This work reports that mutation in a YABBY gene, TOB1, causes pleiotropic phenotypes, such as formation of a cone-shaped organ and premature termination of the meristem in rice spikelets. Molecular genetic analyses show that TOB1 regulates the initiation and growth of the lateral organs in the spikelet and acts non-cell autonomously to maintain activity and proper organization of the meristem.
Abstract
The meristem initiates lateral organs in a regular manner, and proper communication between the meristem and the lateral organs ensures the normal development of plants. Here, we show that mutation of the rice (Oryza sativa) gene TONGARI-BOUSHI1 (TOB1) results in pleiotropic phenotypes in spikelets, such as the formation of a cone-shaped organ instead of the lemma or palea, the development of two florets in a spikelet, or premature termination of the floret meristem, in addition to reduced growth of the lemma or palea and elongation of the awn. These phenotypes seem to result from not only failure in growth of the lateral organs, but also defects in maintenance and organization of the meristem. For example, the cone-shaped organ develops as a ring-like primordium from an initial stage, suggesting that regulation of organ initiation in the meristem may be compromised. TOB1 encodes a YABBY protein, which is closely related to FILAMENTOUS FLOWER in Arabidopsis thaliana, and is expressed in the lateral organ primordia without any patterns of polarization. No TOB1 expression is detected in the meristem, so TOB1 may act non–cell autonomously to maintain proper meristem organization and is therefore likely to play an important role in rice spikelet development.
INTRODUCTION
In plant development, lateral organs, such as the leaves and floral organs, develop from founder cells supplied from stem cells in the meristem. After specification of cell fate for differentiation of each organ, the development of lateral organ primordia is still regulated from the meristem through as yet unidentified signals. For example, isolation of the leaf primordia from the meristem by surgical manipulation or laser ablation results in radially symmetrical abaxialized leaves, suggesting that a signal to specify adaxial identity in the leaf is derived from the meristem (Sussex, 1951, 1954; Reinhardt et al., 2005). Conversely, meristem activity is likely to be affected by a signal from the lateral organs. Recent studies suggest that activities of YABBY genes are involved in this signaling in Arabidopsis thaliana and Antirrhinum majus (Golz et al., 2004; Goldshmidt et al., 2008; Stahle et al., 2009; see below in detail).
The YABBY genes play important roles in the regulation of diverse developmental processes, such as establishment of adaxial–abaxial polarity, lamina expansion, and floral organ development, in eudicots. In Arabidopsis, FILAMENTOUS FLOWER (FIL) and YABBY3 (YAB3) are proposed to promote abaxial cell fate (Sawa et al., 1999b; Siegfried et al., 1999). Consistent with this, FIL and YAB3 are expressed in the abaxial domain of lateral organs (Sawa et al., 1999b; Siegfried et al., 1999; Sarojam et al., 2010). Recent studies using triple (fil yab3 yab5) and quadruple (fil yab3 yab5 yab2) mutants, which show complex loss of abaxial or adaxial identities in radialized leaves, suggest that these YABBY genes are involved in the promotion of leaf expansion, the establishment of leaf polarity, and the general genetic program of lamina formation (Stahle et al., 2009; Sarojam et al., 2010). These YABBY genes also regulate flower development (Chen et al., 1999; Sawa et al., 1999a, 1999b; Siegfried et al., 1999; Stahle et al., 2009; Lugassi et al., 2010; Sarojam et al., 2010). Whereas a fil single mutation results in the formation of filamentous flowers and defects in floral organ morphology, triple and quadruple mutants of these YABBY genes produce radialized filamentous floral organs. GRAMINIFOLIA (GRAM) and PROLONGATA (PROL), orthologs of FIL/YAB3 and YAB5, have a similar function in the development of leaves and flowers in A. majus (Golz et al., 2004; Navarro et al., 2004). Other distantly related Arabidopsis YABBY genes, CRABS CLAW (CRC) and INNER NO OUTER, are responsible for abaxial cell fate in the carpel and specification of nectary and for ovule development, respectively (Bowman and Smyth, 1999; Eshed et al., 1999; Villanueva et al., 1999).
FIL and its related YABBY genes (YAB2, YAB3, and YAB5) are associated with fate and maintenance of the meristems. In fil mutants, phyllotaxy is disorganized, and meristem function is partially compromised (Chen et al., 1999; Sawa et al., 1999b; Siegfried et al., 1999; Lugassi et al., 2010). A fil yab3 double mutant produces ectopic shoot apical meristems (SAMs) and axillary meristems (Kumaran et al., 2002). The expression domain of CLAVATA3 and WUSCHEL is markedly expanded in the SAM of the fil or fil yab3 mutant (Goldshmidt et al., 2008). In the Columbia background, fil shows an enlarged meristem, suggesting its role in meristem organization (Lugassi et al., 2010). In addition, the primary SAM fails to be maintained in the triple and quadruple Yabby mutants (Sarojam et al., 2010). These YABBY genes are not expressed in the meristem, and the mobility of YABBY protein or mRNA is not detected (Sawa et al., 1999b; Siegfried et al., 1999; Goldshmidt et al., 2008; Sarojam et al., 2010). Therefore, YABBY gene activity acts non-cell autonomously on the fate and maintenance of the meristem, suggesting that a YABBY-derived signal produced in a lateral organ acts in a signaling pathway that communicates with the meristem (Goldshmidt et al., 2008). This signal from FIL is partly mediated by the activity of LATERAL SUPPRESSOR (Goldshmidt et al., 2008). The non-cell-autonomous action of a YABBY gene was initially shown as intercellular signaling between abaxial and adaxial domains in the leaf primordia in A. majus (Golz et al., 2004).
The YABBY genes encode putative transcription factors with a zinc-finger and a helix-loop-helix motif (called YABBY domain) similar to the HMG box (Sawa et al., 1999b; Siegfried et al., 1999; Golz et al., 2004). YABBY proteins physically interact with STYLOSA (STY) in A. majus and with LEUNIG (LUG) and LEUNIG_HOMOLOG (LUH) in Arabidopsis (Navarro et al., 2004; Stahle et al., 2009). Both proteins are similar to the GRO/TUP1-like transcriptional corepressor that is involved in a broad range of plant developmental processes, and LUG is well known as a repressor of AGAMOUS in flower development in Arabidopsis (Liu and Meyerowitz, 1995; reviewed in Liu and Karmarkar, 2008). Arabidopsis YABBY proteins also physically interact with SEUSS (SEU), a coregulator working with LUG (Stahle et al., 2009). These findings suggest that YABBY proteins are involved in a repressor complex together with LUG/STY and SEU. Consistent with their biochemical properties, lug/luh, sty, or seu mutations enhance phenotypes caused by loss-of-function mutation of YABBY genes in both A. majus and Arabidopsis (Navarro et al., 2004; Stahle et al., 2009). For example, defects in polarity and lamina expansion in fil yab3 are more severe in a lug or seu mutant background, and the disorganized meristem phenotype in fil is enhanced in a lug luh/+ mutant background (Stahle et al., 2009).
Unlike this progress in understanding of YABBY functions in eudicots, we have only fragmentary information about YABBY genes in monocots. DROOPING LEAF (DL), an ortholog of CRC in rice (Oryza sativa), has been well characterized both genetically and functionally. Loss-of-function mutation of DL causes homeotic transformation of carpels into stamens in the flower and loss of the midrib in the leaf, suggesting that DL regulates carpel specification and midrib formation in rice (Nagasawa et al., 2003; Yamaguchi et al., 2004; Ohmori et al., 2008, 2011). DL is expressed throughout the carpel primordia without any patterns of polarity and the presumptive region of the midrib in leaf primordia (Yamaguchi et al., 2004; Ohmori et al., 2011). It is unlikely that DL expression is regulated by class C genes because DL is still expressed in double mutant of class C genes (Dreni et al., 2011). Identical spatial expression patterns of DL orthologs in both flowers and leaves suggest the function of the DL-related genes is conserved in grasses (Ishikawa et al., 2009). Because there is no report of carpel specification or promotion of midrib formation by CRC/DL-like genes in plants other than grasses, these functions may have been recruited in the lineage of grasses.
Expression analyses in basal angiosperms and a monocot species, Asparagus asparagoides, suggest that abaxial expression in the carpel primordium is an ancestral property of CRC/DL-related genes, suggesting that polarized expression of the DL-related genes may have been lost during grass evolution (Fourquin et al., 2005; Nakayama et al., 2010; Yamada et al., 2011). Conversely, their requirement in nectary development seems to have been acquired in the lineage of eudicots (Lee et al., 2005). Additional functions are suggested in a basal eudicot (Orashakova et al., 2009). Thus, CRC/DL-related YABBY genes have functionally diversified among angiosperms. Although no mutants related to YABBY genes other than dl have been described as yet in grasses, expression patterns also imply that the roles of YABBY genes have diverged in grasses. For example, maize (Zea mays) YABBY genes similar to Arabidopsis FIL are expressed on the adaxial side of the leaf primordia, and rice Os YABBY1, which is similar to Arabidopsis YAB2, is expressed in putative precursor cells of a specific cell type, such as sclerenchyma (Juarez et al., 2004; Toriba et al., 2007). Therefore, the isolation of mutants, followed by functional analysis, would provide clues to understanding YABBY functions characteristic to grasses, in addition to the functional diversification of these genes among angiosperms.
Rice inflorescences are composed of two structural units: a floret and a spikelet. Floral organs, such as carpels, stamens, and lodicules, together with the lemma and palea enclosing them, constitute the floret. In principle, the ABC model applies to rice flower development, except for the crucial contribution of DL to carpel specification (Nagasawa et al., 2003; Yamaguchi et al., 2004, 2006; Hirano et al., 2008). However, our understanding of the development of the lemma and palea is not sufficient, although LEAFY HULL STERILE1 (LHS1) is known to be involved in specification of the identity of both organs (Jeon et al., 2000; Prasad et al., 2005). In this article, we describe a rice mutant, tongari-boushi1 (tob1), which shows unique pleiotropic phenotypes in the spikelet, especially in the lemma and palea. In addition, a prematurely terminated meristem is observed in tob1 spikelets, suggesting that meristem fate is also compromised. Gene isolation revealed that TOB1 encodes a YABBY protein, Os YABBY5, which is closely related to Arabidopsis FIL and A. majus GRAM (Sawa et al., 1999b; Golz et al., 2004; Toriba et al., 2007). TOB1 was found to be expressed in the lateral organs, but not in the meristem. Altogether, we propose that TOB1 is responsible for the initiation and growth of the lemma and palea, and acts non-cell autonomously to maintain the activity and proper organization of the meristem.
RESULTS
tob1 Exhibits Pleiotropic Defects in Spikelet Development
Grass inflorescences are composed of structural units: a spikelet and a floret. In rice, the spikelet has a fertile floret, two sterile lemmas, and two rudimentary glumes. The floret is composed of a lemma, a palea, two lodicules, six stamens, and a pistil (Figures 1A and 1B). The sterile lemma is a tiny flap-like leaf, which is thought to be evolutionarily derived from the lemma of two degenerated lateral florets (Arber, 1934; Yoshida et al., 2009). The rudimentary glume is highly degenerated and detected as a very small organ outside the sterile lemma. The transition from the spikelet meristem to the floret meristem is morphologically unclear in rice because a spikelet forms a single floret.
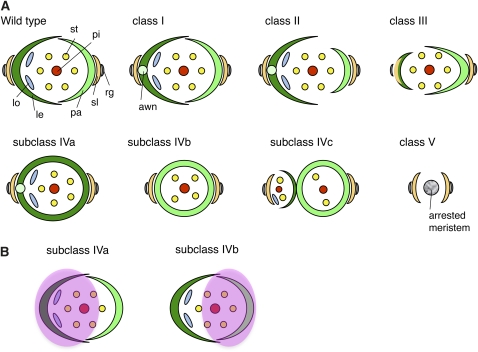
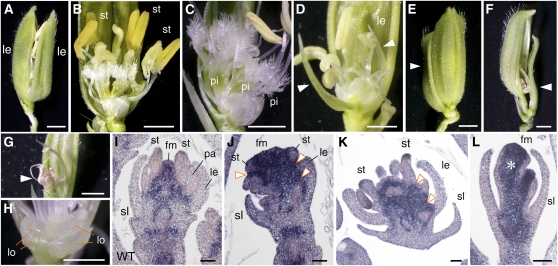
Figure 1.
Phenotypes of Spikelets in the tob1 Mutant.
(A) and (B) A wild-type spikelet.
(C) A tob1 spikelet with an elongated awn (class I).
(D) and (E) A tob1 spikelet with reduced size of the palea (D) and without the palea (E) (class II).
(F) and (G) A tob1 spikelet with an unidentified organ (arrowhead) on the opposite side of the palea (class III). The marginal regions of this unidentified organ are curled inside, similar to those of the lemma.
(H) and (I) A tob1 spikelet in class IV, which forms a cone-shaped organ with ([H]; subclass IVa) and without ([I]; subclass IVb) awn.
(J) and (K) The same tob1 spikelet shown in (I). The distal region of the cone-shaped organ is cut out to show that this organ has no edge (J), and the front of this organ is removed to show the inner organs (K). This spikelet has two stamens and one pistil but no lodicule.
(L) and (M) A tob1 spikelet that has two florets (subclass IVc). The secondary floret (arrowhead) has a pistil, four stamens, and two lodicules inside a lemma/palea-like organ near the cone-shaped organ but lacks the palea or lemma on the opposite side of the organ in (L). Only a palea/lemma-like organ develops as a secondary floret (M).
(N) to (P) Epidermal surface of the lemma (N), palea (O), and sterile lemma (P) in the wild type.
(Q) Epidermal surface of the cone-shaped organ in class IV.
(R) Scanning electron microscopy image of an unidentified organ (close-up view of the unidentified organ indicated with an arrowhead in [G]) in a class III spikelet, which shows a sterile lemma-like surface in the central region and a lemma/palea-like surface in the lateral region.
(S) Close-up view of the epidermal surface at the border region in (R).
(T) and (U) A class IV spikelet with an arrested meristem-like structure (arrowhead) after removal of the cone-shaped organ. (T) is a close-up view of (U).
(V) to (X) tob1 spikelets without organs inside the sterile lemmas (class V). Arrowheads indicate an arrested spikelet after generation of the sterile lemmas (V) and a dome, which is thought to be an arrested meristem (W). A needle-like structure is formed inside the sterile lemmas (X).
(Y) and (Z) Close-up view of the bottom part (Y) of the spikelet, which is indicated by the bottom bracket in (X), and the top part of the needle-like structure (Z), which is indicated by the top bracket in (X).
co, cone-shaped organ; le, lemma; lo, lodicule; pa, palea; pi, pistil; rg, rudimentary glume; rpa, reduced palea size; sl, sterile lemma; st, stamen; WT, wild type. Bars = 1 mm in (A) to (M) and (V) to (X) and 100 μm in (N) to (U), (Y), and (Z).
We identified a recessive mutant, tob1, by a screen of mutants with abnormal morphology in the spikelet. Because tob1 showed pleiotropic phenotypes in spikelets, we categorized these phenotypes into five classes (Figure 1; refer to schematic representations in Figure 8): (I) elongation of the awn, (II) reduction of the palea, (III) reduction of the lemma, (IV) formation of a cone-shaped organ without margin, and (V) loss of the floret. The tob1 phenotype seems to be affected by environmental factors such as temperature because the ratio of these phenotypic classes varied by seasons and culture conditions (see Supplemental Table 1 online). In no case, however, were wild-type spikelets observed. The tob1 mutant had no abnormalities in the vegetative phase.
Figure 8.
Schematic Representation of Spikelet Phenotypes in the Wild Type and the tob1 Mutant.
(A) Spikelet phenotypes. le, lemma; lo, lodicule, pa, palea; pi, pistil; rg, rudimentary glume; sl, sterile lemma; st, stamen.
(B) The domains (pink circles) in the meristem that develop into spikelets in subclass IVa or IVb.
The awn is formed on the top of the lemma in indica rice or wild rice species; by contrast, formation of the awn in japonica rice depends on the cultivar and experimental genetic strain. In strain Taichung 65 (T65), from which the tob1 mutant was isolated, elongation of the awn was completely suppressed (Figures 1A and 1B). In the tob1 mutant, however, most spikelets formed the awn. In class I, the awn was elongated, but no other defect was observed (Figure 1C). In class II, the palea was reduced to various degrees or lost, in addition to awn elongation (Figures 1D and 1E). In both class I and II, floral organs, such as the pistil, stamen, and lodicules, were normal in their morphologies and numbers, suggesting that the effect of tob1 mutation was weak in these spikelets.
In class III, the spikelets lacked an obvious lemma and one of the sterile lemmas; instead, an unidentified organ was observed on the opposite side of the palea (Figures 1F and 1G). This unidentified organ appeared to be a chimera of the lemma (lateral region) and the sterile lemma (central region), as deduced from characteristics of the epidermal surface (Figures 1N to 1P, 1R, and 1S) and morphology of the marginal region (curled inside) (Figures 1F and 1G). Another prominent feature of the spikelets in class III was the loss of lodicules: We could not detect any lodicules in any spikelets examined in this class (Table 1). Furthermore, the number of stamens was reduced, whereas the number of pistils was almost normal.
Table 1.
Distribution of the Number of Floral Organs in tob1 and the Wild Type
| Lodicule |
Stamen |
Carpel |
Arrested Meristem | Other | No. of Spikelets Examined | |||||||||||
| Organ No. | 2 | 1 | 0 | 6 | 5 | 4 | 3 | 2 | 1 | 0 | 2 | 1 | 0 | |||
| Wild type (T65) | 50 | 0 | 0 | 50 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 50 | 0 | 0 | 0 | 50 |
| tob1 class III | 0 | 0 | 20 | 11 | 3 | 3 | 2 | 1 | 0 | 0 | 1 | 19 | 0 | 0 | 0 | 20 |
| tob1 class IV | ||||||||||||||||
| One floret type | ||||||||||||||||
| IVa type floret (with awn) | 5 | 0 | 0 | 3 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 4 | 0 | 0 | 0 | 5 |
| IVb type floret (without awn) | 1 | 0 | 101 | 2 | 2 | 30 | 55 | 11 | 0 | 2 | 1 | 98 | 3 | 2 | 2 | 106 |
| Two floret type | ||||||||||||||||
| IVc type floret (without awn) | 0 | 0 | 9 | 0 | 0 | 0 | 4 | 3 | 2 | 0 | 0 | 6 | 3 | 0 | 0 | 9 |
| 2nd floret | 5 | 0 | 4 | 1 | 0 | 2 | 1 | 1 | 4 | 0 | 0 | 5 | 4 | 0 | 0 | 9 |
In the spikelet consisting of two florets, the cone-shaped organ with awn was not observed.
The class IV spikelets developed a cone-shaped organ, instead of the lemma and palea, inside a pair of sterile lemma (Figures 1H to 1M). The gene name tongari-boushi (meaning “a pointed hat” in Japanese) comes from the cone shape of this organ. Notably, the cone-shaped organ had no edges from the bottom to the top (Figure 1J). The epidermal surface of this organ was similar to that of the lemma and palea of the wild type (Figures 1N, 1O, and 1Q). There were two types of cone-shaped organs: One had the awn at the top of the organ (subclass IVa) and the other did not (subclass IVb). Two normal lodicules were formed in the IVa-type cone, whereas no lodicule was detected inside any IVb-type cone-shaped organ (Table 1). To characterize further the class IV spikelet, we examined the expression of the DL gene, which is expressed in the midrib region of the lemma but not in the palea (Yamaguchi et al., 2004; Ishikawa et al., 2009; Dreni et al., 2011). DL signal was detected only in the class IVa spikelet, which had the lodicules (see Supplemental Figure 1 online). These results suggest that the IVa-type cone-shaped organ is probably derived from the lemma, whereas the IVb-type cone-shaped organ originated from the palea.
The stamen and pistil were formed inside both types of cone-shaped organ, although the number of stamens was reduced and variable among spikelets (Figure 1K, Table 1). In a few extreme cases, no floral organs developed; instead, a terminated meristem-like structure was observed inside the cone-shaped organ (Figures 1T and 1U, Table 1).
Several class IV spikelets were associated with a secondary floret (subclass IVc; Figure 8), which developed variable numbers of floral organs (Figures 1L and 1M, Table 1). In some cases, the secondary floret consisted of only a lemma/palea-like organ (Figure 1M). The development of two florets inside a pair of sterile lemma suggests that the two floret meristems were formed from a spikelet meristem.
Class V showed a marked phenotype. The spikelets in this class formed no florets (Figures 1V to 1X). Inside the sterile lemma, an arrested meristem-like structure was observed (Figure 1W). In another spikelet, a needle-like structure was formed just inside the sterile lemma (Figure 1X). A close-up view showed that this needle-like structure probably developed directly from the meristem, and this structure is similar to a rod-like lemma described by Toriba et al. (2010): The lower part had lemma-like identity with rough surface and trichomes, and the upper part had awn-like identity (Figures 1Y and 1Z).
Scanning Electron Microscopy Analysis Showed Defects in Meristem Maintenance in Addition to Lateral Organ Development in the tob1 Mutant
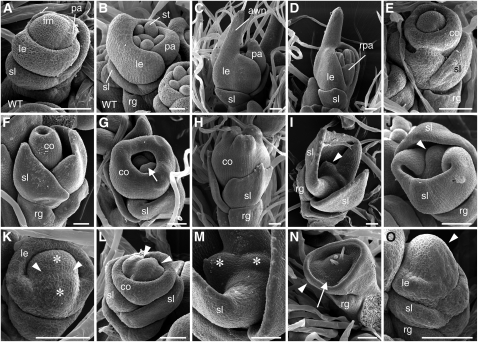
To investigate the abnormalities of tob1 spikelets in detail, we examined early-stage spikelets using scanning electron microscopy. In the wild type, the floret meristem maintained a dome shape until the carpel primordia arose. The lemma and palea primordia developed at the flank of the meristem in an alternate manner and in a synchronized growth pattern (Figures 2A and 2B). In developing tob1 spikelets, a large outgrowth (a putative awn primordium) was observed on the top of the lemma primordium (Figures 2C and 2D). In Figure 2D, growth patterns differed between the lemma and palea, and these primordia were formed at a right angle to the sterile lemma, suggesting that phyllotaxy was partly compromised.
Figure 2.
Spikelets at Early Developmental Stages in the Wild Type and the tob1 Mutant.
(A) and (B) A wild-type spikelet at the stage of lemma and palea initiation (A) and at the stage of stamen initiation (B).
(C) to (O) tob1 spikelets.
(C) A tob1 spikelet with an elongating awn.
(D) A tob1 spikelet with a reduced palea and aberrant phyllotaxy.
(E) to (H) A spikelet developing a cone-shaped organ. This organ initiates as a ring-like primordium (E) and finally fuses at the top (H). Developing stamens (arrow) are observed inside this organ (G).
(I) and (J) Immaturely terminated meristem in a tob1 spikelet, which arrested after the development of a pair of sterile lemma. Arrowheads indicate a terminated meristem.
(K) A large meristem with a groove (arrowheads) in a tob1 spikelet. Asterisks indicate two parts that would split into independent meristems.
(L) A meristem with a few grooves (arrowheads) in a tob1 spikelet.
(M) Close-up view of the prematurely terminated meristems in (I). Putative split meristems are indicated with asterisks.
(N) A meristem with ectopically produced trichome-like structures and a rough epidermal surface (arrow) similar to that of the lemma and palea. Arrowhead indicates a putative sterile lemma primordium.
(O) A spikelet with an abnormally taller meristem (arrowhead).
co, cone-shaped organ; fm, floret meristem; le, lemma; pa, palea; rg, rudimentary glume; rpa, reduced palea size; sl, sterile lemma; st, stamen. Bars = 50 μm.
We found spikelets that initiated a ring-like primordium encompassing the floret meristem (Figure 2E). Subsequently, this ring-like primordium became tube-like and finally fused at the apex to form the cone-shaped organ (Figures 2F to 2H). Stamen-like primordia were detected inside the growing tube-like primordium (Figure 2G). These observations suggest that the cone-shaped organ in class IV may not develop by the fusion of primordia that initiated independently.
Apart from lateral organ development, meristem fate is likely to be compromised in the tob1 mutant. The floret meristems were prematurely terminated after initiation of the sterile lemma primordia (Figures 2I and 2J). Some spikelets had a meristem with a groove crossing the dome structure (Figure 2K). This meristem looked as though it would be subsequently separated by a deeper groove. We found two meristem-like domes adjacent to each other, which might be formed by the split of a meristem (Figure 2M). In another spikelet, the meristem had a few grooves (Figure 2L). More markedly, one spikelet had an arrested meristem within a ring-like primordium inside the rudimentary glume (Figure 2N). In this spikelet, the meristem produced a few trichome-like structures and showed a rough epidermal surface, which was similar to that of the palea/lemma, suggesting that cell fate in the meristem was highly disturbed. We also observed a spikelet that had an abnormally taller meristem, in which a lemma initiated at the lower part of the meristem (Figure 2O). These observations suggest that the maintenance and fate of the meristem are compromised in the tob1 mutant.
Abnormal Expression Patterns of Marker Genes in the Meristem and the Lateral Organs
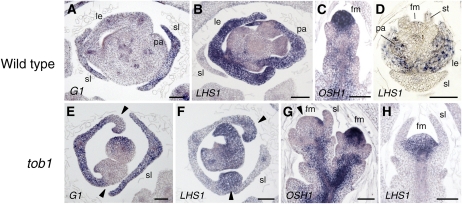
To characterize further the tob1 mutant, we analyzed the spatial expression patterns of a few marker genes in developing tob1 spikelets. First, we examined the expression of the transcripts of LONG STERILE LEMMA (G1) and LHS1 (Prasad et al., 2005; Li et al., 2009; Yoshida et al., 2009). In the wild-type spikelet, G1 was expressed in the sterile lemmas, whereas LHS1 was expressed in the lemma and palea (Figures 3A and 3B). In a cross section of tob1 spikelets, a putative chimeric organ in class III that curled inside in the marginal region was detected at the opposite side of the sterile lemma. G1 was expressed in the central region of the putative chimeric organ of the tob1 spikelet, whereas LHS1 was expressed in the marginal region (Figures 3E and 3F). These results supported the previous morphological observation that the unidentified organ in class III spikelets was a chimera of the lemma and sterile lemma.
Figure 3.
Spatial Expression Patterns of Marker Genes in the Wild Type and the tob1 Mutant.
(A) and (E) In situ localization of G1 transcripts. No expression is observed in the marginal region (arrowheads) of the primordium of the unidentified organ in tob1 spikelets.
(B) and (F) In situ localization of LHS1 transcripts. Expression is observed only in the marginal region (arrowheads) of the primordium of the unidentified organ in tob1 spikelets.
(C) and (G) In situ localization of OSH1 transcripts. Almost no expression is observed in the floret meristem of tob1 spikelets (arrowhead).
(D) and (H) In situ localization of LHS1 transcripts. Ectopic expression is observed in a flattened meristem of tob1 spikelets.
fm, floret meristem; le, lemma; pa, palea; sl, sterile lemma; st, stamen; WT, wild type. Bars = 50 μm.
Next, we examined the expression pattern of the class I KNOX gene ORYZA SATIVA HOMEOBOX1 (OSH1), a marker of indeterminate undifferentiated cells (Sato et al., 1996; Yamaguchi et al., 2004). In the wild type, OSH1 was expressed throughout the floret meristem (Figure 3C). By contrast, we occasionally observed a tob1 spikelet in which OSH1 was not expressed in the meristem (Figure 3G). In addition, we found that LHS1 was ectopically expressed in the abnormal meristem of tob1, although LHS1 transcript was not detected in the meristem in the wild type (Figures 3D and 3H; Li et al., 2009). As above, these observations suggest that the maintenance and fate of the meristem is compromised in some spikelets of the tob1 mutant.
TOB1 Encodes a YABBY Protein
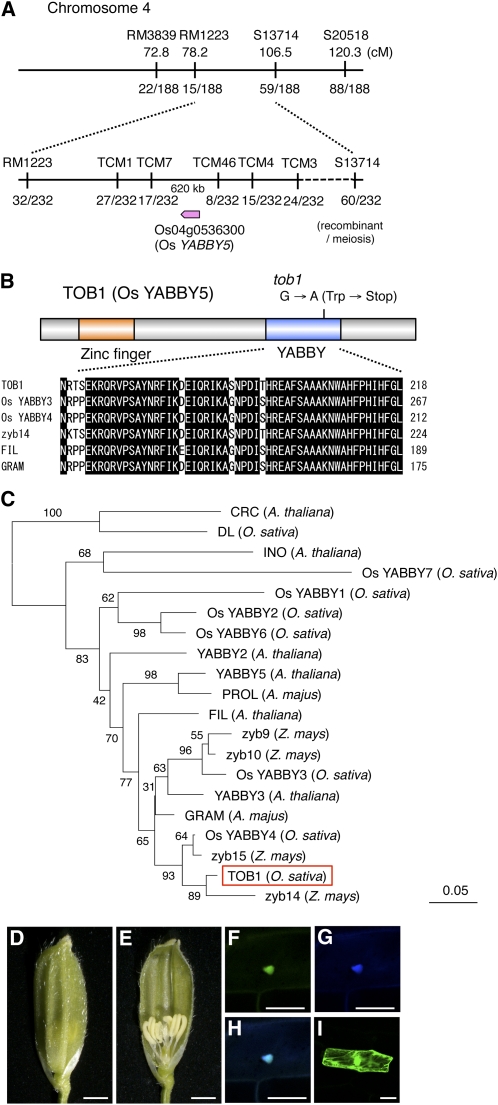
To isolate the gene responsible for the tob1 mutant, we performed map-based cloning. Using F2 plants from a cross between tob1 and Kasalath, TOB1 was mapped to the region between two markers, TCM07 and TCM46, on chromosome 4 (Figure 4A). In this region, we found Os YABBY5 (Os04g0536300), a member of the YABBY gene family that has important roles in plant development (Toriba et al., 2007). By sequencing the genomic DNA, we detected a single nucleotide change that caused a nonsense mutation in the YABBY domain of Os YABBY5 (Figure 4B). Hence, we hypothesized that the tob1 phenotype was caused by loss of function of this gene. We then introduced an 8.6-kb genomic fragment containing the upstream regulatory region and coding region of Os YABBY5 into the tob1 mutant. As a result, the tob1 spikelet phenotype was rescued by this genomic fragment, clearly demonstrating that TOB1 was Os YABBY5 (Figures 4D and 4E).
Figure 4.
Isolation of the TOB1 Gene and Characteristics of the TOB1 Protein.
(A) Map position of the TOB1 locus.
(B) Schematic representation of theTOB1 protein and amino acid alignment of the YABBY domains. The zinc-finger and YABBY domains are indicated by orange and blue boxes, respectively.
(C) Phylogenetic tree of the YABBY proteins. The tree was constructed on the basis of the zinc-finger and YABBY domains by a neighbor-joining method. Numbers above branches indicate the percentage of bootstrap values calculated from 1000 replicates.
(D) and (E) Complementation of the tob1 mutation by a genomic fragment containing the TOB1 locus. No awn is formed on the top of the lemma (D), and normal numbers of inner organs are produced (E). Bars = 1 mm.
(F) to (I) Analysis of subcellular localization of TOB1 protein using onion epidermal tissue. TOB1-GFP fusion protein (F), 4′,6-diamidino-2-phenylindole staining of the nucleus (G), merged image of (F) and (G) in (H), and subcellular localization of GFP (I). Bars = 50 μm.
Rice has eight YABBY genes in the genome (Toriba et al., 2007). We constructed a phylogenetic tree of the YABBY genes including well-characterized members in Arabidopsis, A. majus, and maize (Figure 4C). TOB1 belongs to the YAB1 clade, which includes Arabidopsis FIL, A. majus GRAM, and maize zyb14. The YABBY domains of YABBY proteins in this clade are highly similar to each other (Figure 4B).
To examine the subcellular localization of TOB1, we introduced p35S:TOB1-GFP (for green fluorescent protein) and p35S:GFP-TOB1 into onion epidermal cells. The results clearly showed that both TOB1-GFP and GFP-TOB1 fusion proteins were localized in the nucleus (Figures 4F to 4I; see Supplemental Figure 2 online). These results suggest that TOB1 encodes a nuclear protein, possibly functioning as a transcriptional regulator, similar to well-known YABBY proteins in Arabidopsis.
TOB1 Is Expressed in Lateral Organ Primordia in the Spikelets but Not in the Meristems
First, we examined the organ-specific expression patterns of TOB1 by RT-PCR. TOB1 expression was detected in the vegetative shoot apex, including leaf primordia, and young inflorescences, including developing spikelets (see Supplemental Figure 3 online). By contrast, no expression was detected in fully differentiated leaves and roots.
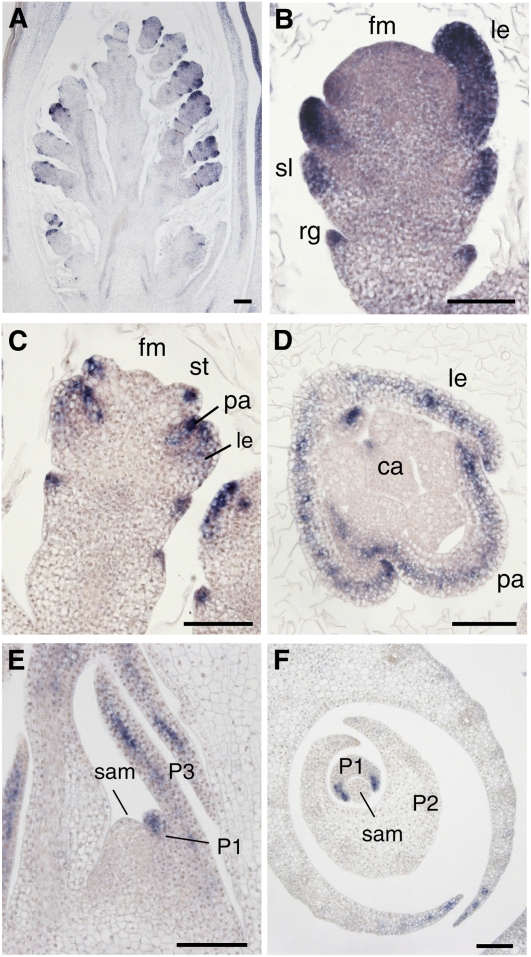
Next, we analyzed the temporal and spatial expression pattern of TOB1 in developing spikelets by in situ hybridization. TOB1 was strongly expressed in the primordia of all lateral organs in the spikelet from their initiation stages (Figure 5A).ca, carpel; fm, floret meristem; le, lemma; pa, palea; P1, P2, and P3, leaf primordium at the indicated stage; rg, rudimentary glume; sl, sterile lemma; st, stamen. Bars = 50 μm. Its expression was detected throughout the primordia: No abaxially or adaxially localized signal was detected (Figures 5B and 5D). The TOB1 signal appeared to be localized to the upper region of the primordia of sterile lemma and rudimentary glume, whereas it was uniformly detected in the lemma and palea primordia (Figures 5B and 5C). In the stamen, in particular, TOB1 signal was restricted to the narrow region of the apical part of the anther primordia (Figure 5C). Notably, no expression was detected in the floret meristem (Figure 5B), raising the possibility that the meristem defect in tob1 was caused by a non-cell-autonomous action of TOB1.
Figure 5.
Temporal and Spatial Expression Pattern of TOB1 during Flower and Leaf Development.
In situ localizations of TOB1 transcripts are shown.
(A) Longitudinal section of wild-type inflorescence at an early stage when lateral organ primordia emerge.
(B) and (C) Longitudinal sections of a wild-type spikelet. A flower at the stage when lemma and palea initiate (B) and at the stage when stamen primordia emerge (C).
(D) Transverse section of a wild-type spikelet.
(E) and (F) Longitudinal (E) and transverse (F) section of a shoot apex of the wild type.
In the vegetative phase, TOB1 was expressed in the apical and peripheral region of the leaf primordia, and no expression was observed in the SAM, as found above for the floret meristem (Figures 5E and 5F).
Overexpression of TOB1 Is Likely to Be Associated with Enhanced Meristem Activity
To examine the function of TOB1 in more detail, we made transgenic rice plants that expressed TOB1 under the control of the rice ACTIN1 promoter, which induces gene expression strongly and constitutively in rice (Suzaki et al., 2008). In transgenic rice overexpressing TOB1, the spikelets were usually unclosed due to an excess number of floral organs (Figure 6A). All floral organs, such as pistils, stamens, and lodicules, were increased in TOB1-overexpressing plants (Figures 6B, 6C, and 6H, Table 2). In particular, the increase in pistil number was marked. The palea was replaced by the lemma, and ectopic palea (or flap)-like organs were formed in the lateral side (Figure 6D). Increases of floral organs and ectopic formation of palea-like organs are observed in fon mutants, which form an enlarged meristem by the failure of meristem maintenance (Suzaki et al., 2004, 2006). In some cases, an extra lemma-like organ was formed outside the floret (Figure 6E). Concerning organ numbers, therefore, TOB1 overexpression resulted in phenotypes that were roughly opposite to those of tob1 mutant. TOB1 overexpression sometimes resulted in disorganized spikelets, with abnormal morphology of the sterile lemma and aborted unidentified organs (Figures 6F and 6G). Although we did not observe obvious phenotypes in the vegetative phase or a reduction in the regeneration rate from calli, effects of TOB1 overexpression will be carefully analyzed in future studies.
Figure 6.
Effects of TOB1 Overexpression.
(A) to (H) Spikelet phenotypes in the transgenic plants overexpressing TOB1.
(A) An unclosed spikelet with an extra lemma instead of the palea.
(B), (C), and (H) Increase in the number of stamens (B), pistils ([B] and [C]), and lodicules (H).
(D) A spikelet that forms ectopic flap-like organs (arrowheads). One of the lemmas is removed to show inner organs.
(E) A spikelet with an extra lemma/palea-like organ (arrowhead) outside the floret.
(F) and (G) A disorganized spikelet with an abnormal sterile lemma (arrowhead in [F]) and aborted unidentified organs (arrowhead in [G]).
(I) to (L) In situ localization of OSH1 transcripts.
(I) In situ localization of OSH1 transcripts in a wild-type spikelet.
(J) to (L) In situ localization of OSH1 transcripts in developing spikelets of TOB1-overexpressing rice. Arrowheads indicate ectopic expression of OSH1. Asterisk indicates a club-shaped structure. The spikelets in (I) and (J) are roughly at the same developmental stage (stamen initiation).
fm, floret meristem; le, lemma; lo, lodicule; pa, palea; pi, pistil; sl, sterile lemma; st, stamen; WT, wild type. Bars = 1 mm in (A) to (H) and 50 μm in (I) to (L).
Table 2.
Distribution of the Number of Floral Organs in TOB1 Overexpression and TOB1-SRDX Lines
| Organ No. | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | No. of Spikelets Examined |
| Overexpression | ||||||||||||||
| Lodicule | 0 | 0 | 16 | 1 | 25 | 17 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 61 |
| Stamen | 0 | 0 | 1 | 0 | 0 | 0 | 13 | 8 | 17 | 11 | 7 | 2 | 2 | 61 |
| Pistil | 6 | 27 | 16 | 6 | 2 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 61 |
| SRDX | ||||||||||||||
| Lodicule | 0 | 0 | 66 | 0 | 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 91 |
| Stamen | 0 | 0 | 0 | 0 | 0 | 8 | 79 | 0 | 4 | 0 | 0 | 0 | 0 | 91 |
| Pistil | 0 | 65 | 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 91 |
The positions of underlined values correspond to the number of each organ in the wild type.
To characterize the effect of TOB1 overexpression, we examined the expression pattern of OSH1. In the wild type, OSH1 expression was observed in the spikelet meristem and was restricted to a narrow region between the stamen primordia just before initiation of the carpel primordia (Figures 3C and 6I). By contrast, the expression domain of OSH1 markedly expanded in the spikelet of TOB1-overexpressing plants, and the expression of OSH1 became much stronger compared with that of the wild type (Figures 6I to 6L). In addition, ectopic expression of OSH1 was observed between the floral organs and in the basal part of the spikelet under the meristem (Figures 6J and 6K). We infrequently observed spikelets with only a club-shaped structure, which appeared to be a large terminated meristem, inside the sterile lemmas (Figure 6L). In these spikelets, OSH1 was strongly expressed in the entire region of the club-shaped structure, except for the lateral epidermal region. No lateral organ was formed from the club-shaped structure, probably due to a failure in the downregulation of OSH1. Taken together, these results suggest that TOB1 overexpression enhances the indeterminate nature of the meristem and increases the number of meristematic cells in the spikelet.
RNA Interference of TOB1 Mimics the tob1 Phenotype but Expression of a Chimeric Repressor Does Not
To get further insight into TOB1 function, we produced transgenic plants in which the endogenous TOB1 (Os YABBY5) gene was suppressed by RNA interference (RNAi) because we failed to identify additional alleles of tob1 in TOS17 or T-DNA insertion lines. About 40% of the RNAi lines (22/53) produced spikelets showing class I and II phenotypes of the tob1 mutant. We focused on two lines in which the level of TOB1 transcript was greatly reduced (see Supplemental Figure 3 online). These RNA suppression lines of TOB1 produced all classes of the spikelets observed in tob1 mutant, including spikelets with severe phenotypes showing the cone-shaped organ or needle-like organ at the expense of the meristem (Figures 7A to 7D). These observations confirmed that class III, IV, and V tob1 spikelets were caused by a severe reduction of TOB1 (Os YABBY5) gene activity.
Figure 7.
Effects of RNAi Suppression of Endogenous TOB1 and Expression of TOB1-SRDX.
(A) to (D) Spikelet phenotypes resulting from RNAi suppression of TOB1.
(A) A spikelet with an elongated awn on the lemma (class I).
(B) A spikelet with a chimeric organ (arrowhead) comprising the sterile lemma and lemma (class III).
(C) A spikelet forming a cone-shaped organ instead of the lemma and palea (class IV)
(D) A spikelet producing a needle-like structure (arrowhead) just inside the sterile lemmas (class V).
(E) and (F) Spikelet phenotypes in transgenic rice expressing TOB1-SRDX.
(E) A spikelet with increased numbers of pistils (arrowheads) and lodicules and with a palea-like organ (arrow) ectopically formed in the lateral side.
(F) A spikelet with an extra lemma instead of the palea.
co, cone-shaped organ; le, lemma; lo, lodicule; pa, palea; sl, sterile lemma. Bars = 1mm.
The rice genome has two YABBY genes, Os YABBY3 and YABBY4, that are closely related to TOB1 (Figures 4B and 4C), suggesting that these three genes have redundant function. To confirm this idea, we employed chimeric repressor silencing technology in which a putative transcription factor is fused to an EAR repression domain (SRDX), resulting in dominant repression of the target genes of the chimeric protein (Hiratsu et al., 2002). This technology is useful for analyzing the function of redundant genes encoding positive transcription factors because the activities of related positive transcription factors are simultaneously inhibited by the silencing method (Hiratsu et al., 2003).
First, we made a construct to produce a fusion protein of GAL4DB-TOB1-SRDX. Transactivation activity of this fusion protein was examined by transient expression assay in Arabidopsis leaves using a luciferase (LUC) reporter construct (Hiratsu et al., 2002). The result indicated that GAL4DB-TOB1-SRDX had reduced reporter activities compared with GAL4DB, suggesting that TOB1-SRDX acts as a repressor (see Supplemental Figure 4 online). Therefore, we hypothesized that, in transgenic rice expressing the TOB1-SRDX fusion protein, the tob1 mutant phenotype might be enhanced by simultaneous repression of TOB1 and its related YABBY genes. Unexpectedly, however, transgenic plants expressing TOB1-SRDX did not show any phenotypes similar to those of tob1 mutant or RNAi suppression lines of TOB1. We could not find any spikelets with an elongated awn, the phenotype most frequently observed in tob1 spikelets (>85%), or any with a cone-shaped organ, a representative phenotype of tob1. Rather, the spikelets in plants expressing TOB1-SRDX partially resembled those of the TOB1-overexpressing plants because the number of floral organs, such as pistils and lodicules, increased (Figure 7E, Table 2), and the palea was replaced by a lemma-like organ in addition to formation of ectopic palea-like organs in the lateral region (cf. Figures 6A and 6D with 7E and 7F). These results indicate that TOB1 is not likely to act as a positive transcription factor but raise the possibility that TOB1 is involved in transcriptional repression.
DISCUSSION
In this article, we have shown that TOB1 regulates spikelet development and meristem fate in rice. A mutation in TOB1 resulted in unique pleiotropic phenotypes, including the formation of a cone-shaped organ without edges, the development of two florets in a spikelet, the formation of chimeric organs, and a reduction in floral organ number. The observation of arrested spikelets with a meristem-like dome or premature termination of the meristem at early stages of spikelet development suggests that cell fate and maintenance of the floret meristem are partially compromised in tob1. TOB1 encodes a YABBY protein, Os YABBY5, which is similar to Arabidopsis FIL and A. majus GRAM. TOB1 is expressed throughout the primordia of spikelet organs, whereas no TOB1 transcript was detected in the meristem. Taken together with meristem defects in tob1, it is suggested that TOB1 acts non-cell autonomously on the meristem.
TOB1 Encodes a YABBY Protein
TOB1 is a YABBY gene, Os YABBY5, which is closely related to Arabidopsis FIL and A. majus GRAM (Sawa et al., 1999b; Golz et al., 2004; Toriba et al., 2007). A mutation resulting in a stop codon in the YABBY domain, indicating loss of function of Os YABBY5, caused the tob1 phenotype. Consistent with this, a severe reduction in the expression level of endogenous Os YABBY5 by RNAi suppression led to spikelet phenotypes characteristic of tob1. Although a few studies concerning YABBY genes in this clade in grasses have been reported, no mutant has been described so far (Juarez et al., 2004; Dai et al., 2007).
TOB1 is expressed in the lateral organ primordia, such as spikelet organs and leaves. It is expressed uniformly: No polar distribution of TOB1 signal was detected. YABBY genes related to TOB1, such as FIL, YAB3, and GRAM, are expressed in the abaxial domain of the leaf primordia in eudicots, whereas they are expressed in the adaxial domain in maize (Sawa et al., 1999b; Siegfried et al., 1999; Golz et al., 2004; Juarez et al., 2004; Goldshmidt et al., 2008; Stahle et al., 2009). Thus, the spatial expression pattern of the TOB1/FIL clade is likely to be highly diverged in the angiosperms. TOB1 was not expressed in the meristem, which is the same as YABBY genes in this clade in other plants. The nuclear localization of TOB1 protein suggests that TOB1 acts as a transcriptional regulator similar to FIL and GRAM (Sawa et al., 1999b; Kanaya et al., 2002; Meister et al., 2002).
The Pleiotropic Phenotypes of the tob1 Spikelet Seem to Be Associated with Defects in Lateral Organ Development and Maintenance of Meristem Organization
In class II and III spikelets, the small sizes of the palea (class II) and the chimeric organ formation in the lemma (class III) indicate that TOB1 is required for normal growth of these organs. This may be a direct consequence of the loss of TOB1 function in the primordia of the lemma or palea, where TOB1 was found to be expressed. By contrast, careful consideration seems to be required to explain the phenotype of class IV spikelets (Figure 8A; see Supplemental Figure 5 online).
In class IV, the cone-shaped organ developed as a ring primordium from the initial stage. Structural features of the epidermal surface, combined with the initiation position of the cone-shaped organ, suggest that this organ is likely to be derived from the lemma or the palea. The IVa-type cone-shaped organ had the awn at the top, and two lodicules developed within this organ (IVa-type floret) (Figure 8A). By contrast, the IVb-type floret had no awn and no lodicule. In a wild-type spikelet, if it has an awn, the awn is formed only at the top of the lemma and the palea never develops the awn. In the wild type, the lodicules develop only at the lemma side with no exception. In addition, DL expression that marks the lemma, but not the palea, was associated with an existence of the lodicule (i.e., IVa-type floret). These observations suggest that the cone-shaped organ in class IVa is derived from the lemma, and the IVa-type floret may be developed from a floret meristem that lacks a domain involving the palea primordium (Figure 8B). Likewise, the IVb-type floret may develop from a floret meristem lacking a domain involving the lemma and lodicule primordia.
It is probable that TOB1 is required for initiation of the lemma or the palea, in addition to their normal growth as discussed above. The field of inhibition model of phyllotaxy hypothesizes that putative inhibitory compounds secreted from organ primordia restrict the position of subsequent primordium formation (Wardlaw, 1949; Steeves and Sussex, 1989; Goldshmidt et al., 2008). According to this hypothesis, it is possible that the failure in palea initiation caused by loss of TOB1 function allows the meristem to differentiate a ring-like primordium originated from the lemma primordium (IVa type floret) (see Supplemental Figure 5 online). Likewise, the failure in lemma initiation may be associated with the formation of the IVb-type floret with the palea-derived cone.
In some spikelets, we observed a large meristem with a groove crossing it and two arrested meristem-like domes in a spikelet. These observations suggest the possibility that the spikelet meristem might split into two floret meristems in the tob1 mutant. Such a split of the meristem may imply that proper meristem organization or zonation along the lemma–plea axis is disturbed, suggesting that TOB1 is required to maintain meristem organization. Class IVc spikelets with two florets are likely to result from this disorganized meristem and subsequent split of the meristem (Figure 8A; see Supplemental Figure 5 online).
TOB1 mutation also caused a mild reduction in the number of floral organs, such as lodicules and stamens. The reduction in the number of stamens is probably due to the size of the floret meristem, whereas the presence or absence of the lodicule may depend on the type of the floret, that is, whether it developed from the lemma hemisphere of the meristem or from the other hemisphere.
Thus, although tob1 showed pleiotropic phenotypes in the spikelet, these defects may be related to failure in initiation or growth of the lemma/palea primordia and disorganization of the meristem along the lemma–palea axis, which is probably associated with reduced communication between these primordia and the meristem.
Most japonica rice cultivars have no awn at present, suggesting that gene activities that promote awn elongation have been eliminated during domestication and subsequent improvements. In the tob1 mutant, all lemma had the long awn, similar to wild ancestral rice species. Therefore, japonica rice strains without the awn seem to have the potential to elongate the awn, but this potential is likely to be suppressed by the activity of TOB1.
TOB1 Is Likely to Act Non-Cell Autonomously on the Meristem
Various defects in the floret meristem were observed in tob1. The floret meristems were prematurely terminated at an early stage in spikelet development. Consistent with this, we found a spikelet in which the expression of OSH1, a maker of meristematic cells, disappeared in the meristem (Sato et al., 1996; Yamaguchi et al., 2004). Some meristems had one or a few grooves, running across the meristem. In some cases, the arrested meristem exhibited an epidermal surface similar to that of the lemma/palea. Whereas LHS1 is predominantly expressed in the primordia of the lemma/palea in the wild type, ectopic expression of LHS1 was detected in this type of arrested meristem in tob1. Even in mature spikelets, we sometimes observed dome-shaped structures, probably derived from an arrested meristem. Thus, maintenance and cell fate of the meristem is likely to be partially compromised in tob1. Because TOB1 was not expressed in the meristem, TOB1 seems to act non-cell autonomously to maintain meristem activity, suggesting again that this gene is involved in communication between the lateral organs and the meristem.
Overexpression of TOB1 resulted in an excess production of floral organs, such as pistils, stamens, and lodicules. The increase in pistil number was the most marked. The domain between the stamen primordia, corresponding to the floret meristem at this stage, was expanded, and greater accumulation of the OSH1 transcript was detected in this region. These phenotypes are similar to those in floral organ number (fon) mutants (fon1 or fon2), which have an enlarged meristem rich in stem cells (Suzaki et al., 2004, 2006). OSH1 was also ectopically expressed between floral organs or, in some cases, the expression domain expanded across the whole spikelet meristem. These results suggest that TOB1 overexpression may enhance the activity of the meristem and promote an indeterminate cell fate in developing spikelets.
Common and Distinct Features of YABBY Genes between Eudicots and Grasses
The spikelet phenotypes of tob1 seem to be similar to the floral phenotypes in fil mutants, such as failure in meristem maintenance, reduction in the floral organ number, and defects in arrangements and identity of floral organs (Chen et al., 1999; Sawa et al., 1999a, 1999b). FIL and related YABBY genes are known to be involved in robust meristem organization and regular phyllotaxy in Arabidopsis (Chen et al., 1999; Sawa et al., 1999a, 1999b; Goldshmidt et al., 2008; Stahle et al., 2009; Lugassi et al., 2010). This YABBY gene activity is non-cell autonomous because these genes are not expressed in the meristem, and movement of the YABBY proteins and RNA has not been detected (Goldshmidt et al., 2008). Thus, these YABBY genes are likely to have a role in short-range signaling from lateral organ primordia to the meristem. A similar mechanism may underlie the function of TOB1 in rice. If so, the contribution of YABBY gene activity in the FIL clade to short-range signaling between lateral organs and the meristem may be conserved in angiosperms.
In Arabidopsis, the expression domains of CLAVATA3 and WUSCHEL expand in a fil single or fil yab3 double mutant, and the meristems are enlarged in a fil mutant (Columbia background), suggesting that FIL and YAB3 negatively regulate stem cell maintenance (Goldshmidt et al., 2008; Lugassi et al., 2010). In triple (fil yab3/5) and quadruple (fil yab2/3/5) mutants, however, the primary SAM fails to be maintained, suggesting that these YABBY genes have a role to promote meristem activity (Sarojam et al., 2010). Thus, actions of the YABBY genes are complicated, and their function seems to depend on the genetic combinations and growth stages. In rice, the tob1 single mutant showed defects in maintenance and organization of the meristem in reproductive phase, suggesting that TOB1 promotes meristem activity and indeterminate cell fate. It will be intriguing to learn how the meristem is affected when the tob1 mutation is combined with other related YABBY genes, such as Os YABBY3 and YABBY4. In addition, further detailed elucidation of the mechanism of FIL/TOB1 signaling, including the final target, in both Arabidopsis and rice will provide clues to understanding YABBY-mediated communication between organ primordia and the meristem and its diverse role depending on plant species.
The effect of tob1 mutation on the meristem seems to result primarily from a failure in the initiation of the lemma or palea primordium, where TOB1 was strongly expressed. Although TOB1 was expressed in the primordia of the sterile lemma and rudimentary glume, no obvious defect was observed in these organs or in the spikelet meristems that initiate these organs. It is probable that closely related YABBY genes, such as Os YABBY3 and YABBY4, function redundantly in these organs (Toriba et al., 2007). Such a redundant function of these related YABBY genes would also explain the lack of abnormalities in the vegetative phase in tob1. It will be of great interest to determine whether TOB1 and its related YABBY genes are involved in lamina expansion, one of the key functions of FIL and its related YABBY genes in Arabidopsis (Siegfried et al., 1999; Goldshmidt et al., 2008; Stahle et al., 2009; Sarojam et al., 2010). TOB1 expression localized to the apical and peripheral region of leaf primordia might imply a distinct role in rice.
YABBY proteins physically interact with LUG/PROL or SEU to form repressor complexes in Arabidopsis and A. majus (Navarro et al., 2004; Stahle et al., 2009). This suggests that YABBY proteins act as negative regulators of transcription. In this study, we showed that the phenotype of TOB1-SRDX was unrelated to that of tob1, excluding the possibility that TOB1 functions as a positive regulator of transcription. Instead, spikelets in TOB1-SRDX plants showed phenotypes similar to those in TOB1-overexpressing plants. Therefore, it is probable that TOB1 is involved in the repression, rather than activation, of target genes.
METHODS
Plant Materials
The tob1 mutant was found in M2 plants of T65 that had been mutagenized with N-methyl-N-nitrosourea. F2 plants from a cross between tob1 (ssp japonica) and Kasalath (ssp indica) were used for map-based cloning of TOB1.
Scanning Electron Microscopy
Young panicles and spikelets were fixed in 4% (w/v) paraformaldehyde and 0.25% glutaraldehyde in 0.1 M sodium phosphate buffer, pH 7.2, at 4°C overnight. Next, they were dehydrated in a graded ethanol series, and 100% ethanol was replaced with 3-methylbutyl acetate. Samples were dried at their critical point, sputter coated with platinum, and observed with a scanning electron microscope (model JSM-820S; JEOL) at an accelerating voltage of 5 kV.
In Situ Hybridization
To generate probes for the TOB1 transcript, partial cDNA fragments were amplified with the primers listed in Supplemental Table 2 online and cloned into a pCRII vector (Invitrogen). Next, RNAs were transcribed with T7 or SP6 RNA polymerase from the above constructs as templates and were labeled with digoxigenin (Roche). For LHS1, the whole coding region was amplified using primers listed in Supplemental Table 2 online and cloned into a pT7 Blue vector (Novagen). After digestion with KpnI, the fragments were inserted into Bluescript SK+ (Stratagene). Next, probes were transcribed and partially hydrolyzed with alkaline solution (60 mM Na2CO3 and 40 mM NaHCO3, pH 10.2) at 60°C for 30 min. G1 and OSH1 probes were prepared as described previously (Sato et al., 1996; Yoshida et al., 2009). Preparation of sections and hybridization experiments were performed as described previously (Suzaki et al., 2004).
Isolation of the TOB1 Gene and Complementation Test
First, the TOB1 locus was mapped to a region between simple sequence repeat marker RM1223 and sequence tagged site marker S13714 on the long arm of chromosome 4 using 94 tob1 homozygotes. Next, the locus was narrowed to a 620-kb region between two closely linked derived cleaved-amplified polymorphic sequence markers, TCM07 and TCM46, which were designed by comparing the genomic sequences of japonica and indica. A database search revealed that a member of the YABBY gene family that plays important roles in plant development was located in this region. We hypothesized that mutation in this YABBY gene, Os YABBY5, caused the tob1 phenotypes, and then determined the genomic sequence of Os YABBY5 by a direct sequencing method. For the complementation test, an 8.6-kb genomic fragment containing the 2-kb Os YABBY5 coding sequence, coupled with the 5-kb upstream and 1.6-kb downstream sequences, was isolated by digestion of a BAC clone OSJNBb 0020O11 (Clemson University Genomics Institute BAC/EST Resource Center) with BalI and cloned into a pENTR 2B vector (Invitrogen). Then, the fragment was transferred into a vector for plant transformation, pBI-Hm12-GW, which contains the Gateway rfC cassette (Invitrogen), by an LR recombination reaction (Yoshida et al., 2009). The recombinant plasmid was introduced into Agrobacterium tumefaciens strain EHA101 and transformed into the tob1 mutant by the method described by Hiei et al. (1994).
Phylogenetic Analysis
Multiple sequence alignments and a phylogenetic tree of the zinc-finger and YABBY domains were generated with MEGA5 (Tamura et al., 2011). The multiple sequence alignment was made with the ClustalW module within MEGA5 using default parameters (gap opening penalty = 10, gap extension penalty = 0.2, protein weight matrix = Gonnet with residue-specific and hydrophylic penalties, gap separation distance = 4, and a 30% delay divergent cutoff). The alignment used to generate the tree is available as Supplemental Data Set 1 online. The tree was constructed using CRC and DL as outgroups by the neighbor-joining method (Saitou and Nei, 1987) within MEGA5 according to a p-distance model with uniform rates among sites and complete deletion of gaps/missing data. Bootstrap values were calculated from 1000 replicates.
RT-PCR Analysis of the TOB1 Transcripts
Total RNAs were isolated from tissues using TRIsure (BIOINE) according to the manufacturer’s protocol. For each sample, first-strand cDNA was synthesized from 5 μg of total RNA using a SuperScript III first-strand synthesis system for RT-PCR (Invitrogen) with the oligo(dT)15 primer. Next, 2 μL of each reverse transcription product was used for PCR using primers listed in Supplemental Table 2 online.
Subcellular Localization of the TOB Protein
The TOB1 coding sequence was amplified using the primers listed in Supplemental Table 2 online and cloned into a pENTR D-TOPO vector (Invitrogen). Next, by an LR recombination reaction, the fragment was transferred into a Gateway binary vector, pGWB5 or pGWB6, to produce a TOB-GFP or GFP-TOB fusion protein (Invitrogen). These constructs were bombarded into onion epidermal cells by a particle-mediated DNA delivery system (PDS1000/He; Bio-Rad). After overnight incubation at 22°C in the dark, the florescence of GFP was detected by a confocal microscanning laser microscope after the cells were stained with 4′,6-diamidino-2-phenylindole (Nikon).
Constructs for RNAi, Overexpression, and the Chimeric Repressor
To make a construct for RNAi, we first amplified a partial sequence of TOB1 cDNA using the primers listed in Supplemental Table 2 online and then cloned it into a pENTR D-TOPO vector (Invitrogen). The fragment was transferred by LR recombination reaction into a modified pANDA vector, pANDA-EG1 (Miki and Shimamoto, 2004; Suzaki et al., 2008). The recombinant plasmid was transformed into the T65 strain via A. tumefaciens (EHA101) by the method described by Hiei et al. (1994). To make constructs for overexpression and a chimeric repressor (SRDX), first the DNA sequence of the reading frame excluding the stop codon of a full-length TOB1 cDNA (accession number AK070205) was amplified by PCR using the primers AK070205Fw and AK070205Rv (see Supplemental Table 2 online), and the resulting PCR product was cloned independently by BP reaction into a Gateway entry vector, pDONR207 (Invitrogen). Next, TOB1 cDNA was transferred by LR reaction into two Gateway destination vectors, derivatives of the pSMAHdN636L-GateA binary vector (Hakata et al., 2010): pSMAHdN638GW for overexpression analysis and pSMAHdN643UGWRD for chimeric repressor analysis. In pSMAHdN638GW, a rice (Oryza sativa) ACTIN1 promoter directed TOB1 cDNA expression. In pSMAHdN643UGWRD, a maize (Zea mays) UBIQUITIN1 promoter directed the expression of the cDNA sequence encoding a chimeric protein of TOB1 and the repression domain (SRDX) of a rice protein (Os05g0286100) that is closely related to Arabidopsis thaliana SUPERMAN. The recombinant plasmids were transformed into a rice cultivar, Nipponbare, via A. tumefaciens (EHA105), according to Toki et al. (2006).
Transactivation Assay
A DNA fragment encoding TOB1-SRDX fusion protein was amplified using the primers listed in Supplemental Table 2 online and cloned into 35S:GAL4DB vector 430T1.2 (Ohta et al., 2000). The resulting construct was bombarded into Arabidopsis leaves with a reporter plasmid, modified Pro35S-GAL4-TATA-LUC (Hiratsu et al., 2002), in which transcriptional terminator of a heat shock gene was used (Pro35S-GAL4-TATA-LUC-HSP; Nagaya et al., 2010). We used 0.8 μg of the reporter plasmid and 0.6 μg of the effector construct for each bombardment. As an internal control, 0.1 μg of plasmid, phRLHSPPTRL, which included modified Renilla LUC gene (Promega) from Renilla under the control of 35S promoter and HSP terminator (Promega), was cobombarded with both reporter and effector plasmids. After bombardment, samples were incubated for 10 h in darkness, and then luciferase activity was determined.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: AB274017 (Os YABBY5), AB274013 (Os YABBY1), AB274014 (Os YABBY2), AB274015 (Os YABBY3), AB274016 (Os YABBY4), AB274018 (Os YABBY6), AB274019 (Os YABBY7), AB106553 (DL), AF136538 (FIL), AF136539 (YAB2), AF136540 (YAB3), AF195047 (INNER NO OUTER), AK119091 (YAB5), AF132606 (CRC), AJ804783 (GRAM), AJ559762 (PROL), AY31390 (zyb9), AY313904 (zyb10), AY313901 (zyb14), and AY313902 (zyb15).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. In Situ Localization of DL Transcripts.
Supplemental Figure 2. Subcellular Localization of TOB1 Protein.
Supplemental Figure 3. Expression Levels of TOB1 mRNA Analyzed by RT-PCR.
Supplemental Figure 4. The Repression Activity of TOB1-SRDX.
Supplemental Figure 5. Models of tob1 Spikelet Development in Each Class.
Supplemental Table 1. Percentage of Observed Phenotypic Classes.
Supplemental Table 2. Primers Used in This Study.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis in Figure 4C.
Acknowledgments
We thank Masaru Fujimoto and Takuya Suzaki for technical support, Mafumi Abiko and Keiko Ohsawa-Yamamoto for technical assistance, and technicians at the Institute for Sustainable Agro-Ecosystem Services at the University of Tokyo for cultivation of rice. This research was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (20380005 and 23012011 to H.-Y.H.), the Program of Basic Research Activities for Innovative Biosciences (to H.-Y.H. and Y.O.), the Global Center of Excellence Program (Integrative Life Science Based on the Study of Biosignaling Mechanisms) from the Ministry of Education, Culture, Sports, Science and Technology (to W.T.), a Research Fellowship for Young Scientists from the Japan Society for the Promotion of Science (to W.T.), and grants from the Ministry of Agriculture, Forestry and Fisheries (Genomics for Agricultural Innovation, AMR0001 and AMR0004, to H.I. and M.O.-T.).
AUTHOR CONTRIBUTIONS
W.T. and H.-Y.H. designed the research. W.T., T.T., Y.O, A.Y., A.K., T.M.-T., H.I., N.M., and M.O.-T. performed the research. W.T. and H.-Y.H. wrote the article.
References
- Arber A. (1934). The Gramineae: A Study of Cereal, Bamboo, and Grasses. (Cambridge, UK: Cambridge University Press) [Google Scholar]
- Bowman J.L., Smyth D.R. (1999). CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126: 2387–2396 [DOI] [PubMed] [Google Scholar]
- Chen Q.Y., Atkinson A., Otsuga D., Christensen T., Reynolds L., Drews G.N. (1999). The Arabidopsis FILAMENTOUS FLOWER gene is required for flower formation. Development 126: 2715–2726 [DOI] [PubMed] [Google Scholar]
- Dai M., Hu Y., Zhao Y., Liu H., Zhou D.-X. (2007). A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development. Plant Physiol. 144: 380–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreni L., Pilatone A., Yun D.P., Erreni S., Pajoro A., Caporali E., Zhang D.B., Kater M.M. (2011). Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy. Plant Cell 23: 2850–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed Y., Baum S.F., Bowman J.L. (1999). Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99: 199–209 [DOI] [PubMed] [Google Scholar]
- Fourquin C., Vinauger-Douard M., Fogliani B., Dumas C., Scutt C.P. (2005). Evidence that CRABS CLAW and TOUSLED have conserved their roles in carpel development since the ancestor of the extant angiosperms. Proc. Natl. Acad. Sci. USA 102: 4649–4654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldshmidt A., Alvarez J.P., Bowman J.L., Eshed Y. (2008). Signals derived from YABBY gene activities in organ primordia regulate growth and partitioning of Arabidopsis shoot apical meristems. Plant Cell 20: 1217–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golz J.F., Roccaro M., Kuzoff R., Hudson A. (2004). GRAMINIFOLIA promotes growth and polarity of Antirrhinum leaves. Development 131: 3661–3670 [DOI] [PubMed] [Google Scholar]
- Hakata M., et al. (2010). Production and characterization of a large population of cDNA-overexpressing transgenic rice plants using Gateway-based full-length cDNA expression libraries. Breed. Sci. 60: 575–585 [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Hirano H.-Y., Hirai A., Sano Y., Sasaki T. (2008). Rice biology in the Genomics Era. (Heidelberg, Germany: Springer) [Google Scholar]
- Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Hiratsu K., Ohta M., Matsui K., Ohme-Takagi M. (2002). The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Lett. 514: 351–354 [DOI] [PubMed] [Google Scholar]
- Ishikawa M., Ohmori Y., Tanaka W., Hirabayashi C., Murai K., Ogihara Y., Yamaguchi T., Hirano H.-Y. (2009). The spatial expression patterns of DROOPING LEAF orthologs suggest a conserved function in grasses. Genes Genet. Syst. 84: 137–146 [DOI] [PubMed] [Google Scholar]
- Jeon J.S., Jang S., Lee S., Nam J., Kim C., Lee S.H., Chung Y.Y., Kim S.R., Lee Y.H., Cho Y.G., An G. (2000). leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12: 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez M.T., Twigg R.W., Timmermans M.C.P. (2004). Specification of adaxial cell fate during maize leaf development. Development 131: 4533–4544 [DOI] [PubMed] [Google Scholar]
- Kanaya E., Nakajima N., Okada K. (2002). Non-sequence-specific DNA binding by the FILAMENTOUS FLOWER protein from Arabidopsis thaliana is reduced by EDTA. J. Biol. Chem. 277: 11957–11964 [DOI] [PubMed] [Google Scholar]
- Kumaran M.K., Bowman J.L., Sundaresan V. (2002). YABBY polarity genes mediate the repression of KNOX homeobox genes in Arabidopsis. Plant Cell 14: 2761–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-Y., Baum S.F., Oh S.-H., Jiang C.-Z., Chen J.-C., Bowman J.L. (2005). Recruitment of CRABS CLAW to promote nectary development within the eudicot clade. Development 132: 5021–5032 [DOI] [PubMed] [Google Scholar]
- Li H., Xue D., Gao Z., Yan M., Xu W., Xing Z., Huang D., Qian Q., Xue Y. (2009). A putative lipase gene EXTRA GLUME1 regulates both empty-glume fate and spikelet development in rice. Plant J. 57: 593–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Karmarkar V. (2008). Groucho/Tup1 family co-repressors in plant development. Trends Plant Sci. 13: 137–144 [DOI] [PubMed] [Google Scholar]
- Liu Z., Meyerowitz E.M. (1995). LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development 121: 975–991 [DOI] [PubMed] [Google Scholar]
- Lugassi N., Nakayama N., Bochnik R., Zik M. (2010). A novel allele of FILAMENTOUS FLOWER reveals new insights on the link between inflorescence and floral meristem organization and flower morphogenesis. BMC Plant Biol. 10: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister R.J., Kotow L.M., Gasser C.S. (2002). SUPERMAN attenuates positive INNER NO OUTER autoregulation to maintain polar development of Arabidopsis ovule outer integuments. Development 129: 4281–4289 [DOI] [PubMed] [Google Scholar]
- Miki D., Shimamoto K. (2004). Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 45: 490–495 [DOI] [PubMed] [Google Scholar]
- Nagasawa N., Miyoshi M., Sano Y., Satoh H., Hirano H.-Y., Sakai H., Nagato Y. (2003). SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130: 705–718 [DOI] [PubMed] [Google Scholar]
- Nagaya S., Kawamura K., Shinmyo A., Kato K. (2010). The HSP terminator of Arabidopsis thaliana increases gene expression in plant cells. Plant Cell Physiol. 51: 328–332 [DOI] [PubMed] [Google Scholar]
- Nakayama H., Yamaguchi T., Tsukaya H. (2010). Expression patterns of AaDL, a CRABS CLAW ortholog in Asparagus asparagoides (Asparagaceae), demonstrate a stepwise evolution of CRC/DL subfamily of YABBY genes. Am. J. Bot. 97: 591–600 [DOI] [PubMed] [Google Scholar]
- Navarro C., Efremova N., Golz J.F., Rubiera R., Kuckenberg M., Castillo R., Tietz O., Saedler H., Schwarz-Sommer Z. (2004). Molecular and genetic interactions between STYLOSA and GRAMINIFOLIA in the control of Antirrhinum vegetative and reproductive development. Development 131: 3649–3659 [DOI] [PubMed] [Google Scholar]
- Ohmori Y., Abiko M., Horibata A., Hirano H.-Y. (2008). A transposon, Ping, is integrated into intron 4 of the DROOPING LEAF gene of rice, weakly reducing its expression and causing a mild drooping leaf phenotype. Plant Cell Physiol. 49: 1176–1184 [DOI] [PubMed] [Google Scholar]
- Ohmori Y., Toriba T., Nakamura H., Ichikawa H., Hirano H.-Y. (2011). Temporal and spatial regulation of DROOPING LEAF gene expression that promotes midrib formation in rice. Plant J. 65: 77–86 [DOI] [PubMed] [Google Scholar]
- Ohta M., Ohme-Takagi M., Shinshi H. (2000). Three ethylene-responsive transcription factors in tobacco with distinct transactivation functions. Plant J. 22: 29–38 [DOI] [PubMed] [Google Scholar]
- Orashakova S., Lange M., Lange S., Wege S., Becker A. (2009). The CRABS CLAW ortholog from California poppy (Eschscholzia californica, Papaveraceae), EcCRC, is involved in floral meristem termination, gynoecium differentiation and ovule initiation. Plant J. 58: 682–693 [DOI] [PubMed] [Google Scholar]
- Prasad K., Parameswaran S., Vijayraghavan U. (2005). OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. Plant J. 43: 915–928 [DOI] [PubMed] [Google Scholar]
- Reinhardt D., Frenz M., Mandel T., Kuhlemeier C. (2005). Microsurgical and laser ablation analysis of leaf positioning and dorsoventral patterning in tomato. Development 132: 15–26 [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sarojam R., Sappl P.G., Goldshmidt A., Efroni I., Floyd S.K., Eshed Y., Bowman J.L. (2010). Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell 22: 2113–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Hong S.K., Tagiri A., Kitano H., Yamamoto N., Nagato Y., Matsuoka M. (1996). A rice homeobox gene, OSH1, is expressed before organ differentiation in a specific region during early embryogenesis. Proc. Natl. Acad. Sci. USA 93: 8117–8122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa S., Ito T., Shimura Y., Okada K. (1999a). FILAMENTOUS FLOWER controls the formation and development of Arabidopsis inflorescences and floral meristems. Plant Cell 11: 69–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa S., Watanabe K., Goto K., Liu Y.G., Shibata D., Kanaya E., Morita E.H., Okada K. (1999b). FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 13: 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried K.R., Eshed Y., Baum S.F., Otsuga D., Drews G.N., Bowman J.L. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126: 4117–4128 [DOI] [PubMed] [Google Scholar]
- Stahle M.I., Kuehlich J., Staron L., von Arnim A.G., Golz J.F. (2009). YABBYs and the transcriptional corepressors LEUNIG and LEUNIG_HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis. Plant Cell 21: 3105–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves T.A., Sussex I.M. (1989). Patterns in Plant Development. (Cambridge, UK: Cambridge University Press) [Google Scholar]
- Sussex I.M. (1951). Experiments on the cause of dorsiventrality in leaves. Nature 167: 651–652 [DOI] [PubMed] [Google Scholar]
- Sussex I.M. (1954). Morphogenesis in Solanum tuberosum L.: experimental investigation of leaf dorsiventrality and orientation in the juvenile shoot. Phytomorphology 5: 286–300 [Google Scholar]
- Suzaki T., Sato M., Ashikari M., Miyoshi M., Nagato Y., Hirano H.-Y. (2004). The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development 131: 5649–5657 [DOI] [PubMed] [Google Scholar]
- Suzaki T., Toriba T., Fujimoto M., Tsutsumi N., Kitano H., Hirano H.-Y. (2006). Conservation and diversification of meristem maintenance mechanism in Oryza sativa: Function of the FLORAL ORGAN NUMBER2 gene. Plant Cell Physiol. 47: 1591–1602 [DOI] [PubMed] [Google Scholar]
- Suzaki T., Yoshida A., Hirano H.-Y. (2008). Functional diversification of CLAVATA3-related CLE proteins in meristem maintenance in rice. Plant Cell 20: 2049–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toki S., Hara N., Ono K., Onodera H., Tagiri A., Oka S., Tanaka H. (2006). Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 47: 969–976 [DOI] [PubMed] [Google Scholar]
- Toriba T., Harada K., Takamura A., Nakamura H., Ichikawa H., Suzaki T., Hirano H.-Y. (2007). Molecular characterization the YABBY gene family in Oryza sativa and expression analysis of OsYABBY1. Mol. Genet. Genomics 277: 457–468 [DOI] [PubMed] [Google Scholar]
- Toriba T., Suzaki T., Yamaguchi T., Ohmori Y., Tsukaya H., Hirano H.-Y. (2010). Distinct regulation of adaxial-abaxial polarity in anther patterning in rice. Plant Cell 22: 1452–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva J.M., Broadhvest J., Hauser B.A., Meister R.J., Schneitz K., Gasser C.S. (1999). INNER NO OUTER regulates abaxial- adaxial patterning in Arabidopsis ovules. Genes Dev. 13: 3160–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardlaw C.W. (1949). Experiments on organogenesis in ferns. Growth 13(suppl.): 93–131 [Google Scholar]
- Yamada T., Yokota S., Hirayama Y., Imaichi R., Kato M., Gasser C.S. (2011). Ancestral expression patterns and evolutionary diversification of YABBY genes in angiosperms. Plant J. 67: 26–36 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T., Lee D.Y., Miyao A., Hirochika H., An G., Hirano H.-Y. (2006). Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa. Plant Cell 18: 15–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Nagasawa N., Kawasaki S., Matsuoka M., Nagato Y., Hirano H.-Y. (2004). The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16: 500–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A., Suzaki T., Tanaka W., Hirano H.-Y. (2009). The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet. Proc. Natl. Acad. Sci. USA 106: 20103–20108 [DOI] [PMC free article] [PubMed] [Google Scholar]