Alignment of cellulose fibers during wall synthesis is crucial for anisotropic plant cell growth. Cortical microtubules guide and align cellulose synthases (CESAs) in the plasma membrane during cellulose synthesis. This study uses genetic and cell biological experiments to show that the CELLULOSE SYNTHASE INTERACTING1/POM2 protein is needed for the guidance of CESAs along cortical microtubules.
Abstract
In plants, regulation of cellulose synthesis is fundamental for morphogenesis and plant growth. Cellulose is synthesized at the plasma membrane, and the orientation of synthesis is guided by cortical microtubules; however, the guiding mechanism is currently unknown. We show that the conditional root elongation pom2 mutants are impaired in cell elongation, fertility, and microtubule-related functions. Map-based cloning of the POM-POM2 locus revealed that it is allelic to CELLULOSE SYNTHASE INTERACTING1 (CSI1). Fluorescently tagged POM2/CSI1s associated with both plasma membrane–located cellulose synthases (CESAs) and post-Golgi CESA-containing compartments. Interestingly, while CESA insertions coincided with cortical microtubules in the pom2/csi1 mutants, the microtubule-defined movement of the CESAs was significantly reduced in the mutant. We propose that POM2/CSI1 provides a scaffold between the CESAs and cortical microtubules that guide cellulose synthesis.
INTRODUCTION
Cell expansion in plants is fundamental for cell morphogenesis and growth. The expansion is driven by vacuolar turgor pressure and is restricted by a strong, yet flexible, cell wall that permits directional cell growth. The cell wall is mainly constituted of polysaccharides, such as cellulose, hemicelluloses, and pectins, and highly glycosylated proteins (Somerville et al., 2004). Cellulose is made of β-1,4-linked glucan chains that form microfibrils by intermolecular hydrogen bonds (Somerville, 2006; Carpita, 2011; Endler and Persson, 2011). These microfibrils provide the main tensile strength to the cell walls and are hence of great importance for directed cell growth (Baskin, 2001; Cosgrove, 2005). Cellulose is believed to be synthesized at the plasma membrane by cellulose synthases (CESAs), which in higher plants have been observed to form hexameric rosette complexes (Kimura et al., 1999; reviewed in Guerriero et al., 2010). The cellulose microfibrils have been observed to align with underlying cortical microtubules (Green, 1962; Ledbetter and Porter, 1963). Consistent with these observations, in vivo studies using fluorescently labeled CESAs and microtubules revealed that the CESAs track along cortical microtubules (Paredez et al., 2006). Furthermore, perturbation of microtubule organization affects the mechanical properties of the cell wall (Baskin, 2001; Bichet et al., 2001; Sugimoto et al., 2003; Baskin et al., 2004; Wasteneys and Fujita, 2006). However, no mechanism for how the guidance of the CESAs by the microtubules occurs has been presented.
Mutant analyses and coimmunoprecipitation studies have shown that three distinct CESA subunits are necessary to form a functional complex (Somerville et al., 2004). Consequently, the primary wall CESA complex requires a CESA1, a CESA3, and a CESA6-related activity to be functional (Desprez et al., 2007; Persson et al., 2007). The CESA complexes are assumed to be assembled in the Golgi and transported to the cell surface where they are inserted into the plasma membrane adjacent to cortical microtubules (Crowell et al., 2009; Gutierrez et al., 2009). Apart from Golgi and plasma membrane localization, the CESAs are found in small post-Golgi compartments referred to as small CESA compartments (smaCCs; Gutierrez et al., 2009) or microtubule-associated CESA compartments (MASCs; Crowell et al., 2009). It has been proposed that the smaCCs/MASCs may be involved in either exo- or endocytosis of the CESAs.
Several components that are important for cell expansion have been identified through genetic screens. Subsequent cloning of the corresponding genes revealed many genes that are associated with cellulose synthesis, such as POM-POM1/CHITINASE-LIKE1 (POM1/CTL1), KORRIGAN/LION’S TAIL1/RADIALLY SWOLLEN2 (KOR/LIT/RSW2), COBRA, KOBITO/ELONGATION DEFECTIVE1, RSW1/CESA1, CONSTITUTIVE EXPRESSION OF VSP1/ECTOPIC LIGNIFICATION1/RSW5/CESA3, and QUILL/PROCUSTE/CESA6 (Hauser et al., 1995; Arioli et al., 1998; Nicol et al., 1998; Fagard et al., 2000; Lane et al., 2001; Schindelman et al., 2001; Pagant et al., 2002; Wang et al., 2006). Mutations in any of these genes result in epidermal cell swelling, restricted root and hypocotyl elongation, and reduced cellulose content. While it is unlikely that all of these proteins partake in the CESA complex, their functions appear crucial for production of the microfibrils. However, only CELLULOSE SYNTHASE INTERACTING1 (CSI1) has so far been identified to interact directly with the primary wall CESAs (Gu et al., 2010). The interaction was identified through a yeast two-hybrid screen using the catalytic domain of CESA6 as bait and was confirmed by dual labeling of the CSI1 and CESAs (Gu et al., 2010). CSI1 is a 2150–amino acid protein that holds between 10 and 20 predicted Armadillo repeats and a C-terminal C2 domain (Gu et al., 2010). Mutations in CSI1 caused reduced movement of CESA particles in the plasma membrane, lower cellulose content, and defective cell elongation.
To gain more knowledge about components that may affect cell morphogenesis and cellulose production, we characterized and cloned the gene responsible for pom2 (Hauser et al., 1995), which displays microtubule and cell elongation related defects. POM2 is allelic to CSI1, and fluorescently labeled POM2/CSI1s are associated with both plasma membrane–located CESAs and smaCCs/MASCs. Intriguingly, while the POM2/CSI1 does not affect insertion of CESAs adjacent to cortical microtubules, the protein is essential for sustained CESA movement along the microtubules. Hence, the POM2/CSI1 protein facilitates a connection between the CESAs and cortical microtubules, either directly or in association with other proteins.
RESULTS
pom2 Has Defects in Cell Elongation and Fertility
pom2-1 and pom2-2 were identified in a genetic screen for root morphogenesis mutants (Hauser et al., 1995). We identified two alleles, designated pom2-3 and pom2-4. All alleles were recessive and developed a conditional expansion phenotype with thick short roots on media supplemented with Suc (see Supplemental Figures 1A and 1B online; Table 1). Whereas wild-type roots grew roughly 3.5 mm/d, pom2 roots typically elongated only 0.5 to 0.7 mm/d under these conditions. On medium without Suc, the pom2 roots also showed reduced growth and cell elongation (see Supplemental Figure 2A online; Table 1). Similar to the root phenotypes on conditions stimulating growth, skotomorphogenic hypocotyls of pom2 seedlings were significantly shorter compared with the wild type (see Supplemental Figure 2B online). However, with the exception of pom2-3, full-grown pom2 mutants were only slightly dwarfed (see Supplemental Figure 2C online). By contrast, floral organs, such as petals, sepals, stamens, and carpels, were also reduced in size (see Supplemental Figure 2D online). The extent of the size reduction of stamens was allele specific and lead to heterostyly in pom2-1 and pom2-2 but not in pom2-3 and pom2-4. These cell expansion phenotypes indicate that POM2 is required for proper cell elongation in distinct organs.
Table 1.
Summary of Quantified Phenotypes of Wild-Type and pom2 Alleles
| Length (mm) |
Etiolated Hypocotyls 4.5% (μm) |
||||||||||
| Roots |
Etiolated Hypocotyls |
Length of Epidermis | Diameter of Palisade Tissue | Pollen Germination (%) | No. per Siliquea |
||||||
| Allele | Accession | 0% | 4.5% | 0% | 4.5% | Diameter | Ovules | Seeds | |||
| pom2-1 | Col | 4.8 ± 2.6 | 2.7 ± 1.0 | 9.2 ± 1.9 | 5.6 ± 2.1 | n.d. | n.d. | 44.3 ± 4.7 | 1.6 | 20.9 ± 3.2 | 0 ± 0b |
| pom2-2 | Col | 3.5 ± 1.7 | 2.3 ± 0.9 | 7.1 ± 1.5 | 4.7 ± 1.8 | 201 ± 47 | 735 ± 120 | 39.8 ± 5.0 | 21.3 | 25.0 ± 6.0 | 0 ± 0b |
| pom2-3 | Ler | 3.4 ± 1.1 | 2.2 ± 0.6 | 5.3 ± 0.9 | 3.4 ± 1.5 | n.d. | n.d. | 36.8 ± 6.1 | 12.0 | 25.5 ± 7.0 | 22.7 ± 5.9 |
| pom2-4 | Col, Nossen | 5.1 ± 2.0 | 3.8 ± 1.1 | 7.3 ± 1.3 | 6.7 ± 1.9 | n.d. | n.d. | 36.5 ± 4.1 | 20.3 | 22.6 ± 3.9 | 18.3 ± 4.9 |
| Wild type | Col | 10.8 ± 2.9 | 16.2 ± 3.3 | 11.9 ± 2.8 | 13.0 ± 3.3 | 389 ± 80 | 302.5 ± 96 | 32.9 ± 4.5 | 83.1 | 36.1 ± 5.0 | 40.0 ± 3.9 |
| Wild type | Ler | 11.0 ± 4.1 | 10.6 ± 2.7 | 11.5 ± 2.3 | 10.6 ± 2.5 | n.d. | n.d. | 39.8 ± 5.0 | 45.4 | 44.0 ± 5.6 | 45.2 ± 6.1 |
Significant differences to the wild type are in bold; 4.5% refers to Suc supplement in MS media. n.d., not determined.
Self-fertilization.
Upon cross-fertilization, up to 20 seeds/siliques developed.
The heterostyly correlated with fertility defects in the different pom2 alleles. All pom2 alleles developed significantly fewer ovules (Table 1). Whereas the wild type held around 40 ovules per gynoecium, only 20 to 30 were visible in the pom2 alleles. pom2-1 and pom2-2 had severely reduced fertility and occasionally displayed one to three seeds in terminal siliques (Table 1). However, if fertilized with wild-type pollen, pom2-1 and pom2-2 set up to 20 seeds/silique (n = 5 for pom2-1; n = 22 for pom2-2). By contrast, all ovules of pom2-3 and pom2-4 were fertilized by their own pollen (Table 1). As self-fertilization of the pom2-1 and pom2-2 was rare, we analyzed the number of pollen released per flower. Whereas wild-type anthers release 326 pollen for Columbia (Col) and 234 for Landsberg erecta (Ler), only 1.6, 5.4, 95, and 113 pollen per flower were harvested from pom2-1, pom2-2, pom2-3, and pom2-4, respectively. In addition, the majority of pollen grains were deformed (see Supplemental Figure 2E online), and pollen tube development was impaired in the pom2 alleles (see Supplemental Figure 2F online; Table 1). Hence, the reduced fertility of the pom2 mutants appears to be a combination of development of smaller gynoecia with fewer ovules, heterostyly, the inability of anthers to release pollen, collapsed pollen grains, and poor pollen germination rate.
pom2/csi1 Mutants Display Microtubule-Related Phenotypes and Mitotic Defects
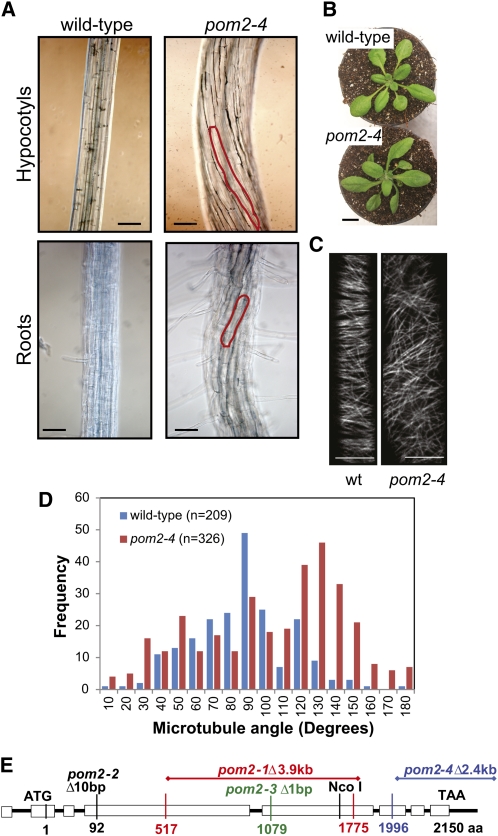
The pom2 mutants also displayed organ and cell twisting (Figures 1A and 1B). This was readily observed in roots and etiolated hypocotyls (left-handed) of seedlings grown on medium with or without added Suc. In addition, twisting was also observed in soil-grown rosette leaves (left-handed; Figures 1A and 1B). Twisting or spiraling of organs occurs in mutants affecting cytoskeleton organization, for example, in the microtubule-associated clasp, lefty1 and 2, and sku6/spr1 (Thitamadee et al., 2002; Sedbrook et al., 2004; Ambrose et al., 2007; Kirik et al., 2007). To investigate whether the microtubule organization was disturbed in pom2, we crossed pom2-4 to the microtubule marker lines Microtubule-Associated Protein4 (MAP4):green fluorescent protein (GFP) or mCherry:Tubulin alpha-5 (TUA5) (Marc et al., 1998; Gutierrez et al., 2009). Rapidly elongating cells in wild-type roots and etiolated hypocotyls typically hold transversely or obliquely orientated cortical microtubule arrays (Sugimoto et al., 2000; Figure 1C). However, the cortical microtubules were less well transversally arranged in rapidly elongating hypocotyl cells (between 2 and 12 mm below the apical hook) in the pom2-4 mutant (Figures 1C and 1D). In addition, microtubule defects were also observed in the other pom2 alleles, for example, in pom2-1 (see Supplemental Figure 3A online). These results suggest that the organization of the microtubule array is impaired in the pom2 mutants. We also investigated whether the microtubule dynamics were altered in pom2/csi1 by selecting instances along kymographs in which contrast changes were unambiguously connected to a single microtubule end. Using this criterion, we found no significant differences in microtubule plus end growth (5.5 ± 2.1 μm/min in pom2/csi1; 4.8 ± 1.3 μm/min in the wild type), plus end shrinkage (15.3 ± 9.2 μm/min in pom2/csi1; 11.5 ± 5.0 μm/min in the wild type), or in minus end shrinkage (2.4 ± 1.4 μm/min in pom2/csi1; 2.2 ± 1.7 μm/min in the wild type) when comparing pom2/csi1 mutant cells and wild-type cells (60 microtubule ends from six cells from six seedlings). These growth and shrinkage rates are in good agreement with previously published results (Shaw and Lucas, 2011).
Figure 1.
Positional Cloning of POM2 and Microtubule-Related Defects in the Mutant.
(A) Six-day-old etiolated pom2-4 hypocotyl displaying twisting of cell files (red outline).
(B) Rosette leaves of 3-week-old pom2-4 plants that spiral.
(C) Microtubule organization in 3-d-old elongating hypocotyl cells of the wild type (wt; left) and pom2-4 (right). Microtubules are labeled by mCherry:TUA5.
(D) Microtubule angle distribution in elongating hypocotyl cells in the wild type and pom2-4. Angles were measured against the growth axis. Seven cells from seven seedlings were used for the analysis.
(E) Mapped pom2 mutations in POM2 (At2g22125). Black line indicates introns and boxed areas exons. aa, amino acids.
Bars = 100 μm in (A), 2 cm in (B), and 10 μm in (C).
Apart from altered microtubule organization, mutations in several microtubule-associated genes also lead to reduced mitotic activity (Ambrose et al., 2007; Kirik et al., 2007; Sunohara et al., 2009). To test whether the pom2 mutants also hold fewer cells that actively divide, we crossed lines expressing the mitotic marker cycB1;1:Cyclin Destruction Box (CDB):β-glucuronidase (GUS) (Hauser and Bauer, 2000) with several pom2 mutant alleles. The activity of the meristem was quantified by histochemical GUS staining. Similar to other microtubule-associated mutants, significantly fewer cells were actively dividing in the pom2 mutants (see Supplemental Figure 3B online). Thus, pom2 mutants are affected both in cell elongation, microtubule organization, and meristematic activity.
POM2 Is Allelic to CSI1 and Contributes the Main CSI Activity
POM2 was isolated by a map-based cloning strategy and was initially mapped in the middle of chromosome II between the cleaved amplified polymorphic sequence markers GPA1 and m429 (Hauser et al., 1995). By analyzing 1022 meiotic events with diverse molecular markers (see Supplemental Table 1 online), POM2 was narrowed down to a 110-kb interval containing 36 candidate genes. This region was covered by two overlapping BACs (see Supplemental Figure 4A online). Allele-specific NcoI and EcoRV restriction fragment length polymorphisms were identified between the Col alleles pom2-1 and pom2-2 using the labeled BAC T16B14 as probe (see Supplemental Figure 4A online), with only a region around At2g22125 with the expected genomic fragment lengths and the detected polymorphism pattern (see Supplemental Figure 4B online). A 14.5-kb binary cosmid clone, containing the full At2g22125 gene, was identified using the (F6/R6) probe and complemented the pom2-3 phenotypes. Mutations in all four alleles were identified through sequencing (outlined in Figure 1E), and all of the lines contained reduced levels of POM2 mRNA (see Supplemental Figure 4C online). Furthermore, T-DNA insertion lines that correspond to At2g22125 (SALK_047252, pom2-5; SALK_051146, pom2-6; SALK_115451, pom2-7; and SALK_136239, pom2-8; all of which are mRNA nulls; Gu et al., 2010) phenocopied the pom2 mutants; hence, POM2 is At2g22125.
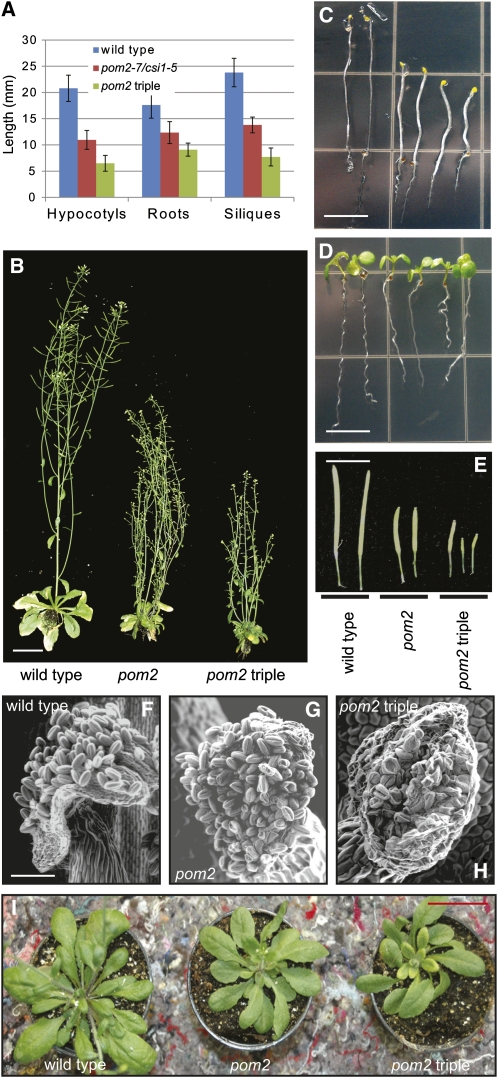
At2g22125 was recently identified as CSI1 (Gu et al., 2010) and is part of a three-member family, which contains POM2/CSI1, CSI2, and CSI3. To assess whether the other members of this family are also of importance for plant development, we obtained homozygous T-DNA insertion mutants for CSI2 (SALK_022491 and SALK_024648) and CSI3 (SALK_009141 and SALK_009157). These mutants were mRNA nulls or contained minute mRNA levels, as assessed via quantitative RT-PCR (qRT-PCR) (see Supplemental Figure 4D online). However, we did not observe any phenotypes in the csi2 and csi3 single mutants or in the double csi2 csi3 (SALK_022491 and SALK_009141) mutants. These data suggest that POM2/CSI1 contributes the main POM2/CSI-related activity in Arabidopsis thaliana. However, pom2-7/csi1-5 csi2 csi3 triple mutants did display some mild synergistic phenotypes (Figure 2), such as dwarfed plant growth, a more dramatic collapsed pollen phenotype, and increased organ twisting, suggesting that some functional redundancy may be contributed by CSI2 and CSI3.
Figure 2.
Triple pom2/csi1 csi2 csi3 Mutant Phenotypes.
(A) Graph displaying length of 5-d-old etiolated hypocotyls, 5-d-old light-grown seedling roots, and mature siliques from 7-week-old soil-grown plants of wild-type, pom2-7/csi1-5, and pom2-7/csi1-5 csi2 csi3 genotypes.
(B) Seven-week-old wild-type, pom2-7/csi1-5, and pom2-7/csi1-5 csi2 csi3 triple mutants. Bar = 5 cm.
(C) Five-day-old etiolated wild-type, pom2-7/csi1-5, and pom2-7/csi1-5 csi2 csi3 triple mutant seedlings. Bar = 5 mm.
(D) Five-day-old light-grown wild-type, pom2-7/csi1-5, and pom2-7/csi1-5 csi2 csi3 triple mutant seedlings. Bar = 5 mm.
(E) Mature siliques from 7-week-old wild-type, pom2-7/csi1-5, and pom2-7/csi1-5 csi2 csi3 triple mutant plants. Bar = 1 cm.
(F) to (H) Environmental scanning electron microscopy images of pollen grains on anthers from wild-type, pom2-7/csi1-5, and pom2-7/csi1-5 csi2 csi3 triple mutant plants. Bar = 70 μm.
(I) Twisting rosette leaves in 4-week-old pom2-7/csi1-5 and pom2-7/csi1-5 csi2 csi3 triple mutant plants. Bar = 5 cm.
POM2/CSI1 Colocalizes with CESAs and CESA-Containing Vesicles
To gain further insight into the function of POM2/CSI1, we made a C-terminal fusion of the protein to cyan fluorescent protein (CFP) under a 35S constitutive promoter. This construct complemented the pom2/csi1 phenotype (pom2-8/csi1-1) and mimicked the published CSI1p:CSI1:red fluorescent protein behavior (Gu et al., 2010). Hence, as previously described, the POM2/CSI1 fluorescent protein was present as distinct punctae at the cell cortex in rapidly elongating hypocotyl cells and displayed bidirectional motility of ~200 to 300 nm/min when imaged with a spinning-disc confocal microscope (see Supplemental Figures 5A and 5B online; Gu et al., 2010). In addition, several of the pom2 alleles could hold truncated proteins based on the location of the insertion and on the qRT-PCR results (Figure 1E; see Supplemental Figure 4C online). We therefore also generated a truncated POM2, in which the C2 domain was removed, and fused it to CFP. This construct did not rescue the pom2 mutants, and the fluorescent pattern was diffuse and distinct from the cortical foci seen using the full-length POM/CSI1:CFP construct (see Supplemental Figure 5C online), suggesting that truncations of the POM2/CSI1 protein render it nonfunctional and that the C-terminal C2 domain contributes to POM2/CSI1 cellular localization.
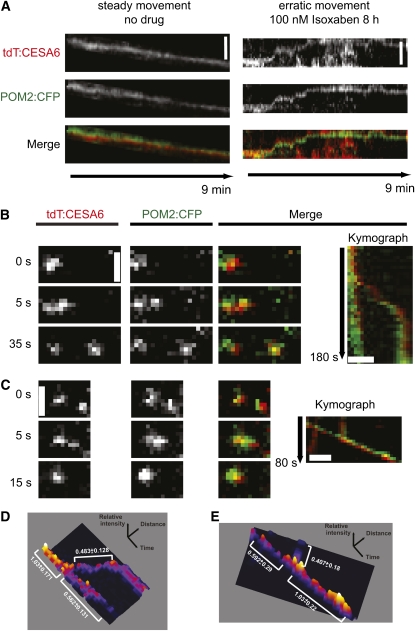
To ensure that the POM2/CSI1:CFP punctae corresponded to plasma membrane–based CESA particles, we crossed the POM2/CSI1:CFP with a tdTomato:CESA6 line. The fluorescent signals from these two markers showed marked coincidence (see Supplemental Figure 5D online). Statistical analysis revealed that the overlap was highly significant compared with randomized signals (see Supplemental Figure 5E online). Furthermore, the CESA and POM2/CSI1 signals co-occurred throughout different cells in etiolated hypocotyls (see Supplemental Figure 5F online). Curiously, we also observed a population of cortical POM2/CSI1s that moved with velocities of well over 1000 nm/min (see Supplemental Figures 5A and 5B online). Such high velocities have not been reported for plasma membrane–bound CESAs, but rather for CESA containing vesicles (i.e., smaCCs/MASCs) at the cell cortex (Gutierrez et al., 2009).
To explore whether the rapidly moving POM2/CSI1s were associated with smaCCs/MASCs, we treated seedlings with the cellulose synthesis inhibitor isoxaben. This drug causes rapid disappearance of the plasma membrane–located CESA complexes and a concomitant accumulation of smaCCs/MASCs tethered at the cell cortex (Gutierrez et al., 2009). After isoxaben treatment, we observed a gradual increase in the number of tethered smaCCs/MASCs, as estimated from their erratic motilities (Gutierrez et al., 2009; Figure 3A). In addition, these vesicles were accompanied by POM2/CSI1 signal, indicating that the POM2/CSI1 can associate with the smaCCs/MASCs (Figure 3A). The smaCCs/MASCs may also bifurcate into smaller compartments or merge into bigger ones (Figure 3; Gutierrez et al., 2009). In agreement with this observation, we also detected merging and bifurcating fluorescent signals of the POM2/CSI1-decorated compartments (Figure 3). These events were corroborated by three-dimensional heat map analyses (i.e., we observed that the sum of the migrating smaCCs/MASCs and POM2/CSI1 branches roughly equaled the trunk of the signal) (Figures 3C and 3D).
Figure 3.
POM2/CSI1-Containing Compartments Can Merge and Bifurcate at the Cell Cortex.
(A) Kymographs from 4-d-old etiolated seedlings expressing tdTomato:CESA6 and POM2:CFP. Kymographs display CESA and POM2/CSI movement at the plasma membrane in nontreated cells (left) and CESA and POM/CSI1 movement at the cell cortex in isoxaben-treated (100 nm for 8 h) cells (right).
(B) and (C) Images from two different time series in 4-d-old etiolated seedlings expressing tdTomato:CESA6 and POM2:CFP. The images show a split of one compartment into two (for both the CESA and POM2/CSI1 fluorescent signal) at 5 s (B), and a merge of the signals at 5 s in another time series (C). Right panel shows kymographs depicting the split of the signal into two (Top) and the merge of two compartments into one (Bottom). Bars = 2 μm.
(D) and (E) Surface plots displayed as heat maps. The mean intensity of the signals above background was measured for (B) and (C) and is displayed as relative values ± sd for all pixels along the indicated trace.
POM2/CSI1 Co-Occurs with Microtubules and Remains at the Cell Cortex after Microtubule Depolymerization
Plasma membrane–located CESA particles track along cortical microtubules (Paredez et al., 2006). To put the POM2/CSI1 in context to the microtubules, we crossed the POM2/CSI1:CFP-expressing line with a plant expressing mCherry:TUA5. Similar to what has been shown for the CESAs, POM2/CSI1 migration occurred along cortical microtubules under control conditions (see Supplemental Figure 6A online). We quantified the coincidence of the two signals using van Steensel’s cross-correlation function algorithm (Bolte and Cordelières, 2006), which confirmed colocalization of the two signals (see Supplemental Figure 6B online). To assess whether the motility and location of POM2/CSI1 depends on microtubules, we treated POM2/CSI1:CFP-expressing seedlings with oryzalin overnight. This treatment completely abolished the microtubule array, and the signal was instead seen as a diffuse cytoplasmic smear (see Supplemental Figure 6E online). However, the POM2/CSI1 signal remained at the cell cortex and followed similar trajectories as the CESAs (see Supplemental Figures 6C and 6D online). The CESA and POM2/CSI1 trajectories were, however, less well defined than in the control-treated cells (Paredez et al., 2006). These results suggest that the interaction between CESAs and POM2/CSI1 is sustained in the absence of microtubules.
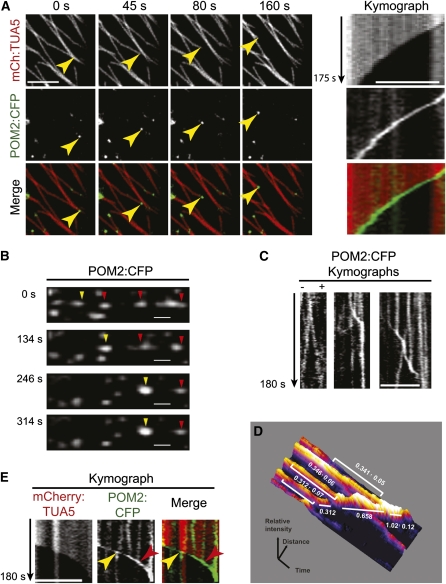
POM2/CSI1 Co-Occurs with Microtubule Depolymerizing Ends and smaCCs/MASCs upon Isoxaben Treatment
Exposure of cells to isoxaben causes, in addition to the accumulation of smaCCs/MASCs, tethering of the vesicles to cortical microtubules and co-occurrence of the smaCCs/MASCs with microtubule depolymerizing ends (Gutierrez et al., 2009). Similar isoxaben treatments also resulted in the co-occurrence of POM2/CSI1 foci with microtubule depolymerizing ends (Figure 4A; see Supplemental Movie 1 online). We observed POM2/CSI1 on 33 out of 67 microtubule depolymerizing plus and minus ends (three cells from three seedlings) under these conditions. These POM2/CSI1s displayed rapid movement interspersed with infrequent stops, which followed the dynamics of the microtubule ends (Figure 4A). Furthermore, multiple POM2/CSI1s that populated the same microtubule were collected by the depolymerizing end–associated POM2/CSI1s (Figures 4B to 4D; see Supplemental Movie 2 online). Hence, aggregation of the POM2/CSI1s occurred in a stepwise fashion, each step adding an additional POM2/CSI1 (Figures 4C and 4D). This behavior is similar to what has been described for smaCCs/MASCs (Gutierrez et al., 2009). The POM2/CSI1 particles also exclusively co-occurred with microtubule depolymerizing ends and not with growing ends (see Supplemental Figure 7A and Supplemental Movie 3 online), which also was reported for smaCCs/MASCs (Gutierrez et al., 2009). Interestingly, the association of POM2/CSI1 with microtubule depolymerizing ends may be connected with a bifurcation mechanism. Bifurcation of microtubule end–associated POM2/CSI1 signals occurred when depolymerization progressed across another adjacent microtubule (Figure 4D). Hence, the retracting microtubule ends may aid the bifurcation, perhaps facilitating redistribution of POM2/CSI1 and, therefore, possibly also smaCCs/MASCs, to other cellular locations.
Figure 4.
POM2/CSI1 Fluorescent Signals Associate with Microtubule Depolymerizing Ends.
(A) Images from time series (see Supplemental Movie 1 online) of cells in 4-d-old hypocotyls expressing mCherry:TUA5 and POM2:CFP treated with 100 nM isoxaben for 2 h. Yellow arrowheads indicate POM2/CSI1 fluorescent signal associated with a depolymerizing microtubule end. Right panel displays kymographs, which show that the POM2/CSI1 signal follows the microtubule retracting end.
(B) Time series (see Supplemental Movie 2 online) of POM2/CSI1 collecting other POM2/CSI1s. Yellow arrowheads indicate collecting POM2/CSI1 and red arrowheads collected POM2/CSI1s.
(C) Kymograph displaying POM2/CSI1 bidirectional migration (left), accelerating and stopping POM2/CSI1 (middle), and collecting POM2/CSI1 (right).
(D) Surface plot displayed as a heat map. The mean intensity of the signals above background was measured for the right kymograph in (C) and is displayed as relative values ± sd.
(E) Kymograph showing bifurcation and merge of POM2/CSI1 signal that is associated with a retracting microtubule end. When the end crosses another microtubule, the POM2/CSI1 signal bifurcates (yellow arrowheads). Merging events of two POM2/CSI1 signals happen along retracting microtubules that contain laterally associated POM2/CSI1 (red arrowheads).
Bars = 5 μm (A), (C), and (E) and 1 μM in (B).
POM2/CSI1 Does Not Affect Insertion Rates or the Spatial Distribution of CESA Deliveries
To assess how the POM2/CSI1 punctae are established at the cell cortex, we bleached the existing POM2/CSI1:CFP signal and observed its repopulation. The POM2/CFP fluorescence first dimly reemerged evenly across the bleached area (see Supplemental Figure 7B online), as expected from a cytosolic protein due to rapid cytoplasmic streaming. After ~60 s, distinct punctae appeared at the cell cortex that moved with constant velocities of ~200 to 300 nm/min (see Supplemental Figure 7B online). The POM2/CSI1 punctae continued to appear during the time of recovery, and after ~120 s, the bleached area was repopulated by the fluorescent foci. The establishment of distinct POM2/CSI1-related punctae at the plasma membrane could occur either through codelivery together with CESA particles or through the gradual association of cytosolic POM2/CSI1 with newly delivered CESAs that subsequently amass to a brighter puncta. To investigate this, we bleached both fluorescent signals in the dual-labeled POM2/CSI1:CFP and tdTomato:CESA6 line. We observed that the appearance of a CESA particle at the plasma membrane was accompanied by the appearance of a POM2/CSI1 fluorescent puncta at the cell cortex in 16 out of 69 (or 23%) observed delivery events (three cells from three seedlings; see Supplemental Figure 7C online). The appearance of the remaining CESA punctae occurred independently from any clear POM2/CSI1 signal at the plasma membrane (see Supplemental Figure 7C online). Bleaching of smaller areas did not appear to change the estimate of codelivery events. The co-occurrence of the two fluorescent signals observed in some of the delivery events suggests that POM2/CSI1 can accumulate with CESAs prior to plasma membrane delivery (e.g., with the smaCCs/MASCs, as discussed above). Consequently, POM2/CSI1 could have a role in the delivery of CESAs to the plasma membrane. To assess this, we generated GFP:CESA3 and mCherry:TUA5 dual-labeled lines in the pom2 mutant background. To quantify the rate, as well as the spatial distribution, of CESA insertions, we bleached the GFP channel and measured the appearance of new CESAs at the plasma membrane. The criteria used to identify delivery events were identical to what was described by Gutierrez et al. (2009) (i.e., a CESA particle appeared in the focal plane and showed a brief period of wobbling motility, which after a short period of time reverted to a steady movement of ~200 to 400 nm/min along a linear track). We subsequently mapped these events on images of mCherry:TUA5. In the wild type, we observed an insertion rate of ~0.06 CESAs μm−2 min−1, which corresponds to 3.1 (±0.6) insertion events μm−2 h−1 (three cells from three seedlings). This observation is in close agreement with previously reported insertion rates (4.8 insertion events μm−2 h−1; Gutierrez et al., 2009). However, we observed similar insertion rates in the pom2 mutants (4.2 ± 1.4 in pom2-4; 3.1 ± 1.4 in pom2-5; three cells from three seedlings); therefore, the overall rate of CESA insertion does not appear to be affected in the pom2 mutants. Furthermore, 283 out of 324 CESA insertions, or 87%, occurred at sites occupied by microtubules in the pom2 mutants (three cells from three seedlings for pom2-4 and pom2-5, respectively; Figures 5A and 5B). This was considerably more than expected by chance (54% based on microtubule image coverage); hence, CESA insertion coincides with microtubules also in the pom2 mutants.
Figure 5.
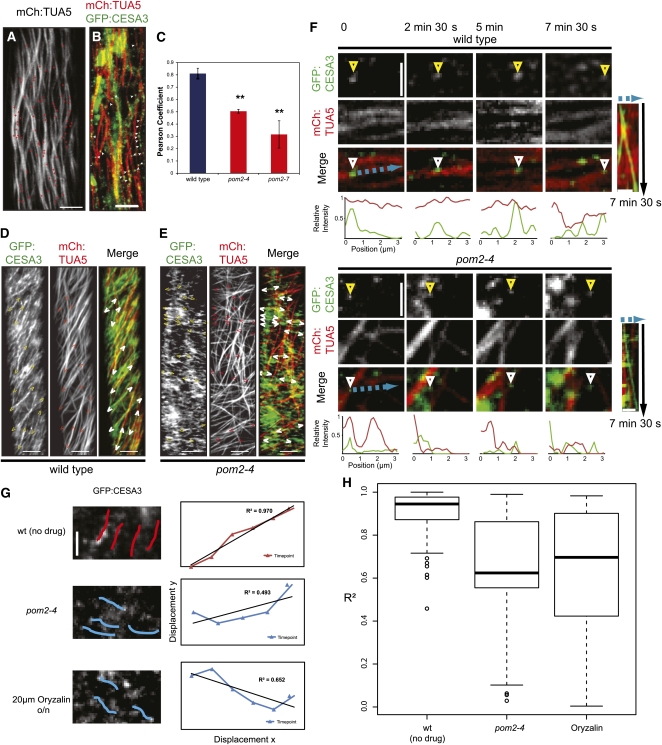
POM2/CSI1 Is Necessary for Coalignment of CESAs and Microtubules at the Cell Cortex.
(A) Map of CESA complex delivery sites in an elongating pom2-4 mutant cell from a 4-d-old etiolated hypocotyl. The sites were mapped on a 10-min average projection of microtubule signal. Red plus symbols indicate CESA insertion sites.
(B) Single image frame showing CESA insertions (green channel; white arrowheads) on microtubules (red channel) in a pom2-4 mutant cell from a 4-d-old etiolated hypocotyl. Bars = 5 μm in (A) and (B).
(C) Quantification of colocalization between GFP:CESA3 and mCherry:TUA5 trajectories in 500-s time averages using Pearson correlation (n = 9 cells from 9 seedlings). Error bars indicate sd. Asterisks indicate significance (**P value < 0.01).
(D) and (E) Average projections of 53 frames in elongating control (D) and pom2-4 (E) cells in 4-d-old etiolated hypocotyls expressing mCherry:TUA5 and GFP:CESA3. Brackets indicate CESA migration trajectories. Bars = 5 μm.
(F) Time frames of individual plasma membrane–located CESAs in the wild type (Top) and pom2-4 (Bottom) in relation to cortical microtubules. CESAs (arrowheads) track along microtubules in the wild type (Top), but the tracking of CESAs in pom2-4 is independent of the microtubule orientation (Bottom). Kymographs of the observations are supplied to the right of the respective time frames. The intensity of both channels was plotted along the cyan trace.
(G) and (H) Degree of linearity of moving CESAs over time. The displacement of individual CESA particles (145 particles from nine seedlings; between 45 and 50 particles per treatment and line) was traced over 8 min (100 frames). The left panel shows examples of individual traces. x and y coordinates at different time points were plotted ([G], right panel) and fitted against a linear curve. Deviations from linearity result in a low R2 value; ideal linear traces are represented with an R2 of 1. The distribution of R2 values (right panel) shows a significant reduction in linear CESA movement in pom2/csi1 mutants and in oryzalin-treated cells (t test: the wild type (wt) versus pom2-4 and/or oryzalin treatment; P < 0.01, pom2 versus oryzalin treatment; P > 0.5).
POM2/CSI1 Is Essential for Coalignment of Microtubules and CESAs
After insertion, the CESAs begin to move with a steady rate along trajectories defined by the underlying microtubules (Gutierrez et al., 2009). To assess whether POM2/CSI1 has a role in this process, we analyzed the tracking behavior of the CESAs and compared the coincidence of CESA molecules and microtubules over time using Pearson correlation coefficients (Figure 5C). To do the latter, we compared the coincidence of fluorescent signals in time averages from 100 single images (500-s total imaging time) of wild-type and pom2 cells expressing both GFP:CESA3 and mCherry:TUA5. While we observed a high correlation between CESA and microtubule fluorescence in the wild-type background, the correlation between the two channels was significantly lower in the pom2 mutants (Figure 5C). We further measured the amount of CESAs that maintained trajectories associated with cortical microtubules in the wild type and pom2 mutants. Only ~10% of the CESAs were associated with microtubules in the pom2 mutants; however, 94% of the CESAs maintained trajectories along microtubules in the wild-type background (284 particles from three cells from three seedlings in the wild type, pom2-4, and pom2-7, respectively). These analyses were corroborated by visual inspection of consecutive and averaged time frame images, which revealed that the CESA trajectories moved independently of the direction of the underlying cortical microtubules in the pom2 mutants (cf. Figures 5D to 5F; see Supplemental Movie 4 online). Thus, POM2/CSI1 is important for the microtubule-based guidance of the CESAs.
Loss of the microtubule array does not affect the velocity of CESA particles localized to the plasma membrane (Paredez et al., 2006; Chen et al., 2010; Bischoff et al., 2011). However, plasma membrane–located CESAs in the pom2/csi1 mutants showed a two- to threefold reduction in speed compared with the wild-type background (Gu et al., 2010). One possible explanation for this is that POM2/CSI1, in addition to its role in coaligning the CESAs and microtubules, also influences the activity of the CESAs as determined by the decreased motility. Another possibility is that POM2/CSI1 contributes to a currently not recognized guiding mechanism for the CESAs independently of the microtubule array. To test the latter, we tracked individual plasma membrane located CESA particles over 100 single images (500-s total imaging time) and assessed the degree of linearity of migrating CESAs over time. We subsequently estimated the deviations from linear trajectories in the pom2-4 mutant and the wild type by plotting the displacement in the x and y direction over time and calculating the linear regression coefficients (Figure 5G). The CESAs deviated significantly more from linear tracks in the pom2-4 compared with the wild type (Figure 5G). However, similar deviations were also observed in oryzalin-treated wild-type cells (Figure 5G). It is therefore likely that the reduced CESA velocities observed in the pom2 mutants correspond to an alteration in CESA activity and not an alternative guiding mechanism by POM2/CSI1. Hence, it is evident that the POM2/CSI1 not only provides for an association between the cortical microtubules and the CESAs but also is important for the maintenance of an active CESA complex. These results further show that the linear movement of the plasma membrane located CESAs is stabilized by the cortical microtubules.
DISCUSSION
Turgor-driven expansion, coupled with cell wall reorganization, is essential for anisotropic cell development and plant growth. Several genetic screens have revealed mutants with defects in cell elongation, some of which affect cellulose production. We show that the conditional root expansion mutant pom2 affects CSI1 (Gu et al., 2010) and that the function of the protein is essential for the guidance of the CESA complexes along the cortical microtubules.
Already in the early 1960s, Paul Green and others observed that cellulose microfibrils were disrupted by the antispindle fiber drug colchicine and proposed that the spindle fibers, later termed microtubules (Ledbetter and Porter, 1963), may have a role in the orientation of microfibril deposition. This hypothesis was corroborated by live-cell imaging (Paredez et al., 2006), which showed that cortical microtubules define the trajectories of plasma membrane–based CESAs. The tight spatial and temporal link between the CESA trajectories and microtubules indicated an intimate contact between the two arrays (Baskin, 2001; Paredez et al., 2006). We show that mutations in POM2/CSI1 substantially reduce coalignment of the CESAs and cortical microtubules, suggesting that POM2/CSI1 facilitates a link between the CESAs and the microtubules during cellulose synthesis. POM2/CSI1 contains many Armadillo repeats, which are present in several microtubule binding proteins (reviewed in Tewari et al., 2010). One possible scenario could therefore be that POM2/CSI1 contributes a direct link between the CESAs and microtubules. Another scenario may involve additional components (e.g., proteins that could promote motility along the microtubules). For instance, mutations in a kinesin-like protein caused altered deposition of secondary wall cellulose microfibrils in Arabidopsis (Zhong et al., 2002). Similarly, mutations in the rice (Oryza sativa) kinesin-4 also led to deficiencies in secondary wall cellulose production (Zhang et al., 2010). While these mutants affect secondary wall cellulose synthesis, it is possible that kinesin-related activity also could play a role in primary wall cellulose production, perhaps by supporting the means of motility for POM2/CSI1 and, hence, the CESAs, along the microtubules. Interestingly, an Armadillo repeat protein, KAP3, associates with kinesins and is important for cargo movement along microtubules in neuronal axons (reviewed in Manning and Snyder, 2010).
While it is possible that active microtubule-associated machinery could be involved in promoting migration of the CESA complex along the microtubule, it is perhaps more likely that polymerization of the cellulose microfibril drives CESA motility. For example, both CESA and POM2/CSI1 motility can be observed in the absence of a microtubule array (see Supplemental Figure 6C online; Paredez et al., 2006). Providing that this hypothesis is correct, and also that the main POM2/CSI1 activity is related to guidance of the CESAs along cortical microtubules, we would expect the CESA complexes to migrate with a similar speed in pom2/csi1 mutant cells as in wild-type cells. However, this is not the case. The rate of CESA migration is reduced two- to threefold in the pom2/csi1 background (Gu et al., 2010). This raises some interesting questions regarding the function of POM2/CSI1 and regarding the role of microtubules in the maintenance of CESA migration. Two recent reports show that disruption of the microtubule array can promote CESA migration (Chen et al., 2010; Bischoff et al., 2011), which further corroborates that the CESA activity supports CESA motility. For example, one of these studies showed that the migration of CESA5 was reduced in etiolated prc1-1 seedlings compared with the wild type (Bischoff et al., 2011). However, removal of the microtubules using oryzalin restored CESA5 migration to speeds observed in wild-type seedlings. These data suggest that the removal of microtubules should result in CESA migratory rates comparable to control seedlings. Yet, the abolished coalignment of the CESAs and cortical microtubules in the pom2/csi1 mutants still result in reduced CESA velocities. Several scenarios may explain this observation. For example, it is possible that POM2/CSI1 may provide an additional layer of guidance to the CESA migration, perhaps via the putative C2 domain that is present in the C terminus of POM2/CSI1 that may interact with plasma membrane lipids. Such interactions could provide directionality of the CESA complex based on the lipid environment. If this hypothesis is true, one would expect a different CESA complex directionality in wild-type cells without microtubules and cells from pom2/csi1 mutants. However, we observed similar deviations in linear tracking behavior of CESAs in oryzalin-treated wild-type cells and in pom2/csi1 cells (Figure 5G). These results support an important function of POM2/CSI1 in microtubule-based tracking of the CESAs. The reduced CESA motility in the pom2/csi1 mutants could instead be due to an important function of POM2/CSI1 for CESA activity, perhaps by modulating the CESA active sites with which POM2/CSI1 appears to interact (Gu et al., 2010). In support of this, mutations in several components that affect cellulose synthesis, such as KOR and CESA6, lead to reduced CESA motility (Paredez et al., 2008; Bischoff et al., 2011). It is also possible that POM2/CSI1 facilitates a docking site for other proteins necessary for CESA activity (e.g., for Suc synthase) that may provide substrate for cellulose synthesis.
While mutations in POM2/CSI1 result in phenotypes that typically are observed in cellulose-deficient mutants, such as cell elongation deficiency, collapsed pollen grains, and reduced cellulose levels (Gu et al., 2010), the pom2/csi1 mutants also displayed defects in microtubule organization, cell file spiraling, and organ twisting. Spiraling, or twisting, phenotypes have been reported for mutations in several microtubule-associated proteins, such as SKU6/SPIRAL1, LEFTY1 and 2, and CLASP (Thitamadee et al., 2002; Sedbrook et al., 2004; Kirik et al., 2007), and perturbation of these proteins also lead to microtubule defects. Although cellulose-deficient mutants (i.e., mutants that have reduced CESA velocities and cellulose content) typically display defects in microtubule organization (Paredez et al., 2008), no spiraling phenotypes have been reported. Moreover, clasp and other microtubule-related mutants also affect mitotic activity, presumably by disturbances in the mitotic spindle formation (Ambrose et al., 2007; Kirik et al., 2007; Sunohara et al., 2009). Interestingly, pom2/csi1 mutants also held a reduced number of cells that actively divided in the root meristem. Considering that no alterations in mitotic activity were observed in the cellulose-deficient mutant ctl1/pom1 (Hermans et al., 2011), it is apparently not the reduction in cellulose per se that caused the impaired mitotic activity. Rather, certain defects in the microtubule stability or arrangement affect the mitotic activity. Although POM2/CSI1 appears to be more strongly connected to the CESAs than the microtubules, it is clear that the protein also executes important microtubule related functions. Hence, the microtubule-related phenotypes displayed by the pom2/csi1 mutants corroborate the role of POM2/CSI1 in microtubule-related processes, which support that the protein facilitates a direct or indirect connection between the CESAs and microtubules.
Delivery of CESA particles to the plasma membrane may be facilitated either via the Golgi or via the post-Golgi compartments, which are referred to as smaCCs/MASCs (Crowell et al., 2009; Gutierrez et al., 2009). The latter compartments are induced by treating cells with isoxaben or osmotic stress (Crowell et al., 2009; Gutierrez et al., 2009). Interestingly, in addition to the close colocalization of CESA and POM2/CSI1 at the plasma membrane, we also observed pronounced colocalization of POM2/CSI1 signal with smaCCs/MASCs. Hence, POM2/CSI1 can associate with CESAs, or CESA-containing compartments, also at other locations than the plasma membrane. While the function of the smaCCs/MASCs is unclear, it has been proposed that the vesicles may hold CESA complexes that are in transit to the plasma membrane (Gutierrez et al., 2009). Considering that POM2/CSI1s can associate with the smaCCs/MASCs, one possible scenario would be that CESAs and POM2/CSI1s are codelivered to the plasma membrane. However, we only observed codelivery of a minor part of the observed delivery events. One reason for this observation may be that POM2/CSI1s are codelivered with CESAs from the smaCCs/MASCs only and not with CESAs that are inserted directly from Golgi-derived compartments. For example, it is possible that either the cortical microtubules or the plasma membrane composition could provide an advantageous environment for POM2/CSI1s to interact with CESAs at the cell cortex. Consistent with this, we did not observe any convincing colocalization of POM2/CSI1 and CESAs associated with rapidly moving Golgi. We therefore favor the occurrence of two types of delivery events: one in which POM2/CSI1 molecules already are associated with CESAs prior to insertion (possibly defined by the smaCCs/MASCs) and another in which POM2/CSI1 molecules continuously associate with the already delivered CESAs (presumably delivered directly from the Golgi). While some POM2/CSI1s were codelivered with the CESAs, we did not observe an alteration in CESA delivery rate in the pom2/csi1 mutants compared with the wild type. CESA insertions preferentially coincide with cortical microtubules (Gutierrez et al., 2009). Considering the importance of POM2/CSI1 for coalignment of the microtubules and CESAs, it appeared plausible that POM2/CSI1 could establish an initial contact point between the CESAs and microtubules prior to insertion (e.g., via the smaCCs/MASCs association). Nevertheless, we observed that the CESA insertion coincided with microtubules also in the absence of POM2/CSI1. In addition, the absence of POM2/CSI1 also did not affect the behavior of the microtubule-tethered smaCCs/MASCs (see Supplemental Figure 8 online). We therefore propose that the mechanisms governing the insertion of the CESAs and of the motility patterns of the smaCCS/MASCs are different from the mechanism of microtubule-based guidance of the plasma membrane–located CESAs.
In summary, our results show that POM2/CSI1 can associate with CESA molecules that are actively synthesizing cellulose and CESA-containing compartments at the cell cortex. This association does not contribute to the rate of CESA insertions or to the localized insertions of CESAs adjacent to cortical microtubules. Rather, POM2/CSI1 is required for the guidance of CESAs along microtubules during cellulose synthesis.
METHODS
Plant Material
pom2-1 and pom2-2 were previously isolated in a screen for root morphogenesis mutants (Hauser et al., 1995). pom2-3 was a donation of T. Wada and was isolated from an ethyl methanesulfonate–mutagenized population in Ler. pom2-4 was isolated from a T-DNA–mutagenized progeny of a cross between Col and Nossen kindly provided by Christian Luschnig. pom2-5 (SALK_047252), pom2-6 (SALK_051146), pom2-7 (SALK_115451), pom2-8 (SALK_136239), csi2 (SALK_022491 and SALK_024648), and csi3 (SALK_009141 and SALK_009157) are T-DNA insertion lines from the Arabidopsis Stock Center (http://www.Arabidopsis.org/; Alonso et al., 2003). The marker line expressing cycB1;1:CDB:GUS was a gift from John Celenza. Col-0 seeds containing the constructs mCherry:TUA5 and GFP:CESA3 as well as tdTomato:CESA6 and MAP4:GFP (Desprez et al., 2007; Gutierrez et al., 2009) were kindly donated by Ryan Gutierrez, David Ehrhardt, and Herman Höfte.
Growth Conditions and Phenotypic Characterization
Seedlings were cultivated after stratification for at least 2 d at 4°C vertically on sterile nutrient plates according to Hauser et al. (1995). Plants were propagated on soil with a 16-h-light/8-h-dark cycle according to Hauser et al. (1995). Homozygous plants of pom2-1, pom2-2, and pom2-4 were used as female parents for crosses. Reciprocal crosses were only successful with pom2-3. For hypocotyl measurements under skotomorphogenic conditions, nutrient agar plates with sterile seeds were wrapped in aluminum foil. After 4 d at 4°C of stratification, the covered plates were transferred to 22°C and incubated for another 7 d. Cell walls were visualized with propidium iodine (10 μg/mL). Cell and organ sizes were measured using differential interference contrast microscopy on chloral hydrate–cleared preparations as described by Müller et al. (2002) on whole seedling or plant organ levels. Alternatively, seeds were sterilized and germinated on nutrient plates after stratification for at least 2 d at 4°C and measured after 5 d of growth at 22°C. For confocal microscopy of hypocotyls expressing diverse marker lines, seedlings were cultivated on Murashige and Skoog (MS) media in light (16-h photoperiod) or dark conditions at 21°C for 3 or 4 d as described by Sampathkumar et al. (2011).
Drug Treatments
Drug treatments were performed essentially as described by Sampathkumar et al. (2011). Seedlings were immersed in 2 mL of solution with drugs or a control solution in 12-well cell culture plates in the dark and were subsequently imaged. A stock solution of oryzalin was dissolved in DMSO, and working stocks were made fresh by further dilution in water (stock solutions; 20 mM oryzalin in DMSO and 20 μM isoxaben in water).
Meristem Activity and Pollen Germination
Meristem activity was quantified in the GUS-stained seedlings of crosses between different pom2 mutants and the cycB1;1:CDB:GUS according to Hauser and Bauer (2000). Histochemical GUS staining was done as described by Hauser and Bauer (2000). For pollen germination assays (Azarov et al., 1990), anthers of soil-grown plants were harvested at different developmental stages. Pollen was applied to a microscope slide covered with a thin layer of pollen germination medium (3% [w/v] agar, 25% [w/v] Suc, 0.7 mM CaCl2, 1 mM MgSO4, 2 mM Ca[NO3]2, 0.25 mM K2HPO4, 1 mM K2SO4, and 8 mM H3BO3 adjusted to pH 7.4). After incubation in a humid chamber at 24°C for ~17 h, pollen grains were considered as germinated if the pollen tube length exceeded one grain diameter.
Mounting of Seedlings for Confocal Microscopy
Seedlings were mounted in half-strength MS medium supplemented with 1% (w/v) Suc and buffered with 10 mM MES, pH 5.7, or between a cover glass and a 1-mm thick 1% agar pad affixed on a circular cover slip (Roth), thus stabilizing the sample and preventing it from compression and mechanical damage.
Genetic Mapping and Chromosomal Walking toward the pom2 Locus
Mapping was performed with the F2 progeny of a cross between pom2-3 and Wassilewskija (Ws). The phenotypes of >2000 F2 seedlings were scored on nutrient agar plates. A total of 511 pom2 seedlings were used for fine mapping with the microsatellite marker F14M13 and the duplex analysis markers F7D8_91, T16B14_20, T26C19_19, and T26C19_43 (Hauser et al., 1998; see Supplemental Table 1 online). Genomic DNA isolation and PCR analyses were performed as described by Hauser et al. (1998). Allele-specific polymorphisms were detected with DNA gel blot analysis of NcoI- and EcoRV-digested genomic DNA and probed with the digoxigenin-labeled BAC clone T16B14 as described by Adhami et al. (1999). Overlapping PCR fragments of the genomic region around the polymorphic NcoI site from wild-type Col, Ler, pom2-1, pom2-2, pom2-3, and pom2-4 and of the cDNA clones RZL02E12, APZL03F11, and RZL15D2 were sequenced on the ABI-Prism 310 genetic analyzer using the BigDye terminator cycle sequencing chemistry (Perkin-Elmer, Applied Biosystems, and Amersham). Ambiguous exon/intron borders were confirmed by RT-PCR (for primers, see Supplemental Table 2 online) and sequencing. pom2-1 has a 3.9-kb deletion from amino acid 517 in exon IV to amino acid 1755 in exon V, causing a predicted protein of 912 amino acids; pom2-2 has a short AATGCGTGAG deletion in exon IV that causes a frame shift and leads to a stop codon at amino acid 119; pom2-3 has a single base pair deletion at the beginning of exon V leading to a frame shift and a premature stop codon at amino acid 1108; pom2-4 has a 2446-bp deletion starting in exon IV and ending in the adjacent gene (see Figure 1E for schematic view).
Cosmid Library Screen, Transformations, and Constructs
A cosmid carrying a 14.5-kb genomic region of the POM2 locus was isolated from a genomic Ler library constructed in pBIC20 (Meyer et al., 1996) and used to transform the pom2-3 allele and wild-type controls by the floral dipping method of Clough and Bent (1998). T1 transformants were selected on MS plates containing 100 μg mL−1 kanamycin. The segregation of the resistance and the phenotype was scored in 13 independent T2 lines. POM2/CSI1cDNA was amplified from the full-length cDNA containing clone RZL02e12 (obtained from the Kazusa DNA Research Institute, Japan; see Supplemental Table 3 online) and transferred to the vector pENTRY/SD/D Topo (Invitrogen), followed by homologous recombination to the destination vector pGBPGWC using Gateway technology (Invitrogen). Col-0 plants expressing POM2:CFP were generated by Agrobacterium tumefaciens–mediated transformation (Clough and Bent, 1998). T1 transgenic plants were selected on kanamycin and resistant progeny was used for spinning disc confocal microscopy as described by Sampathkumar et al. (2011).
Microscopy
Light microscopy was performed using Leica stereomicroscopes (MZ12.5 or MZFLIII and the digital camera DFC420 or DC500). Microtubules were visualized in the F2 progeny of crosses between pom2-1, pom2-3, pom2-4, and the MAP4:GFP marker line with a confocal laser scanning microscope (Leica TCS-SP2). Seedlings expressing mCherry:TUA5, POM2:CFP, and dual-labeled lines of GFP:CESA3 and mCherry:TUA5, POM2:CFP, and mCherry:TUA5 as well as tdTomato:CESA6 and POM2:CFP were imaged on a confocal microscope equipped with a CSU-X1 Yokogawa spinning disc head fitted to a Nikon Ti-E inverted microscope, a CFI APO TIRF 3100 oil immersion objective (numerical aperture of 1.49), an Evolve charge-coupled device camera (Photometrics Technology), and a 31.2 lens between the spinning disc and camera. GFP was excited at 491 nm, mCherry at 561 nm, and CFP at 405 nm using a multichannel dichroic and an ET455/50M, ET525/50M, or ET595/50M band-pass emission filter (Chroma Technology) for CFP, GFP, and mCherry/tdTomato, respectively. Image acquisitions were performed using Metamorph online premier, version 7.5. Typical exposure times were 600 ms for GFP and 300 ms for mCherry. Environmental scanning electron microscopy was performed as described by Persson et al. (2007).
General Image Processing and Analysis
All images were processed using ImageJ software (W.S. Rasband; National Institutes of Health). Background correction was performed using the “subtract background” tool (rolling ball radius 30 to 40 pixels), and StackReg was used to correct focus drift. Linear adjustments in pixel values were made when measuring signal intensities. Microtubule angles were measured with regard to the growth axis and always in a clockwise direction using the angle tool in ImageJ.
Colocalization Analysis
All colocalization analyses were done with the Jacop plugin for ImageJ (Bolte and Cordelières, 2006). Pearson coefficients were calculated for the time-averaged signals of GFP:CESA3 and TUA5:mCherry as well as POM2:CFP and GFP:CESA3 and compared with randomized images using Costes randomization and automatic threshold (Costes et al., 2004). Colocalization analysis of POM2:CFP and mCherry:TUA5, as well as POM2:CFP and tdTomato:CESA6, was performed using the van Steensel’s cross-correlation function mechanism, which determined the degree of co-occurrence of the fluorescent signals of two channels by shifting the corresponding channels in relation to each other in the x direction (shift = 20 pixels; van Steensel et al., 1996). Correlation maxima at a shift of 0 pixels indicate that colocalization of both channels is apparent for most fluorescent foci.
Analysis of Fluorescent Particle and Microtubule Dynamics Using Kymographs and Three-Dimensional Heat Maps
Kymographs were made using the multiple Kymograph plugin for ImageJ (http://www.embl.de/eamnet/html/body_kymograph.html), applying a line width of three pixels. Slopes of the kymographs were measured with the angle tool in ImageJ. POM2 and CESA particle velocities as well as microtubule growth and shrink velocities were calculated from the slopes of the kymographs as follows:
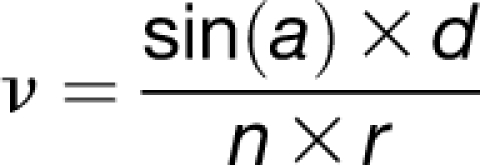 |
where ν = velocity (μm/min), a = angle between fluorescent trace and orthogonal axis (time axis) of kymograph (rad), d = distance of movement (μm), n = number of frames, and r = frame rate (min). Three-dimensional heat maps of kymographs were generated using the Interactive 3D Surface Plots plugin (http://rsbweb.nih.gov/ij/plugins/surface-plot-3d.html). CESA tracks were defined as trajectories of CESAs during steady movement (seen as lines in time average images). CESA particles were considered to be associated with microtubules if they tracked (with steady motion) along microtubule fluorescent signal over a time frame of 120 frames (5-s intervals) = 10 min. Microtubule angles were measured as described by Sampathkumar et al. (2011). The ImageJ angle tool was used to determine the angle between the microtubule and the longitudinal axis of the cell.
Analysis of CESA Delivery Sites at Cortical Microtubules
Delivery events of CESAs were analyzed as described by Gutierrez et al. (2009). Wild-type plants and mutant plants for pom2-4 and -5 expressing GFP:CESA3 and mCherry:TUA5 were bleached with a 2-s, 405-nm laser pulse, followed by image acquisition in 5-s time intervals over 300 frames. Photobleaching was performed by a fluorescence recovery after photobleaching/photo ablation system (Roper Scientific) integrated into the setup above.
GFP:CESA3 and POM2:CFP Density Measurements and Delivery Rates
Particle numbers of tdTomato:CESA6 and POM2:CFP were determined using the particle tracker plugin for ImageJ (Sbalzarini and Koumoutsakos, 2005) combined with counting by eye. In the case of CESA density quantification, areas covered with background fluorescence caused by Golgi vesicles were not taken into consideration. CESA insertion rates were determined by calculating the slope of CESA density (particles/μm2) plotted against bleach recovery time (min) for at least three biological replicates through regression of the linear phase of recovery.
qRT-PCR Analysis of T-DNA Insertion Lines
The qRT-PCR reaction was performed using the primers presented in Supplemental Tables 2 and 3 online. Power SYBRGreen (Agilent) was used as the detector. Reactions were run on an ABI Prism 7900 HT fast real-time PCR system (Applied Biosystems). mRNA from Arabidopsis thaliana leaves was extracted using the RNAeasy extraction kit (Qiagen) following the supplier’s instructions. cDNA synthesis was performed using the Superscript III reverse transcription kit (Invitrogen). Cycle threshold values were normalized to the housekeeping gene UBI10 (Czechowski et al., 2005). Alternatively, total RNA of seedlings and organs was isolated according to the manufacturers’ protocols using TRI reagent (Molecular Research Center). After DNase I (Roche) treatment, 2.5 to 5 μg total RNA was reverse transcribed with Superscript I (Invitrogen) and 2 μM oligo(dT)18 primers (Karsai et al., 2002). Absolute and relative copy numbers of individual mRNA species were calculated with standard curves of known molar concentrations for each of the genes. The expression was normalized to the housekeeping gene TUB9. At least three different RNA isolations and cDNA synthesis were used for quantification, and each cDNA was measured in triplicate. Primers, gene name, and the size of genomic and cDNA amplicons are listed in Supplemental Tables 1 and 2 online. In silico expression analysis was done using the Web-based program Spot History with two clones on the AFGC chip (ID132C7T7 and ID192M4T7) and one clone on the Affimetrix chip (SpotID 418556 as At2g22125).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: POM2/CSI1 (At2g22125), CSI2 (At1g44120), CSI3 (At1G77460), CESA6 (At5g64740), CESA3 (At5g05170), TUA5 (At5g19780), T26C19 (AF076243), and T16B14 (AC007232).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phenotypes of pom2 and Wild-Type Seedlings after Germination on Medium Supplemented with 4.5% Suc.
Supplemental Figure 2. pom2 Mutant Phenotypes.
Supplemental Figure 3. pom2 Mutants Are Impaired in Meristematic Activity and Microtubule Organization.
Supplemental Figure 4. Schematic Representation of the Positional Cloning Strategy of POM2 and qPCR Analyses.
Supplemental Figure 5. POM2/CSI1:CFP Behavior and Colocalization with CESA Complexes.
Supplemental Figure 6. Co-Occurrence of POM2/CSI1 and Microtubules at the Cell Cortex in Etiolated Hypocotyls.
Supplemental Figure 7. POM2/CSI1 Only Moves Together with Depolymerizing Microtubule Ends, and Delivery of CESAs and POM2/CSI1 to the Plasma Membrane.
Supplemental Figure 8. Mutations in POM2/CSI1 Have No Visible Effect on the Attachment of smaCCs/MASCs to Microtubules.
Supplemental Table 1. Summary of the Molecular Markers Used for Fine Mapping the POM2 Locus.
Supplemental Table 2. Primers Used for Sequencing and Real-Time PCR Analyses.
Supplemental Table 3. Primers for POM2 Cloning, Genotyping, and qRT-PCR Analysis.
Supplemental Movie 1. POM2/CSI1 Can Track along Depolymerizing Microtubule Ends.
Supplemental Movie 2. POM2/CSI1 Collecting Other POM2/CSI1s.
Supplemental Movie 3. POM2/CSI1 Is Only Associated with Depolymerizing Microtubule Ends.
Supplemental Movie 4. Coalignment of CESAs and Cortical Microtubules Is Lost in pom2/csi1 Mutants.
Acknowledgments
We thank Takuji Wada for the pom2-3 allele, Christian Luschnig for the pom2-4 allele, Ryan Gutierrez and Dave Ehrhardt for tdTomato:CESA6, and John Celenza for the cycB1;1:CDB:GUS marker line. We also thank Nicole Schlager, Gernot Resch, Norma Funke, and Anja Froehlich for technical assistance, Thomas Nedoma for the initial mapping, Albert Karsai for T-DNA segregation analysis, and the other members of the lab for advice and patience. The cosmid library was a kind gift of Erwin Grill and Andreas Bachmair. We thank Thomas Herter and Yi Zhang for experimental support. We thank the Kazusa DNA Research Institute (RIKEN) for providing the cDNA clones of POM2. E.L. was supported by the Asia-Europe Forestry Exchange Program and Austria Science Foundation (FWF) Project P13353. T.K. was financed through the AKTION Österreich-Tschechische Republik Project 21p8. This project was further supported by the FWF Project P14477 to M.-T.H. M.B., A.S., and S.P. were funded through the Max-Planck Gesellschaft and through Deutsche Forschungsgemeinshaft Grant PE1642/5-1. This project was further supported by FWF Projects P14477 and F03707 to M.-T.H.
AUTHOR CONTRIBUTIONS
M.B., M.-T.H., and S.P. designed the research. M.B., E.L., A.S., T.K., M.-T.H., and S.P. performed the research. M.B., E.L., A.S., M.-T.H., and S.P. analyzed data. M.B., A.S., M.-T.H., and S.P. wrote the article.
References
- Adhami F., Müller S., Hauser M.-T. (1999). Nonradioactive labeling of large DNA fragments for genome walking, RFLP and northern blot analysis. Biotechniques 27: 314–320 [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Ambrose J.C., Shoji T., Kotzer A.M., Pighin J.A., Wasteneys G.O. (2007). The Arabidopsis CLASP gene encodes a microtubule-associated protein involved in cell expansion and division. Plant Cell 19: 2763–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arioli T., et al. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279: 717–720 [DOI] [PubMed] [Google Scholar]
- Azarov A.S., Tokarev B.I., Netchepurenko A.E. (1990). Effect of sodium chloride on pollen germination and pollen tube growth in vitro in Arabidopsis thaliana (L.) Heynh. Arabidopsis Inf. Serv. 27. http://www.arabidopsis.org/ais/1990/azaro-1990-aacdq.html
- Baskin T.I. (2001). On the alignment of cellulose microfibrils by cortical microtubules: A review and a model. Protoplasma 215: 150–171 [DOI] [PubMed] [Google Scholar]
- Baskin T.I., Beemster G.T.S., Judy-March J.E., Marga F. (2004). Disorganization of cortical microtubules stimulates tangential expansion and reduces the uniformity of cellulose microfibril alignment among cells in the root of Arabidopsis. Plant Physiol. 135: 2279–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bichet A., Desnos T., Turner S., Grandjean O., Höfte H. (2001). BOTERO1 is required for normal orientation of cortical microtubules and anisotropic cell expansion in Arabidopsis. Plant J. 25: 137–148 [DOI] [PubMed] [Google Scholar]
- Bischoff V., Desprez T., Mouille G., Vernhettes S., Gonneau M., Höfte H. (2011). Phytochrome regulation of cellulose synthesis in Arabidopsis. Curr. Biol. 21: 1822–1827 [DOI] [PubMed] [Google Scholar]
- Bolte S., Cordelières F.P. (2006). A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224: 213–232 [DOI] [PubMed] [Google Scholar]
- Carpita N.C. (2011). Update on mechanisms of plant cell wall biosynthesis: How plants make cellulose and other (1->4)-β-D-glycans. Plant Physiol. 155: 171–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Ehrhardt D.W., Somerville C.R. (2010). Mutations of cellulose synthase (CESA1) phosphorylation sites modulate anisotropic cell expansion and bidirectional mobility of cellulose synthase. Proc. Natl. Acad. Sci. USA 107: 17188–17193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cosgrove D.J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Costes S.V., Daelemans D., Cho E.H., Dobbin Z., Pavlakis G., Lockett S. (2004). Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys. J. 86: 3993–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell E.F., Bischoff V., Desprez T., Rolland A., Stierhof Y.D., Schumacher K., Gonneau M., Höfte H., Vernhettes S. (2009). Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21: 1141–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T., Juraniec M., Crowell E.F., Jouy H., Pochylova Z., Parcy F., Höfte H., Gonneau M., Vernhettes S. (2007). Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler A., Persson S. (2011). Cellulose synthases and synthesis in Arabidopsis. Mol. Plant 4: 199–211 [DOI] [PubMed] [Google Scholar]
- Fagard M., Desnos T., Desprez T., Goubet F., Refregier G., Mouille G., McCann M., Rayon C., Vernhettes S., Höfte H. (2000). PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12: 2409–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P.B. (1962). Mechanism for plant cellular morphogenesis. Science 138: 1404–1405 [DOI] [PubMed] [Google Scholar]
- Gu Y., Kaplinsky N., Bringmann M., Cobb A., Carroll A., Sampathkumar A., Baskin T.I., Persson S., Somerville C.R. (2010). Identification of a cellulose synthase-associated protein required for cellulose biosynthesis. Proc. Natl. Acad. Sci. USA 107: 12866–12871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerriero G., Fugelstad J., Bulone V. (2010). What do we really know about cellulose biosynthesis in higher plants? J. Integr. Plant Biol. 52: 161–175 [DOI] [PubMed] [Google Scholar]
- Gutierrez R., Lindeboom J.J., Paredez A.R., Emons A.M., Ehrhardt D.W. (2009). Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11: 797–806 [DOI] [PubMed] [Google Scholar]
- Hauser M.T., Adhami F., Dorner M., Fuchs E., Glössl J. (1998). Generation of co-dominant PCR-based markers by duplex analysis on high resolution gels. Plant J. 16: 117–125 [DOI] [PubMed] [Google Scholar]
- Hauser M.T., Bauer E. (2000). Histochemical analysis of root meristem activity in Arabidopsis thaliana using a cyclin:GUS (β-glucuronidase) marker line. Plant Soil 226: 1–10 [Google Scholar]
- Hauser M.T., Morikami A., Benfey P.N. (1995). Conditional root expansion mutants of Arabidopsis. Development 121: 1237–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans C., Porco S., Vandenbussche F., Gille S., De Pessemier J., Van Der Straeten D., Verbruggen N., Bush D.R. (2011). Dissecting the role of CHITINASE-LIKE1 in nitrate-dependent changes in root architecture. Plant Physiol. 157: 1313–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsai A., Müller S., Platz S., Hauser M.T. (2002). Evaluation of a homemade SYBR green I reaction mixture for real-time PCR quantification of gene expression. Biotechniques 32: 790–792, 794–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S., Laosinchai W., Itoh T., Cui X., Linder C.R., Brown R.M., Jr (1999). Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant Vigna angularis. Plant Cell 11: 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V., Herrmann U., Parupalli C., Sedbrook J.C., Ehrhardt D.W., Hülskamp M. (2007). CLASP localizes in two discrete patterns on cortical microtubules and is required for cell morphogenesis and cell division in Arabidopsis. J. Cell Sci. 120: 4416–4425 [DOI] [PubMed] [Google Scholar]
- Lane D.R., et al. (2001). Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-beta-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol. 126: 278–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledbetter M.C., Porter K.R. (1963). A “microtubule” in plant cell fine structure. J. Cell Biol. 19: 239–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning B.D., Snyder M. (2010). Drivers and passengers wanted! The role of kinesin-associated proteins. Trends Cell Biol. 10: 281–289 [DOI] [PubMed] [Google Scholar]
- Marc J., Granger C.L., Brincat J., Fisher D.D., Kao Th., McCubbin A.G., Cyr R.J. (1998). A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 10: 1927–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K., Benning G., Grill E. (1996). Cloning of plant genes based on the genetic map position. In Genome Mapping in Plants, A.H. Patterns, ed (New York: Academic Press), pp. 137–154 [Google Scholar]
- Müller S., Fuchs E., Ovecka M., Wysocka-Diller J., Benfey P.N., Hauser M.T. (2002). Two new loci, PLEIADE and HYADE, implicate organ-specific regulation of cytokinesis in Arabidopsis. Plant Physiol. 130: 312–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol F., His I., Jauneau A., Vernhettes S., Canut H., Höfte H. (1998). A plasma membrane-bound putative endo-1,4-beta-D-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 17: 5563–5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagant S., Bichet A., Sugimoto K., Lerouxel O., Desprez T., McCann M., Lerouge P., Vernhettes S., Höfte H. (2002). KOBITO1 encodes a novel plasma membrane protein necessary for normal synthesis of cellulose during cell expansion in Arabidopsis. Plant Cell 14: 2001–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez A.R., Persson S., Ehrhardt D.W., Somerville C.R. (2008). Genetic evidence that cellulose synthase activity influences microtubule cortical array organization. Plant Physiol. 147: 1723–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez A.R., Somerville C.R., Ehrhardt D.W. (2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495 [DOI] [PubMed] [Google Scholar]
- Persson S., Paredez A., Carroll A., Palsdottir H., Doblin M., Poindexter P., Khitrov N., Auer M., Somerville C.R. (2007). Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 15566–15571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampathkumar A., Lindeboom J.J., Debolt S., Gutierrez R., Ehrhardt D.W., Ketelaar T., Persson S. (2011). Live cell imaging reveals structural associations between the actin and microtubule cytoskeleton in Arabidopsis. Plant Cell 23: 2302–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbalzarini I.F., Koumoutsakos P. (2005). Feature point tracking and trajectory analysis for video imaging in cell biology. J. Struct. Biol. 151: 182–195 [DOI] [PubMed] [Google Scholar]
- Schindelman G., Morikami A., Jung J., Baskin T.I., Carpita N.C., Derbyshire P., McCann M.C., Benfey P.N. (2001). COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 15: 1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook J.C., Ehrhardt D.W., Fisher S.E., Scheible W.R., Somerville C.R. (2004). The Arabidopsis sku6/spiral1 gene encodes a plus end-localized microtubule-interacting protein involved in directional cell expansion. Plant Cell 16: 1506–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw S.L., Lucas J. (2011). Intrabundle microtubule dynamics in the Arabidopsis cortical array. Cytoskeleton (Hoboken) 68: 56–67 [DOI] [PubMed] [Google Scholar]
- Somerville C. (2006). Cellulose synthesis in higher plants. Annu. Rev. Cell Dev. Biol. 22: 53–78 [DOI] [PubMed] [Google Scholar]
- Somerville C., Bauer S., Brininstool G., Facette M., Hamann T., Milne J., Osborne E., Paredez A., Persson S., Raab T., Vorwerk S., Youngs H. (2004). Toward a systems approach to understanding plant cell walls. Science 306: 2206–2211 [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Himmelspach R., Williamson R.E., Wasteneys G.O. (2003). Mutation or drug-dependent microtubule disruption causes radial swelling without altering parallel cellulose microfibril deposition in Arabidopsis root cells. Plant Cell 15: 1414–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Williamson R.E., Wasteneys G.O. (2000). New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant Physiol. 124: 1493–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunohara H., Kawai T., Shimizu-Sato S., Sato Y., Sato K., Kitano H. (2009). A dominant mutation of TWISTED DWARF 1 encoding an alpha-tubulin protein causes severe dwarfism and right helical growth in rice. Genes Genet. Syst. 84: 209–218 [DOI] [PubMed] [Google Scholar]
- Tewari R., Bailes E., Bunting K.A., Coates J.C. (2010). Armadillo-repeat protein functions: Questions for little creatures. Trends Cell Biol. 20: 470–481 [DOI] [PubMed] [Google Scholar]
- Thitamadee S., Tuchihara K., Hashimoto T. (2002). Microtubule basis for left-handed helical growth in Arabidopsis. Nature 417: 193–196 [DOI] [PubMed] [Google Scholar]
- van Steensel B., van Binnendijk E.P., Hornsby C.D., van der Voort H.T., Krozowski Z.S., de Kloet E.R., van Driel R. (1996). Partial colocalization of glucocorticoid and mineralocorticoid receptors in discrete compartments in nuclei of rat hippocampus neurons. J. Cell Sci. 109: 787–792 [DOI] [PubMed] [Google Scholar]
- Wang J., Howles P.A., Cork A.H., Birch R.J., Williamson R.E. (2006). Chimeric proteins suggest that the catalytic and/or C-terminal domains give CesA1 and CesA3 access to their specific sites in the cellulose synthase of primary walls. Plant Physiol. 142: 685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasteneys G.O., Fujita M. (2006). Establishing and maintaining axial growth: wall mechanical properties and the cytoskeleton. J. Plant Res. 119: 5–10 [DOI] [PubMed] [Google Scholar]
- Zhang M., Zhang B., Qian Q., Yu Y., Li R., Zhang J., Liu X., Zeng D., Li J., Zhou Y. (2010). Brittle Culm 12, a dual-targeting kinesin-4 protein, controls cell-cycle progression and wall properties in rice. Plant J. 63: 312–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Burk D.H., Morrison W.H., III, Ye Z.H. (2002). A kinesin-like protein is essential for oriented deposition of cellulose microfibrils and cell wall strength. Plant Cell 14: 3101–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]