates containing different concentrations of ABA ranging from 0 to 1.5 μM (see Methods), and radical emergence was examined 2 d after removal from a 4°C chamber. Full radical emergence (100%) was observed in seeds germinated on MS+SUC plates without ABA (Figure 9D; also depicted in left panel of Figure 9C). With the increase of ABA concentration in the medium to 0.75 μM and higher, a significant reduction in radical emergence of ATI-KD seeds was observed in comparison to wild-type seeds and a significant enhancement in radical emergence of ATI1-OE seeds was observed in comparison to wild-type seeds (Figure 9D; also depicted in Figure 9C). Those observations indicate that exogenous ABA can alter the rate of germination (as demonstrated by radical emergence) in a concentration-dependent manner and that ATI1/2 gene expression levels affect the germination ability of seeds in presence of exogenous ABA.
Figure 9.
ATI Expression Levels Alter Seed Radical Emergence Ability in the Presence of Exogenous ABA.
(A) Analysis of the ATI1 gene expression in wild-type and ATI-KD plants. mRNA was extracted and cDNA was synthesized from three independent wild-type (WT) and ATI1-KD plants (SAIL_404_D09). RT-PCR was used to amplify the ATI1 full-length coding sequence (780 bp) using specific primers spanning the 5′ and 3′ ends of the ATI1 cDNA. CYCLOPHILIN was used for cDNA quality control and as a reference gene. No ATI1 expression was detected in ATI1-KD plants.
(B) Analysis of the ATI2 gene expression in wild-type and ATI-KD plants. mRNA was extracted and cDNA was synthesized from four independent wild-type and ATI2 RNAi plants (CATMA4a_00420). Real-time PCR was used to amplify a partial ATI2 sequence from all samples. UBI-C was used as an internal standard. The average relative ATI2 mRNA level in ATI-KD plants as quantified by the real-time PCR was significantly lower (marked with an asterisk, P < 0.05) than the ATI2 mRNA level in wild-type plants. Error bars indicate se values.
(C) Radical emergence images of wild-type, ATI1-OE, and ATI-KD seeds in three different concentrations of ABA (0, 0.75, and 1.5 μM) 2 d after removal from 4°C (2 d after germination). Each line is represented by three seedlings depicting the common phenotype.
(D) Quantification of seed radical emergence percent of the genotypes described in (C) observed 2 d after germination. Each of five ABA treatments was repeated twice, and in each treatment, ~100 seeds of every one of the three genotypes were examined. Values in table are averages of both experimental repeats. Samples were statistically analyzed using a two-way analysis of variance. A significant F score for both factors as well as their interaction (P < 0.01) was calculated. A least square means difference Tukey test was performed to find groups of samples with significant differences in which α = 0.05. Letters (a to g) represent different statistically significant groups within the experimental samples. Error bars represent se for each experiment. conc, concentration.
[See online article for color version of this figure.]
The Expression Level of ATI1 and ATI2 Does Not Influence the Steady State Number of the Bodies Containing Them
Since the function of ATIs on the ATI1 bodies is unknown, we wished to determine whether ATIs affect the number of ATI1 bodies in the cell. To address this issue, we subjected seedlings of the wild-type genotype as well as ATI1-OE and ATI-KD to carbon starvation and counted the number of ATI1 bodies accumulating in their hypocotyl epidermis cells. As shown in Supplemental Figure 12 online, no significant difference in the number of bodies was observed between the three genotypes, indicating that the expression level of the ATI proteins does not influence the steady state number of these bodies, but rather presumably influences their functions.
DISCUSSION
ATI1 and ATI2 Are Newly Identified Plant-Specific Atg8-Interacting Proteins
A number of multifunctional proteins that specifically bind to Atg8 proteins, but themselves are unrelated to the core autophagy machinery, have been described in mammalian cells (Johansen and Lamark, 2011). It was generally hypothesized that the interaction of these proteins with Atg8 is used for their selective turnover by the autophagy machinery, although in some cases, binding to Atg8 also resulted in the modulation of the function of these proteins (Johansen and Lamark, 2011). In contrast with mammals, there are only several very recent reports describing plant Atg8 binding proteins: At-TSPO, which binds the toxic metabolite heme and apparently regulates its selective turnover inside the vacuoles when heme is present in excess (Vanhee et al., 2011); and At-NBR1/Nt-Joka2, a plant functional hybrid protein of the mammalian autophagic adapters p62 and NBR1, which is a selective autophagy substrate degraded in the vacuole (Svenning et al., 2011; Zientara-Rytter et al., 2011).
In this report, we describe two closely related Arabidopsis proteins (ATI1 and ATI2) that bind to the autophagy-associated Atg8f protein (Atg8f was used as a representative in this study) but do not belong to the core autophagy machinery. We confirmed the interaction of ATI1 and ATI2 with Atg8f by two independent approaches, namely, the yeast two-hybrid method and the in vivo BiFC method (Bracha-Drori et al., 2004). Both ATI1 and ATI2 proteins contain two putative AIMs (Noda et al., 2010), located on both sides of a predicted transmembrane domain (Figure 1). Interestingly, as opposed to the proteins of the core plant autophagy machinery, which are homologous to their counterparts in other organisms, ATI1 and ATI2 are specific to plants and possess no homologs outside the plant kingdom. Homologs of ATI1 and ATI2 are present in both dicotyledonous and monocotyledonous plants, and a distant homolog of these proteins also exists in the moss P. patens (see Supplemental Figure 1 online). Thus, ATI1 and ATI2 apparently serve common, yet unknown, functions that are of special importance to plants. In the following discussion, we focus on ATI1 as a representative of the closely homologous ATI1 and ATI2 proteins, displaying similar amino acid sequence, expression, and in situ localization patterns.
ATI1 Is Located on Spherical Bodies That Move Dynamically along the ER Network, but Apparently Do Not Contain Luminal ER Resident Proteins
Our results showed that under favorable growth conditions, ATI1 is generally associated with ER membranes, being in close proximity to the ER marker mCherry-HDEL (Figure 2). Upon exposure to carbon starvation, GFP fluorescence of ATI1-GFP becomes associated particularly with the surface of previously undescribed spherical bodies, which apparently do not contain ER lumen, as determined by the lack of mCherry-HDEL or GFP-HDEL fluorescence inside them (Figure 3; see Supplemental Figure 10 online). The stress-induced newly identified spherical bodies that were visualized in the transmission image of Figures 3 and 4 and Supplemental Figure 7 online were also detected in carbon starved wild-type plants, indicating that these bodies appear naturally and are not a result of our transgenic approach (see Supplemental Figure 8 online). In addition, these ATI1-GFP–containing bodies, named ATI1 bodies, dynamically move on the ER network and occasionally tend to cluster with each other, move together on the ER membrane, and then separate from each other (see Supplemental Movies 2 and 3 online). Using organelle marker proteins, we also showed that ATI1 bodies are not related to the Golgi, mitochondria, or peroxisomes (Figure 6). ATI1 bodies are clearly distinct from the previously described ER-derived spindle-shaped bodies (Matsushima et al., 2003) by their differential morphology and also by the fact that the ATI1 bodies, in contrast with the ER bodies, are not labeled by a fluorescent ER lumen marker containing a C-terminal HDEL signal (Figure 3; see Supplemental Figures 8 and 10 online). These observations render ATI1 bodies as newly identified ER-associated bodies that are induced by carbon starvation, a stress causing energy deprivation. Although ATI1 bodies are principally induced by carbon starvation, in some cases we observed relatively small amounts of these bodies without exposure to darkness. Statistical quantification of this phenomenon in transgenic ATI1-GFP plants has shown that the cellular ATI1 body content during carbon starvation is significantly increased in comparison to their content during favorable growth conditions (see Supplemental Figure 9 online). It is possible that existence of minor amounts of ATI1 bodies under favorable conditions is needed for housekeeping duties, as is also the case with autophagosomes (Inoue et al., 2006; Yano et al., 2007).
Potential Organization of ATI1 on the Membranes of the ER and the ATI1 Bodies
Since ATI1 contains two putative AIMs, on both sides of the predicted transmembrane domain, it is likely that one of these AIMs is located inside and the second located outside ATI1 bodies. Based on a previous report describing the canonical AIM sequence (Noda et al., 2010), the putative AIM located upstream of the predicted transmembrane domain toward the N terminus of ATI1 appears to be a more canonical AIM in terms of its sequence. Furthermore, the positive interaction of ATI1 with Atg8f in the BiFC assay was observed with a construct possessing an in-frame fusion of the N-terminal part of ATI1 to the split YFP vector. Thus, these results imply that the more canonical N-terminal AIM sequence of ATI1, facing outside the ER and the ATI1 bodies, could possibly be used in vivo for the interaction with Atg8f. The localization of ATI1-GFP on the surface of ATI1 bodies upon exposure to carbon starvation (Figure 5; see Supplemental Figure 7 online) agrees with the presence of a putative transmembrane domain in ATI1 and ATI2 (Figure 1; see Supplemental Figure 2 online). However, electron microscopy analysis showed that some of the gold-labeled molecules are detected inside the bodies and not only on their surface (Figure 5). We assume that a certain degree of ATI1-GFP proteolysis occurs, causing a cleavage of the GFP moiety and its internalization to the bodies, probably due to high proteolytic activity within the bodies.
ATI1 Bodies Are Distinct from Autophagosomes
Since ATI1 bodies are clearly visible in a transmission image of the confocal microscope, we were able to analyze their potential colocalization with classical autophagosomes that are labeled by either GFP-Atg8f or mRFP-Atg8f marker proteins. In plants exposed to carbon starvation, the majority of the ATI1 bodies moving on the ER network and visualized in the transmission image were not colocalized with autophagosomes, labeled with the GFP-Atg8f autophagosome marker (Figure 6D). In addition, under carbon starvation conditions, only a very small fraction of these bodies, if any, appear to be colocalized with mRFP-Atg8f autophagosome marker in the cytosol, as determined by the rare identification of bodies that are labeled with both mRFP-Atg8f and ATI1-GFP in the confocal images (Figures 7A to 7C). Moreover, under carbon starvation, a number of small bodies labeled with the ATI1-GFP protein appear inside the vacuole (Figure 8), implying that the ATI1 bodies are transported into the vacuole. Taken together, these results imply that ATI1 bodies may be gradually associated with Atg8f either on the surface of the ER or when detached from the ER into the cytoplasm and that the interaction with Atg8f also may trigger the transport of the ATI1 bodies from the cytoplasm into the vacuole. Such a process may be analogous to the Atg8-mediated selective autophagy, which has been proven to transport specific cellular proteins for degradation in the lysosome of mammalian cells and in the vacuole of plant cells (Johansen and Lamark, 2011; Svenning et al., 2011; Vanhee et al., 2011; Zientara-Rytter et al., 2011).
Potential Roles of ATI1 and ATI2 during Energy Deprivation and in Selective Turnover of Germination-Inhibiting ABA-Associated Macromolecules during Seed Germination
Like many other proteins that bind Atg8 but do not belong to the core autophagy machinery (Mohrlüder et al., 2007a, 2007b; Pankiv et al., 2007), ATI1 and ATI2 apparently do not belong to the plant core autophagy machinery. This is supported by the fact that the core autophagy machinery is conserved between plants and animals, while ATI1 and ATI2 are plant-specific proteins. The exact function of ATI1 and ATI2 in plants and the reason for their interaction with Atg8f are still unknown. Yet, since the expression of ATI1 and ATI2 is induced by exposure to darkness (carbon starvation) and also to various other abiotic stresses that cause energy deprivation and may also trigger autophagy (see Supplemental Figure 3 online), we hypothesize that the function of ATI1 and ATI2 may be related to the selective disposal of macromolecules that are not needed from the ER in the vacuole. Moreover, since ATI1 contains a predicted transmembrane domain and since ATI1 bodies appear in the confocal microscope to move along the ER membranes, it may also be possible that ATI1 functions in the disposal of ER membrane proteins. The phenomenon of direct movement from ER to the vacuole in plant cells was previously reported regarding precursor-accumulating vesicles delivering storage protein precursors in pumpkin (Cucurbita sp cv Kurokawa Amakuri Nankin) seeds (Hara-Nishimura et al., 1998) and regarding the ER-to-vacuole movement of Cys proteases carrying the ER retention signal H/KDEL (Toyooka et al., 2000). In both cases, the macromolecules were transferred to the vacuole for storage. However, it might as well be that such strategy is used by the plants for massive disposal of macromolecules that are not needed as well. This kind of massive disposal may be generally required upon switching from one physiological or developmental state to another, such as exposure to stresses causing energy deprivation or the transition from seed development to seed germination. Exposure to stress may cause extensive misfolding of nascent proteins formed in the ER due to the lack of energy required for their folding (Vitale and Boston, 2008; Liu and Howell, 2010), and such proteins may be transported and degraded in the vacuole. Elimination of such proteins generally occurs by ER-associated degradation machineries that target the proteins for degradation either by the proteasome or by proteases in the vacuole (Pedrazzini et al., 1997; Frigerio et al., 2001; Pimpl et al., 2006; Foresti et al., 2008; Liu and Bassham, 2010). It is possible that ATI1 and ATI2 participate in these processes. We showed that ATI1-GFP changes its cellular localization after exposure of young seedlings to relative short carbon starvation to newly identified spherical bodies, which increase significantly in number as a result of the stress. Those bodies might function as a newly identified transport system that is based, according to our data, on movement on the ER network, probably assisted by actino-myosin motors, similarly to other ER-associated transport systems in plants (Sparkes et al., 2009). We believe that the actual presence of the bodies under favorable conditions demonstrates that the mechanism of ATI1 bodies is probably constitutively active at basal levels and by that it resembles autophagy that was shown to be a steady state process active in favorable conditions and increased during stress and starvation (Inoue et al., 2006; Yano et al., 2007). However, in contrast with autophagy, the ATI1 body disposal mechanism seems to be unique to plants and by that may function in plant-unique situations like shortage of energy, which requires suitable adaptations, or seed development and germination, which requires massive degradation procedures. The transition from seed development to seed germination requires the selective turnover of germination-inhibiting ABA-associated proteins accumulating during seed development, apparently by transporting them for degradation in the vacuole. Indeed, our observations show a positive correlation between the expression level of ATI1 and ATI2 and seed germination on a medium containing the ABA hormone, with seeds of the ATI1-GFP–overexpressing plants germinating at a higher frequency than wild-type plants and seeds with suppressed expression of ATI1 and ATI2 germinating at a lower frequency than wild-type plants, supporting our hypothesis. These observations also support the functionality of the ATI1 and ATI2 proteins in these biological processes. In addition, the fact that altered expression level of ATI1 and ATI2 does not affect the steady state number of ATI1 bodies (see Supplemental Figure 12 online) may indicate that ATIs do not function in the formation of ATI1 bodies, but rather participate in other processes, such as cargo selection. Further studies are needed to identify the cargo(s) of ATI1 bodies and decipher their functions.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana ecotype Colombia was used in this study. ATI-KD plants were generated from a cross between homozygous ATI1 T-DNA knockout plants (SAIL_404_D09, obtained from the ABRC seed collection, Ohio State University [Alonso et al., 2003]; http://abrc.osu.edu) and homozygous ATI2-RNAi plants (CATMA4a_00420 seed line 2762, obtained from the AGRIKOLA collection [Hilson et al., 2004]; http://www.agrikola.org). F3 progeny was analyzed by RT-PCR and real-time PCR to quantify the ATI1 and ATI2 gene expression level, respectively. The F3 line demonstrating the lowest expression level was used for the physiological analysis.
Several marker lines were used in the experiments: the ER lumen markers mCherry-HDEL and GFP-HDEL (Nelson et al., 2007), the Golgi marker GmMan1-mCherry (Nelson et al., 2007), the mitochondrial marker ScCOX4-mCherry (Nelson et al., 2007), the peroxisome marker AtPEX5-CFP (Tian et al., 2004), and the autophagosome marker GFP-Atg8f (Sláviková et al., 2005).
Seeds of relevant genotypes and various cellular marker lines were surface sterilized using bleach/HCl mixture, sown on 1MS+SUC plates (full-strength MS medium including vitamins and 1% Suc) with the proper antibiotic selection, incubated at 4°C for 48 h, and put in a long-day growth chamber (16 h light/8 h dark, 25°C, 40% humidity).
Carbon Starvation and ABA Treatments
For carbon starvation, wild-type and transgenic seeds were sown on 1MS medium lacking Suc (1MS-SUC) plates and grown for 6 d in the growth chamber. A day before microscopy analysis, the 1MS-SUC plates were covered by double layer of aluminum foil and left in the same growth chamber for additional 24 h. All plants were microscopically analyzed 7 d after germination. For ABA treatments, the wild type and relevant transgenic genotypes were sown in 0.5MS+SUC plates without ABA or supplemented with 0.5, 0.75, 1, or 1.5 μM ABA (diluted from a 10 mM ABA stock solution) and incubated at 4°C as described above. The plants were vertically grown, and radicle emergence was analyzed 2 d after plate transfer to a long-day growth chamber.
Protein Interaction Verification Using the Yeast Two-Hybrid and BiFC Methods
The yeast two-hybrid analysis was performed using the MATCHMAKER (Clontech) GAL4 two-hybrid system. The Atg8f coding DNA region was cloned in frame downstream to the GAL4 DNA binding domain in the yeast bait vector pGBKT7 (Clontech). This plasmid was cointroduced with a pACT2 vector (containing a GAL4 activation domain), harboring a cDNA expression library prepared from a dark-grown Arabidopsis cell suspension (Németh et al., 1998), to the yeast AH109 strain containing the GAL4-responsive genes ADE2, HIS3, and lacZ (Clontech). These reporter genes are expressed when in vivo two-hybrid interaction combines the GAL4 binding and activation domains (AD+BD). pACT2 clones capable of activating the ADE2 and lacZ reporters genes were rescued and their cDNA inserts were sequenced. lacZ, ADE2, and HIS3 activation was rechecked several times by one-on-one yeast two-hybrid interaction for all the positive clones to eliminate false positive interactions.
For analyzing in planta interactions between Atg8f and two of the positive clones identified in the yeast two-hybrid screen (ATI1 and ATI2 proteins encoded by the At2g45980 and At4g00355 genes, respectively), the coding sequence of Atg8f was cloned into a pSY736 vector (Bracha-Drori et al., 2004) in frame and downstream to the first half of the YFP coding sequence (YN), while the coding sequences of ATI1 or ATI2 were each cloned into the pSY735 vector (Bracha-Drori et al., 2004) in frame and downstream to the second half of the YFP coding sequence (YC). The YN-Atg8f, YC-ATI1, and YC-ATI2 expression cassettes were each transferred to a binary pCAMBIA1300 vector, which was further individually transformed into GV3101 strain Agrobacterium tumefaciens (Koncz and Schell, 1986) using electroporation. For verification of the interaction of Atg8f with ATI1 or ATI2, Agrobacterium strains harboring the pCAMBIA-YN-Atg8f or pCAMBIA-YC-ATI vector were transiently coexpressed either in Arabidopsis cotyledons or in Nicotiana benthamiana leaves (see below). pCAMBIA2300-YC-FTB was used as a negative control vector for this BiFC interactions because FTB lacks the consensus AIM motif and thus was not expected to interact with the Atg8f protein in the cytosol. FTB and FTA are two subunits reconstructing the Arabidopsis-soluble cytoplasmic PFT (Bracha-Drori et al., 2004); hence, pCAMBIA2300-YC-FTB and pCAMBIA2300-YN-FTA (donated by Shaul Yalovsky, Tel-Aviv University, Israel) were both used for split-YFP interaction positive control.
Plasmid Construction
All primers used for amplification of different genes are listed in Supplemental Table 1 online.
Construction of ATI1-GFP, ATI2-GFP, and ATI2-mRFP Fusion Proteins
To construct the ATI1-GFP and ATI2-GFP translational fusion proteins, the coding sequences of ATI1 (780 bp) and ATI2 (810 bp) were amplified from Arabidopsis wild-type cDNA. Both ATI1 and ATI2 coding DNA sequences were cloned in frame with the coding sequence of GFP to generate ATI1-GFP and ATI2-GFP in the vector pART7 (Gleave, 1992). The Pro35S:ATI1-GFP and Pro35S:ATI2-GFP expression cassettes were further transferred into the plant binary vector pART27 (Gleave, 1992). To construct the ATI2-mRFP translational fusion protein, the coding sequence of mRFP (681 bp) was amplified from pART7-RD21-mRFP vector (kindly donated by Olga Davidov, The Weizmann Institute). Next, the GFP coding sequence was removed from the pART7-ATI2-GFP vector and replaced with the amplified mRFP coding sequence, in frame with the ATI2 coding sequence. As a result of this fusion, a C-terminal mRFP-tagged ATI2 construct was created. The Pro35S:ATI2-mRFP cassette was further transferred into the plant binary vector pMLBART (Gleave, 1992).
Construction of GFP-Atg8f and mRFP-Atg8f Fusion Proteins
To construct the GFP-Atg8f-3HA translational fusion protein, the coding sequence of Atg8f (363 bp) was amplified from Arabidopsis cDNA and cloned in frame with the coding sequence of GFP in the vector pART7. A six-Ala linker bridged between GFP and Atg8f coding sequences. Three repeats of the hemaglutation (HA) tag were also cloned to this vector, downstream to the Atg8f coding sequence. As a result of those fusions, an N-terminal GFP-tagged and C-terminal HA-tagged Atg8f translational fusion was created. The 3HA tag is cleaved prior to incorporation of the Atg8f to the autophagosome membrane (Sláviková et al., 2005). The Pro35S:GFP-Atg8f-3HA cassette was further transferred into plant binary vector pART27. To construct the mRFP-Atg8f-3HA translational fusion protein, the coding sequence of mRFP (681 bp) was amplified from pART7-RD21-mRFP vector as described above. The GFP coding sequence in pART7- Pro35S:GFP-Atg8f-3HA was then replaced with the mRFP coding sequence fragment. The Pro35S:mRFP-Atg8f-3HA cassette was further transferred into plant binary vector pMLBART. All binary vectors were introduced into GV3101 Agrobacterium strain as described above.
RNA Extraction, cDNA Synthesis, RT-PCR, and Real-Time PCR
RNA was extracted from 100 mg of frozen seedling tissues using a standard Tri-reagent extraction method. cDNA was synthesized using the high-capacity cDNA reverse transcription kit (Applied Biosystems), which includes random primers and multiscribe reverse transcriptase. RT-PCR reactions were performed with primers amplifying the full-length ATI1 coding sequence. CYCLOPHILIN (At2g36130) was used as reference gene. Real-time PCR experiments for quantifying ATI2 expression level were performed using the power SYBR green kit (Applied Biosystems) and the 7300 real-time PCR system (Applied Biosystems) according to the manufacturer’s instructions. The UBIQUITIN-CONJUGATING 21 gene (Ubi-C; At5g25760) was used as an internal standard. Real-time primers for ATI2 and Ubi-C genes were designed using primer express 3.0 software (Applied Biosystems). Real-time PCR experiments were repeated twice using cDNA templates prepared from four independent batches of plants, and every reaction was set up in triplicates. All RT-PCR and real-time PCR primer sequences are listed in Supplemental Table 1 online.
ConA Treatment of Arabidopsis Seedlings
ConA (Fluka 27689; 100 μM stock in DMSO) was generally used according to a previously described protocol (Ishida et al., 2008) with some modifications. Arabidopsis seedlings were grown in 1MS-SUC Petri plates for 6 d. Then, three to five whole seedlings of each of the examined line (Pro35S:ATI1-GFP) were transferred into Eppendorf tubes containing 1 μM ConA diluted in incubation buffer (10 mM MES-NaOH, pH 5.5). As a control, the same number of seedlings was subjected to the same treatment using DMSO (used to dissolve ConA) without ConA. The seedlings were incubated in the liquid media for 20 h, at 23°C, in the dark. Afterwards, the seedlings were directly examined by a confocal microscope.
Transient Expression in N. benthamiana Leaves and in Arabidopsis Cotyledons
Transient expression of proteins in N. benthamiana leaves was performed according to Sparkes et al. (2006). Transient expression of proteins in Arabidopsis cotyledons was modified from a published protocol (Li et al., 2009) as follows: Wild-type Arabidopsis seeds were surface sterilized and sown (10 to 15 seeds in each well) in a sterile 24-well plate containing solid quarter-strength MS medium (pH 6, 1% Suc). The plates were incubated for 48 h at 4°C and afterwards transferred to a growth chamber (16 h light/8 h dark, 25°C, 40% humidity) and grown for 4 d until the cocultivation procedure. The Agrobacterium culture preparation was done as previously described (Li et al., 2009). Finally, 100 μL of the Agrobacterium-containing cocultivation medium were dripped on top of the seedlings in the wells using a pipette. The plates were sealed again with Parafilm, covered with aluminum foil, and were put back to the growth chamber, causing a mild, short-term carbon starvation. Forty-eight hours after the cocultivation process, the plant cotyledons were scanned for positive fluorescence expressing cells using an Olympus SZX12 fluorescent stereoscope and then examined by a confocal microscope.
Confocal Microscopy
An Olympus Fluoview 1000 IX81 confocal microscope system was used in this study. For Arabidopsis seedling analysis, 7-d-old whole seedlings were put between two microscope glass cover slips (24 × 40mm, No. 1 thickness) in an aqueous environment. For transient expression assays, the cotyledon epidermis cells were analyzed. For examination of stable transgenic plants under favorable conditions or under carbon and nitrogen starvation, hypocotyl epidermis cells were analyzed. For ConA treatments, both hypocotyl and root cells were analyzed. For N. benthamiana transient expression, a small piece (0.5 cm2) of the injected leaf was placed between two glass cover slips as described above, and the epidermis cells were analyzed. All images were taken from single focal plane (no Z slices were used). For all image acquisition, the UPlanSApo ×60 oil objective (numerical aperture of 1.35) was used. GFP fluorescence images were taken using a 488-nm argon laser. mCherry and mRFP images were taken using a 559-nm diode laser. YFP images were taken using a 515-nm argon laser. Transmittance images were taken using the differential interference contrast method. Time-lapse images were all composed from 30 to 50 images taken using line sequence. All acquired images were converted to TIFF (for images) and AVI (for movies) formats using the Olympus Fluoview 1000 viewer.
In Situ Quantification of the ATI1 Bodies and Statistical Analysis
For quantification of ATI1 bodies under favorable and carbon starvation conditions, ATI1-GFP 7-d-old seedlings were used. Plants were either carbon starved or grown under favorable conditions (as described above). Images of hypocotyl epidermal cells from different plants of each growth condition (starved and nonstarved) were acquired using confocal microscopy. The cells chosen for analysis were approximately of similar size, and seven sample cells were taken from each treatment. ATI1 bodies were identified using both transmittance and GFP fluorescence. The average number of bodies per cell was calculated for each condition, and statistical significance was determined using a Student's t test (P < 0.01).
For quantification of ATI1 bodies in lines expressing varying levels of ATI1 and ATI2, ATI1-OE, ATI-KD, and wild-type 7-d-old seedlings were used. Plants were carbon starved as described above. Images of hypocotyl epidermal cells from different plants were acquired using confocal microscopy. The cells chosen for analysis were approximately of similar size, and 10 sample cells were taken from line. ATI1 bodies were identified using transmittance images. The average number of bodies per cell was calculated for each condition and statistical significance was determined using one-way analysis of variance.
Electron Microscopy
Sample Preparation for Electron Microscopy
The proximal 2-mm regions of the hypocotyls of transgenic ATI1-GFP 7-d-old seedlings, grown under carbon starvation as described above, were removed under a dissection microscope and immediately suspended in a fixative solution consisting of 1.5% paraformaldehyde and 0.25% glutaraldehyde in 50 mM phosphate buffer, pH 7.4. The hypocotyls were maintained during fixation for 60 min under moderate vacuum, to remove air, and then were transferred to 4°C for overnight. After washing in cold 50 mM phosphate buffer and in double distilled water, the samples were dehydrated in a graded ethanol series for 1 h at each concentration of 15, 30, 50, 70, 85, and 95%, and three times in absolute ethanol stored over molecular sieve as a drying agent. After dehydration, the samples were embedded in Lowicryl HM20 (EMS) according to the following conditions: 5 h or overnight in each of a series of Lowicryl/ethanol mixture of 10, 20, 30, 40, 50, 65, 80, 90, and 95% and then in 100% Lowicryl for 3 × 5 h or overnight. All dehydration and embedding procedures were performed at 4°C. Polymerization was performed at −20°C under UV light for 72 h in an AFS2 freeze substitution device (Leica Mycrosystems).
Immunolocalization of GFP
Formvar-coated 300 mesh nickel grids carrying ultrathin sections (90 mM) of Lowicryl-embedded transgenic Arabidopsis hypocotyls were washed in phosphate buffer containing 0.2% Gly for 2 × 3 min and then blocked with a blocking solution consisting of 0.5% gelatin, 0.5% BSA, 0.2% Gly, and 1% Tween 20 in PBS, pH 7.4, for 30 min. The sections were then treated with the primary antibody rabbit polyclonal anti-GFP (ab-6556 abCam) diluted 1:25 and 1:50 in blocking solution for 2 h in a wet chamber. The grids were washed 5 × 1min in PBS containing 0.2% Gly and blocked again for 5 min. They were transferred to a droplet of Goat anti-rabbit IgG conjugated to 10 nM gold particles diluted 1:20 in blocking solution for 30 min. Labeled samples were washed 5 × 1min in PBS containing 0.2% Gly and then in double distilled water for 3 × 1min. The grids were poststained with 2% uranyl acetate dissolved in 50% ethanol, rinsed in lead citrate, and then examined at 120 kV in a Tecnai Spirit transmission electron microscope (FEI). Images were recorded using an Eagle 2K × 2K charge-coupled device camera (FEI).
Sequence and Bioinformatic Analysis
ATI1 and ATI2 coding sequences and protein sequences were retrieved from The Arabidopsis Information Resource (http://www.Arabidopsis.org). Protein sequences of ATI orthologs from different plant species were retrieved from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) using the BLASTP tool. Multiple alignment of ATI protein sequences (available as Supplemental Data Set 1 online) was done by ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html), and phylogenetic tree buildup was done using MEGA software (http://www.megasoftware.net). A neighbor-joining tree of protein sequences was constructed using the default settings (Poisson model of substitution, uniform rates among sites, homogeneous pattern among lineages, and missing data treatment of complete deletion). The test of phylogeny was performed using the bootstrap method with 1000 bootstrap replications. Gene expression analysis in different Arabidopsis tissues and developmental stages and under different environmental stimuli was displayed using GENEVESTIGATOR (https://www.genevestigator.com) and BAR (http://bar.utoronto.ca/interactions/) public microarray databases. Transmembrane domain prediction display was done by the ARAMEMNON plant membrane protein database (http://aramemnon.botanik.uni-koeln.de). Hydrophobicity plots were created using the subcellular proteomic database (SUBA; http://www.plantenergy.uwa.edu.au/applications/suba2). Coexpressed gene network analysis was done using the ATTED-II coregulated gene database (http://atted.jp). The overrepresentation analysis of the ATI’s coexpressed genes was done using the PAGEMAN multiparallel experiments overview tool (http://mapman.mpimp-golm.mpg.de/general/ora/ora.shtml).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under accession numbers At2g45980 (At-ATI1), At4g00355 (At-ATI2), and At4g16520 (At-Atg8f).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogenetic Analysis Shows That ATI1 and ATI2 from Arabidopsis Have Ortholog Proteins in Different Plant Species.
Supplemental Figure 2. Hydrophobicity Index of ATI1 and ATI2 Predicts One Transmembrane Domain in the Sequence of Each Protein.
Supplemental Figure 3. Microarray Data Analysis of ATI1, ATI2, and Atg8f Gene Expression Shows Increased Expression under Different Stress and Starvation Conditions.
Supplemental Figure 4. ATI1, ATI2, and Atg8f Show Similar Gene Expression Patterns in Various Arabidopsis Tissues.
Supplemental Figure 5. ATTED-II Coexpression Predictions of the ATI1 and ATI2 Gene Network Relate Them to Protein Degradation-Associated Genes.
Supplemental Figure 6. ATI2-mRFP Is Extensively Colocalized with ATI1-GFP and Partially Localized with the ER Marker mCherry-HDEL.
Supplemental Figure 7. ATI1-GFP Becomes Associated with Spherical Intracellular Bodies upon Exposure to Carbon Starvation.
Supplemental Figure 8. Newly Identified Bodies Are Found in Hypocotyl and Cotyledon Tissues of Wild-Type Arabidopsis under Carbon Starvation Conditions.
Supplemental Figure 9. ATI1 Bodies Are Seen Also under Favorable Growth Conditions in ATI1-GFP Plants, but Significantly Induced by Carbon Starvation.
Supplemental Figure 10. ATI1 Bodies Are Clearly Distinct in Structure from Spindle-Shaped ER Bodies and Do Not Colocalize with the ER Lumen Marker GFP-HDEL.
Supplemental Figure 11. Microarray Data Analysis of ATI1 and ATI2 Gene Expression during Seed Development and Germination.
Supplemental Figure 12. ATI Expression Levels Do Not Affect the Steady State Number of ATI1 Bodies during Carbon Starvation.
Supplemental Table 1. Primer List.
Supplemental Movie 1. Dynamic Movement of Newly Identified Spherical-Shaped Bodies in Comparison to Classic ER Body Movement in Arabidopsis Hypocotyl Epidermis Cells under Carbon Starvation.
Supplemental Movie 2. Dynamic Movement of ATI1 Bodies (Detected by ATI1-GFP) on the ER Membrane Network in Arabidopsis Hypocotyl Epidermis Cell under Carbon Starvation.
Supplemental Movie 3. Dynamic Movement of ATI1 Bodies on the ER Membrane Network in Arabidopsis Hypocotyl Epidermis Cell under Carbon Starvation.
Supplemental Movie 4. Dynamic Movement of ATI1-GFP–Containing Particles within the Vacuolar Lumen of ConA-Treated Carbon-Starved Arabidopsis Cotyledon Cell Expressing the ATI1-GFP Polypeptide.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis in Supplemental Figure 1.
Acknowledgments
We thank Zvulun Elazar and Hilla Weidberg for critical reading of the manuscript and helpful comments, Daniela Ben-Tov for her excellent assistance in the scientific experiments, Vladimir Kiss for his invaluable help with confocal microscope image acquisition, Hanna Levanony and Eyal Shimoni for performing the transmission electron microscopy sample preparations and analysis, Csaba Konz and Aviah Zilberstein for providing the yeast two-hybrid system, and Shaul Yalovsky for providing the cloning and control vectors for BiFC analysis. Our research was supported by grants from The Israel Science Foundation (Grant 764/07), the J & R Center for Scientific Research at The Weizmann Institute of Science, and the Israeli Ministry of Agriculture. G.G. is an incumbent of the Bronfman Chair of Plant Science at The Weizmann Institute of Science.
AUTHOR CONTRIBUTIONS
A.H. and G.G. designed the research. A.H., T.A.-W., and S.U. each performed different aspects of the research and analyzed the data. A.H., T.A.-W. and G.G. contributed to the writing of the article.
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Baena-González E., Sheen J. (2008). Convergent energy and stress signaling. Trends Plant Sci. 13: 474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham D.C. (2007). Plant autophagy—More than a starvation response. Curr. Opin. Plant Biol. 10: 587–593 [DOI] [PubMed] [Google Scholar]
- Bassham D.C. (2009). Function and regulation of macroautophagy in plants. Biochim. Biophys. Acta 1793: 1397–1403 [DOI] [PubMed] [Google Scholar]
- Bewley J.D. (1997). Seed germination and dormancy. Plant Cell 9: 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha-Drori K., Shichrur K., Katz A., Oliva M., Angelovici R., Yalovsky S., Ohad N. (2004). Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40: 419–427 [DOI] [PubMed] [Google Scholar]
- Braybrook S.A., Harada J.J. (2008). LECs go crazy in embryo development. Trends Plant Sci. 13: 624–630 [DOI] [PubMed] [Google Scholar]
- Contento A.L., Xiong Y., Bassham D.C. (2005). Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J. 42: 598–608 [DOI] [PubMed] [Google Scholar]
- Dröse S., Boddien C., Gassel M., Ingenhorst G., Zeeck A., Altendorf K. (2001). Semisynthetic derivatives of concanamycin A and C, as inhibitors of V- and P-type ATPases: Structure-activity investigations and developments of photoaffinity probes. Biochemistry 40: 2816–2825 [DOI] [PubMed] [Google Scholar]
- Foresti O., De Marchis F., de Virgilio M., Klein E.M., Arcioni S., Bellucci M., Vitale A. (2008). Protein domains involved in assembly in the endoplasmic reticulum promote vacuolar delivery when fused to secretory GFP, indicating a protein quality control pathway for degradation in the plant vacuole. Mol. Plant 1: 1067–1076 [DOI] [PubMed] [Google Scholar]
- Frigerio L., Pastres A., Prada A., Vitale A. (2001). Influence of KDEL on the fate of trimeric or assembly-defective phaseolin: Selective use of an alternative route to vacuoles. Plant Cell 13: 1109–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J., Parcy F., Bertauche N., Gosti F., Leung J., Morris P.C., Bouvier-Durand M., Vartanian N. (1994). Current advances in abscisic acid action and signalling. Plant Mol. Biol. 26: 1557–1577 [DOI] [PubMed] [Google Scholar]
- Gleave A.P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Hara-Nishimura I.I., I, Shimada T., Hatano K., Takeuchi Y., Nishimura M. (1998). Transport of storage proteins to protein storage vacuoles is mediated by large precursor-accumulating vesicles. Plant Cell 10: 825–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawes C., Saint-Jore C., Martin B., Zheng H.Q. (2001). ER confirmed as the location of mystery organelles in Arabidopsis plants expressing GFP! Trends Plant Sci. 6: 245–246 [DOI] [PubMed] [Google Scholar]
- Hayward A.P., Dinesh-Kumar S.P. (2010). What can plant autophagy do for an innate immune response? Annu. Rev. Phytopathol. 49: 4.1–4.20 [DOI] [PubMed] [Google Scholar]
- Hilson P., et al. (2004). Versatile gene-specific sequence tags for Arabidopsis functional genomics: Transcript profiling and reverse genetics applications. Genome Res. 14(10B): 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y., Komatsu M. (2010). Selective degradation of p62 by autophagy. Semin. Immunopathol. 32: 431–436 [DOI] [PubMed] [Google Scholar]
- Inoue Y., Suzuki T., Hattori M., Yoshimoto K., Ohsumi Y., Moriyasu Y. (2006). AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol. 47: 1641–1652 [DOI] [PubMed] [Google Scholar]
- Ishida H., Yoshimoto K., Izumi M., Reisen D., Yano Y., Makino A., Ohsumi Y., Hanson M.R., Mae T. (2008). Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol. 148: 142–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T., Lamark T. (2011). Selective autophagy mediated by autophagic adapter proteins. Autophagy 7: 279–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V., McEwan D.G., Novak I., Dikic I. (2009). A role for ubiquitin in selective autophagy. Mol. Cell 34: 259–269 [DOI] [PubMed] [Google Scholar]
- Koncz C., Schell J. (1986). The promoter of Tl-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204: 383–396 [Google Scholar]
- Li J.F., Park E., von Arnim A.G., Nebenführ A. (2009). The FAST technique: A simplified Agrobacterium-based transformation method for transient gene expression analysis in seedlings of Arabidopsis and other plant species. Plant Methods 5: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.X., Howell S.H. (2010). Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 22: 2930–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.M., Bassham D.C. (2010). TOR is a negative regulator of autophagy in Arabidopsis thaliana. PLoS ONE 5: e11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C., Daniel-Vedele F., Dechorgnat J., Chardon F., Gaufichon L., Suzuki A. (2010). Nitrogen uptake, assimilation and remobilization in plants: Challenges for sustainable and productive agriculture. Ann. Bot. (Lond.) 105: 1141–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima R., Hayashi Y., Kondo M., Shimada T., Nishimura M., Hara-Nishimura I. (2002). An endoplasmic reticulum-derived structure that is induced under stress conditions in Arabidopsis. Plant Physiol. 130: 1807–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima R., Hayashi Y., Yamada K., Shimada T., Nishimura M., Hara-Nishimura I. (2003). The ER body, a novel endoplasmic reticulum-derived structure in Arabidopsis. Plant Cell Physiol. 44: 661–666 [DOI] [PubMed] [Google Scholar]
- Meijer W.H., van der Klei I.J., Veenhuis M., Kiel J.A. (2007). ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy 3: 106–116 [DOI] [PubMed] [Google Scholar]
- Mittler R., Blumwald E. (2010). Genetic engineering for modern agriculture: Challenges and perspectives. Annu. Rev. Plant Biol. 61: 443–462 [DOI] [PubMed] [Google Scholar]
- Mohrlüder J., Hoffmann Y., Stangler T., Hänel K., Willbold D. (2007a). Identification of clathrin heavy chain as a direct interaction partner for the gamma-aminobutyric acid type A receptor associated protein. Biochemistry 46: 14537–14543 [DOI] [PubMed] [Google Scholar]
- Mohrlüder J., Stangler T., Hoffmann Y., Wiesehan K., Mataruga A., Willbold D. (2007b). Identification of calreticulin as a ligand of GABARAP by phage display screening of a peptide library. FEBS J. 274: 5543–5555 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H., Ichimura Y., Ohsumi Y. (2007). Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130: 165–178 [DOI] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Németh K., et al. (1998). Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev. 12: 3059–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda N.N., Ohsumi Y., Inagaki F. (2010). Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 584: 1379–1385 [DOI] [PubMed] [Google Scholar]
- Noda N.N., Kumeta H., Nakatogawa H., Satoo K., Adachi W., Ishii J., Fujioka Y., Ohsumi Y., Inagaki F. (2008). Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells 13: 1211–1218 [DOI] [PubMed] [Google Scholar]
- Novak I., et al. (2010). Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 11: 45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K., Kondo-Okamoto N., Ohsumi Y. (2009). Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell 17: 87–97 [DOI] [PubMed] [Google Scholar]
- Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. (2007). p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282: 24131–24145 [DOI] [PubMed] [Google Scholar]
- Pedrazzini E., Giovinazzo G., Bielli A., de Virgilio M., Frigerio L., Pesca M., Faoro F., Bollini R., Ceriotti A., Vitale A. (1997). Protein quality control along the route to the plant vacuole. Plant Cell 9: 1869–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimpl P., Taylor J.P., Snowden C., Hillmer S., Robinson D.G., Denecke J. (2006). Golgi-mediated vacuolar sorting of the endoplasmic reticulum chaperone BiP may play an active role in quality control within the secretory pathway. Plant Cell 18: 198–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sláviková S., Shy G., Yao Y.L., Glozman R., Levanony H., Pietrokovski S., Elazar Z., Galili G. (2005). The autophagy-associated Atg8 gene family operates both under favourable growth conditions and under starvation stresses in Arabidopsis plants. J. Exp. Bot. 56: 2839–2849 [DOI] [PubMed] [Google Scholar]
- Slavikova S., Ufaz S., Avin-Wittenberg T., Levanony H., Galili G. (2008). An autophagy-associated Atg8 protein is involved in the responses of Arabidopsis seedlings to hormonal controls and abiotic stresses. J. Exp. Bot. 59: 4029–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I.A., Frigerio L., Tolley N., Hawes C. (2009). The plant endoplasmic reticulum: A cell-wide web. Biochem. J. 423: 145–155 [DOI] [PubMed] [Google Scholar]
- Sparkes I.A., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Svenning S., Lamark T., Krause K., Johansen T. (2011). Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 7: 993–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Shimada T., Ono E., Tanaka Y., Nagatani A., Higashi S.I., Watanabe M., Nishimura M., Hara-Nishimura I. (2003). Why green fluorescent fusion proteins have not been observed in the vacuoles of higher plants. Plant J. 35: 545–555 [DOI] [PubMed] [Google Scholar]
- Thompson A.R., Doelling J.H., Suttangkakul A., Vierstra R.D. (2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138: 2097–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.R., Vierstra R.D. (2005). Autophagic recycling: Lessons from yeast help define the process in plants. Curr. Opin. Plant Biol. 8: 165–173 [DOI] [PubMed] [Google Scholar]
- Tian G.W., et al. (2004). High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol. 135: 25–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka K., Okamoto T., Minamikawa T. (2000). Mass transport of proform of a KDEL-tailed cysteine proteinase (SH-EP) to protein storage vacuoles by endoplasmic reticulum-derived vesicle is involved in protein mobilization in germinating seeds. J. Cell Biol. 148: 453–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhee C., Zapotoczny G., Masquelier D., Ghislain M., Batoko H. (2011). The Arabidopsis multistress regulator TSPO is a heme binding membrane protein and a potential scavenger of porphyrins via an autophagy-dependent degradation mechanism. Plant Cell 23: 785–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale A., Boston R.S. (2008). Endoplasmic reticulum quality control and the unfolded protein response: Insights from plants. Traffic 9: 1581–1588 [DOI] [PubMed] [Google Scholar]
- Weidberg H., Shvets E., Shpilka T., Shimron F., Shinder V., Elazar Z. (2010). LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 29: 1792–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Contento A.L., Nguyen P.Q., Bassham D.C. (2007). Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 143: 291–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K., Suzuki T., Moriyasu Y. (2007). Constitutive autophagy in plant root cells. Autophagy 3: 360–362 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K. (2010). Plant autophagy puts the brakes on cell death by controlling salicylic acid signaling. Autophagy 6: 192–193 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K., Hanaoka H., Sato S., Kato T., Tabata S., Noda T., Ohsumi Y. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zientara-Rytter K., Lukomska J., Moniuszko G., Gwozdecki R., Surowiecki P., Lewandowska M., Liszewska F., Wawrzyńska A., Sirko A. (2011). Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors. Autophagy 7: 1145–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]