Leguminous plants have mutualistic symbiotic relationships with rhizobial bacteria and arbuscular mycorrhizal fungi. These two distinct symbioses share calcium calmodulin–dependent protein kinase (CCaMK), a key regulator for accommodation of bacterial and fungal symbionts. This article demonstrates that the two symbioses are distinguished by differential regulation of CCaMK by calmodulin binding.
Abstract
Ca2+/calmodulin (CaM)–dependent protein kinase (CCaMK) is a key regulator of root nodule and arbuscular mycorrhizal symbioses and is believed to be a decoder for Ca2+ signals induced by microbial symbionts. However, it is unclear how CCaMK is activated by these microbes. Here, we investigated in vivo activation of CCaMK in symbiotic signaling, focusing mainly on the significance of and epistatic relationships among functional domains of CCaMK. Loss-of-function mutations in EF-hand motifs revealed the critical importance of the third EF hand for CCaMK activation to promote infection of endosymbionts. However, a gain-of-function mutation (T265D) in the kinase domain compensated for these loss-of-function mutations in the EF hands. Mutation of the CaM binding domain abolished CaM binding and suppressed CCaMKT265D activity in rhizobial infection, but not in mycorrhization, indicating that the requirement for CaM binding to CCaMK differs between root nodule and arbuscular mycorrhizal symbioses. Homology modeling and mutagenesis studies showed that the hydrogen bond network including Thr265 has an important role in the regulation of CCaMK. Based on these genetic, biochemical, and structural studies, we propose an activation mechanism of CCaMK in which root nodule and arbuscular mycorrhizal symbioses are distinguished by differential regulation of CCaMK by CaM binding.
INTRODUCTION
Legumes have developed a unique mechanism of nitrogen fixation by symbiosis with soil bacteria that are collectively termed rhizobia. The symbiotic interaction between legumes and rhizobia is initiated by the recognition of plant-derived flavonoids by rhizobia, which induces the biosynthesis and secretion of lipochito-oligosaccharide signal molecules, Nod-factors (NFs), from rhizobia. NFs trigger the rhizobial infection process through root hairs and trigger cortical cell division in compatible host legumes, leading to the formation of root nodules (RNs), in which the symbiotic rhizobia reside and fix atmospheric nitrogen (Kouchi et al., 2010).
In addition to the nitrogen-fixing RN symbiosis, legumes are also able to establish endosymbiotic associations with arbuscular mycorrhizal (AM) fungi, which are found in more than 80% of land plants. Plant-derived strigolactones stimulate spores of AM fungi to germinate and fungal hyphae to branch and contact host roots (Akiyama et al., 2005). AM fungi secrete a mixture of sulfated and nonsulfated lipochito-oligosaccharide signals (LCOs), so-called Myc-LCOs, to stimulate formation of AM in host roots (Maillet et al., 2011). Inside the roots, the fungi penetrate the cortex and develop an intracellular hyphal network and branched fungal structures that are termed arbuscules, where nutrient exchange takes place between the fungi and the host plant (Bonfante and Genre, 2010).
Molecular genetic studies using two model legumes, Lotus japonicus and Medicago truncatula, have led to the identification of many genes that are essential for NF perception and subsequent signal transduction, leading to nodulation (Oldroyd and Downie, 2008; Kouchi et al., 2010). Lys motif (LysM) receptor–like kinases are believed to be NF receptors. In L. japonicus, two LysM receptor–like kinases, NFR1 and NFR5, have been proven to be responsible for the perception of NFs secreted by Mesorhizobium loti (Madsen et al., 2003; Radutoiu et al., 2003, 2007). Among the components involved in NF signaling, some are also required for symbiosis with AM fungi, indicating the presence of a common symbiosis pathway that is shared between RN and AM symbioses. Thus far, the following components of the common symbiosis pathway have been identified: SYMRK/DMI2 (Endre et al., 2002; Stracke et al., 2002), CASTOR and POLLUX/DMI1 (Ané et al., 2004; Imaizumi-Anraku et al., 2005), NUP133 (Kanamori et al., 2006), NUP85 (Saito et al., 2007), NENA (Groth et al., 2010), CCaMK/DMI3 (Lévy et al., 2004; Tirichine et al., 2006), and CYCLOPS/IPD3 (Messinese et al., 2007; Yano et al., 2008).
Ca2+ spiking, which involves an oscillation in cytoplasmic Ca2+ concentrations in the perinuclear region, is one of the earliest physiological responses of legumes upon the perception of NFs and Myc factors and has been shown to be important for both RN and AM symbioses (Ehrhardt et al., 1996; Miwa et al., 2006; Kosuta et al., 2008; Chabaud et al., 2011; Maillet et al., 2011). Among the components of the common symbiosis pathway identified thus far from L. japonicus, SYMRK, CASTOR, POLLUX, NUP133, and NUP85 are positioned upstream of Ca2+ spiking and are believed to be required for generating Ca2+ spiking. By contrast, Ca2+/calmodulin (CaM)–dependent protein kinase (CCaMK) and CYCLOPS are positioned downstream of the Ca2+ spiking (Miwa et al., 2006). Loss-of-function mutants of CCaMK are completely defective in both RN and AM symbioses (Lévy et al., 2004; Tirichine et al., 2006), whereas gain-of-function mutation results in spontaneous nodule formation in the absence of rhizobia (Gleason et al., 2006; Tirichine et al., 2006). These findings, together with recent genetic studies using gain-of-function CCaMK in various symbiosis-defective mutant backgrounds (Hayashi et al., 2010; Madsen et al., 2010), indicate that CCaMK is a key regulator of nodule formation and infection by symbiotic microbes. Because CCaMK shows structural similarity to the animal Ca2+/CaM-dependent protein kinase II (CaMKII), which is activated by cytosolic Ca2+ oscillation in a frequency-dependent manner (De Koninck and Schulman, 1998), CCaMK therefore seems to be a strong candidate as the decoder of microsymbiont-induced Ca2+ signals.
CCaMK was first isolated from lily (Lilium longiflorum) as a chimeric Ca2+/CaM-dependent protein kinase that is found only in plants (Patil et al., 1995; Yang and Poovaiah, 2003). Its biochemical properties in relation to Ca2+ and CaM have been studied in detail (Patil et al., 1995; Takezawa et al., 1996; Ramachandiran et al., 1997; Sathyanarayanan et al., 2000). Based on the characterization of CCaMK, including in vitro phosphorylation and ligand binding assays with a CCaMK deletion series, a working model for CCaMK activation has been proposed (Sathyanarayanan et al., 2000). Additional studies have been made of the biochemical and expression properties of CCaMK proteins in nonlegumes (Liu et al., 1998; Poovaiah et al., 1999; Pandey and Sopory, 2001); however, the detailed mechanism of CCaMK activation in response to cytosolic Ca2+ signals remains unclear.
In this study, we performed detailed complementation analyses of the ccamk loss-of-function mutant of L. japonicus with various kinds of mutated CCaMK to clarify the in vivo activation mechanism of CCaMK in the signaling pathway of legumes, leading to RN and AM symbioses. Based on our results, we propose a model for CCaMK activation in symbiotic signal transduction with a special emphasis on the significance of CCaMK functional domains.
RESULTS
Contributions of the EF-Hand Motifs and the Kinase Domain in Rhizobial Infection
CCaMK proteins contain a catalytic (Ser/Thr kinase) domain, a CaM binding regulatory domain (CaMBD) with strong similarity to the CaMBD of animal CaMKII, and a visinin-like Ca2+ binding domain that includes three canonical EF-hand motifs (Patil et al., 1995; Gleason et al., 2006; Tirichine et al., 2006). It is generally accepted that the CaMBD in CCaMK overlaps an autoinhibitory domain that keeps CCaMK inactive as its ground state (Sathyanarayanan et al., 2000). To investigate the roles of these functional domains in the activation of CCaMK, we created various deletion constructs of L. japonicus CCaMK that lacked one to three of the EF-hands (1-471, 1-429, and 1-340) and construct 1-314, which retains only the kinase domain (Figure 1). These constructs were placed under the control of the cauliflower mosaic virus 35S promoter and transformed into ccamk-3 mutant plants that were defective in both rhizobial and AM fungal infection (Tirichine et al., 2006) by Agrobacterium rhizogenes–mediated hairy root transformation.
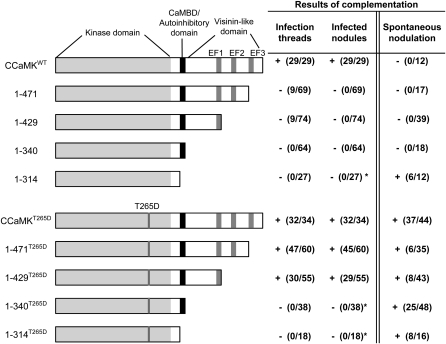
Figure 1.
Complementation of Nodulation on the ccamk-3 Mutant by a Deletion Series of CCaMK Constructs.
Illustrations of the structure of the wild type and deletion series of CCaMK constructs, with the names of the deletion constructs assigned according to the amino acid region of CCaMK. The numbers in the table indicate the number of plants that showed infection threads and the formation of infected nodules after M. loti inoculation, followed by the total number of plants tested. The number of plants that formed spontaneous nodules in the absence of M. loti is also shown. * indicates that empty nodules without rhizobial invasion were formed.
In the complementation tests, fully mature nodules were formed only in roots that expressed the wild-type CCaMK (CCaMKWT); none of the deletion constructs could sustain rhizobial infection, because we observed no penetration of an infection thread into the cortex, except in rare cases (Figure 1). In the roots transformed with the kinase-only construct (1-314), however, nodule organogenesis was induced regardless of the presence or absence of rhizobia. By contrast, the construct with the kinase domain and CaMBD (1-340) did not induce spontaneous nodule formation, although the truncated 1-340 protein was expressed (Figure 1; see Supplemental Figure 1 online). These results indicate that the lack of CaMBD releases CCaMK from autoinhibition, thereby allowing nodule organogenesis on the roots of ccamk-3/1-314, and that the EF-hand motifs of CCaMK are indispensable for rhizobial infection.
We have previously reported that substitution of the putative autophosphorylation site, Thr (T), at position 265 of L. japonicus CCaMK by Asp (D) confers a gain-of-function mutation in CCaMK and can induce spontaneous nodule formation in L. japonicus (Banba et al., 2008; Hayashi et al., 2010). To obtain more insight into the mechanisms of CCaMK activation, we introduced this T265D substitution into the deletion series of CCaMK constructs and used them for complementation of ccamk-3. In contrast with the deletion series of CCaMKWT described above, the introduction of 1-429T265D and 1-471T265D restored the formation of mature nodules filled with endosymbiotic rhizobia, whereas the ccamk-3 roots transformed with 1-314T265D and 1-340T265D showed no bacterial invasion and consequently formed empty nodules (Figure 1). In the absence of symbiotic rhizobia, all the deletion constructs that contained the T265D mutation induced spontaneous nodule formation. These results indicate that the T265D mutation can compensate for the lack of the second and third EF-hand motifs in regard to rhizobial infection. In the case of nodule organogenesis, however, the lack of CaMBD and visinin-like domain motifs can be suppressed by the T265D mutation.
The Third EF-Hand Motif Predominantly Contributes to a Ca2+-Dependent Conformational Change in CCaMK
It is generally accepted that Ca2+ signal transmission is mediated by a conformational change that results when Ca2+ binds to EF-hand motifs (Lewit-Bentley and Réty, 2000; Grabarek, 2006). To verify whether Ca2+ binding to the EF-hand motifs leads to a conformational change in CCaMK, we analyzed the Ca2+-dependent mobility shift (Takezawa et al., 1996; Lin et al., 2002; Franz et al., 2011) of CCaMKWT and site-directed mutants of the visinin-like domain (amino acids 334 to 518). Among the four plant CCaMK proteins shown in Figure 2A, the amino acid sequences of the EF-hand motifs are highly conserved, especially in the 12-amino-acid stretch of the Ca2+ binding loops (Figure 2A, gray boxes). Glu (E) in the 12th (-Z) position (Figure 2A, arrowheads) is known to be critical for Ca2+ chelation, and indeed, a mutation in this residue has been proven to reduce the Ca2+ binding capacity of the EF hand (Babu et al., 1992; Busch et al., 2000; Ogasawara et al., 2008). Therefore, we introduced a mutation of Glu (E) to Gln (Q) at this position into each EF-hand motif of L. japonicus CCaMK in an attempt to abolish its Ca2+-dependent CCaMK function. This mutation is henceforth denoted “EQ”.
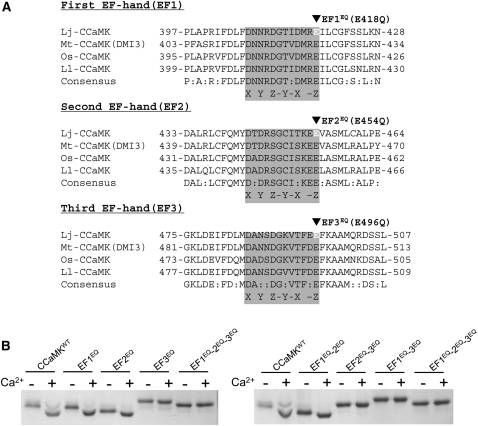
Figure 2.
Amino-Acid Alignments of the Three Canonical EF-Hand Motifs and Ca2+-Dependent Conformational Changes in the Visinin-Like Domain of CCaMK.
(A) Amino-acid alignments of the EF-hand motifs of four plant CCaMK proteins. Lj, L. japonicus; Mt, M. truncatula; Os, O. sativa; Ll, L. longiflorum. The residues responsible for Ca2+ binding (the Ca2+ binding loop) in the first, second, and third EF-hand motifs are shown in gray boxes, with the position of consensus coordinating Ca2+-ligating residues of the first (X), third (Y), fifth (Z), seventh (-Y), ninth (-X), and 12th (-Z). Black arrowheads indicate the residues in which the EQ mutation, from Glu (E) to Gln (Q), was introduced.
(B) The Ca2+-dependent mobility shift of the visinin-like domain proteins. The left gel shows the mobility shifts of the wild type and mutant visinin-like domain proteins harboring the EFEQ mutation(s) in single and triple combinations. The right gel shows the mobility shift of double and triple EFEQ mutations. Mobility shifts of the visinin-like domain proteins were analyzed by means of SDS-PAGE in the presence (+) and absence (-) of Ca2+ (i.e., in the presence of the chelator EGTA).
In the presence of Ca2+, the Escherichia coli-expressed recombinant visinin-like domain protein of CCaMKWT was shifted toward a lower molecular weight than in the presence of a Ca2+ chelator (EGTA), indicating that the visinin-like domain protein of CCaMKWT is responsible for the Ca2+-dependent conformational change (Figure 2B). Mutant proteins of either the first (EF1EQ) or the second EF-hand motif (EF2EQ) showed the same Ca2+-dependent change in mobility. However, the mobility shift was lost in the mutant harboring a mutation in the third EF-hand motif (EF3EQ) (Figure 2B). Furthermore, the Ca2+-dependent mobility shift was also lost in the double- and triple-mutant proteins that included a mutation in the third EF-hand motif (EF2EQ-3EQ, EF1EQ-3EQ, and EF1EQ-2EQ-3EQ) (Figure 2B). These results indicate that the third EF-hand motif predominantly contributes to the Ca2+-dependent conformational change of the visinin-like domain of CCaMK.
Point Mutations in the EF-Hand Motifs Abolish the Ability of CCaMK to Accommodate Rhizobia
To evaluate the effects of the EF-hand motifs on the ability of CCaMK to accommodate rhizobia, we introduced mutated CCaMK constructs that contained every possible combination of the EFEQ mutations. Transformation of constructs containing a single mutation (EF1EQ, EF2EQ, or EF3EQ) into ccamk-3 resulted in the development of normal infection threads and the formation of mature nodules (see Supplemental Figure 2 and Supplemental Table 1 online). This suggests that a mutation in a single EF-hand motif does not affect the ability of CCaMK to accommodate rhizobia.
We next introduced CCaMK constructs with two mutated EF-hand motifs. Defects in rhizobial infection in the ccamk-3 mutant were restored by EF1EQ-2EQ (Figures 3B, 3G, 3L, and 3Q), whereas EF2EQ-3EQ and EF1EQ-3EQ did not complement ccamk-3; neither infection threads nor bump-like structures formed (Figure 3). As expected, in the case of EF1EQ-2EQ-3EQ, rhizobial infection and nodulation did not occur (Figure 3), even though the protein was expressed stably in the transformed roots as well as CCaMKWT (see Supplemental Figure 1 online). Together with the results of the mobility-shift assay for CCaMK, the binding of Ca2+ to the third EF-hand seems to be crucial for CCaMK activation that leads to the accommodation of rhizobia.
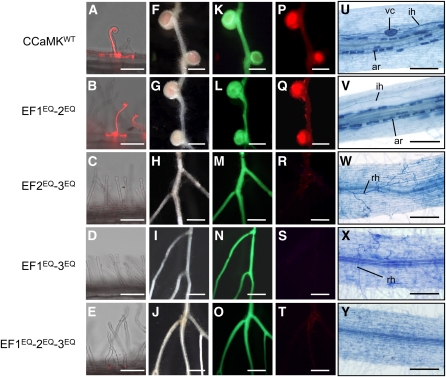
Figure 3.
Complementation of Nodulation and Mycorrhization of ccamk-3 by CCaMK Variants Containing Point Mutations in the EF-Hand Motifs.
(A) to (E) Root hairs of ccamk-3/CCaMKWT (A) and ccamk-3/CCaMK EFEQ mutants ([B] to [E]), shown as merged images of bright-field and fluorescence of DsRed.
(F) to (T) Nodulation of CCaMK-transformed hairy roots was observed using bright images ([F] to [J]) and fluorescence images ([K] to [T]) of GFP ([K] to [O]) and DsRed ([P] to [T]) as markers of transformed hairy roots and infected rhizobial bacteria, respectively.
(U) to (Y) Colonization of the roots by AM fungi in ccamk-3/CCaMKWT (U) and ccamk-3/CCaMK EFEQ mutants ([V] to [Y]), visualized using trypan blue staining. ar, arbuscule; ih, intracellular hyphae; rh, running hyphae; vc, vesicles.
Bars in (A) to (E) = 100 μm; bars in (F) to (T) = 1 mm; bars in (U) to (Y) = 200 μm.
Mutations in the EF-Hand Motifs of CCaMK Adversely Affect Mycorrhization
CCaMK is one of the components of a common symbiosis pathway that is essential for both RN and AM symbiosis. To investigate the significance of the EF-hand motifs of CCaMK in its activation of AM symbiosis, we examined the ability of a series of CCaMK mutations to regulate AM fungi infection. Upon inoculation with Glomus intraradices, ccamk-3/CCaMKWT roots were densely colonized by AM fungi, including the development of intracellular hyphae, arbuscules, and vesicles (Figure 3U). The functional relevance of the EF hands for AM fungi infection was examined using the 1-340, EF2EQ-3EQ, EF1EQ-3EQ, or EF1EQ-2EQ-3EQ constructs, which encode loss-of-function mutations in the EF-hand motifs. On the roots of ccamk-3/1-340, many running hyphae were observed, but they could not penetrate the root tissues (see Supplemental Figure 3A online). Similarly, transformation of EF2EQ-3EQ, EF1EQ-3EQ, and EF1EQ-2EQ-3EQ into ccamk-3 resulted in a failure to restore AM fungi infection (Figures 3W to 3Y). By contrast, EF1EQ-2EQ restored both mycorrhization (Figure 3V) and rhizobial infection (Figure 3). These results indicate that the EF-hand motifs are also important for the function of CCaMK in promoting AM fungi infection.
The CCaMK EF-Hand Motifs Pair to Form Two EF-Hand Domains, Both of Which Are Responsible for Rhizobial Infection
EF-hand motifs mostly appear in pairs, and the adjacent motifs constitute a “two-EF-hand domain” that transduces Ca2+ signals into cell responses (Grabarek, 2006; Gifford et al., 2007). To identify functional pairs of EF-hand motifs in CCaMK, we performed computational modeling of the visinin-like domain of CCaMK with chicken myristoylated guanylate cyclase–activating protein1 (myrGCAP1) with Ca2+ bound, as a reference (PDB 2R2I.A) (Stephen et al., 2007). myrGCAP1 possesses four EF-hand motifs, one of which (EF1) is unable to bind Ca2+. In the predicted structure of CCaMK, the EF2 and EF3 motifs formed a typical EF-hand domain structure, with two EF-loops located close together. In addition to the EF-hand domain consisting of the EF2 and EF3 motifs, our molecular modeling also suggested that the EF1 motif formed another EF-hand domain with a neighboring upstream region, which includes a noncanonical EF-hand motif designated EF0 (Figures 4A and 4B). In general, interaction between the short antiparallel β-sheets formed in each EF-hand motif is important for stabilization of an EF-hand domain (Grabarek, 2006; Gifford et al., 2007). In the case of a typical EF-hand protein, Troponin C, substitution of conserved nonpolar amino acid at the eighth position with polar Gln led to decreased Ca2+ affinity and decreased cooperativity between the first and second EF-hand motifs, likely because of conformational changes in the Ca2+ binding loops (Tikunova et al., 2002). In our model, Ala378 (A) in the seventh position of EF0 and Ile414 (I) in the eighth position of EF1, both of which are nonpolar amino acids, were predicted to be positioned in the β-sheets of each EF-hand motif (Figure 4B, closed arrowheads). To test cooperative interaction between EF0 and EF1 motifs, we created constructs containing mutations of A378Q and I414Q, both of which are expected to disrupt the conformation of the EF0-EF1 domain. The ccamk-3 roots that expressed a single mutation (EF0AQ or EF1IQ) showed normal infection by rhizobia (see Supplemental Figure 2 online). On the roots of ccamk-3/EF0AQ-1IQ, however, many bumps with infection threads in the epidermis were formed, but most of the bumps failed to develop into mature nodules (Figures 4C to 4F), supporting our hypothesis that the EF0 and EF1 motifs form an EF-hand domain that also contributes to the biological activity of CCaMK. Our results demonstrate that two EF-hand domains of CCaMK (i.e., EF0-EF1 and EF2-EF3) function in rhizobial infection.
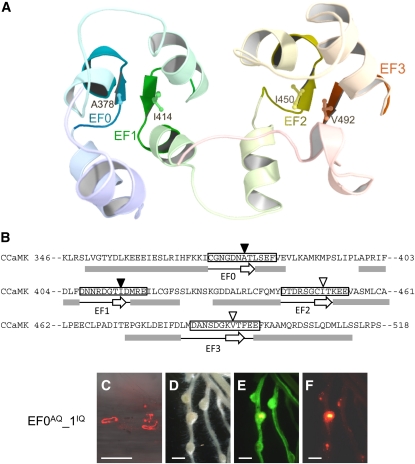
Figure 4.
Structural Model of the Visinin-Like Domain of CCaMK and the Adverse Effect on Rhizobial Infection Caused by the EF0AQ-EF1IQ Double Mutation.
(A) Two EF-hand domains, with EF0-EF1 and EF2-EF3 pairs. Hydrophobic amino acids, Ala378 in EF0, Ile414 in EF1, Ile450 in EF2, and Val492 in EF3, positioned in β-sheets, are highlighted.
(B) The amino-acid sequence of the visinin-like domain (amino acids 346 to 518) of CCaMK. Gray boxes and open arrows below the sequence represent the residues that encode the α-helix and β-sheet structures, respectively, predicted by the MOE software. The Ca2+ binding loop of each EF-hand motif is enclosed in a black box. Closed and open arrowheads indicate the hydrophobic amino acid residues.
(C) to (F) Nodulation phenotype observed in the roots of ccamk-3/EF0AQ-1IQ. Infection threads are shown as (C) merged images of bright field and fluorescent field of DsRed. Nodulation phenotypes were observed using bright-field images (D) and fluorescent field images of GFP (E) and DsRed (F).
Bar in (C) = 100 μm; bars in (D) to (F) = 1 mm.
Mutation in the CaMBD/Autoinhibitory Domain Alters CCaMK Activity
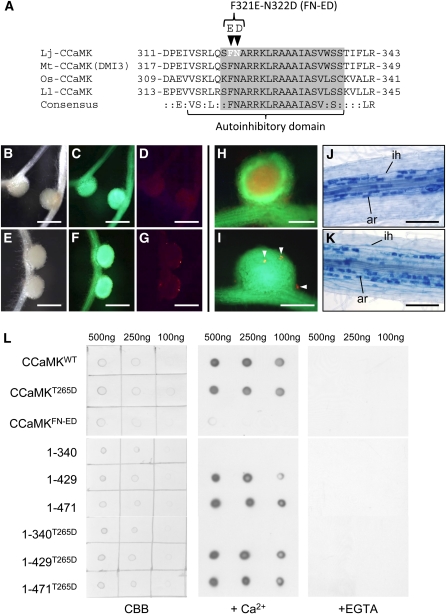
The CaMBD of lily CCaMK was identified by Takezawa et al. (1996), and examination of deletion mutants of lily CCaMK revealed the presence of an autoinhibitory domain of lily CCaMK that overlaps the CaMBD (Ramachandiran et al., 1997). Amino acid sequences around the CaMBD/autoinhibitory domain of several CCaMK proteins are highly conserved (Figure 5A); thus, we hypothesized that the 315 to 338 amino acid region of L. japonicus CCaMK would have a regulatory effect on its kinase domain.
Figure 5.
Biological Properties of CCaMKFN-ED and the CaM Binding Assay Using Mutated CCaMK Proteins.
(A) Amino-acid alignment of the CaMBD/autoinhibitory domain of four plant CCaMK proteins. Lj, L. japonicus; Mt, M. truncatula; Os, O. sativa; Ll, L. longiflorum. The CaMBD and autoinhibitory domains assigned by Ramachandiran et al. (1997) are indicated by the gray box and underline, respectively. Arrowheads above the alignment indicate the residues in which mutations (F321E and N322D) were introduced.
(B) to (G) Nodulation of ccamk-3/CCaMKFN-ED roots in the absence ([B] to [D]) and presence ([E] to [G]) of M. loti, observed as bright-field images (B) and (E) and fluorescence images of GFP ([C] and [F]) and DsRed ([D] and [G]).
(H) and (I) RN formed on ccamk-3/CCaMKWT (H) and ccamk-3/CCaMKFN-ED roots (I), shown as merged images of fluorescent field of GFP and DsRed. Arrowheads indicate microcolonies formed on ccamk-3/CCaMKFN-ED nodule.
(J) and (K) Mycorrhization of ccamk-3 roots transformed with CCaMKWT (J) or CCaMKFN-ED (K). ar, arbuscule; ih, intracellular hyphae.
(L) CaM binding assay of CCaMKWT, CCaMKT265D, CCaMKFN-ED (Top) and a deletion series of CCaMK proteins (Bottom). Images are (left) staining of spotted CCaMK proteins with Coomassie brilliant blue (CBB) and immunodetection of the binding of biotin-labeled CaM in the presence (middle) and absence (right) of Ca2+ (+EGTA). The concentrations of the CCaMK proteins spotted on membrane are shown above the images.
Bars in (B) to (G) = 1 mm; bars in (H) and (I) = 500 μm; bars in (J) and (K) = 200 μm.
Analyses of the biochemical properties of the CaMBD in CaMKII by means of site-directed mutagenesis revealed that substitutions of several amino acid residues conserved within CaMBD rendered Ca2+-independent activity to CaMKII (Yang and Schulman, 1999). Based on these findings, we introduced amino acid substitutions into the CaMBD of CCaMK, in which the residues Phe321 (F) and Asn322 (N) were replaced with Glu (E) and Asp (D), respectively, to create the FN-ED mutant (Figure 5A, arrowheads). On the roots of ccamk-3/CCaMKFN-ED, spontaneous nodule formation was induced (Figures 5B to 5D). Upon M. loti inoculation, however, rhizobial infection was not restored, and empty nodules formed (Figures 5E to 5G). Microcolonies and short infection threads were observed on the surface of roots or nodules at a low frequency (Figures 5H and 5I). These results suggest that the replacement of FN with ED in CaMBD confers Ca2+-independent activity on CCaMK that is sufficient for nodule organogenesis but not for rhizobial infection.
We next evaluated the functional significance of the CaMBD in AM symbiosis. Interestingly, CCaMKFN-ED in the ccamk-3 roots allowed successful infection by AM fungi (Figures 5J and 5K), that is, G. intraradices passed the epidermal cell layer in both intra- and intercellular fashion and formed fully developed arbuscules as well as those in the roots of ccamk-3/CCaMKWT (see Supplemental Figure 4 online). Thus, the FN-ED mutation in CaMBD does not seem to affect the ability of CCaMK to induce infection by AM fungi.
Mycorrhization Does Not Necessarily Require CaM Binding to CCaMK, But Rhizobial Infection Does Require CaM Binding
To address whether the interaction of CaM with CCaMK is required to establish rhizobial infection, we purified E. coli-expressed recombinant CCaMK variant proteins and analyzed their CaM binding ability. In the presence of Ca2+, both CCaMKWT and CCaMKT265D bound to CaM, but we did not detect CaM binding to CCaMKFN-ED (Figure 5L, top), indicating that the FN-ED mutation in the CaMBD leads to a loss of the ability of CCaMK to bind with CaM. We also analyzed the CaM binding activity in a CCaMK deletion series. The recombinant proteins 1-429/1-429T265D and 1-471/1-471T265D, which retain EF1 and EF1-EF2, respectively, bound to CaM, whereas 1-340/1-340T265D, which lacks the whole visinin-like domain, did not bind to CaM, even in the presence of Ca2+ (Figure 5L, bottom). The failure of 1-340 and 1-340T265D to bind with CaM demonstrates that the region of 341 to 429 amino acids, which contains noncanonical EF0 and canonical EF1 motifs, is essential for the CaM binding activity of CCaMK.
As described above, mycorrhization occurred normally in the roots of ccamk-3/CCaMKFN-ED (Figure 5K) and also in ccamk-3/1-340T265D roots (see Supplemental Figure 3B online), whereas infection by rhizobia was aborted on those roots (Figures 1 and 5E to 5G). Collectively, these results demonstrate that the differential regulation of CCaMK by CaM binding is a prerequisite for successful infection by rhizobia but not for mycorrhization.
The in Vitro Kinase Activity of CCaMK Variants and Their Ability to Restore the Symbiotic Defects of ccamk-3
To examine the correlation between the kinase activity of the mutated CCaMK proteins and their ability to restore symbiotic defects of ccamk-3, we performed an in vitro kinase assay using the recombinant CCaMK proteins. Following autophosphorylation in response to the addition of Ca2+, CCaMKWT showed increased substrate (myelin basic protein [MBP]) phosphorylation activity by addition of CaM (see Supplemental Figure 5A online). When we compared the kinase activity of CCaMKWT and CCaMKT265D, both the autophosphorylation and substrate phosphorylation activities of CCaMKT265D were clearly lower than those of CCaMKWT (see Supplemental Figure 5B online). In addition, the kinase activity of CCaMK proteins that lacked any of the EF-hand domains was significantly reduced compared with that of CCaMKWT (see Supplemental Figure 5B online), suggesting that the visinin-like domain is indispensable for kinase activity. By contrast, CCaMK proteins containing point mutations in any two of the three EF-hand motifs retained their kinase activity (see Supplemental Figure 5C online). In the case of CCaMKFN-ED, we detected no autophosphorylation or substrate phosphorylation activity (see Supplemental Figure 5C online).
To further assess the importance of the kinase activity of CCaMK, we analyzed the kinase activity of four recombinant CCaMK proteins that contained previously reported loss-of-function mutant alleles of CCaMK (Murray et al., 2006; Tirichine et al., 2006). As shown in Supplemental Figure 5D online, CCaMKG30E (a mutation in ccamk-3) and CCaMKG124D (a mutation in ccamk-5) did not phosphorylate the substrate, whereas CCaMKS25F (a mutation in ccamk-4) and CCaMKG204R (a mutation in ccamk-6) showed substrate phosphorylation activity comparable with that of CCaMKT265D (see Supplemental Figure 5D online).
CYCLOPS, one of the components of the common signaling pathway, has been identified as a putative in vivo phosphorylation target of CCaMK and seems to act together with CCaMK as a signaling complex in symbiosis (Yano et al., 2008). We analyzed the phosphorylation of recombinant CYCLOPS protein by various mutated CCaMK proteins (see Supplemental Figures 6 and 7 online). The results of the in vitro kinase assays with CYCLOPS were nearly identical to those with MBP, except that CCaMKT265D showed reduced phosphorylation of CYCLOPS compared with S25F and G204R (see Supplemental Figure 7A online).
We also analyzed in vitro interactions between CYCLOPS and mutated CCaMK proteins by yeast two-hybrid analysis. Indeed, in vitro interactions were essentially the same as the kinase activities (see Supplemental Figures 5 to 7 online). Thus, the in vitro kinase activities of the mutant CCaMK proteins and in vitro interactions with CYCLOPS do not seem to correlate well with the biological activities of the mutated CCaMK proteins.
Phosphorylation and Gain-of-Function Mutations Disrupt a Hydrogen Bond Network That Acts as a “Molecular Brake” on CCaMK
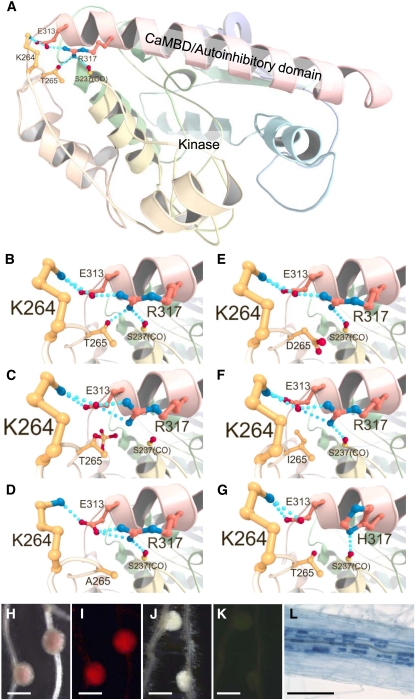
In addition to phosphomimetic CCaMKT265D, several mutated CCaMKs, such as CCaMKT265A (see Supplemental Figure 8 online) (Gleason et al., 2006), CCaMKT265I (Tirichine et al., 2006), and CCaMKFN-ED (Figure 5), showed gain-of-function activity in terms of spontaneous nodule formation. These mutated CCaMKs also have decreased kinase activity compared with CCaMKWT (see Supplemental Figure 8 online) (Tirichine et al., 2006), implying that the gain-of-function activity of the mutated CCaMKs is not defined by the presence of kinase activity. To obtain more insights into the mechanism by which these point mutations alter CCaMK function, we performed homology modeling of the CCaMK N-terminal domains (amino acids 9 to 340). Based on the similarity of amino acid sequence and the predicted secondary structure by Jpred3 (Cole et al., 2008), we selected the x-ray structure of Caenorhabditis elegans CaMKII (PDB 2BDW) (Rosenberg et al., 2005) as a reference. Because 2BDW represents an unphosphorylated, inactive form of CaMKII, this model is suitable for homology modeling of CCaMK to understand the structural differences between the unphosphorylated (inactive) and autophosphorylated (active) statuses of CCaMK. As shown in Figure 6A, the CCaMK N-terminal domains consist of a kinase domain followed by a long α helix in which the CaMBD/autoinhibitory domain is located (Figure 6A, in pink). In the unphosphorylated CCaMK model structure, a backbone atom of Ser237, side chains of Lys264, Thr265, Glu313, and Arg317 interact with one another through a network of hydrogen bonds (Figure 6B, dotted line). This network is predicted to be disrupted by autophosphorylation of Thr265 (Figure 6C). In addition, the disruption of the network is also predicted in all gain-of-function mutants, CCaMKT265A, CCaMKT265D, and CCaMKT265I (Figures 6D to 6F), suggesting that this hydrogen bond network among four residues acts as a molecular brake by which CCaMK is maintained in an inactive state. To further test the importance of this hydrogen bond network for CCaMK activation, we constructed a novel mutant, CCaMKR317H. In our model structure, Arg317 is engaged in hydrogen bonding with both Thr265 and Glu313, thus the replacement of Arg317 by His is expected to disrupt the hydrogen bond network (Figure 6G). Spontaneous nodule formation and rhizobial and AM fungi infections were observed on the roots of ccamk-3/CCaMKR317H (Figure 6), supporting our hypothesis that the hydrogen network is crucial for CCaMK regulation.
Figure 6.
Structural Models of the CCaMK N-Terminal Domain and Hydrogen Bond Network around Thr265.
(A) Ribbon diagram of unphosphorylated-CCaMKWT. CaMBD within a long α helix is shown in pink. Hydrogen bonds among Ser237(CO), Lys264, Thr265, Glu313, and Arg317 are indicated by dotted lines.
(B) to (G) Close-up views around the position 265 residue of the CCaMK models. (B) The hydrogen bond network of unphosphorylated-CCaMKWT. (C) Phosphorylation of CCaMK at Thr265 (CCaMKP-T265) resulted in the disruption of the hydrogen bond network. The disruptions of the hydrogen bond network are also predicted in CCaMKT265A (D), CCaMKT265D (E), and CCaMKT265I (F). A gain-of-function mutant CCaMKR317H in which the hydrogen bond network is predicted to be disrupted (G).
(H) to (L) Symbiotic phenotypes of ccamk-3/CCaMKR317H, with (H) and (I) or without (J) and (K) DsRed-labeled M. loti inoculation (visualized with bright field [H] and [J] and fluorescence of DsRed [I] and [K]). Upon G. intraradices inoculation, normal mycorrhization occurred (L).
Bars in (H) to (K) = 1 mm; bar in (L) = 200 μm.
The disruption of the hydrogen bond network was not predicted in the homology model of CCaMKFN-ED; however, Jpred3 predicted differences in secondary structures between CCaMKWT and CCaMKFN-ED. In the CCaMKWT, the starting position of CaMBD helix was Ile314, but FN to ED mutations resulted in a shift of the starting position to Glu321 (see Supplemental Figure 9 online). Because Arg317 occupies an intermediate position between Ile314 and Glu321, the hydrogen network seems to be disrupted in CCaMKFN-ED as a consequence of the improper helical conformation of CaMBD. In addition, the calmodulin target database program (Yap et al., 2000) predicted the region Gln319-Thr339 of CCaMKWT as a CaM binding motif, whereas the corresponding region of CCaMKFN-ED was not predicted as a CaM binding motif (see Supplemental Figure 9B online). Thus, the FN-ED mutation is assumed to cause both disruption of the molecular brake and the loss of CaM binding ability of CCaMKFN-ED.
Epistatic Relationships among the CCaMK Functional Domains
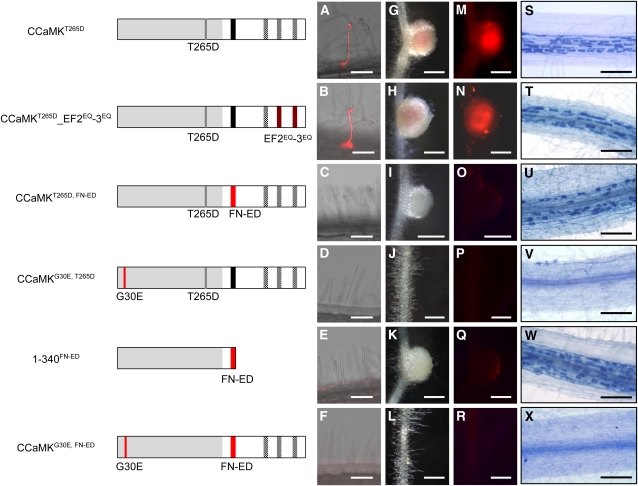
Thus far, we have shown the functional roles of the domains of CCaMK in RN and AM symbioses. To further understand the in vivo activation processes of CCaMK, it is necessary to analyze the functional relationship between the two CCaMK domains. To evaluate epistasis of the domains in RN symbiosis, we tested the activities of CCaMK containing combinations of the mutations described above. On the assumption that T265D substitution in the kinase domain mimics the autophosphorylation state of CCaMK, we constructed mutated CCaMK proteins that contained both T265D in the kinase domain and mutations in the other functional domains. In comparison with the EF2EQ-3EQ mutations alone, in which nodule formation and rhizobial infection did not occur (Figure 3), nodule formation accompanied by rhizobial infection occurred with a combination of T265D and the EF2EQ-3EQ mutation (CCaMKT265D_EF2EQ-3EQ) (Figure 7), indicating that T265D suppresses the loss of function of the EF-hand motifs. In addition, CCaMKT265D_EF1EQ-3EQ also restored the ability of ccamk-3 to form mature nodules (see Supplemental Table 1 online). Because the loss-of-function effect of the EF-hand motif mutations was suppressed by T265D, autophosphorylation of CCaMK, mimicked by T265D, seems to be epistatic to Ca2+ binding to the EF-hand motifs.
Figure 7.
Epistasis of the CCaMK Functional Domains.
(A) to (R) Illustrations of the structure of the CCaMK constructs used for the epistasis analyses. Complementation of rhizobial infection was evaluated based on the development of infection threads ([A] to [F]; shown as merged images of bright-field and DsRed fluorescence, a marker of infected rhizobial bacteria) and infected nodules ([G] to [R]; visualized with bright field ([G] to [L]) and DsRed fluorescence ([M] to [R]).
(S) to (X) Infection by AM fungi was analyzed by staining the intracellular arbuscules with trypan blue.
Bars in (A) to (F) = 100 μm; bars in (G) to (R) = 500 μm; bars in (S) to (X) = 200 μm.
With the combination of T265D and FN-ED (CCaMKT265D, FN-ED) in full-length CCaMK, neither infection threads nor infected nodules were observed upon M. loti inoculation, although nodule organogenesis was restored (Figure 7). These results indicate that the CaMBD suppresses the role of T265D in restoring rhizobial infection. The FN-ED mutation in 1-340 (1-340FN-ED), which lacks the entire visinin-like domain, resulted in phenotypes (Figure 7; see Supplemental Table 1 online) similar to those of ccamk-3/CCaMKFN-ED (Figure 5). These results indicate that the CaMBD is epistatic to the EF-hand domains in nodule organogenesis.
Recombinant CCaMK with the G30E mutation (CCaMKG30E) lacks kinase activity in vitro (see Supplemental Figures 5D and 7A online) (Tirichine et al., 2006). Expression of the construct that carries both the G30E and the T265D mutations (CCaMKG30E, T265D) in ccamk-3 could not complement defects in either rhizobial infection or nodule organogenesis (Figure 7). Similarly, the introduction of CCaMKG30E, FN-ED into ccamk-3 rescued neither M. loti infection nor nodule organogenesis (Figure 7). These results indicate that the gain-of-function mutations caused by T265D and FN-ED could not overcome the loss-of-function mutation caused by G30E in the kinase domain.
To understand whether infection by AM fungi is regulated by the same CCaMK activation process as that of rhizobia, we also examined epistatic relationships among the domains in AM symbiosis. When the T265D mutation was introduced in combination with mutations of the EF-hand motifs (CCaMKT265D_EF2EQ-3EQ and CCaMKT265D_EF1EQ-3EQ), G. intraradices infection was restored, as was the case for rhizobial infection (Figure 7T; see Supplemental Figure 10 online). Correspondingly, constructs carrying double mutations of G30E with either T265D (CCaMKG30E, T265D) or FN-ED (CCaMKG30E, FN-ED) did not restore AM fungi infection in ccamk-3 (Figures 7V and 7X), suggesting that the loss of function caused by the G30E mutation also adversely affects infection by AM fungi, regardless of the gain-of-function mutations conferred by T265D and FN-ED. In ccamk-3/CCaMKT265D, FN-ED roots, in which rhizobial infection was not restored, AM fungi could penetrate and develop arbuscules normally (Figure 7U). Furthermore, the introduction of FN-ED into CCaMK1-340, which could not itself restore AM fungi infection (see Supplemental Figure 3A online), resulted in normal mycorrhization (Figure 7W). Similar results were obtained for the ccamk-3/1-340T265D roots (see Supplemental Figure 3B online). Taken together, these results suggest that CCaMKFN-ED acts as a gain-of-function mutation for infection by AM fungi, even though it acts as a loss-of-function mutation in rhizobial infections (Figures 5 and 7).
DISCUSSION
To date, CCaMK proteins have been identified in several plant species, including bryophytes, monocots, and dicots (Patil et al., 1995; Liu et al., 1998; Godfroy et al., 2006). Because of its unique structural features, which enable CCaMK to bind to both CaM and Ca2+, the biochemical characteristics of CCaMK proteins have been analyzed (Patil et al., 1995; Takezawa et al., 1996; Liu et al., 1998; Sathyanarayanan et al., 2000). Over the past decade, however, the lack of CCaMK mutants has made it difficult to determine the function of CCaMK in specific biological processes as well as its activation mechanisms in vivo. This situation changed as a result of the isolation of ccamk mutants in the model legumes L. japonicus and M. truncatula (Lévy et al., 2004; Tirichine et al., 2006). The identification of both loss- and gain-of-function mutants of CCaMK allowed us to investigate CCaMK function in great detail and to build an activation model of CCaMK in response to symbiotic interactions.
Significance of the EF-Hand Domains in CCaMK Function
Complementation of the L. japonicus ccamk mutant by the CCaMK deletion series revealed that the EF-hand motifs are indispensable for the infection of rhizobia (Figure 1). Similar results have been previously described in complementation analyses for the dmi3 mutant of M. truncatula, in which EF-hand deletion variants of DMI3 (M. truncatula CCaMK) could not induce the formation of infected nodules (Gleason et al., 2006). In addition to the EF-hand deletion variants, we also confirmed the significance of the EF-hand domains using full-length CCaMKs that contain point mutations in the EF-hand motifs. Double mutation in two EF-hand motifs (EF2EQ-3EQ and EF1EQ-3EQ) of CCaMK abolished its ability to rescue symbiotic defects of ccamk-3 (Figure 3). Taken together with the results of the Ca2+-dependent mobility-shift assay (Figure 2B), these data suggest that the loss of function caused by the EF-hand motif mutants results from the loss of a Ca2+-induced conformational change in the visinin-like domain. The Ca2+-dependent mobility shift assay also reveals that the third EF-hand motif accounts for the dominant contribution to the conformational change of CCaMK in response to Ca2+ binding. Therefore, Ca2+ binding and the resultant conformational change in the visinin-like domain are likely to be critical for CCaMK activation to induce intracellular accommodation of endosymbionts. However, a point mutation in any one of the three EF-hand motifs alone did not affect the function of CCaMK (see Supplemental Figure 2 and Supplemental Table 1 online). When these CCaMK variants were expressed by the native promoter of CCaMK, they were also able to complement M. loti infection in the ccamk-3 roots (see Supplemental Table 1 online). Thus, it seems that overexpression of these variants by the cauliflower mosaic virus 35S promoter is not the cause of the rescue of the ccamk-3 phenotype.
The EF-hand motifs tend to occur in pairs that form an EF-hand domain. It is commonly accepted that this pairing is important for Ca2+ binding as well as for stability of the EF-hand domain (Grabarek, 2006; Gifford et al., 2007). Indeed, our modeling of the visinin-like domain of CCaMK shows that the EF2 and EF3 motifs together form an EF-hand domain (Figure 4A). Accordingly, the loss of symbiotic activity in CCaMK EF2EQ-3EQ probably results from the loss of synergistic effects caused by the double mutation in EF2EQ and EF3EQ.
CCaMK proteins contain three canonical EF-hand motifs in their C terminus, unlike the EF-hand proteins, which commonly contain an even number of EF-hand motifs (Grabarek, 2006). Homology modeling of CCaMK raised the possibility that the EF1 motif forms a pair with EF0, a neighboring upstream noncanonical EF-hand motif (Figures 4A and 4B). Indeed, the introduction of a double mutation (A378Q in EF0 and I414Q in EF1) led to defects in CCaMK function; formation of fully effective nodules were disrupted in the roots of ccamk-3/CCaMK EF0AQ-1IQ (Figures 4C to 4F). In addition to the formation of an EF-hand domain between two adjacent EF-hand motifs, synergistic effects between the EF-hand domains have been previously reported in animal visinin-like proteins and some other EF-hand proteins, in which mutations in multiple EF-hand motifs decrease the Ca2+ binding ability or conformational stability at levels greater than those caused by a single EF-hand motif mutation (Dotson and Putkey, 1993; Lin et al., 2002). Taking these previous findings into account, it seems that the ccamk-3/CCaMK EF1EQ-3EQ phenotype (Figure 3) can be interpreted as a loss of synergistic effects between the first (EF0-EF1) and second (EF2-EF3) EF-hand domains of CCaMK.
Contrasting Roles of CaM Binding for Endosymbiont Accommodation
CCaMK has a CaMBD in addition to its EF-hand domains, and both domains respond to Ca2+ signals. As typified by mammalian CaMKII, the CaMBD of CCaMK overlaps an autoinhibitory region. The autoinhibitory effect of this region on the symbiotic activity of CCaMK was previously suggested for M. truncatula DMI3; the roots of dmi3 plants transformed with kinase-only DMI3 (amino acids 1 to 311), which lacks both CaMBD and EF-hand motifs, could induce spontaneous nodule formation (Gleason et al., 2006). This study provides further insights into the significance of CaMBD, especially in RN symbiosis. Among the CCaMK variants we examined, 1-314 (which lacks the CaMBD and a visinin-like domain) and CCaMKFN-ED both had a gain-of-function effect on nodule organogenesis, but neither could accommodate M. loti (Figures 1 and 5E to 5G). Our CaM binding assay showed that CCaMKFN-ED had an impaired ability to bind to CaM (Figure 5L). In addition to 1-314 and CCaMKFN-ED, deletion of the whole visinin-like domain of CCaMK abolished its ability to bind to CaM; for example, 1-340, 1-340T265D, and 1-340FN-ED belonged to this category (Figure 5L). Although both 1-340T265D and 1-340FN-ED could induce nodule organogenesis, they did not restore the ability to accommodate rhizobia, as was also the case in 1-314 and CCaMKFN-ED. These results reveal the importance of CaM binding to CCaMK to establish rhizobial infection. By contrast, mycorrhization was not affected by these four mutated CCaMK proteins, 1-314 (Takeda et al., 2012), CCaMKFN-ED (Figure 5K; see Supplemental Figure 4 online), 1-340T265D (see Supplemental Figure 3 online), and 1-340FN-ED (Figure 7W). Collectively, our findings clearly demonstrate that deregulation of the autoinhibition by these mutations is sufficient for nodule organogenesis and mycorrhization but that binding of CaM to CCaMK is subsequently required for rhizobial infection.
Dispensability of CaM binding to CCaMK in AM symbiosis is somewhat controversial, because all CCaMKs identified in nonleguminous plants retain the CaMBD (Patil et al., 1995; Liu et al., 1998; Pandey and Sopory, 2001; Godfroy et al., 2006). Furthermore, lily and rice (Oryza sativa) CCaMK could complement the nodulation defective phenotype of dmi3 mutant in M. truncatula (Gleason et al., 2006) and of ccamk-3 mutant in L. japonicus (Banba et al., 2008), respectively. In our study, gain-of-function mutated CCaMKs, T265D and FN-ED, are likely to mimic the activated status caused by Ca2+ binding to EF-hand domains. However, it is possible that this gain of function also confers additional functions to CCaMK (i.e., a decreased requirement for CaM binding). In our experiment, rhizobial infection showed an absolute requirement for CaM binding in the gain-of-function mutants, but AM fungi infection did not. In this view, comparative studies of wild-type CCaMK and CaM dynamics during RN and AM symbioses will give us important clues for understanding the action of CCaMK in planta.
Takezawa et al. (1996) showed that Ca2+-dependent autophosphorylation of lily CCaMK was inhibited by CaM, suggesting that Ca2+ and CaM have opposite effects on CCaMK regulation. This result supports the view that CaM binding to CaMBD is required for the negative regulation of CCaMK. It has also been reported that lily and tobacco (Nicotiana tabacum) CCaMKs are expressed in anthers (Patil et al., 1995; Liu et al., 1998; Poovaiah et al., 1999), and increased expression of both CCaMK and CaM were detected in pollen cells of tobacco (Poovaiah et al., 1999). These results imply the possibility that nonleguminous CCaMKs are also involved in reproductive stages in which CCaMK and CaM interaction is required.
Biological Activity of CCaMK Is Not Solely Defined by in Vitro Kinase Activity
In this study, we defined the biological (in vivo) activity of the mutated CCaMK variants by their ability to complement the nodulation and infection-defective ccamk-3 mutant. We also defined the kinase activity of CCaMK in terms of autophosphorylation and substrate phosphorylation, both of which were examined by in vitro biochemical assays. In agreement with previous studies, CCaMKWT was activated by dual regulation in response to Ca2+ and CaM (see Supplemental Figure 5A online) (Sathyanarayanan et al., 2000; Tirichine et al., 2006). After Ca2+-stimulated autophosphorylation, substrate phosphorylation was promoted by the addition of CaM.
The deletion series of CCaMK proteins showed relatively low autophosphorylation and substrate phosphorylation activities compared with CCaMKWT (see Supplemental Figures 5B and 6A online). This reinforces the importance of the EF-hand motif for the kinase activity of CCaMK. In addition, the CCaMKT265D deletion series also showed decreased kinase activity (see Supplemental Figures 5B and 6A online), even though 1-429T265D and 1-471T265D could mediate M. loti infection and spontaneous nodule formation in the ccamk-3 mutant (Figure 1). In contrast with deletion mutations for the EF-hand motifs, CCaMK proteins containing point mutations in any two of the three EF-hand motifs (EF1EQ-2EQ, EF2EQ-3EQ, and EF1EQ-3EQ) showed kinase activity regardless of their ability to accommodate endosymbionts (see Supplemental Figures 5C and 6B online). S25F and G204R, both of which were identified as loss-of-function mutations in CCaMK, showed low but significant substrate phosphorylation activities (see Supplemental Figures 5D and 7A online). These results suggest that the degrees of autophosphorylation and substrate phosphorylation activity do not directly determine the ability of CCaMK to induce endosymbiotic interactions. In addition, yeast two-hybrid analysis indicated that protein interactions between mutated CCaMKs and CYCLOPS correlate with the presence of kinase activity of CCaMK (see Supplemental Figure 7 online), although these interactions do not seem to correspond to the biological activity of the mutated CCaMKs. Further analysis will be required to elucidate the functional significance of CYCLOPS phosphorylation by CCaMK in endosymbioses.
In fact, there are several protein kinases whose biological activities do not correspond to their in vitro kinase activities (Schwartzberg et al., 1997; Hayashi and Yamaguchi, 1999; Rathjen et al., 1999; Charette et al., 2001; Hu et al., 2001; Hüser et al., 2001; Hojjati et al., 2007; Chen et al., 2008). Hojjati et al. (2007) reported that the autophosphorylation and substrate phosphorylation activities of animal αCaMKII are not required for it to function in the modulation of short-term presynaptic plasticity, suggesting that αCaMKII plays a structural role rather than an enzymatic role (Hojjati et al., 2007). Kinase-inactive Raf-1, a mouse Ser/Thr protein kinase, can activate the downstream signaling cascade (the MAPK/ERK pathway) normally, leading to cell division (Hüser et al., 2001). In plants, Rathjen et al. (1999) reported that a mutant tomato (Solanum lycopersicum) Pto kinase (PtoY207D), which shows no detectable autophosphorylation activity, nonetheless constitutively induces hypersensitive cell death in the absence of AvrPto (Rathjen et al., 1999). Considering these previous reports, our results support the notion that the biological activities of protein kinases are not necessarily defined by their kinase activities examined using in vitro biochemical assays.
As a common feature, gain-of-function CCaMK proteins—CCaMKT265A (see Supplemental Figure 8 online) (Gleason et al., 2006), CCaMKT265D (Gleason et al., 2006), CCaMKT265I (Tirichine et al., 2006), and CCaMKFN-ED (this article)—showed decreased phosphorylation activities (see Supplemental Figure 8 online), even though they could induce spontaneous nodule formation. Homology modeling suggested a potentially interesting difference among the CCaMKWT, gain-of-function CCaMK, and CCaMKP-T265 proteins. Thr265 was predicted to form a hydrogen bond network with the backbone atom of Ser237, side chains of Lys264, Glu313, and Arg317 in unphosphorylated CCaMKWT (Figure 6B), whereas gain-of-function mutations (CCaMKT265A, CCaMKT265D, and CCaMKT265I) were expected to destabilize this hydrogen bond network (Figures 6D to 6F), as well as autophosphorylated CCaMKP-T265 (Figure 6C). Based on the structural comparison, we created a novel gain-of-function CCaMK (CCaMKR317H, Figure 6G) and showed that the hydrogen bond network including Thr265 and Arg317 is crucial for the regulation of CCaMK activity.
It has been reported that the hydrogen bond network through the triad of residues Glu565, Gln549, and Lys641 of the fibroblast growth factor receptor2 Tyr kinase (FGFR2K) acts as a molecular brake that keeps FGFR2K in an inactive state (Chen et al., 2007). Phosphorylation of Tyr residue or gain-of-function mutations in FGFR2K resulted in destabilization of the hydrogen bond network, thereby allowing FGFR2K to be activated. In the case of CCaMK, disruption of the hydrogen bond network caused by autophosphorylation at Thr265 or gain-of-function mutations in Thr265 or Arg317 seems to reduce interaction of the kinase domain with the CaMBD/autoinhibitory, thereby allowing CCaMK to be deregulated. In contrast with gain-of-function mutations at Thr265 or Arg317, FN to ED mutation was predicted to cause a conformational change of CaMBD (see Supplemental Figure 9 online). Pleiotropic characteristics of CCaMKFN-ED (i.e., gain-of-function for nodule organogenesis and loss-of-function in CaM binding) are assumed to be caused by this conformational change. Determination of crystal structures of inactivated and Ca2+-activated CCaMK will allow us to further understand the mechanism of CCaMK activation by Ca2+/CaM.
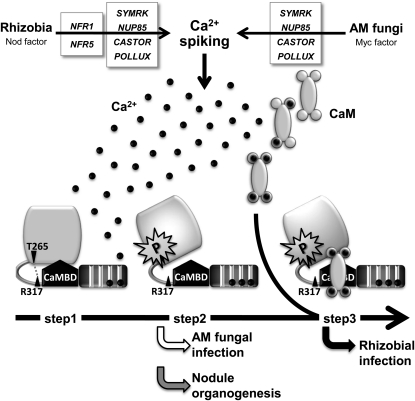
Mechanism of CCaMK Activation during RN and AM Symbioses
To evaluate the functional relationships among the respective domains, we performed epistasis analysis with CCaMK proteins containing multiple mutations. Deletion and point mutations in the EF-hand motifs abolished the ability of CCaMK to induce both rhizobial and AM fungal infection (Figures 1 and 3), and many of these loss-of-function mutations were suppressed by the T265D mutation (Figures 1 and 7). Although T265D is fully functional in both RN and AM symbioses, FN-ED suppresses rhizobial infection in the T265D background, indicating that CaM binding is epistatic to autophosphorylation of the T265 residue that is mimicked by T265D. In addition, neither T265D nor FN-ED could overcome the effect of the G30E mutation (Figure 7). Taken together, these results suggest a potential activation mechanism for CCaMK in the infection by rhizobia and AM fungi (Figure 8): The initial step involves the binding of Ca2+ to the EF-hand domains (step 1). This is followed by autophosphorylation of the kinase domain (step 2). As shown in Supplemental Figure 4 online, weak but significant substrate phosphorylation was detected in CCaMKWT with Ca2+, suggesting that autophosphorylation of CCaMK occurs by an intermolecular mechanism (Sathyanarayanan et al., 2000; Tirichine et al., 2006). The disruption of the hydrogen bond network including Thr265 and Arg317, which is caused by autophosphorylation at Thr265 residue, results in deregulating the activity of CCaMK. This stage of activation is sufficient for the induction of nodule organogenesis and for mycorrhization. After autophosphorylation, the binding of CaM to the CaMBD (step 3) stimulates the activity of CCaMK, which is required to accommodate rhizobia inside the roots. This activation model of CCaMK is consistent with the previously proposed model for lily CCaMK based on in vitro biochemical analyses (Sathyanarayanan et al., 2000).
Figure 8.
A Model for the Activation of CCaMK in RN and AM Symbiosis.
A Ca2+ signal that is induced in response to symbiotic signals from rhizobia or AM fungi is perceived by the EF-hand domains (step 1). Binding of Ca2+ to the EF-hand domains results in autophosphorylation of the kinase domain, which causes the disruption of hydrogen bond network including Thr265 and Arg317, thereby inducing deregulation of CCaMK (step 2). The disruption of hydrogen bond network results in release of CCaMK from autoinhibition of the kinase by the autoinhibitory domain. Binding of Ca2+/CaM to the deregulated CCaMK confers the ability to accommodate rhizobia (step 3).
Different Symbionts Induce Different Levels of CCaMK Activity
Two distinct symbiotic relationships, RN and AM symbioses, are mediated by a common symbiosis pathway that is responsible for generating Ca2+ spiking in response to NFs derived from rhizobia and Myc factors derived from AM fungi (Kosuta et al., 2008; Maillet et al., 2011). Legumes can distinguish the two types of symbionts and properly regulate downstream pathways specific to RN or AM symbioses. In this study, we found the cases in which identical CCaMK constructs led to different results for rhizobial and AM fungal infection, such as in the cases of CCaMKFN-ED, CCaMKT265D, FN-ED, 1-340T265D, and 1-340FN-ED (Figures 5 and 7; see Supplemental Figure 3 online). These differences caused by the identical CCaMK constructs seem to reflect the complexity and different regulation of the CCaMK activity by the respective endosymbionts.
It has been previously demonstrated that Ca2+ spiking has different signatures depending on RN or AM symbiosis, and the difference seems to be a factor in determining the specificity of the respective symbioses (Kosuta et al., 2008). Recently, we have shown that CCaMKT265D suppresses the loss-of-function mutations of common symbiosis genes required for the generation of Ca2+ spiking (Hayashi et al., 2010), which suggests that the gain-of-function status conferred by CCaMKT265D represents the activation status of autophosphorylated CCaMK in response to a Ca2+ spiking. However, the introduction of CCaMKT265D into nfr1 or nfr5 mutants resulted in the failure of intracellular infection by rhizobia, indicating that CCaMK activation by Ca2+ spiking alone is not sufficient to accommodate rhizobia. Our findings in this study, combined with previous reports, led us to hypothesize that rhizobial and AM fungal infections are regulated by qualitatively different Ca2+ signals elicited by the symbionts. Ca2+ signals of different quality are reflected in the differential activation status of CCaMK. As described above, the failure of rhizobial infections in the roots of both nfr1/CCaMKT265D and nfr5/CCaMKT265D indicated that the existence of another signaling pathway, which is directly derived from NFR1/NFR5 and distinct from the common symbiosis pathway, is essential for intracellular rhizobial accommodation. Our previous results also suggest that these two signaling pathways converge on CCaMK (Hayashi et al., 2010). In RN symbiosis, Ca2+ influx has been reported as a NF-induced Ca2+ signal derived from NFR1/NFR5 (Miwa et al., 2006). It remains unknown whether the Ca2+ influx corresponds to another signal derived from NFR1/NFR5, although it seems likely that the Ca2+ influx is required to form a Ca2+ signature that is specific to rhizobial infection. It has also been reported that Ca2+-stimulated autophosphorylation elevates the CaM binding affinity of CCaMK (Sathyanarayanan et al., 2000). In this view, it seems likely that NF-induced Ca2+ spiking and a Ca2+ influx enable CCaMK to bind CaM more stably, thereby promoting CCaMK activity and increasing the ability to accommodate rhizobia. In addition to Ca2+ signature, differential expression of CaM genes might be a cause for differential regulation of two symbioses through CCaMK activation. The difference in the biochemical nature of the activation status of CCaMK between RN and AM symbiosis is still an open question, and elucidation of CCaMK dynamics in response to symbiont-induced Ca2+ and CaM signals will improve our understanding of the ability of CCaMK to activate downstream events specific to RN or AM symbiosis.
METHODS
Plant and Microbial Materials
For complementation analyses of the ccamk mutant, we used mutants of Lotus japonicus B-129 “Gifu” background ccamk-3 (sym72-1) lines that were described previously (Tirichine et al., 2006). Mesorhizobium loti strain MAFF303099 expressing red fluorescent protein from Discosoma sp (DsRed) (Maekawa et al., 2009) was used in our nodulation tests. For mycorrhization, Glomus intraradices DAOM 197198 (Premier Technologies) was used.
Plasmid Constructions
For complementation of the ccamk mutant, the full-length protein coding region of CCaMK of L. japonicus (CCaMK) was amplified by PCR with primers (caccCCaMK_Fw and CCaMK_Rv; see Supplemental Table 2 online) and cDNA synthesized from total RNA of mature plant roots. The amplified fragment was then cloned into a pENTR/D-TOPO vector (Invitrogen), and the resulting entry clone (ENT_CCaMK) was confirmed by sequencing. The insert in ENT_CCaMK was transferred to a Gateway-compatible binary vector of P35S:green fluorescent protein (GFP)-gw (Banba et al., 2008) by LR recombinase (Invitrogen) reaction. For construction of deletion CCaMK variants, corresponding regions were obtained by PCR with ENT_CCaMK as a PCR template, and the amplified fragments were cloned individually into a pENTR/D-TOPO vector. For point mutation, the PrimeSTAR mutagenesis kit (Takara Bio) was used with point mutation primers (see Supplemental Table 2 online) and ENT_CCaMK as a PCR template. The resulting entry clones of deletion and point mutation CCaMKs were transferred to P35S:GFP-gw by LR recombinase reaction.
Complementation Tests on the ccamk Mutant
The constructs carrying the wild type and mutated CCaMK variants were introduced into ccamk-3 mutant by means of hairy root transformation with Agrobacterium rhizogenes LBA1334 (Offringa et al., 1986) as described by Díaz et al. (2005). Plants with GFP-positive hairy roots were transplanted in vermiculite pots supplied with B and D medium (Broughton and Dilworth, 1971) supplemented with 0.5 mM ammonium nitrate. For nodulation, M. loti strain MAFF303099 expressing DsRed was inoculated at 3 d after transplanting. For spontaneous nodule formation, plants with GFP-positive hairy roots were grown in vermiculite pots supplied with B and D medium supplemented with 0.5 mM KNO3. The plants were grown in a growth chamber with a 16 h/8 h night cycle at 24°C for 4 weeks for nodulation and 6 weeks for spontaneous nodule formation. M. loti infection and nodule formation were examined using a MZFLIII stereomicroscope (Leica) and BioLebo 9000 (KEYENCE).
For mycorrhization, plants with GFP-positive hairy roots were grown in an autoclaved 1:1:1 (v/v/v) mixture of vermiculite, commercial soil (Shibametuchi; Sunbellex), and river sand. After inoculation of G. intraradices, plants were grown for 6 weeks in a growth chamber with a 16 h/8 h night cycle at 24°C and a one-half-strength Hoagland solution supplemented with 100 μM KH2PO4. Mycorrhizal colonization was evaluated by the same procedure as described in Banba et al. (2008). Root colonization was estimated by measuring the arbuscular-colonized region per total length of randomly selected mycorrhizal roots.
Expression and Purification of Recombinant CCaMK and CYCLOPS Protein
DNA fragments of the wild type and CCaMK variants were amplified by PCR with primers containing restriction sites for NdeI or XhoI (see Supplemental Table 2 online). After digestion with NdeI and XhoI, the amplified fragment was inserted into NdeI and XhoI site of pCold-GST, which was a pCold I cold-shock vector (Takara Bio) modified by introducing a glutathione S-transferase (GST) tag in front of a 6× His tag. GST-tagged CCaMK proteins were induced in Escherichia coli Rosetta cells by adding 1 mM isopropyl-β-d-thiogalactopyranoside and were cultured at 15°C for 20 h. After sonication, expressed CCaMK proteins were purified using a GST spin trap purification column (GE Healthcare). Recombinant CYCLOPS protein was expressed and purified by the same procedures as above, except that the protein was induced by adding 0.5 mM isopropyl-β-d-thiogalactopyranoside and was cultured at 15°C for 12 h. Protein amounts were determined by the Bradford method, using BSA as a standard and adjusted according to Coomassie staining visualization in a SDS polyacrylamide gel.
Mobility Shift Assay
The carboxyl terminal visinin-like domain (amino acids 334 to 518) of the wild type and mutated CCaMK were cloned into pCold-GST and expressed and purified by the same procedure as described above. A total of 1 μg of purified visinin-like domain proteins were incubated with 0.25 mM CaCl2 or 5 mM EGTA for 30 min and were then analyzed for mobility shift by 12% SDS polyacrylamide gel.
In Vitro Kinase Assay
Kinase assays were performed using 0.2 μM CCaMK protein per assay in total 25 μL reaction for 30 min at 25°C in the presence of 50 mM HEPES (pH 7.4) containing 10 mM MgCl2, 1 mM DTT, 200 μM ATP, 10 μCi [γ-32P] ATP, 0.1 mM CaCl2, and 1 μM bovine brain CaM (Sigma-Aldrich). A total of 2 μg of MBP (Sigma-Aldrich) or 1 μg of CYCLOPS was used as a substrate. Kinase reactions were stopped by adding SDS-PAGE sample buffer and boiling for 5 min. Two-thirds aliquots of samples (20 μL) were separated by SDS-PAGE. Radioactive images were obtained by exposing SDS-PAGE gel to imaging plate sheet for 12 h (for MBP phosphorylation) or 24 h (for CYCLOPS phosphorylation) and were analyzed by STORM860 (GE Healthcare).
CaM Binding Assay
CaM from bovine brain (Sigma-Aldrich) was labeled with biotin using ECL Protein Biotinylation Module (GE Healthcare). Purified CCaMK proteins were spotted on polyvinylidene difluoride membrane (Immobilon-P; Millipore), and the membrane was blocked in Tris-buffered saline containing 0.1% Tween 20 and 5% skim milk at room temperature for 1 h. The membrane was then incubated with 100 ng/mL biotinylated CaM in binding buffer (Tris-buffered saline containing 1% w/v BSA and 1 mM CaCl2 or 5 mM EGTA) at 4°C for overnight. After several washes in Tris-buffered saline containing 0.1% Tween 20 and 1 mM CaCl2 or 5 mM EGTA, the membrane was incubated with streptavidin-conjugated horseradish peroxidase in binding buffer at room temperature for 1 h. Binding of biotinylated CaM to CCaMK was detected by ECL Plus Western Blotting Detection Reagents (GE Healthcare).
Homology Modeling of CCaMK
Homology modeling of the visinin-like domain and N-terminal part of CCaMK was performed using Molecular Operating Environment (MOE) software (MOE version 2009; Chemical Computing Group, http://www.chemcomp.com/software.htm) (Shirakawa et al., 2008). Based on the homology search in the Swiss-Model (http://swissmodel.expasy.org/), including gapped BLAST and HHsearch template library searches, chicken myristoylated guanylate cyclase-activating protein1, chain A (PDB 2R2I.A) (Stephen et al., 2007) and Caenorhabditis elegans CaMKII (PDB 2BDW.B) (Rosenberg et al., 2005) were selected as templates for the prediction of visinin-like domain and N-terminal domain of CCaMK, respectively.
Immunodetection of CCaMK Proteins
For immunodetection of CCaMK proteins, triple c-Myc tags were fused to CCaMKWT, 1-340, and EF1EQ-2EQ-3EQ at their N termini. These tagged CCaMKs were transferred to Gateway-compatible binary vector of P35S:GFP-gw and were introduced into ccamk-3 mutant by means of hairy root transformation as described above. Approximately 0.5 g of transformed hairy roots were ground in liquid nitrogen and incubated in extraction buffer (25 mM MOPS, 0.5 mM EDTA, 100 mM NaCl, 1% Triton X-100, and 0.4 mM phenylmethylsulfonyl fluoride) for 1 h on ice. Samples were then filtered through four layers of Miracloth, and flow-through was centrifuged at 12,000 rpm for 5 min. The supernatant was collected, and its protein concentration was measured by the Bradford method. Twenty micrograms of total protein was separated by SDS-PAGE and transferred to polyvinylidene difluoride membrane (Immobilon-P, Millipore). Immunodetection was performed using ECL Plus Western Blotting Detection Reagents (GE Healthcare) with c-Myc rabbit polyclonal IgG primary antibody (Santa Cruz) and ECL peroxidase-labeled anti-rabbit secondary antibody (GE Healthcare).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: Lj-CCaMK (AM230793), Mt-CCaMK (AY496049), Os-CCaMK (AK070533), and Ll-CCaMK (U24188).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Immunodetection of Myc-tagged CCaMK Protein Variants on ccamk-3 Roots.
Supplemental Figure 2. Complementation of Nodulation of ccamk-3 Mutant by CCaMK Variants Containing Point Mutation in Single EF-Hand Motif.
Supplemental Figure 3. Mycorrhization on ccamk-3/1-340 and ccamk-3/1-340T265D Roots.
Supplemental Figure 4. G. intraradices Infection on ccamk-3/CCaMKWT and ccamk-3/CCaMKFN-ED Roots.
Supplemental Figure 5. In Vitro Kinase Activity of Recombinant CCaMK Proteins with MBP as a Substrate.
Supplemental Figure 6. In Vitro Kinase Activity of Recombinant CCaMK Proteins with CYCLOPS as a Substrate.
Supplemental Figure 7. In Vitro Kinase Assay with CCaMK Variants and Their Interaction with CYCLOPS.
Supplemental Figure 8. Complementation of ccamk-3 Mutant by CCaMKT265A and in Vitro Kinase Activity of Gain-of-Function CCaMK Proteins.
Supplemental Figure 9. Comparison of Predicted Protein Structure of CCaMKWT and CCaMKFN-ED.
Supplemental Figure 10. Mycorrhization on ccamk-3/CCaMKG30E and ccamk-3/CCaMKT265D_EF1EQ-3EQ.
Supplemental Table 1. Results of Complementation Analysis of ccamk-3 Mutant by Point Mutated CCaMKs.
Supplemental Table 2. Sequences of Primers Used in This Study.
Acknowledgments
We thank Satoko Yoshida (RIKEN) for providing experimental information for the in vitro kinase assay of CCaMK. We thank Hiroshi Kouchi (National Institute of Agrobiological Sciences) for critical reading and discussion of the manuscript. We thank Yu Tanokura (University of Tokyo) and Hidetaka Kaya and Kazuyuki Kuchitsu (Tokyo University of Science) for providing information for the EF-hand analysis. This article was supported by the Program of Basic Research Activities for Innovative Biosciences (H.I.-A. and Y.S.) and by the Funding Program for Next Generation World-Leading Researchers from Japan Society for the Promotion of Science (M.H.).
AUTHOR CONTRIBUTIONS
Y.S., M.H., and H.I.-A. designed the research. Y.S., L.H., and H.I.-A. performed research. Y.S., T.Y., R.S., and H.I.-A. analyzed data. Y.S., M.H., and H.I.-A. wrote the article.
References
- Akiyama K., Matsuzaki K., Hayashi H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827 [DOI] [PubMed] [Google Scholar]
- Ané J.M., et al. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303: 1364–1367 [DOI] [PubMed] [Google Scholar]
- Babu A., Su H., Ryu Y., Gulati J. (1992). Determination of residue specificity in the EF-hand of troponin C for Ca2+ coordination, by genetic engineering. J. Biol. Chem. 267: 15469–15474 [PubMed] [Google Scholar]
- Banba M., Gutjahr C., Miyao A., Hirochika H., Paszkowski U., Kouchi H., Imaizumi-Anraku H. (2008). Divergence of evolutionary ways among common sym genes: CASTOR and CCaMK show functional conservation between two symbiosis systems and constitute the root of a common signaling pathway. Plant Cell Physiol. 49: 1659–1671 [DOI] [PubMed] [Google Scholar]
- Bonfante P., Genre A. (July 27, 2010). Mechanisms underlying beneficial plant-fungus interactions in mycorrhizal symbiosis. Nature Commun. 1 (online), doi10.1038/ncomms1046 [DOI] [PubMed] [Google Scholar]
- Broughton W.J., Dilworth M.J. (1971). Control of leghaemoglobin synthesis in snake beans. Biochem. J. 125: 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch E., Hohenester E., Timpl R., Paulsson M., Maurer P. (2000). Calcium affinity, cooperativity, and domain interactions of extracellular EF-hands present in BM-40. J. Biol. Chem. 275: 25508–25515 [DOI] [PubMed] [Google Scholar]
- Chabaud M., Genre A., Sieberer B.J., Faccio A., Fournier J., Novero M., Barker D.G., Bonfante P. (2011). Arbuscular mycorrhizal hyphopodia and germinated spore exudates trigger Ca2+ spiking in the legume and nonlegume root epidermis. New Phytol. 189: 347–355 [DOI] [PubMed] [Google Scholar]
- Charette S.J., Lambert H., Landry J. (2001). A kinase-independent function of Ask1 in caspase-independent cell death. J. Biol. Chem. 276: 36071–36074 [DOI] [PubMed] [Google Scholar]
- Chen L., Huynh L., Apgar J., Tang L., Rassenti L., Weiss A., Kipps T.J. (2008). ZAP-70 enhances IgM signaling independent of its kinase activity in chronic lymphocytic leukemia. Blood 111: 2685–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Ma J., Li W., Eliseenkova A.V., Xu C., Neubert T.A., Miller W.T., Mohammadi M. (2007). A molecular brake in the kinase hinge region regulates the activity of receptor tyrosine kinases. Mol. Cell 27: 717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C., Barber J.D., Barton G.J. (2008). The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36(Web Server issue): W197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Koninck P., Schulman H. (1998). Sensitivity of CaM kinase II to the frequency of Ca2+ oscillations. Science 279: 227–230 [DOI] [PubMed] [Google Scholar]
- Díaz C.L., Grønlund M., Schlaman H.R.M., Spaink H.P. (2005). Induction of hairy roots for symbiotic gene expression studies. Lotus japonicus Handbook, Márquez A.J., (Dordrecht, The Netherlands: Springer; ), pp. 261–277 [Google Scholar]
- Dotson D.G., Putkey J.A. (1993). Differential recovery of Ca2+ binding activity in mutated EF-hands of cardiac troponin C. J. Biol. Chem. 268: 24067–24073 [PubMed] [Google Scholar]
- Ehrhardt D.W., Wais R., Long S.R. (1996). Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85: 673–681 [DOI] [PubMed] [Google Scholar]
- Endre G., Kereszt A., Kevei Z., Mihacea S., Kaló P., Kiss G.B. (2002). A receptor kinase gene regulating symbiotic nodule development. Nature 417: 962–966 [DOI] [PubMed] [Google Scholar]
- Franz S., Ehlert B., Liese A., Kurth J., Cazalé A.C., Romeis T. (2011). Calcium-dependent protein kinase CPK21 functions in abiotic stress response in Arabidopsis thaliana. Mol. Plant 4: 83–96 [DOI] [PubMed] [Google Scholar]
- Gifford J.L., Walsh M.P., Vogel H.J. (2007). Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem. J. 405: 199–221 [DOI] [PubMed] [Google Scholar]
- Gleason C., Chaudhuri S., Yang T., Muñoz A., Poovaiah B.W., Oldroyd G.E. (2006). Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441: 1149–1152 [DOI] [PubMed] [Google Scholar]
- Godfroy O., Debellé F., Timmers T., Rosenberg C. (2006). A rice calcium- and calmodulin-dependent protein kinase restores nodulation to a legume mutant. Mol. Plant Microbe Interact. 19: 495–501 [DOI] [PubMed] [Google Scholar]
- Grabarek Z. (2006). Structural basis for diversity of the EF-hand calcium-binding proteins. J. Mol. Biol. 359: 509–525 [DOI] [PubMed] [Google Scholar]
- Groth M., Takeda N., Perry J., Uchida H., Dräxl S., Brachmann A., Sato S., Tabata S., Kawaguchi M., Wang T.L., Parniske M. (2010). NENA, a Lotus japonicus homolog of Sec13, is required for rhizodermal infection by arbuscular mycorrhiza fungi and rhizobia but dispensable for cortical endosymbiotic development. Plant Cell 22: 2509–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Banba M., Shimoda Y., Kouchi H., Hayashi M., Imaizumi-Anraku H. (2010). A dominant function of CCaMK in intracellular accommodation of bacterial and fungal endosymbionts. Plant J. 63: 141–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S., Yamaguchi M. (1999). Kinase-independent activity of Cdc2/cyclin A prevents the S phase in the Drosophila cell cycle. Genes Cells 4: 111–122 [DOI] [PubMed] [Google Scholar]
- Hojjati M.R., van Woerden G.M., Tyler W.J., Giese K.P., Silva A.J., Pozzo-Miller L., Elgersma Y. (2007). Kinase activity is not required for alphaCaMKII-dependent presynaptic plasticity at CA3-CA1 synapses. Nat. Neurosci. 10: 1125–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Baud V., Oga T., Kim K.I., Yoshida K., Karin M. (2001). IKKalpha controls formation of the epidermis independently of NF-kappaB. Nature 410: 710–714 [DOI] [PubMed] [Google Scholar]
- Hüser M., Luckett J., Chiloeches A., Mercer K., Iwobi M., Giblett S., Sun X.M., Brown J., Marais R., Pritchard C. (2001). MEK kinase activity is not necessary for Raf-1 function. EMBO J. 20: 1940–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi-Anraku H., et al. (2005). Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature 433: 527–531 [DOI] [PubMed] [Google Scholar]
- Kanamori N., et al. (2006). A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc. Natl. Acad. Sci. USA 103: 359–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuta S., Hazledine S., Sun J., Miwa H., Morris R.J., Downie J.A., Oldroyd G.E. (2008). Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proc. Natl. Acad. Sci. USA 105: 9823–9828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouchi H., Imaizumi-Anraku H., Hayashi M., Hakoyama T., Nakagawa T., Umehara Y., Suganuma N., Kawaguchi M. (2010). How many peas in a pod? Legume genes responsible for mutualistic symbioses underground. Plant Cell Physiol. 51: 1381–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévy J., et al. (2004). A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303: 1361–1364 [DOI] [PubMed] [Google Scholar]
- Lewit-Bentley A., Réty S. (2000). EF-hand calcium-binding proteins. Curr. Opin. Struct. Biol. 10: 637–643 [DOI] [PubMed] [Google Scholar]
- Lin L., Braunewell K.H., Gundelfinger E.D., Anand R. (2002). Functional analysis of calcium-binding EF-hand motifs of visinin-like protein-1. Biochem. Biophys. Res. Commun. 296: 827–832 [DOI] [PubMed] [Google Scholar]
- Liu Z., Xia M., Poovaiah B.W. (1998). Chimeric calcium/calmodulin-dependent protein kinase in tobacco: Differential regulation by calmodulin isoforms. Plant Mol. Biol. 38: 889–897 [DOI] [PubMed] [Google Scholar]
- Madsen E.B., Madsen L.H., Radutoiu S., Olbryt M., Rakwalska M., Szczyglowski K., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640 [DOI] [PubMed] [Google Scholar]
- Madsen L.H., Tirichine L., Jurkiewicz A., Sullivan J.T., Heckmann A.B., Bek A.S., Ronson C.W., James E.K., Stougaard J. (April 12, 2010). The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nature Commun. 1 (online), doi/10.1038/ncomms1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T., Maekawa-Yoshikawa M., Takeda N., Imaizumi-Anraku H., Murooka Y., Hayashi M. (2009). Gibberellin controls the nodulation signaling pathway in Lotus japonicus. Plant J. 58: 183–194 [DOI] [PubMed] [Google Scholar]
- Maillet F., et al. (2011). Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469: 58–63 [DOI] [PubMed] [Google Scholar]
- Messinese E., Mun J.H., Yeun L.H., Jayaraman D., Rougé P., Barre A., Lougnon G., Schornack S., Bono J.J., Cook D.R., Ané J.M. (2007). A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula. Mol. Plant Microbe Interact. 20: 912–921 [DOI] [PubMed] [Google Scholar]
- Miwa H., Sun J., Oldroyd G.E., Downie J.A. (2006). Analysis of Nod-factor-induced calcium signaling in root hairs of symbiotically defective mutants of Lotus japonicus. Mol. Plant Microbe Interact. 19: 914–923 [DOI] [PubMed] [Google Scholar]
- Murray J., et al. (2006). Genetic suppressors of the Lotus japonicus har1-1 hypernodulation phenotype. Mol. Plant Microbe Interact. 19: 1082–1091 [DOI] [PubMed] [Google Scholar]
- Offringa I.A., Melchers L.S., Regensburg-Tuink A.J., Costantino P., Schilperoort R.A., Hooykaas P.J. (1986). Complementation of Agrobacterium tumefaciens tumor-inducing aux mutants by genes from the T(R)-region of the Ri plasmid of Agrobacterium rhizogenes. Proc. Natl. Acad. Sci. USA 83: 6935–6939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara Y., et al. (2008). Synergistic activation of the Arabidopsis NADPH oxidase AtrbohD by Ca2+ and phosphorylation. J. Biol. Chem. 283: 8885–8892 [DOI] [PubMed] [Google Scholar]
- Oldroyd G.E., Downie J.A. (2008). Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59: 519–546 [DOI] [PubMed] [Google Scholar]
- Pandey S., Sopory S.K. (2001). Zea mays CCaMK: Autophosphorylation-dependent substrate phosphorylation and down-regulation by red light. J. Exp. Bot. 52: 691–700 [DOI] [PubMed] [Google Scholar]
- Patil S., Takezawa D., Poovaiah B.W. (1995). Chimeric plant calcium/calmodulin-dependent protein kinase gene with a neural visinin-like calcium-binding domain. Proc. Natl. Acad. Sci. USA 92: 4897–4901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poovaiah B.W., Xia M., Liu Z., Wang W., Yang T., Sathyanarayanan P.V., Franceschi V.R. (1999). Developmental regulation of the gene for chimeric calcium/calmodulin-dependent protein kinase in anthers. Planta 209: 161–171 [DOI] [PubMed] [Google Scholar]
- Radutoiu S., Madsen L.H., Madsen E.B., Felle H.H., Umehara Y., Grønlund M., Sato S., Nakamura Y., Tabata S., Sandal N., Stougaard J. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592 [DOI] [PubMed] [Google Scholar]
- Radutoiu S., Madsen L.H., Madsen E.B., Jurkiewicz A., Fukai E., Quistgaard E.M., Albrektsen A.S., James E.K., Thirup S., Stougaard J. (2007). LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J. 26: 3923–3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandiran S., Takezawa D., Wang W., Poovaiah B.W. (1997). Functional domains of plant chimeric calcium/calmodulin-dependent protein kinase: Regulation by autoinhibitory and visinin-like domains. J. Biochem. 121: 984–990 [DOI] [PubMed] [Google Scholar]
- Rathjen J.P., Chang J.H., Staskawicz B.J., Michelmore R.W. (1999). Constitutively active Pto induces a Prf-dependent hypersensitive response in the absence of avrPto. EMBO J. 18: 3232–3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg O.S., Deindl S., Sung R.J., Nairn A.C., Kuriyan J. (2005). Structure of the autoinhibited kinase domain of CaMKII and SAXS analysis of the holoenzyme. Cell 123: 849–860 [DOI] [PubMed] [Google Scholar]
- Saito K., et al. (2007). NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell 19: 610–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyanarayanan P.V., Cremo C.R., Poovaiah B.W. (2000). Plant chimeric Ca2+/Calmodulin-dependent protein kinase. Role of the neural visinin-like domain in regulating autophosphorylation and calmodulin affinity. J. Biol. Chem. 275: 30417–30422 [DOI] [PubMed] [Google Scholar]
- Schwartzberg P.L., Xing L., Hoffmann O., Lowell C.A., Garrett L., Boyce B.F., Varmus H.E. (1997). Rescue of osteoclast function by transgenic expression of kinase-deficient Src in src-/- mutant mice. Genes Dev. 11: 2835–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakawa K., Takaori-Kondo A., Yokoyama M., Izumi T., Matsui M., Io K., Sato T., Sato H., Uchiyama T. (2008). Phosphorylation of APOBEC3G by protein kinase A regulates its interaction with HIV-1 Vif. Nat. Struct. Mol. Biol. 15: 1184–1191 [DOI] [PubMed] [Google Scholar]
- Stephen R., Bereta G., Golczak M., Palczewski K., Sousa M.C. (2007). Stabilizing function for myristoyl group revealed by the crystal structure of a neuronal calcium sensor, guanylate cyclase-activating protein 1. Structure 15: 1392–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke S., Kistner C., Yoshida S., Mulder L., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J., Szczyglowski K., Parniske M. (2002). A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417: 959–962 [DOI] [PubMed] [Google Scholar]
- Takeda N., Maekawa T., Hayashi M. (February 14, 2012) Nuclear-localized and deregulated calcium- and calmodulin-dependent protein kinase activates rhizobial and mycorrhizal responses in Lotus japonicus. Plant Cell http://dx.doi.org/10.1105/tpc.111.091827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takezawa D., Ramachandiran S., Paranjape V., Poovaiah B.W. (1996). Dual regulation of a chimeric plant serine/threonine kinase by calcium and calcium/calmodulin. J. Biol. Chem. 271: 8126–8132 [DOI] [PubMed] [Google Scholar]
- Tikunova S.B., Rall J.A., Davis J.P. (2002). Effect of hydrophobic residue substitutions with glutamine on Ca2+ binding and exchange with the N-domain of troponin C. Biochemistry 41: 6697–6705 [DOI] [PubMed] [Google Scholar]
- Tirichine L., et al. (2006). Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441: 1153–1156 [DOI] [PubMed] [Google Scholar]
- Yang E., Schulman H. (1999). Structural examination of autoregulation of multifunctional calcium/calmodulin-dependent protein kinase II. J. Biol. Chem. 274: 26199–26208 [DOI] [PubMed] [Google Scholar]
- Yang T., Poovaiah B.W. (2003). Calcium/calmodulin-mediated signal network in plants. Trends Plant Sci. 8: 505–512 [DOI] [PubMed] [Google Scholar]
- Yano K., et al. (2008). CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci. USA 105: 20540–20545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap K.L., Kim J., Truong K., Sherman M., Yuan T., Ikura M. (2000). Calmodulin target database. J. Struct. Funct. Genomics 1: 8–14 [DOI] [PubMed] [Google Scholar]