The L-shaped skeleton of the chloroplast NADH dehydrogenase-like complex is highly conserved among the related complexes. An assembly model of its stroma-protruding arm is proposed. Three intermediates are assembled in the stroma in a stepwise manner via specific factors that facilitate the biogenesis of subunits or mediate their integration into the complex.
Abstract
Chloroplast NADH dehydrogenase-like complex (NDH) mediates photosystem I cyclic electron transport and chlororespiration in thylakoids. Recently, substantial progress has been made in understanding the structure of NDH, but our knowledge of its assembly has been limited. In this study, a series of interactive proteomic analyses identified several stroma-localized factors required for the assembly of a stroma-protruding arm of NDH (subcomplex A). In addition to further characterization of the previously identified CHLORORESPIRATORY REDUCTION1 (CRR1), CRR6, and CRR7, two novel stromal proteins, CRR41 and CRR42, were discovered. Arabidopsis thaliana mutants lacking these proteins are specifically defective in the accumulation of subcomplex A. A total of 10 mutants lacking subcomplex A, including crr27/cpn60β4, which is specifically defective in the folding of NdhH, and four mutants lacking NdhL–NdhO subunits, were extensively characterized. We propose a model for subcomplex A assembly: CRR41, NdhO, and native NdhH, as well as unknown factors, are first assembled to form an NDH subcomplex A assembly intermediate (NAI500). Subsequently, NdhJ, NdhM, NdhK, and NdhI are incorporated into NAI500 to form NAI400. CRR1, CRR6, and CRR42 are involved in this process. CRR7 is likely to be involved in the final step, in which the fully assembled NAI, including NdhN, is inserted into thylakoids.
INTRODUCTION
In higher plants, the chloroplast NADH dehydrogenase-like complex (NDH) is a multisubunit complex embedded in stroma thylakoids. Studies of mutant phenotypes revealed that chloroplast NDH is involved in cyclic electron transport around photosystem I (PSI) and chlororespiration (Shikanai, 2007). Chloroplast NDH recycles electrons from ferredoxin (Fd) to plastoquinone (PQ) and subsequently to PSI through the cytochrome (Cyt) b6f complex. Both the NDH complex and the Cyt b6f complex contribute to the formation of ΔpH across thylakoid membranes, but the rate of NDH-dependent electron transport is low and is unlikely to contribute substantially to ATP synthesis during steady state photosynthesis (Okegawa et al., 2008). However, there is some evidence that chloroplast NDH is required to alleviate stromal overreduction under stress conditions (Endo et al., 1999; Munekage et al., 2004).
Eleven subunits of chloroplast NDH are homologous to the subunits of mitochondrial complex I (NADH dehydrogenase) and eubacterial NDH-1 (Suorsa et al., 2009; Peng et al., 2011b), suggesting that all of the NDH-related complexes have similar structures and enzyme activities. The Escherichia coli NDH-1 complex has an L-shaped structure and comprises two major parts, the membrane and peripheral segments. Recently, the crystal structure of the peripheral and membrane segments of NDH-1 was solved in the thermophilic bacterium Thermus thermophilus and E. coli, respectively (Sazanov and Hinchliffe, 2006; Efremov et al., 2010; Efremov and Sazanov, 2011). The membrane segment consists of six hydrophobic subunits (which correspond to the chloroplast NDH membrane subunits NdhB–NdhG) with 55 transmembrane helices, and it is required for proton translocation (Efremov et al., 2010; Efremov and Sazanov, 2011). The peripheral segment of E. coli NDH-1 protrudes into the bacterial cytosol and contains other seven hydrophilic subunits (four of which are homologs to chloroplast NdhH–NdhK) carrying redox centers, flavin mononucleotide, and Fe-S clusters (Sazanov and Hinchliffe, 2006). During the last decade, however, dozens of subunits specific to chloroplast NDH have been identified using extensive genetic, proteomic, and bioinformatic approaches (Suorsa et al., 2009; Ifuku et al., 2010; Peng et al., 2011b; Yamamoto et al., 2011). On the basis of structural information on eubacterial NDH-1, as well as the biochemical and genetic characterization of Arabidopsis thaliana mutants lacking NDH subunits, we divided chloroplast NDH into four subcomplexes: membrane, lumen, and stroma-exposed A and B subcomplexes (Peng et al., 2009). Whereas the membrane subcomplex is composed of seven plastid-encoded subunits, NdhA–NdhG, subcomplex A contains four plastid-encoded subunits, NdhH–NdhK, and four nucleus-encoded subunits, NdhL–NdhO (Peng et al., 2009). Subunits in the B and lumen subcomplexes are specific to higher plants (Peng et al., 2009). Recently, a unified nomenclature for nucleus-encoded subunits was proposed (Ifuku et al., 2011), and in this article, we use the new names as well as the old names. We also identified three subunits that are localized to the putative catalytic subcomplex of chloroplast NDH (Yamamoto et al., 2011). Among them, the peripheral subunit NdhS (CHLORORESPIRATORY REDUCTION31 [CRR31]) forms an Fd binding site, indicating that chloroplast NDH accepts electrons from Fd rather than from NAD(P)H (Yamamoto et al., 2011). Our studies also revealed that chloroplast NDH interacts with at least two copies of PSI to form the NDH-PSI supercomplex, which is required for stabilization of NDH, especially under strong light conditions (Peng et al., 2008, 2009; Peng and Shikanai, 2011). Two minor light-harvesting complex I proteins, Lhca5 and Lhca6, are required for the interaction between NDH and PSI (Peng et al., 2009).
Despite the progress made in revealing the structure of chloroplast NDH, few reports have focused on the assembly of this complex. As NDH subunits are encoded by both the nuclear and the plastid genome, assembly of NDH must require a concerted interplay between plastid- and nucleus-encoded products, and it is likely that NDH assembly occurs in a stepwise manner, as do the processes of assembly of other photosynthetic complexes (Rochaix, 2011). Because of the low abundance and fragile nature of chloroplast NDH, it is hard to apply biochemical approaches, such as pulse-chase labeling, to investigate the NDH assembly process. In a complementary approach to biochemistry, genetics has identified a large number of assembly factors specific to an individual photosynthetic complex, including photosystem II (PSII) and PSI (Nixon et al., 2010; Rochaix, 2011), resulting in the accumulation of knowledge on the assembly machinery and on the dynamics that regulate the process. Mutant screening focusing on NDH activity has identified many nonsubunit factors that are required for the accumulation of NDH (Hashimoto et al., 2003). One series of mutants is defective in genes encoding the members of the pentatricopeptide repeat family; these RNA binding proteins are specifically required for the expression of plastid genes encoding NDH subunits (Suorsa et al., 2009). However, plastid gene expression is not affected in the crr6 and crr7 mutants (Peng et al., 2010). CRR6 and CRR7 are localized to the chloroplast stroma and are required for the accumulation of NDH subcomplex A. CRR6 was copurified with an NDH subunit, NdhH. CRR1 is also a stromal nonsubunit factor required for the accumulation of NDH and shows weak similarity to dihydrodipicolinate reductase (DHPR), which is involved in Lys biosynthesis (Shimizu and Shikanai, 2007). NDH-DEPENDENT CYCLIC ELECTRON FLOW5 (NDF5) was discovered by in silico coexpression analysis (Ishida et al., 2009). Although NDF5 shows weak sequence similarity to the NDH subunit PnsB2 (NDF2/NDH45), it is not an NDH subunit and may be involved in the assembly of both the A and the B subcomplex of NDH (Ishida et al., 2009).
Subcomplex A of chloroplast NDH is composed of eight subunits, and its assembly is independent of other NDH subcomplexes (Peng et al., 2011b). Three assembly intermediate complexes containing NdhH, with molecular masses of ~800, ~500, and ~400 kD, respectively, have been found in the chloroplast stroma, suggesting that assembly of subcomplex A occurs there (Peng et al., 2010). Accumulation of the 500- and 400-kD assembly intermediates is impaired in crr27 mutants defective in a gene encoding a minor chaperonin subunit, Cpn60β4, which is specifically required for the folding of NdhH (Peng et al., 2011a). These findings imply that only properly folded NdhH can be incorporated into the 500- and 400-kD assembly intermediates.
Although several nonsubunit factors have been discovered, our knowledge of the assembly process has been limited. In this study, we extensively analyzed 10 Arabidopsis mutants specifically defective in the accumulation of subcomplex A and also performed a series of interactive proteomics analyses. Although direct biochemical evidence of the function of each assembly component is still required, we were able to figure out the entire assembly process of subcomplex A and place some factors in our model on the basis of the mutant phenotypes. Surprisingly, many factors are preferentially required for specific points in the process.
RESULTS
Mass Spectrometry Analysis of the Proteins Associated with CRR6 and CRR7
We previously demonstrated that the stromal proteins CRR6 and CRR7 are required for the assembly of subcomplex A and that NdhH is copurified with CRR6 (Peng et al., 2010). To investigate the components associated with CRR6 and CRR7 more comprehensively, their interacting proteins were affinity purified from the crr6 and crr7-1 mutants complemented by the introduction of CRR6-hemagglutinin (HA) and CRR7-HA, respectively. Because the HA tag in the C terminus does not affect the function of CRR6 and CRR7 (Peng et al., 2010), the proteins associated with these proteins might also be copurified. The purified proteins were separated by SDS-PAGE and further analyzed by liquid chromatography–tandem mass spectrometry (LC-MS/MS) using the linear ion-trap triple quadrupole (LTQ)-Orbitrap XL-HTC-PAL system (Fujiwara et al., 2009). Neither NDH subunits nor nonsubunit factors were purified from wild-type plants, which were used as negative controls (see Supplemental Data Set 1 online), thus excluding the possibility that subcomplex A subunits were bound nonspecifically to the magnetic beads used in the affinity purification. Consistent with the previous results (Peng et al., 2010), NdhH was detected in a sample purified from CRR6-HA plants (Table 1; see Supplemental Data Set 1 online), confirming that CRR6 associates with NdhH or the assembly intermediate(s) containing NdhH. Besides NdhH, subcomplex A subunits NdhI, NdhJ, NdhK, NdhM, and NdhO were also identified specifically in the CRR6-immunoprecipitate (IP) (Table 1), suggesting that the majority of subcomplex A subunits are already assembled in the stroma before being inserted into the thylakoid membranes. NDH subunits in other subcomplexes have been preferentially detected in the thylakoid membranes (Peng et al., 2009); the assembly of subcomplex A therefore seems to occur in the chloroplast stroma, and the process is independent of the other parts of NDH. By contrast, none of the NDH subunits were found in fractions isolated from CRR7-HA plants (see Supplemental Data Set 1 online).
Table 1.
Summary of the NDH Subunits and Biogenesis Factors Identified from LTQ-Orbitrap Mass Analysis of the CRR6-, CRR41-, and CR42-IP Samples
| Mascot Score |
||||
| AGI Code | Protein Name | CRR6-IP | CRR41-IP | CRR42-IP |
| ATCG01110 | NdhH | 1272 | 1015 | 270 |
| ATCG00430 | NdhK | 471 | 233 | 154 |
| ATCG01090 | NdhI | 429 | 274 | 96 |
| AT4G37925 | NdhM | 364 | 155 | 67 |
| ATCG00420 | NdhJ | 284 | 410 | 147 |
| AT1G74880 | NdhO | 88 | 179 | 44 |
| AT5G58260 | NdhN | – | 52 | – |
| AT2G47910 | CRR6 | 2821 | 114 | 34 |
| AT3G24430 | HCF101 | 430 | – | – |
| AT1G51100 | CRR41 | 303 | 648 | 20 |
| AT5G20935 | CRR42 | 60 | 106 | 428 |
| AT5G52100 | CRR1 | – | 59 | – |
The complete list of proteins can be found in Supplemental Data Set 1 online. Number indicates the Mascot score, which is a measure of the significance of protein identification. AGI, Arabidopsis Genome Initiative; -, proteins were not detected in the samples.
In addition to subunits of subcomplex A, HIGH CHLOROPHYLL FLUORESCENCE101 (HCF101) was copurified with CRR6 (Table 1). This stromal protein has been proposed to function as a scaffold for assembly of the [4Fe-4S] cluster in chloroplasts (Schwenkert et al., 2010). Examination of the crystal structure of the hydrophilic domain of NDH-1 from T. thermophilus showed that Nqo9 and Nqo6, which correspond to NdhI and NdhK, respectively, in chloroplast NDH, bind three [4Fe-4S] clusters (Sazanov and Hinchliffe, 2006). Copurification of HCF101 and CRR6 as well as subcomplex A subunits suggested that HCF101 is required for the formation of [4Fe-4S] clusters in NdhI and/or NdhK.
CRR41 and CRR42 Are Novel Factors Specifically Required for the Accumulation of Subcomplex A
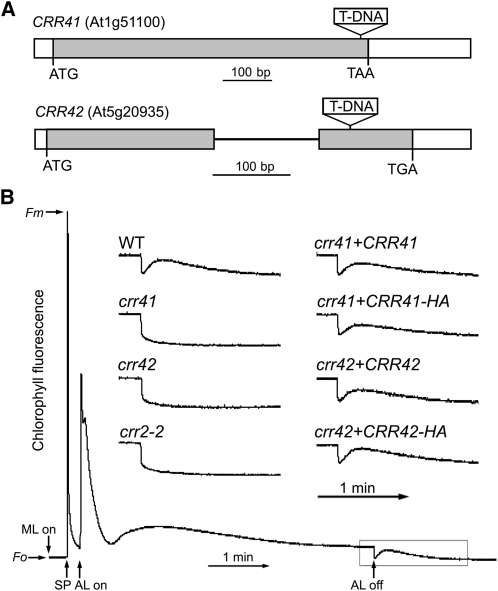
Besides CRR6, HCF101, and subunits of subcomplex A, numerous proteins with unknown function were detected in the CRR6-IP (see Supplemental Data Set 1 online). On the basis of our examination of a transcriptome database of Arabidopsis, ATTED-II (Obayashi et al., 2009), we found that two genes, At1g51100 and At5g20935, were coexpressed with genes encoding NDH subunits and nonsubunit factors (see Supplemental Table 1 online), suggesting their involvement in the assembly of subcomplex A. To clarify the functions of these two genes, we characterized homozygous T-DNA insertion lines obtained from the ABRC (Figure 1A). Under standard growth conditions, like other crr mutants specifically defective in NDH activity, neither mutant showed any visible difference in phenotype. Chloroplast NDH activity can be monitored as a transient increase in chlorophyll fluorescence after turning off actinic light; this reflects NDH-dependent PQ reduction in darkness (Shikanai et al., 1998) (Figure 1B). Neither mutant exhibited this postillumination increase in chlorophyll fluorescence, thus indicating that NDH activity was impaired (Figure 1B). Because both mutants had the typical phenotypes observed in crr mutants, we designated them crr41 and crr42, respectively (Figure 1). The wild-type genomic sequences of At1g51100 and At5g20935 were introduced into crr41 and crr42, respectively. NDH activity was fully rescued in the complementation lines (Figure 1B), confirming that disruption of the CRR41 and CRR42 genes is responsible for the absence of NDH activity. We also fused the sequence encoding the influenza HA protein epitope tag to the C-terminal region of CRR41 and CRR42 cDNA and overexpressed them in their corresponding mutants under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The transformation fully restored NDH activity (Figure 1B), suggesting that the additional C-terminal HA tag did not affect the function of CRR41 and CRR42.
Figure 1.
Characterization of crr41 and crr42 Mutants.
(A) Schematic presentation of CRR41 and CRR42 genes and T-DNA insertion sites. White boxes, gray boxes, and a black line indicate untranslated regions, exons, and intron, respectively.
(B) Determination of NDH activity using chlorophyll fluorescence. The bottom curve shows a typical trace of chlorophyll fluorescence in wild-type (WT) plants. Leaves were exposed to actinic light (AL; 50 μmol photons m–2 s–1) for 5 min. After illumination, the subsequent transient increase in fluorescence ascribed to NDH activity was monitored. Fluorescence levels were normalized against Fm (maximum fluorescence at closed PSII center in the dark) levels. Insets are magnified traces from the boxed area. crr41+CRR41 and crr42+CRR42 represent crr41 and crr42 transformed with CRR41 and CRR42 cDNAs, respectively, expressed under the control of the CaMV 35S promoter. crr41+CRR41-HA and crr42+CRR42-HA: crr41 and crr42 complemented by the introduction of CRR41 and CRR42 cDNA, respectively, fused to the sequence encoding the HA tag under the control of the CaMV 35S promoter. Fo, minimum fluorescence at open PSII center in the dark; ML, measuring light; SP, saturating light pulse.
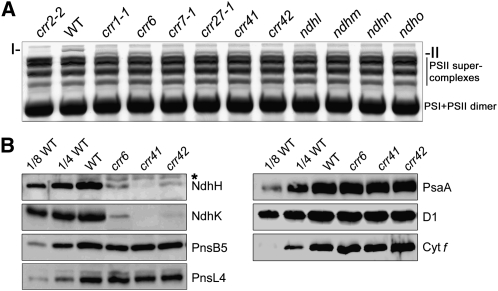
To investigate further the changes in NDH-PSI supercomplex formation in crr41 and crr42, thylakoid membranes isolated from these two mutants as well as wild-type and crr6 plants were solubilized in 1% n-dodecyl-β-d-maltoside and then the thylakoid protein complexes were separated by blue native (BN)-PAGE (Figure 2A; see Supplemental Figure 1 online). The NDH-PSI supercomplex corresponding to band I was visible at the top of the BN-PAGE gel in the lane containing wild-type thylakoids (Figure 2A; Peng et al., 2008). Consistent with our previous reports (Peng et al., 2008, 2010), band I was replaced by band II in crr6 (Figure 2A). Previous mass spectrometry and biochemical analyses revealed that only subcomplex A, which is composed of four plastid-encoded subunits (NdhH–NdhK) and four nucleus-encoded subunits (NdhL–NdhO), was absent in the band II subsupercomplex (Peng et al., 2009). As in crr6, band I was replaced by band II in crr41 and crr42 (Figure 2B), suggesting that subcomplex A was absent in both mutants and that the accumulation of other parts of the NDH-PSI supercomplex was not affected. Immunoblot analysis further confirmed that the NdhH level was dramatically reduced in crr41 and crr42 (Figure 2B). We also produced an antibody against a subcomplex A subunit, NdhK, and the levels of NdhK were also substantially decreased in crr6, crr41, and crr42 (Figure 2B). By contrast, subunits of the other NDH subcomplexes, PnsB5 (NDH18) and PnsL4 (FKBP16-2), were almost unaffected (Figure 2B). Identical levels of D1 (PSII), PsaA (PSI), and Cyt f (Cyt b6f) were detected in crr41, crr42, and wild-type plants (Figure 2B). Taken together, our results suggest that, like CRR6 and CRR7, CRR41 and CRR42 are specifically required for the accumulation of subcomplex A of chloroplast NDH.
Figure 2.
Analysis of the NDH Complex in Different NDH Mutant Backgrounds.
(A) BN-PAGE analysis of thylakoid protein complexes isolated from wild-type (WT) plants and the indicated mutants. Magnification of the top part of the BN-PAGE gel in Supplemental Figure 1 is shown. After electrophoresis, the gel was stained with Coomassie blue. I, NDH-PSI supercomplex detected in the wild type; II, sub-NDH-PSI supercomplex lacking subcomplex A.
(B) Immunoblot analysis of thylakoid proteins, with the indicated antibodies. Thylakoid proteins were loaded on an equal chlorophyll basis, and the series of dilutions is indicated. An asterisk indicates nonspecific signals.
CRR41 and CRR42 Are Localized to the Chloroplast Stroma
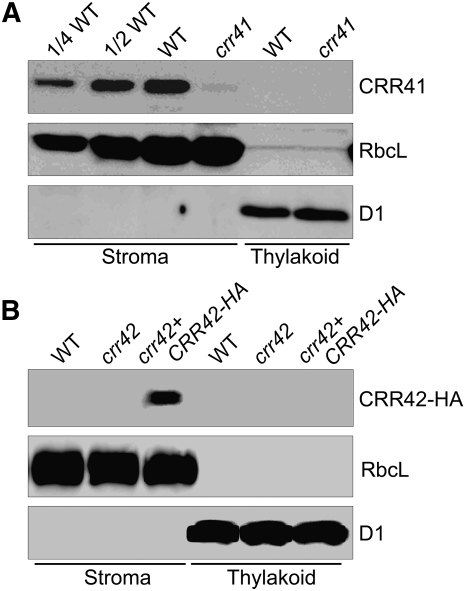
CRR41 encodes a 211–amino acid protein without any known motif suggesting its function (see Supplemental Figure 2 online). The N-terminal 38 amino acids of CRR41 were predicted to be the plastid-targeting signal (ChloroP; http://www.cbs.dtu.dk/services/ChloroP/). CRR41 is unlikely to contain any transmembrane helix (predicted by TMHMM; http://www.cbs.dtu.dk/services/TMHMM-2.0/); this is consistent with the fact that CRR41 was copurified with CRR6 from the chloroplast stroma. To confirm the localization of CRR41, we produced a specific antibody against the recombinant CRR41 protein. In accordance with the theoretical molecular mass of mature CRR41 (~19 kD), immunoblot analysis detected a signal with a molecular mass slightly less than 20 kD in the chloroplast stromal fraction isolated from wild-type plants, but the signal was barely detected in the stromal fraction isolated from crr41 plants (Figure 3A). CRR41 was absent in the thylakoid membranes isolated from both wild-type and crr41 plants (Figure 3A), supporting the conclusion that CRR41 is a stromal protein. A BLAST database search revealed that CRR41 is conserved in angiosperms, but no homolog was found in Chlamydomonas reinhardtii or cyanobacteria (see Supplemental Figure 2 online). A hypothetical protein exhibiting low similarity (38% amino acid identity) with the C-terminal half of Arabidopsis CRR41 was found in Physcomitrella patens (see Supplemental Figure 2 online). Terrestrial plants may have acquired the novel protein CRR41 that is specifically required for the biogenesis of NDH subcomplex A during the evolution of land plants.
Figure 3.
Immunodetection of CRR41 and CRR42.
Freshly isolated chloroplasts from various genetic backgrounds were further fractionated into membrane and stromal fractions. Immunoblot analysis was performed with antibodies against CRR41 (A) and HA tag (B). Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (RbcL) and D1 were detected as loading and fractionation controls, respectively. Stromal proteins were loaded by equal protein content, and the series of dilutions is indicated. crr42+CRR42-HA: crr42 transformed with CRR42 cDNA fused to the sequence encoding the HA tag expressed under the control of the CaMV 35S promoter. WT, wild type.
CRR42 is predicated to encode a small chloroplast protein (108 amino acids) included in the DUF (domain of unknown function) 3148 superfamily. CRR42 is also not predicted to have any transmembrane domain by TMHMM (see Supplemental Figure 3 online). Attempts to produce a specific antibody against CRR42 have been unsuccessful. To determine the localization of CRR42 in chloroplasts, we fractionized chloroplasts isolated from CRR42-HA plants into the stromal and membrane fractions. A signal with a molecular mass of between 10 and 15 kD was detected in the stroma using a monoclonal antibody against the HA tag (Figure 3B); this was consistent with the molecular mass of the putative mature CRR42 (11.1 kD) plus the linker and the HA tag (1.6 kD). This protein was absent in thylakoid membranes isolated from wild-type, crr42, and crr42-HA plants and also absent in the wild-type and crr42 stromal fraction (Figure 3B). Recent proteomic analysis has also shown that the homolog of CRR42 in maize (Zea mays; protein name: GRMZM2G175019_P01) is present in the chloroplast stroma (Plant Proteome Database, http://ppdb.tc.cornell.edu/; Friso et al., 2010). From these findings, we conclude that CRR42 is also a stromal protein.
CRR1 Is Required for the Accumulation of Subcomplex A
To study the function of CRR41 and CRR42, proteins associated with them were immunoprecipitated from transgenic plants expressing CRR41-HA and CRR42-HA, respectively, and then subjected to a second mass spectrometric analysis (see Supplementary Data Set 1 online). The NDH subunits NdhH–NdhK, NdhM, and NdhO, as well as CRR6, CRR41, and CRR42, were identified in both IPs (Table 1). Furthermore, NdhN was found in the CRR41-IP sample (Table 1), indicating that all of the hydrophilic subunits of subcomplex A were copurified with CRR41.
CRR1, which was discovered in our genetic screening (Shimizu and Shikanai, 2007), was also detected in the CRR41-IP sample, indicating that CRR1 is associated with the assembly intermediates of subcomplex A. CRR1 may also be involved in the assembly of subcomplex A. This idea is consistent with the results of the BN-PAGE analysis, in which a subsupercomplex lacking subcomplex A, corresponding to band II, was detected in crr1-1 (Figure 2A). To reveal the exact role of CRR1 in the biogenesis of subcomplex A, we also overexpressed CRR1-HA in crr1-1 plants, and a third mass spectrometric analysis was performed using HA-tagged CRR1. However, no NDH subunits or nonsubunit factors were found in the sample (see Supplementary Data Set 1 online). In crr1-1 transformed with CRR1-HA driven by the CaMV 35S promoter, the NDH activity (detected as chlorophyll fluorescence change) was only partially rescued (see Supplemental Figure 4 online). Addition of the HA tag may have partly impaired the function of CRR1. It is also possible that the HA tag is hidden inside the CRR1-containing complex and that the free form of CRR1-HA protein was trapped by the HA antibody in our affinity chromatography analysis (see Supplementary Data Set 1 online).
Stability of the Stroma-Localized NDH Subunits and Biogenesis Factors under the NDH-Less Mutant Backgrounds
Besides crr1, crr6, crr7, crr41, and crr42, five mutants, ndhl, ndhm, ndhn, ndho, and crr27, specifically lack subcomplex A in thylakoids (Figure 2A). As reported previously (Peng et al., 2008, 2009), NdhL–NdhO are nucleus-encoded subunits of subcomplex A, and the absence of any of these subunits destabilizes the entire subcomplex A (Figure 2A). CRR27/Cpn60β4 is required for the folding of NdhH to the native form, and plants failed to accumulate subcomplex A in the absence of native NdhH (Figure 2A; Peng et al., 2011a). Thus, we have 10 mutants that are specifically defective in the accumulation of subcomplex A. Because the absence of any subunits of subcomplex A totally destabilizes subcomplex A in thylakoids, resulting in the detection of band II in the BN gel, no more information on the assembly of subcomplex A is obtained from the analysis of thylakoid-localized subunits. However, as in crr6 and crr7 (Peng et al., 2010), subcomplex A subunits may be partly stable in the assembly intermediates in the stroma. Comprehensive characterization of the stromal assembly intermediates may provide new insights into subcomplex A biogenesis.
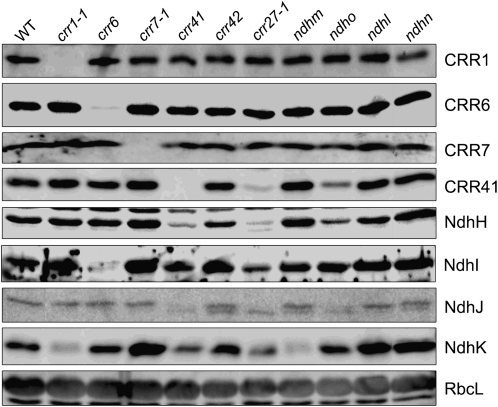
Copurification of the hydrophilic subunits of subcomplex A with CRR6, CRR41, and CRR42 suggests that these subunits are present in the chloroplast stroma (Table 1). This idea was further confirmed by the detection of four plastid-encoded subunits, NdhH–NdhK, in the wild-type stromal fraction (Figure 4). To determine whether accumulation of the stroma-localized subunits was dependent on nonsubunit factors or other subcomplex A subunits, we isolated stromal fractions from 11 genotypes and performed a matrix analysis of immunoblotting using four antibodies against NdhH–NdhK (Figure 4). The levels of the nonsubunit factors, CRR1, CRR6, CRR7, and CRR41, were also investigated in these mutants. Besides CRR1, the NdhK level was reduced in crr1-1 compared with that in the wild type, whereas those of other subunits or nonsubunit factors were not affected (Figure 4). In ndhm, the NdhK signal was also barely detected (Figure 4), suggesting that NdhM is required for the accumulation of NdhK in the chloroplast stroma. Because we did not have an antibody against NdhM, we could not evaluate the accumulation of NdhM in crr1-1. We therefore could not determine whether CRR1 was directly required for the accumulation of NdhK or was required for the accumulation of NdhM and consequently NdhK. By contrast, accumulation of NdhI was impaired in the stroma of crr6, suggesting that CRR6 is specifically involved in the maturation and/or assembly of NdhI.
Figure 4.
Accumulation of Stroma-Localized NDH Subunits and Nonsubunit Factors.
Stromal proteins were isolated from the wild type (WT) and various mutants, and protein blot analyses were performed with the indicated antibodies. Stromal proteins were loaded by equal protein content, and RbcL was detected as the loading control.
In crr41, the levels of CRR41 and NdhH were drastically reduced, and the NdhK level was also lower than that in wild-type plants (Figure 4). A similar accumulation pattern was found in the crr27-1 and ndho mutants, although the effect was milder in ndho than in other mutants (Figure 4). These results indicate that CRR41, NdhO, and native NdhH depend on each other for their accumulation in the chloroplast stroma and that NdhK accumulation is partially dependent on CRR41, NdhO, and native NdhH. In crr41, crr27-1, and ndho, the level of NdhJ was also reduced (Figure 4). Instead, a weak band with slightly higher mobility in the gel was detected in these mutants. Both the original NdhJ and lower bands were detected in crr42, ndhm, and ndhn. This lower band may represent a degradation product of NdhJ. It is also possible that the upper band detected in the wild type corresponds to phosphorylated NdhJ. In both cases, our results imply that accumulation of functional NdhJ is dependent on CRR41, NdhO, and native NdhH. Although accumulation of the nonsubunit factors CRR1, CRR6, and CRR7 did not depend on subcomplex A subunits, CRR41 levels were drastically and partly reduced in crr27-1 and ndho, respectively (Figure 4), suggesting that the accumulation of CRR41 depends on native NdhH and NdhO. These results imply that there is a protein interaction network between the nonsubunit factor CRR41 and the subunits of subcomplex A, NdhH and NdhO, in the stroma.
In crr7-1, crr42, ndhl, and ndhn, however, no NDH subunits and nonsubunit factors investigated were missing (Figure 4). However, the levels of NdhH, NdhI, and NdhK were higher in the stroma of these mutants than in the wild type (Figure 4), suggesting that these proteins are involved in insertion of the subcomplex A assembly intermediate into thylakoids.
Components of the NDH Subcomplex A Assembly Intermediates
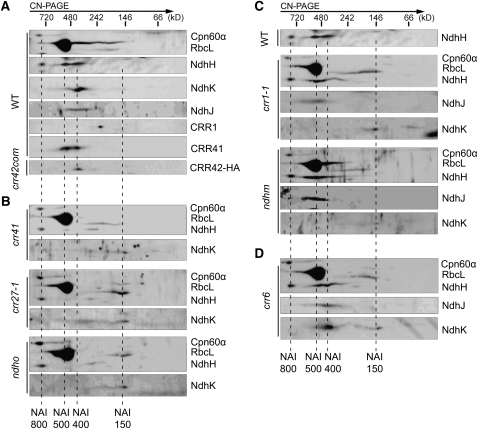
Three complexes containing NdhH, representing NDH subcomplex A assembly intermediates (NAIs) (Figure 5; Peng et al., 2010), were detected in the chloroplast stroma. On the basis of their apparent molecular masses, we designated them as NAI800, NAI500, and NAI400, respectively (Figure 5). To study the components of these NAIs, we performed clear native (CN)-PAGE and subsequent immunoblot analyses using the stromal fraction isolated from wild-type plants. Whereas CRR41 comigrated with NAI500 and NAI400, NdhK, NdhJ, and CRR42 were preferentially localized to NAI400 (Figure 5A). For some unknown reason, we could not detect NdhI in the two-dimensional (2D) CN/SDS-PAGE, although it can be detected in one-dimensional SDS-PAGE (Figure 4). Because CRR42-HA was preferentially associated with NAI400 (Figure 5A), NdhH–NdhK, NdhM, and NdhO, which were detected in the CRR42-IP sample by interactive proteomic analysis, represent the components of NAI400 (Table 1). These results suggest that NAI400 contains the majority of hydrophilic subunits of subcomplex A. Fewer NDH subunits were detected in NAI800 and NAI500 than in NAI400 (Figure 5A), suggesting that NAI400 is closer to the fully assembled form of subcomplex A and that formation of NAI800 and NAI500 occurs before NAI400 formation (Figure 6).
Figure 5.
Formation of NAIs in the Chloroplast Stroma of Various Mutants.
Stromal protein complexes isolated from the wild type (WT) crr42 overexpressing the CRR42-HA protein (crr42com) (A), crr41, crr27-1, and ndho (B), crr1-1 and ndhm (C), and crr6 (D) plants were separated by CN-PAGE followed by 2D SDS-PAGE. Proteins were immunodetected with three antibodies against Cpn60α, RbcL, and NdhH or the specific antibodies indicated. Dashed lines indicate the positions of NAIs.
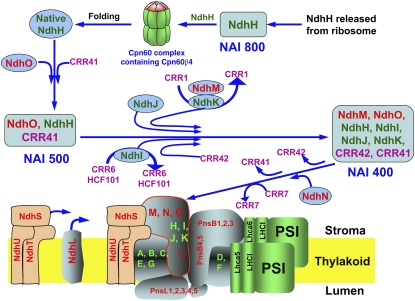
Figure 6.
A Schematic Model of the Chloroplast NDH Subcomplex A Assembly.
The nature of NAI800 is unclear. This complex appears to deliver nonnative NdhH to the chaperonin complex containing Cpn60β4 for subsequent protein folding. NdhO, CRR41, and native NdhH, as well as some unknown factors, are first assembled together to form NAI500. Subsequently, NdhJ, NdhK, NdhM, and NdhI are assembled to form NAI400, which accepts NdhN in the final assembly step and then, together with the membrane subunit NdhL and the catalytic subcomplex including NdhS–NdhT, is incorporated into the thylakoids to form the functional NDH-PSI supercomplex. This process is facilitated by CRR41, CRR1, CRR6, HCF101, CRR42, and CRR7 at the various assembly steps indicated. On the basis of their molecular masses, the NAI complexes are expected to contain additional unknown factors that are not depicted in the model. Nonsubunit factors, nucleus-encoded NDH subunits, and chloroplast-encoded NDH subunits are shown in pink, red, and green fonts, respectively.
Previously, we showed that CRR6 and CRR7 are not components of NAIs (Peng et al., 2010). Here, we showed that CRR1 is present in a complex with a molecular mass of ~240 kD (Figure 5A); CRR1 is also not a component of NAIs. However, the copurification of CRR1 and CRR41, as well as the hydrophilic subunits of subcomplex A, suggests that CRR1 transiently interacts with NAI(s) (Table 1). Because the molecular masses of NAIs are larger than those of the mature NDH subcomplex A in thylakoids, some other unknown proteins may be included in NAIs; these unknown factors were probably detected in our proteomic analysis (see Supplemental Data Set 1 online). Two chaperone proteins, cpHSC70-1 and cpHSC70-2, were detected in the CRR41-IP sample (see Supplemental Data Set 1 online). However, 2D CN/SDS-PAGE and the immunoblot analysis detected three complexes containing cpHSC70 in the chloroplast stroma, and none of them comigrated with NAIs (see Supplemental Figure 5 online), suggesting that the interaction is transient, as in the case of CRR1, CRR6, and CRR7. Because cpHSC70s are required for the translocation of chloroplast proteins across the envelopes (Su and Li, 2010), it is possible that the precursors of nucleus-encoded subunits or nonsubunit factors are still associated with cpHSC70s in the stroma.
Analyses of NAI Complex Formation under Mutant Backgrounds
As mentioned above, we had 10 mutants that were defective in the accumulation of subcomplex A in thylakoids, and six of them showed specific defects in the accumulation of NDH subunits in the stroma (Figures 2 and 4). To obtain further information on NAI formation, we performed 2D CN/SDS-PAGE and subsequent immunoblot analyses using the mutants. NAI800 was present in all of the mutants investigated, and this complex did not include NdhK and NdhJ (Figure 5), suggesting that formation of NAI800 including NdhH occurs before the incorporation of NdhK and NdhJ into NAIs and is independent of other subunits and nonsubunit factors. Moreover, NAI800 was detected in crr27-1 (Figure 5B), indicating that formation of NAI800 also occurs before the folding of NdhH (Peng et al., 2011a). Consistently, NAI800 did not comigrate with Cpn60α (Figure 5A); thus, NAI800 is unlikely to be the Cpn60 complex associated with NdhH. It is therefore likely that NAI800 includes nonnative NdhH and that its formation initiates the steps of subcomplex A assembly in the stroma. The components of NAI800, with the exception of NdhH, are largely unknown, and further investigations are required.
In crr41, crr27-1, and ndho, formation of NAI500 and NAI400 was almost completely impaired, and NdhH protein was detected mainly in NAI800 (Figure 5B), implying that CRR41, native NdhH, and NdhO are required for the formation of NAI500 and NAI400. The 2D CN/SDS-PAGE and immunoblot analyses confirmed the idea that CRR41 and NdhH are components of the NAI500 complex (Figure 5A). Levels of CRR41 and NdhH were reduced in the ndho stroma (Figure 4), and NAI500 was undetectable in ndho in CN-PAGE (Figure 5B), implying that NAI500 is unstable in the absence of NdhO in CN-PAGE and also in vivo. These findings suggest that NdhO is also a component of NAI500 (Figure 6), although we have no antibody against NdhO to confirm this idea directly. CRR41 was unstable in the absence of Cpn60β4 and NdhO, and NdhH was also unstable in the absence of CRR41 (Figure 4), suggesting that interaction among CRR41, NdhO, and folded NdhH is necessary for stabilization of NAI500 (Figure 6). Because the molecular mass of NAI500 is larger than that of NAI400, some nonsubunit factors must be released from NAI500 during the transition to NAI400. The identity of these unknown nonsubunit factors is currently unclear. From these results, we propose that CRR41, native NdhH, and NdhO are first assembled into NAI500 (Figure 6).
NAI400 was also absent in crr27, crr41, and ndho (Figure 5B). Furthermore, the level of NdhK was reduced in the stroma of these three mutants (Figure 4), and a trace level of NdhK was detected in the 150-kD complex (NAI150) (Figure 5B), suggesting that NdhK could not be incorporated into NAI400 in the absence of NAI500 (Figure 5B). The 2D CN/SDS-PAGE and immunoblot analyses showed that formation of NAI400, but not NAI500, was impaired in crr1-1 and ndhm (Figure 5C), in which stroma-localized NdhK was barely detected (Figures 4 and 5C), implying that NdhK and NdhM were integrated into NAI500 to produce NAI400 (Figure 6). These results also support the idea that formation of NAI500 is a prerequisite for the formation of NAI400 (Figure 6).
Stroma-localized NdhJ was missing in crr41, crr27-1, and ndhO (Figure 4), indicating that NdhJ is stabilized by NAI500 in the stroma. Although NdhJ was detected in NAI400 in the wild type, it was preferentially detected in NAI500 in both crr1-1 and ndhm (Figures 5A and 5C), suggesting that NdhJ can be assembled into NAI500 in the absence of NdhK and/or NdhM. Because we do not have an ndhJ-defective mutant, it is unclear whether NdhJ is required for the incorporation of NdhK and NdhM. However, our results suggest that NdhJ is incorporated into NAI500 before, or independently of, the incorporation of NdhK and NdhM during the formation of NAI400 from NAI500 (Figure 6).
Although stroma-localized NdhI was specifically missing in crr6 (Figure 4), NAI formation was not affected (Figure 5D). NdhJ and NdhK were found in a complex with a molecular mass similar to that of NAI400 detected in the wild type, although a trace level of NdhK was detected in the position of NAI150 (Figure 5D). This result indicates that these two subunits could be assembled into NAI500 to form a complex resembling NAI400 in the wild type in the absence of NdhI. The molecular mass of this complex in crr6 may differ slightly from that of NAI400 in the wild type, but CN-PAGE could not distinguish this minor difference (Figure 5D). This contrasts with the finding that NdhM is essential for the transition from NAI500 to NAI400 (Figure 5C). We also investigated NAI formation in the crr42 mutants; no difference was found between this mutant and wild-type plants (see Supplemental Figure 6 online). CRR42 was preferentially detected in NAI400 (Figure 5A), indicating that it is a component of this assembly intermediate. NdhI was found in the CRR42-IP samples (Table 1), implying that NdhI is also a component of NAI400 (Figure 6). These results lead to the conclusion that NdhI and CRR42 are incorporated into NAI during the transition from NAI500 into NAI400, as are NdhJ, NdhK, and NdhM (Figure 6).
Accumulation of NAI complexes was not impaired in ndhn (see Supplemental Figure 6 online). Consistently, NdhN was found in the CRR41-IP but not in the CRR42-IP (Table 1), indicating that NdhN was not included in NAI400. These results suggest that integration of NdhN into NAI400 occurs after the release of CRR42 but before the release of CRR41 (Figure 6). We also investigated NAI formation in crr7; no difference was found between crr7 and the wild type (see Supplemental Figure 6 online). Furthermore, CRR7 was detected in neither the CRR41-IP nor the CRR42-IP sample analysis (see Supplemental Data Set 1 online), suggesting that CRR7 is involved in the later steps of subcomplex A assembly (Figure 6). CRR7 may be involved in the recruitment of fully assembled NAI to the thylakoids. It is also possible that CRR7 is indirectly involved in the process and does not interact with any NAIs.
Absence of subcomplex A does not affect any of the membrane subunits, except for NdhL (Shimizu et al., 2008). NdhL contains three transmembrane helices and was not detected in the stromal fraction (Peng et al., 2009), and accumulation of stroma-localized NDH subunits was not affected in ndhl (Figure 4), suggesting that NdhL serves as a receptor for the fully assembled NAI and that subcomplex A, including NdhL, finally docks with the other parts of the NDH-PSI supercomplex in the thylakoid membranes (Figure 6). We recently reported three NDH subunits, NdhS (CRR31), NdhT (CRRJ), and NdhU (CRRL), and NdhS forms the Fd binding site of NDH (Yamamoto et al., 2011). The subunits accumulate partly independently of subcomplex A and were not detected in any NAIs. NdhT and NdhU each contain a transmembrane domain (Yamamoto et al., 2011), and the subunits may interact with fully assembled NDH in the thylakoid membranes.
DISCUSSION
A Model for the Assembly of NDH Subcomplex A
The levels of stromal subcomplex A subunits were comparable to those in the thylakoids (Peng et al., 2010) and were not affected in the mutants severely lacking subcomplex A in their thylakoids (Figure 4). These results suggest that NAIs are assembly intermediates rather than degradation products dissociated from thylakoids. Three complexes (NAI800, NAI500, and NAI400) contain NdhH in the stroma (Figure 5A). Formation of NAI800 occurs at the beginning of subcomplex A assembly (Figure 6). In angiosperms, folding of NdhH requires the Cpn60 complex containing Cpn60β4, and Cpn60β4 accumulates at an extremely low level compared with other major Cpn60β subunits (Peng et al., 2011a). Plants may have a mechanism for efficiently delivering nonnative NdhH to the specific Cpn60 complex, and NAI800 may be involved in this process (Figure 6).
After proper folding, the native NdhH, NdhO, CRR41, and other unknown proteins form NAI500. Our results suggest that NAI500 provides a scaffold for the assembly of other subcomplex A subunits, NdhI, NdhJ, NdhK, and NdhM, to form NAI400 and finally NdhN (Figure 6). Ycf4 is a thylakoid protein required for PSI assembly in cyanobacteria and Chlamydomonas (Wilde et al., 1995; Boudreau et al., 1997). It has been shown that Ycf4 forms a 1500-kD complex with the newly synthesized PSI subunits PsaA–PsaF and some unknown factors, and this large multiprotein complex acts as a scaffold for PSI assembly (Ozawa et al., 2009). CRR41 is required for the formation and/or stabilization of NAI500 (Figure 5B), and in NDH subcomplex A assembly, NAI500 may play a role equivalent to that of the Ycf4 complex in PSI assembly.
In our model, NdhJ, NdhK, NdhM, and NdhI are integrated into NAI500 to produce NAI400, although we still cannot determine their exact assembly order in the wild type. As discussed above, NdhJ is assembled into NAI500 before, or independently of, the incorporation of NdhK and NdhM. Consistent with this fact, Nqo5 (corresponding to chloroplast NdhJ) closely interacts with Nqo4 (corresponding to chloroplast NdhH) in the three-dimensional structure of the hydrophilic domain of complex I from T. thermophilus (Sazanov and Hinchliffe, 2006). Similarly, NdhJ and NdhK can be incorporated into NAI500 even in the absence of NdhI in crr6 (Figure 5D); this is consistent with the fact that Nqo6 (corresponding to chloroplast NdhK) also directly interacts with Nqo4 (Sazanov and Hinchliffe, 2006). Because Nqo9 (corresponding to chloroplast NdhI) interacts with Nqo6 in T. thermophilus (Sazanov and Hinchliffe, 2006), NdhI stability may depend on NdhK. Although we could not test this possibility in 2D CN/SDS-PAGE because of unknown technical reasons, in one-dimensional SDS-PAGE, we detected a higher level of NdhI in crr1-1, which is defective in NdhK accumulation, than in the wild type (Figure 4). NdhO, which is absent in T. thermophilus complex I, may interact with NdhI and stabilize it in the absence of NdhK in chloroplasts.
Function and Evolution of Assembly Factors
Besides the assembly factors CRR1, CRR6, and CRR7, which were originally identified in our forward genetics research (Shimizu and Shikanai, 2007; Peng et al., 2010), two novel assembly factors, CRR41 and CRR42, were discovered in this study as components of NAIs. All of these factors are chloroplast stromal proteins (Figure 3; Shimizu and Shikanai, 2007; Peng et al., 2010), and this is consistent with the conclusion that assembly of subcomplex A occurs in the stroma. The exact function of CRR42 remains unclear because NAI formation was not affected in crr42 (see Supplemental Figure 6 online). However, this finding suggests that incorporation of NdhI–NdhK and NdhM into NAI occurs before, or independently of, incorporation of CRR42 into NAI. Because CRR42 was specifically detected in NAI400, insertion of CRR42 may take place after the decrease in the molecular mass of NAI500. Recently, SipA, a CRR42 homolog in Synechococcus sp PCC 7942, was shown to interact with the His kinase (Hik) NblS (Espinosa et al., 2006). The homolog of CRR42 in Synechocystis sp PCC 6803, Ssl3451, is also likely to activate Hik33 (Sakayori et al., 2009). No homologs of NblS/Hik33 have been found in land plants (Sakayori et al., 2009); this is consistent with the fact CRR42 is a component of NAI400 and is specifically required for the assembly of subcomplex A. Because crr42 accumulated NAI400 (see Supplemental Figure 6 online), CRR42 may endow NAI400 with the ability to incorporate NdhN or enable fully assembled NAI400 to be inserted into thylakoids.
Although CRR1 is weakly homologous to DHPR, which functions in Lys biosynthesis (Shimizu and Shikanai, 2007), it is specifically required for the accumulation of subcomplex A (Figure 2A). The DHP binding motif is not conserved in CRR1, and this may explain the distinctive function of CRR1 compared with those of two other paralogs functioning as genuine DHPRs in Arabidopsis (Hudson et al., 2005). Our results suggest that CRR1 is required for the accumulation of NdhK and/or NdhM in the chloroplast stroma (Figure 4). Because CRR1 interacts with NAI500 and/or NAI400, it is unlikely to be involved in transcription or translation of the ndhK gene in chloroplasts. Furthermore, in mutants defective in NAI500 formation (crr27, crr41, and ndho), the level of stroma-localized NdhK was higher than in crr1-1 but lower than in the wild type (Figure 4). CRR1 may be directly involved in the maturation of NdhK and/or NdhM, rather than assisting with their incorporation into NAI500. Although CRR1 is unlikely to be DHPR, its NAD(P)H binding motif is conserved (Shimizu and Shikanai, 2007). CRR1 may be involved in the formation of prosthetic groups of NdhK as reductase, although the substrate is unknown.
CRR6 is essential for the accumulation of stroma-localized NdhI (Figure 4). The NdhI level was more drastically affected in crr6 than in the crr27, crr41, and ndho mutants defective in the accumulation of NAI500 (Figure 4), suggesting that CRR6 is required for the biogenesis of NdhI. However, the majority of the subcomplex A subunits were detected in our interactive proteomic analysis of the CRR6-IP sample, and vice versa, CRR6 was found in the CRR41- and CRR42-IP samples, implying that CRR6 transiently interacts with NAIs, including NdhI (Table 1). These findings suggest that CRR6 is also required for the incorporation of NdhI into NAI after NdhI maturation. CRR6 may have dual functions. Similar examples have been reported in LOW PSII ACCUMULATION1 and PHOTOSYNTHESIS AFFECTED MUTANT68, which interact with PSII core subunit D1, probably assisting with the proper folding and integration of D1 into thylakoid membranes (Peng et al., 2006; Armbruster et al., 2010).
HCF101 has been proposed to be involved in [4Fe-4S] cluster assembly in chloroplasts (Schwenkert et al., 2010), and it was also copurified with CRR6 (Table 1), indicating the involvement of HCF101 in the incorporation of [4Fe-4S] clusters into NdhI. In contrast with the specific phenotype of crr6 in NDH activity, the hcf101 mutant has a pleiotropic defect in chloroplast function (Lezhneva et al., 2004), suggesting that HCF101 is a general factor required for [4Fe-4S] cluster assembly. The [4Fe-4S] cluster is also included in NdhK, and its assembly may require HCF101 because Nqo6 (corresponding to chloroplast NdhK) and Nqo9 (corresponding to chloroplast NdhI) are closely localized in T. thermophilus complex I (Sazanov and Hinchliffe, 2006). However, HCF101 was not copurified with CRR41, implying that HCF101 only weakly interacts with NdhI.
Q Module Corresponding to Subcomplex A Is Assembled in the Matrices of Mammalian Mitochondria
Complexes related to chloroplast NDH are present in nonphototrophic and phototrophic bacteria, mitochondria, and chloroplasts, and they probably have the same origin. We elucidated several of the assembly factors specific to chloroplast NDH, and the question is now how the assembly step is conserved among organisms. In mammalian mitochondria, it has been proposed that an early subassembly of the Q module, which corresponds to subcomplex A in chloroplast NDH, is anchored to the membrane by other membrane-embedded subunits, forming an ~400-kD subcomplex (Lazarou et al., 2009). This assembly step is followed by further association with an ~460-kD subcomplex containing the P module (corresponding to the membrane part of chloroplast NDH) to form a stable ~830-kD intermediate. In the last stage of complex I assembly, the N module, which is responsible for binding and oxidizing NADH, is assembled into the 830-kD complex to form the functional enzyme (Lazarou et al., 2009). Because the N module is replaced by the catalytic subcomplex, including NdhS–NdhU, conferring different enzyme activity (Yamamoto et al., 2011), and NdhM–NdhO are specific to phototrophs, it is not surprising that the assembly factors of chloroplast NDH (CRR1, CRR6, CRR7, CRR41, and CRR42) are not conserved in nonphototrophs. Despite the structural diversity of the complex and the different assembly machinery, the connecting arm that links the catalytic domain to the membrane domain is assembled via the soluble intermediate in both mitochondrial complex I (Q module) and chloroplast NDH (subcomplex A), suggesting that the frame of the complex assembly is conserved among organisms. This may reflect the findings that the L-shaped skeleton consisting of NdhA–NdhK in chloroplast NDH is highly conserved among other NADH dehydrogenase-related complexes and that the skeleton is involved in the fundamental functions of the complexes; in chloroplast NDH, that function is electron transport from the hemes present in NdhI to PQ, probably coupled with proton pumping across the membrane subunits (Efremov et al., 2010).
Assembly of complex I seems to occur via a dynamic process in which a subunit or an assembly intermediate interacts with preexisting subunits or intermediates, resulting in dynamic transitions between fully assembled complex I and its intermediates (Lazarou et al., 2009). Our work suggests that assembly of chloroplast NDH also occurs via a dynamic cycling between fully assembled NDH and its intermediates. This idea is supported by the fact that a substantial quantity of subcomplex A subunits were detected in the chloroplast stroma (Peng et al., 2010) and that NDH subsupercomplex lacking subcomplex A is stable in thylakoids (Peng et al., 2009). On the basis of this dynamic assembly mechanism, the damaged subcomplex A in thylakoids can be rapidly replaced by another without de novo synthesis of the membrane parts of NDH.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana (ecotype Columbia-0) plants used in this work were grown in soil in a growth chamber (50 μmol photons m–2 s–1, 16-h photoperiod, 23°C) for 3 to 4 weeks. SAIL T-DNA lines for CRR41 (SAIL_697_B12) and for CRR42 (SAIL_889_G06) were obtained from the ABRC. Insertion sites of these two mutants were confirmed by direct sequencing of the PCR products. For the complementation of crr41 and crr42, cDNA fragments of wild-type At1g51100 and At5g20935 were subcloned into the pBI121 vector under the control of the CaMV 35S promoter. The cDNA sequences used for the complementation were also modified to be followed by the sequences encoding a linker (SRGG) and the HA tag (YPYDVPDYAG) and then subcloned into pBI121. The vectors were transferred into Agrobacterium tumefaciens C58C by electroporation, and the bacteria were used to transform crr41 or crr42 respectively, by floral dipping (Clough and Bent, 1998).
Chlorophyll Fluorescence Analysis
The transient increase in chlorophyll fluorescence after actinic light was turned off was monitored as previously described (Shikanai et al., 1998) using a MINI-PAM portable chlorophyll fluorometer (Waltz).
Chloroplast Preparation, BN-PAGE, CN-PAGE, and Immunoblot Analysis
Chloroplasts were osmotically ruptured in buffer containing 20 mM HEPES-KOH, pH 7.6, 5 mM MgCl2, and 2.5 mM EDTA (Munekage et al., 2004). Thylakoid membranes were separated from the stromal fraction by centrifugation (15,000g for 10 min at 4°C). The concentration of the stromal protein was determined with a Protein Assay Kit (Bio-Rad). The chlorophyll content was determined by the method of Porra et al. (1989). BN-PAGE was performed as described (Peng et al., 2008). CN-PAGE was performed according to the method of Peltier et al. (2004), with the following modifications. Intact chloroplasts were osmotically broken in cold buffer containing 50 mM BisTris-HCl, pH 7.0, 5 mM MgCl2, and protease inhibitors (Complete Mini; Roche) and then centrifuged at 15,000g for 10 min at 4°C. Supernatants were collected as a stromal soluble fraction. A total of 10 μg of stromal proteins was mixed with one-quarter volume of sample buffer (40 mM BisTris-HCl, pH 7.0, 0.008% Ponceau S, 200 mM 6-aminon-caproic acid, and 60% glycerol [w/v]). Stromal protein complexes were separated by 5 to 12% gradient CN-PAGE in 0.75-mm-thick gels connected to a circulating cooler. For 2D PAGE/immunoblot analysis, excised CN-PAGE lanes were soaked for 30 min at 25°C in SDS sample buffer (100 mM Tris-HCl, pH 6.8, 2% SDS, and 15% glycerol) containing 2.5% β-mercaptoethanol and then layered onto 1-mm-thick SDS-PAGE gels or Tricine-SDS-PAGE gels (for CRR7 and CRR42 immunodetection). After electrophoresis, the proteins were transferred to nitrocellulose membranes and probed with specific antibodies. The signal was visualized with the LAS-3000 luminescent image analyzer (Fuji Film).
Affinity Chromatography of Proteins Associated with Assembly Factors
Intact chloroplasts isolated from CRR6-HA, CRR7-HA, CRR1-HA, CRR41-HA, and CRR42-HA, as well as from wild-type plants, were osmotically ruptured in the lysis buffer (50 mM Tris-HCl, pH 8.0, 0.01% Tween 20, and 10 mM MgCl2) plus protease inhibitors (Complete Mini; Roche). Thylakoid membranes were pelleted by centrifugation at 20,000g for 10 min at 4°C, and the supernatants were transferred to new tubes. NaCl and Nonidet P-40 were added to the supernatants to final concentrations of 150 mM and 1%, respectively. Samples were generally mixed with 50 μL anti-HA MicroBeads (Miltenyi Biotec). After incubation of the mixture for 2 h at 4°C, the beads were transferred to columns placed in a magnetic field. Columns were rinsed four times with 200 μL washing buffer I (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1 mM EDTA, and 1% Nonidet P-40). After final washing with 200 μL washing buffer II (25 mM Tris-HCl, pH 7.5), total proteins were eluted with elution buffer (50 mM Tris-HCl, pH 6.8, 50 mM DTT, 1% SDS, 1 mM EDTA, 0.005% bromophenol blue, and 10% glycerol). The proteins were separated on 12.5% SDS-PAGE gels (Perfect NT gel; DRC) and stained with Coomassie Brilliant Blue. SDS-PAGE lanes were cut into several slices and analyzed by LC-MS/MS.
Peptide Preparation for MS/MS Analysis
Peptide preparation and LC-MS/MS analyses were performed as described previously (Fujiwara et al., 2009). The excised SDS-PAGE lanes were cut into several slices and then treated twice with 25 mM ammonium bicarbonate in 30% (v/v) acetonitrile for 10 min and in 100% (v/v) acetonitrile for 15 min. After being dried in a vacuum concentrator, the gel pieces were treated with 0.01 mg/mL trypsin (sequence grade; Promega) in 50 mM ammonium bicarbonate and incubated at 37°C for 16 h. The digested peptides in the gel pieces were recovered twice with 20 mL 5% (v/v) formic acid/50% (v/v) acetonitrile. The extracted peptides were combined and then dried in a vacuum concentrator.
MS Analysis and Database Search
LC-MS/MS analyses were performed on an LTQ-Orbitrap XL-HTC-PAL system. Trypsin-digested peptides were loaded onto the column (diameter 0.1 mm, 150 mm; L-Column; CERI) using a Paradigm MS4 HPLC pump (Michrom BioResources) and an HTC-PAL autosampler (CTC Analytics) and were eluted with a gradient of 5 to 45% (v/v) acetonitrile in 0.1% (v/v) formic acid over a period of 26 min. The eluted peptides were introduced directly into the LTQ-Orbitrap XL MS at a flow rate of 500 nL/min and a spray voltage of 2.0 kV. The range of the MS scan was mass-to-charge ratio of 450 to 1500, and the top three peaks were analyzed by MS/MS analysis. MS/MS spectra were compared using the Mascot server (version 2.3.2) against TAIR8 (The Arabidopsis Information Resource), with the following search parameters: set-off threshold at 0.05 in the ion score cutoff; peptide tolerance, 10 ppm; MS/MS tolerance, 0.8 D; peptide charge, 2+ or 3+; trypsin as enzyme allowing up to one missed cleavage; carboxymethylation on Cys residues as a fixed modification; and oxidation of Met as a variable modification.
Antibody Production
Nucleotide sequences encoding the last 173 amino acids of CRR41 and the entire NdhK protein were amplified from cDNA using synthetic oligonucleotides (see Supplemental Table 2 online). The fragments were cloned into the pET30 expression vector (Novagen, now part of Merck). Expression of the recombinant proteins was induced by 1 mM isopropylthio-β-galactoside at 37°C for 2 h in the host Escherichia coli strain BL21 (DE3), and recombinant proteins were purified from the inclusion bodies in Ni2+-nitrilotriacetic acid columns (Qiagen) under denaturing conditions. Recombinant proteins CRR42 and NdhK were used to raise polyclonal antiserum in rabbits.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At CRR41 (Arabidopsis, At1g51100), At CRR42 (At5g20935), Vv CRR41 (Vitis vinifera, XP_002279843), Rc CRR41 (Ricinus communis, XP_002522837), Pt CRR41 (Populus trichocarpa, XP_002316975), Os CRR41 (Oryza sativa, Os01g0676200), Pp CRR41 (Physcomitrella patens, XP_001770273), Os CRR42 (Os07g0164200), Pt CRR42 (XP_002312864), Pp CRR42 (XP_001776311), and Sy CRR42/Ss l3451 (Synechocystis sp PCC 6803, BAA17314).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. BN-PAGE Analysis of the Thylakoid Protein Complexes in the Wild Type and Various Mutants.
Supplemental Figure 2. Amino Acid Sequence Alignment of CRR41.
Supplemental Figure 3. Amino Acid Sequence Alignment of CRR42.
Supplemental Figure 4. Determination of NDH Activity by Chlorophyll Fluorescence Analysis.
Supplemental Figure 5. Analysis of the Stromal Protein Complexes in the Wild-Type Plants.
Supplemental Figure 6. Analysis of the NDH Assembly Intermediates Formation in the crr42, crr7, and ndhn Mutants.
Supplemental Table 1.r Value between CRR41 and CRR42 with NDH Complex-Related Genes.
Supplemental Table 2. Primers Used in This Work.
Supplemental Data Set 1. Total Proteins Detected in the WT-IP, CRR6-IP, CRR7-IP, CRR41-IP, CRR42-IP, and CRR1-IP Fractions.
Acknowledgments
We thank Tsuyoshi Endo, Hualing Mi, Masato Nakai, Peter J. Nixon, and Amane Makino for giving us antibodies. T.S. was supported by Grants 22114509 and 22247005 from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Genomics for Agricultural Innovation; GPN0008). L.P. was supported by the Hundred Talents Program of the Chinese Academy of Sciences.
AUTHOR CONTRIBUTIONS
L.P. and T.S. conceived and designed the experiments. L.P., Y.F., and M.F. performed the experiments. L.P., Y.F., M.F., and T.S. analyzed the data. L.P. and T.S. wrote the article.
References
- Armbruster U., Zühlke J., Rengstl B., Kreller R., Makarenko E., Rühle T., Schünemann D., Jahns P., Weisshaar B., Nickelsen J., Leister D. (2010). The Arabidopsis thylakoid protein PAM68 is required for efficient D1 biogenesis and photosystem II assembly. Plant Cell 22: 3439–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau E., Takahashi Y., Lemieux C., Turmel M., Rochaix J.-D. (1997). The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J. 16: 6095–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Efremov R.G., Baradaran R., Sazanov L.A. (2010). The architecture of respiratory complex I. Nature 465: 441–445 [DOI] [PubMed] [Google Scholar]
- Efremov R.G., Sazanov L.A. (2011). Structure of the membrane domain of respiratory complex I. Nature 476: 414–420 [DOI] [PubMed] [Google Scholar]
- Endo T., Shikanai T., Takabayashi A., Asada K., Sato F. (1999). The role of chloroplastic NAD(P)H dehydrogenase in photoprotection. FEBS Lett. 457: 5–8 [DOI] [PubMed] [Google Scholar]
- Espinosa J., Fuentes I., Burillo S., Rodríguez-Mateos F., Contreras A. (2006). SipA, a novel type of protein from Synechococcus sp. PCC 7942, binds to the kinase domain of NblS. FEMS Microbiol. Lett. 254: 41–47 [DOI] [PubMed] [Google Scholar]
- Friso G., Majeran W., Huang M., Sun Q., van Wijk K.J. (2010). Reconstruction of metabolic pathways, protein expression, and homeostasis machineries across maize bundle sheath and mesophyll chloroplasts: Large-scale quantitative proteomics using the first maize genome assembly. Plant Physiol. 152: 1219–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M., Hamada S., Hiratsuka M., Fukao Y., Kawasaki T., Shimamoto K. (2009). Proteome analysis of detergent-resistant membranes (DRMs) associated with OsRac1-mediated innate immunity in rice. Plant Cell Physiol. 50: 1191–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M., Endo T., Peltier G., Tasaka M., Shikanai T. (2003). A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J. 36: 541–549 [DOI] [PubMed] [Google Scholar]
- Hudson A.O., Bless C., Macedo P., Chatterjee S.P., Singh B.K., Gilvarg C., Leustek T. (2005). Biosynthesis of lysine in plants: Evidence for a variant of the known bacterial pathways. Biochim. Biophys. Acta 1721: 27–36 [DOI] [PubMed] [Google Scholar]
- Ifuku K., Endo T., Shikanai T., Aro E.-M. (2011). Structure of the chloroplast NADH dehydrogenase-like complex: Nomenclature for nuclear-encoded subunits. Plant Cell Physiol. 52: 1560–1568 [DOI] [PubMed] [Google Scholar]
- Ifuku K., Ishihara S., Sato F. (2010). Molecular functions of oxygen-evolving complex family proteins in photosynthetic electron flow. J. Integr. Plant Biol. 52: 723–734 [DOI] [PubMed] [Google Scholar]
- Ishida S., Takabayashi A., Ishikawa N., Hano Y., Endo T., Sato F. (2009). A novel nuclear-encoded protein, NDH-dependent cyclic electron flow 5, is essential for the accumulation of chloroplast NAD(P)H dehydrogenase complexes. Plant Cell Physiol. 50: 383–393 [DOI] [PubMed] [Google Scholar]
- Lazarou M., Thorburn D.R., Ryan M.T., McKenzie M. (2009). Assembly of mitochondrial complex I and defects in disease. Biochim. Biophys. Acta 1793: 78–88 [DOI] [PubMed] [Google Scholar]
- Lezhneva L., Amann K., Meurer J. (2004). The universally conserved HCF101 protein is involved in assembly of [4Fe-4S]-cluster-containing complexes in Arabidopsis thaliana chloroplasts. Plant J. 37: 174–185 [DOI] [PubMed] [Google Scholar]
- Munekage Y., Hashimoto M., Miyake C., Tomizawa K., Endo T., Tasaka M., Shikanai T. (2004). Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 429: 579–582 [DOI] [PubMed] [Google Scholar]
- Nixon P.J., Michoux F., Yu J., Boehm M., Komenda J. (2010). Recent advances in understanding the assembly and repair of photosystem II. Ann. Bot. (Lond.) 106: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T., Hayashi S., Saeki M., Ohta H., Kinoshita K. (2009). ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 37(Database issue): D987–D991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okegawa Y., Kagawa Y., Kobayashi Y., Shikanai T. (2008). Characterization of factors affecting the activity of photosystem I cyclic electron transport in chloroplasts. Plant Cell Physiol. 49: 825–834 [DOI] [PubMed] [Google Scholar]
- Ozawa S., Nield J., Terao A., Stauber E.J., Hippler M., Koike H., Rochaix J.-D., Takahashi Y. (2009). Biochemical and structural studies of the large Ycf4-photosystem I assembly complex of the green alga Chlamydomonas reinhardtii. Plant Cell 21: 2424–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier J.B., Ripoll D.R., Friso G., Rudella A., Cai Y., Ytterberg J., Giacomelli L., Pillardy J., van Wijk K.J. (2004). Clp protease complexes from photosynthetic and non-photosynthetic plastids and mitochondria of plants, their predicted three-dimensional structures, and functional implications. J. Biol. Chem. 279: 4768–4781 [DOI] [PubMed] [Google Scholar]
- Peng L., Cai W., Shikanai T. (2010). Chloroplast stromal proteins, CRR6 and CRR7, are required for assembly of the NAD(P)H dehydrogenase subcomplex A in Arabidopsis. Plant J. 63: 203–211 [DOI] [PubMed] [Google Scholar]
- Peng L., Fukao Y., Fujiwara M., Takami T., Shikanai T. (2009). Efficient operation of NAD(P)H dehydrogenase requires supercomplex formation with photosystem I via minor LHCI in Arabidopsis. Plant Cell 21: 3623–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Fukao Y., Myouga F., Motohashi R., Shinozaki K., Shikanai T. (2011a). A chaperonin subunit with unique structures is essential for folding of a specific substrate. PLoS Biol. 9: e1001040. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Peng L., Ma J., Chi W., Guo J., Zhu S., Lu Q., Lu C., Zhang L. (2006). LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18: 955–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Shikanai T. (2011). Supercomplex formation with photosystem I is required for the stabilization of the chloroplast NADH dehydrogenase-like complex in Arabidopsis. Plant Physiol. 155: 1629–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Shimizu H., Shikanai T. (2008). The chloroplast NAD(P)H dehydrogenase complex interacts with photosystem I in Arabidopsis. J. Biol. Chem. 283: 34873–34879 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Peng L., Yamamoto H., Shikanai T. (2011b). Structure and biogenesis of the chloroplast NAD(P)H dehydrogenase complex. Biochim. Biophys. Acta 1807: 945–953 [DOI] [PubMed] [Google Scholar]
- Porra R.J., Thompson W.A., Kriedemann P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectrometry. Biochim. Biophys. Acta 975: 384–394 [Google Scholar]
- Rochaix J.-D. (2011). Assembly of the photosynthetic apparatus. Plant Physiol. 155: 1493–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakayori T., Shiraiwa Y., Suzuki I. (2009). A Synechocystis homolog of SipA protein, Ssl3451, enhances the activity of the histidine kinase Hik33. Plant Cell Physiol. 50: 1439–1448 [DOI] [PubMed] [Google Scholar]
- Sazanov L.A., Hinchliffe P. (2006). Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science 311: 1430–1436 [DOI] [PubMed] [Google Scholar]
- Schwenkert S., Netz D.J.A., Frazzon J., Pierik A.J., Bill E., Gross J., Lill R., Meurer J. (2010). Chloroplast HCF101 is a scaffold protein for [4Fe-4S] cluster assembly. Biochem. J. 425: 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T. (2007). Cyclic electron transport around photosystem I: Genetic approaches. Annu. Rev. Plant Biol. 58: 199–217 [DOI] [PubMed] [Google Scholar]
- Shikanai T., Endo T., Hashimoto T., Yamada Y., Asada K., Yokota A. (1998). Directed disruption of the tobacco ndhB gene impairs cyclic electron flow around photosystem I. Proc. Natl. Acad. Sci. USA 95: 9705–9709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu H., Peng L., Myouga F., Motohashi R., Shinozaki K., Shikanai T. (2008). CRR23/NdhL is a subunit of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant Cell Physiol. 49: 835–842 [DOI] [PubMed] [Google Scholar]
- Shimizu H., Shikanai T. (2007). Dihydrodipicolinate reductase-like protein, CRR1, is essential for chloroplast NAD(P)H dehydrogenase in Arabidopsis. Plant J. 52: 539–547 [DOI] [PubMed] [Google Scholar]
- Su P.-H., Li H.M. (2010). Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 22: 1516–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suorsa M., Sirpiö S., Aro E.-M. (2009). Towards characterization of the chloroplast NAD(P)H dehydrogenase complex. Mol. Plant 2: 1127–1140 [DOI] [PubMed] [Google Scholar]
- Wilde A., Härtel H., Hübschmann T., Hoffmann P., Shestakov S.V., Börner T. (1995). Inactivation of a Synechocystis sp strain PCC 6803 gene with homology to conserved chloroplast open reading frame 184 increases the photosystem II-to-photosystem I ratio. Plant Cell 7: 649–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H., Peng L., Fukao Y., Shikanai T. (2011). An Src homology 3 domain-like fold protein forms a ferredoxin binding site for the chloroplast NADH dehydrogenase-like complex in Arabidopsis. Plant Cell 23: 1480–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]