This article examines the role of cyclin-dependent kinase (Cdk) activity in pairing of homoeologous chromosomes in wheat (Triticum aestivum), finding that the Ph1 locus suppresses Cdk2-type activity, thus affecting replication and histone H1 phosphorylation.
Abstract
Despite possessing multiple sets of related (homoeologous) chromosomes, hexaploid wheat (Triticum aestivum) restricts pairing to just true homologs at meiosis. Deletion of a single major locus, Pairing homoeologous1 (Ph1), allows pairing of homoeologs. How can the same chromosomes be processed as homologs instead of being treated as nonhomologs? Ph1 was recently defined to a cluster of defective cyclin-dependent kinase (Cdk)-like genes showing some similarity to mammalian Cdk2. We reasoned that the cluster might suppress Cdk2-type activity and therefore affect replication and histone H1 phosphorylation. Our study does indeed reveal such effects, suggesting that Cdk2-type phosphorylation has a major role in determining chromosome specificity during meiosis.
INTRODUCTION
Organisms exhibiting sexual reproduction carry two copies (homologs) of each chromosome. Before meiosis, each homolog is replicated, forming two sister chromatids that remain linked together. At the start of meiosis, each chromosome must recognize its homolog from among all of the chromosomes present in the nucleus. The homologs generally then become intimately aligned or paired along their entire lengths. A proteinaceous structure known as the synaptonemal complex (SC) is assembled between them in a process termed synapsis (reviewed in Zickler and Kleckner, 1998). This process is preceded by clustering of the chromosome telomeres to form a bouquet at the onset of meiosis. Synapsis of the chromosomes is initiated in the telomere regions. Meiotic recombination occurs through the generation of double strand breaks (DSBs), which are then repaired after synapsis using the homologs. Thus, meiotic recombination forms chiasmata, physical links that hold the homologs together after the disassembly of the SC. The homologs are then segregated, so that each gamete carries only a single copy of each chromosome.
In contrast with mammals, many plant species are polyploid. Despite possessing multiple genomes, only true homologous chromosomes pair during meiosis in these species. This correct pairing involves suppressing associations between the multiple sets of related chromosomes. Such diploid-like chromosome pairing in hexaploid wheat (Triticum aestivum) and tetraploid wheat (Triticum turgidum) is under the genetic control of a single major dominant locus, the Pairing homoeologous1 (Ph1) locus, located on chromosome 5B (Okamoto, 1957; Riley and Chapman, 1958; Sears and Okamoto, 1958). Wheat lacking Ph1 accumulates extensive chromosome rearrangements and eventually becomes infertile (Sánchez-Morán et al., 2001). In wheat, therefore, Ph1 stabilizes the polyploid genome, resulting in the high fertility of this crop. Sexual hybridization between wheat and a wild relative generally produces an interspecific hybrid containing a haploid set of only related chromosomes (homoeologs), in which chromosome pairing is largely prevented as a result of the presence of Ph1 (Dhaliwal et al., 1977). Therefore, understanding the basis for this pairing suppression will have the important practical application of enabling researchers to switch the pairing on and off, thereby enhancing breeding strategies (Able and Langridge, 2006).
Molecular analysis of the relevant region of chromosome 5B has defined the Ph1 locus to a region containing a cluster of cyclin-dependent kinase (Cdk)-like genes, which share some sequence similarity to mammalian Cdk2 (Griffiths et al., 2006; Al-Kaff et al., 2008; Yousafzai et al., 2010a). Cdks are a group of Ser/Thr protein kinases that control the progression through the various phases of the cell cycle and subsequent cell divisions in eukaryotic cells. Cdk2 has been shown to participate in the transition from G1 into S phase and also to affect DNA replication (Nasmyth, 1996; Thomson et al., 2010). Surprisingly, although the 5B Cdk-like genes are transcribed, they all seem to be defective copies (see Methods). Thus, the hypothesis is that these defective Cdk-like genes are responsible for reducing Cdk-type activity, and that this leads to the Ph1 effect. If this is the case, then increased Cdk-type activity should phenocopy the effect of deleting the Ph1 locus. Treatment with okadaic acid, a Ser-Thr phosphatase inhibitor, increases Cdk-type activity (Yamashita et al., 1990; Ajiro et al., 1996). A recent study has shown that okadaic acid treatment of wheat–rye (Secale cereale) interspecific hybrids does indeed induce homoeologous chromosome pairing even in the presence of Ph1, thereby phenocopying the effect of deleting Ph1 (Knight et al., 2010). This implies that Cdk-type phosphorylation affects chromosome pairing. This raises the question of whether it is possible to demonstrate that Ph1 actually suppresses Cdk-type activity, and, if so, whether this suppression is specifically of Cdk2-type activity. Mammalian Cdk2 phosphorylates many substrates, including histone H1, at specific Cdk2 consensus sites (Garcia et al., 2004). We therefore investigated whether we could carry out phosphoproteomics on limited numbers of staged wheat meiocytes. This would reveal whether wheat histone H1 is phosphorylated at similar Cdk2-type consensus sites to mammalian histone H1, and whether Ph1 alters the level of phosphorylation of these sites; this would suggest that Ph1 acts by affecting Cdk2-type activity. We also investigated whether Ph1 alters premeiotic DNA replication. Our study does indeed reveal both increased Cdk2-type phosphorylation of histone H1 in the absence of Ph1 and alterations in the course of replication.
RESULTS AND DISCUSSION
Ph1 Affects Premeiotic Replication
If the Cdk-like activity regulated by Ph1 is similar to mammalian Cdk2, then it should alter the replication process. To assess whether this is the case, we used wheat–rye hybrids either carrying the Ph1 locus (Ph1+) or carrying the Ph1 deletion (Ph1−) in the wheat genome. These hybrids have a haploid set of 21 wheat chromosomes and seven rye chromosomes, and replication of the rye heterochromatin knobs can be easily visualized in these lines. The pattern of replication in wheat–rye anthers was originally analyzed by diffusing bromodeoxyuridine into live excised wheat tillers. However we repeated the entire study using 5-ethynyl-2′-deoxyuridine (EdU) (a thymidine analog) when this technology became available, because of better sensitivity and structural preservation and the ease of dual labeling with either centromere or telomere probes. After EdU diffusion for 4 h, the anthers were fixed, and the labeling patterns in premeiotic anther sections were imaged using three-dimensional (3D) fluorescence microscopy. Therefore, in these experiments, any DNA that is replicated within the 4-h period before fixation will potentially incorporate EdU. When the anthers are fixed and the labeling visualized, all chromosome structures replicating within the preceding 4-h period will potentially appear as labeled, even if they individually replicate at different times within the 4-h period. We have analyzed a minimum of 50 anther sections for each of the replication stages that we categorized, in the presence and absence of Ph1. These whole anther sections typically contain between four and eight meiocytes, surrounded by the tapetal cell layer. The total number of meiocytes scored using 3D images stacks within these anther sections was 611 in the presence of Ph1 and 398 in the absence of Ph1 (Table 1). The analysis showed that either in the presence or absence of Ph1, premeiotic S phase commences in the meiocytes with the replication of dispersed chromatin in the same images as, and thus within 4 h of, the replication of tapetal cells surrounding the meiocytes (Figure 1). After the tapetal cell replication finishes, the dispersed chromatin is still observed in full replication (i.e., throughout the nucleus) in the meiocytes. The rye heterochromatin knob replication and full replication of the dispersed chromatin are observed in the same images, and thus occur within 4 h of each other, in the presence of Ph1. In the absence of Ph1, the rye heterochromatin knob replication is rarely observed in meiocytes with the dispersed chromatin in full replication. The pericentromeric regions and rye heterochromatin knobs finish replicating, in the presence and absence of Ph1, within the 4-h period before the full telomere bouquet formation at the onset of meiosis (Figures 1E, 1J,2D, and 3F). Thus, either with or without Ph1, dispersed chromatin replication is initiated in the meiocytes as the tapetal cells are finishing their replication, and all meiocyte replication is completed within the 4-h period before the telomeres form the bouquet.
Table 1.
Analysis of the Replication Progression in Meiocytes in the Presence (Ph1+) and Absence (Ph1−) of the Ph1 Locus
| Category | Ph1 Locus | No. of Cells | Proportion of Cells | se | Student’s t Test Probability |
| 1) Dispersed chromatin only | Ph1+ | 343 | 0.56 | 0.03 | P = 0.065 |
| Ph1− | 252 | 0.65 | 0.04 | ||
| 2) Dispersed chromatin and heterochromatin knobs | Ph1+ | 102 | 0.17 | 0.02 | P < 0.001 |
| Ph1− | 8 | 0.02 | 0.01 | ||
| 3) Only heterochromatin knobs | Ph1+ | 166 | 0.27 | 0.02 | P = 0.034 |
| Ph1− | 138 | 0.35 | 0.03 |
Each nucleus was independently classified as in category 1) (dispersed chromatin only), category 2) (dispersed chromatin + heterochromatin knobs), or category 3) (only heterochromatin knobs) from either Ph1+ or Ph1−. A multinomial regression in a general linear model was then performed to conduct an analysis of deviance, and the means were then compared using Student’s t test probabilities within the regression model. The difference was highly significant between Ph1+ and Ph1− in category 2) but of marginal significance between categories 1) and 2).
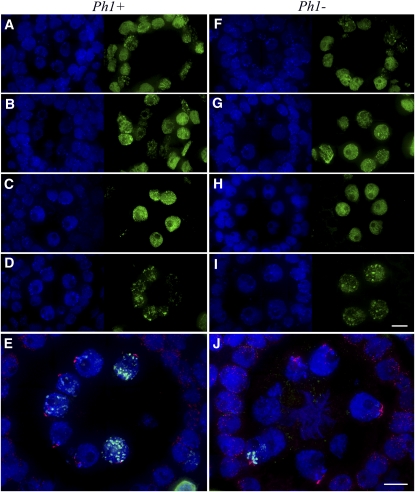
Figure 1.
Whole Anther Sections Showing Tapetal Cell and Meiocyte Replication in the Presence and Absence of the Ph1 Locus.
EdU incorporation, indicating active DNA synthesis, is shown in green, and telomeres (E) and (J) are shown in magenta. All sections were counterstained with DAPI (blue).
(A) and (F) Prior to meiocyte replication, the tapetal and middle layer cells replicate asynchronously.
(B) Meiocytes with dispersed chromatin replication are seen in the same images as tapetal and middle cell replication.
(G) Middle layer cells are seen replicating in the same images as meiocytes replicating.
(C) and (H) After tapetal cell and middle layer replication has finished, the dispersed chromatin in the meiocytes is still in full replication.
(D) and (I) The rye heterochromatin knobs are predominantly replicating along with the pericentromeric regions in the meiocytes.
(E) and (J) Merged images showing that replication of heterochromatin knobs is seen in the same images as the full telomere bouquet in the meiocytes. In (J), replication in most meiocytes showing a tight telomere bouquet formation has finished, apart from one meiocyte with knobs still replicating.
Bar = 10 μm.
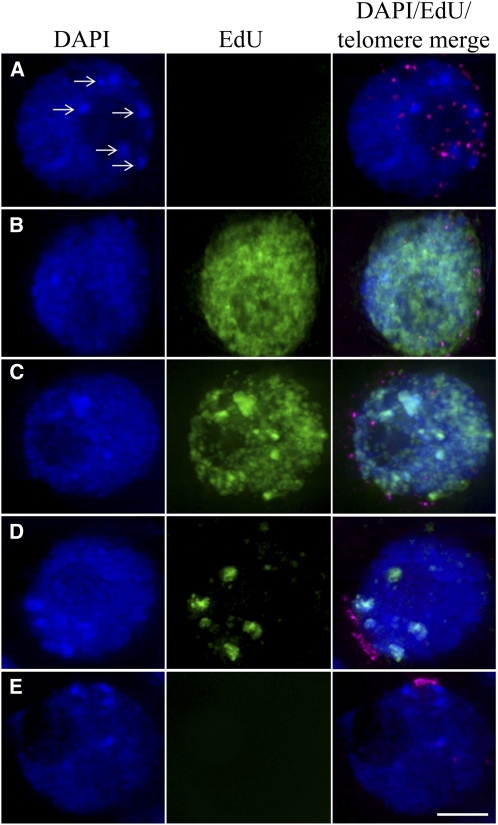
Figure 2.
Progression of DNA Replication within Meiocytes in Relation to Telomere Dynamics.
These images are derived from cross-sections through entire anther locules and show the central premeiotic cells at different stages in their development. Telomeres are shown in magenta in the merged panel. EdU incorporation, indicating active DNA synthesis, is shown in green. DNA, stained by DAPI, is shown in blue.
(A) Prior to replication, with the telomeres dispersed around approximately one-half of the nuclear periphery.
(B) Dispersed chromatin in full replication, with the telomeres beginning to cluster together on the nuclear membrane.
(C) The rye subtelomeric heterochromatin knobs (the intensely DAPI-stained regions indicated by the arrows in [A]) replicating, with the dispersed chromatin in full replication as the telomeres still clustering.
(D) The rye heterochromatin knobs replicating, with the telomeres close to a full bouquet.
(E) All replication finished, with the telomeres as a tight cluster or bouquet on the nuclear membrane.
Bar = 5 μm.
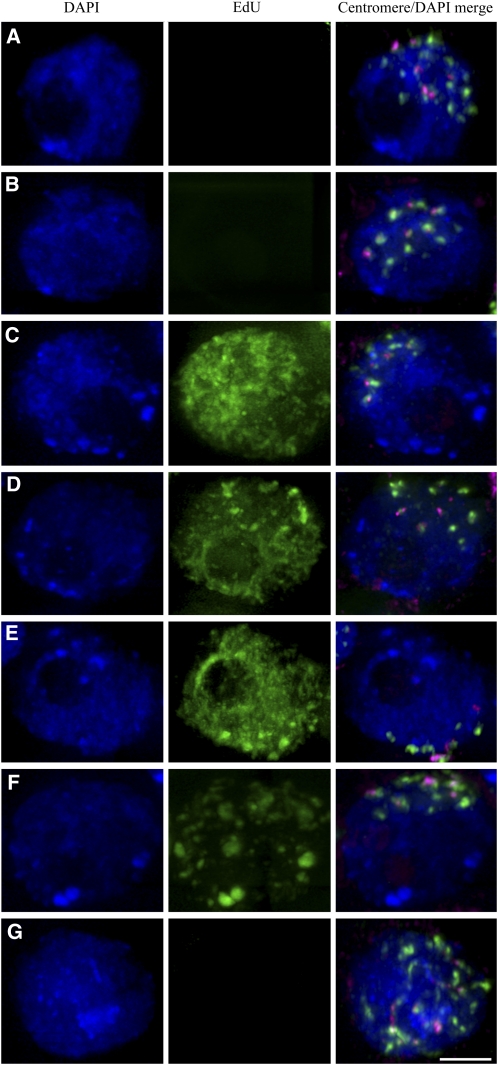
Figure 3.
The Dynamics of Wheat and Rye Centromeres throughout Premeiotic Replication.
These images are derived from cross-sections through entire anther locules and show the central premeiotic cells at different stages in their development. DNA, stained by DAPI, is shown in blue, and EdU incorporation, indicating active DNA synthesis, is shown in green in the central panels. In the merged panel at right, wheat centromeres are shown in green, and rye centromeres are shown in magenta, with DNA shown in blue. Prior to replication, the centromeres are unpaired (A). Just before replication begins, the centromeres associate as an average of 10 sites (B). As replication commences, the centromeric associations persist, maintaining between 14 and seven centromeric sites. The centromeres replicate within 4 h of the dispersed chromatin replication (C) to (E). During the later stages of meiocyte replication, up to 4 h after the rye subtelomeric heterochromatin knob replication, the centromeres begin to separate (F). As replication finishes, the centromeres are unpaired (G) and begin to remodel.
Bar = 5 μm.
There is, however, a difference in how replication progresses between Ph1+ and Ph1−. Premeiotic S phase can be split into three stages: 1) dispersed chromatin replication only (corresponding to the images in Figure 2B), 2) dispersed chromatin and rye heterochromatin knob replication (corresponding to the images in Figure 2C), and 3) predominantly rye heterochromatin knob replication (corresponding to the images in Figure 2D). Meiocyte nuclei (Ph1+ and Ph1−) were assigned to each category by visual observation. Analysis of these data are shown in Table 1 and demonstrated that the difference between Ph1+ and Ph1− samples in the proportion of nuclei in categories 1) and 3) was of marginal significance. However, there were significantly fewer Ph1− nuclei in category 2) than Ph1+ nuclei (Table 1). Cdk2 has been shown to activate origins of replication (Krasinska et al., 2008), and one explanation for our observations is that the absence of Ph1 (hence higher Cdk-like activity) increases the activation of origins and hence the rate of replication of the dispersed chromatin. An alternative explanation is that the initiation of heterochromatin replication is delayed in the absence of Ph1. Okadaic acid treatment, which mimics the effect of deleting Ph1 by inducing chromosome pairing in wheat–rye hybrids in the presence of Ph1 (Knight et al., 2010), also delays heterochromatin replication (Polit and Kazmiercazk, 2007). Therefore, as with the okadaic acid treatment, Ph1 could be altering heterochromatin structure and replication.
The chromosomes are intimately aligned while at the telomere bouquet stage, although some homologs in wheat can be paired via their telomeres prior to the telomere bouquet being formed (Prieto et al., 2004; Colas et al., 2008). Previous studies revealed that centromeres start associating during floral development and decrease to seven sites at the onset of meiosis, and Ph1 promotes homologous centromere association at this stage (Martinez-Perez et al., 2001; Martinez-Perez et al., 2003). Analysis of the premeiotic S phase period reveals that the 21 wheat centromeres (representing seven centromeres from each of the three wheat genomes) and the seven rye centromeres are associated as ~10 sites at the beginning of replication (Figure 3; see Supplemental Figure 1 online). The mean number of sites visible then decreases during premeiotic S phase, but the number is not fixed, varying between seven and 14 sites. This suggests a dynamic process in which the centromeres are continually associating and disassociating throughout the premeiotic S phase. Initially, the telomeres are dispersed across one-half of the nuclear membrane before the start of replication (Figure 2A). Telomeres are observed to begin to cluster in images where dispersed replication is visible (Figures 2B and 2C) and are nearly fully clustered in images where only the late rye heterochromatin is being replicated. This suggests that the telomeres start reorganizing themselves to form the bouquet during premeiotic S phase; that is, that the pairing process is initiated before the completion of replication.
Ph1 Alters Histone H1 Phosphorylation
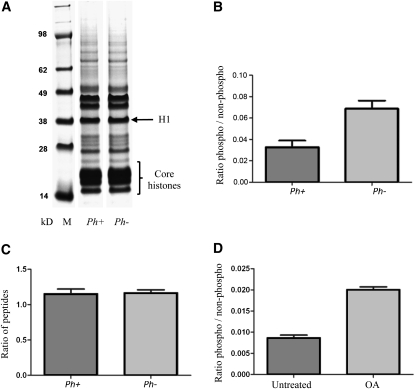
Many proteins are known to be phosphorylated by Cdk2 on specific consensus sites (Garcia et al., 2004). If Ph1 affects Cdk2-type activity, then it would be expected to affect the phosphorylation of similar sites. Histone H1 is one of the best-characterized Cdk2 substrates and was selected for study because it could be relatively easily purified from small amounts of meiotic tissue. In wheat florets, the three anthers and the meiocytes within them are highly synchronized in development, so that one anther can be used for staging by microscopy, leaving the other two anthers for biochemical analysis. In histone fractions purified from wheat–rye anthers staged in early meiotic prophase, there is no visible difference between Ph1+ and Ph1− samples in the proportions of the different histones (Figure 4A). Detailed proteomic analysis of the histone H1 fraction (arrow in Figure 4A) after digestion with trypsin and endoproteinase AspN and mass spectrometry showed the occurrence of phosphorylation at SPAK, TPKK, and TPAAK sites (Figure 5). These sites are very similar to those reported for consensus Cdk2 phosphorylation sites, (S/T) –P-X-Z, from studies of mammalian histones (Garcia et al., 2004). The peptide containing the TPAAK site was observed frequently enough to make a statistically reliable estimate of the relative level of phosphorylation between Ph1+ and Ph1− samples. This showed that the TPAAK site is consistently twofold more phosphorylated in Ph1− samples than in Ph1+ samples (Figure 4B; see Supplemental Table 1 online) (A two-tailed unpaired t test on the Ph1+ and Ph1− biological samples gives a P value of 0.003). As a control, levels of two regularly observed peptides from histone H1 (SGSTIAIGK and ILLTQIK) were compared in the same way, giving a nonsignificant difference (P = 0.9465) (Figure 4C; see Supplemental Table 2 online). This shows that Cdk2-type activity is indeed increased when the Ph1 Cdk-like cluster is deleted. As mentioned earlier, a previous study showed that okadaic acid treatment of wheat–rye hybrids prior to meiosis induced pairing between the homoeologs even in the presence of Ph1 (Knight et al., 2010). Moreover, it has been shown that this drug treatment increases histone H1 phosphorylation (Ajiro et al., 1996). Proteomic analysis was therefore also performed on histone fractions prepared from okadaic acid treated wheat–rye anthers with Ph1 present to assess whether phosphorylation of Ph1 TPAAK site was affected. The analysis did indeed reveal that this same TPAAK site, which showed twofold phosphorylation in the absence of Ph1, also showed twofold phosphorylation with okadaic acid treatment (P = 0.0064) (Figure 4D; see Supplemental Table 3 online). This confirms that okadaic acid treatment increases Cdk2-type activity and phosphorylation of histone H1 in Ph1+, as well as phenocopying the Ph1− pairing phenotype. In many species, increased histone H1 phosphorylation has been reported to reduce heterochromatin protein1 binding, leading to a more open chromatin structure (Hale et al., 2006). This in turn leads to premature condensation, which has been proposed to occur because of greater accessibility of the chromatin to condensing proteins (Roth and Allis, 1992). The appearance of heterochromatin in premeiotic and early wheat–rye meiotic nuclei in the presence and absence of Ph1 is very similar when stained with 4′,6-diamidino-2-phenylindole (DAPI). However, DAPI shows a bias toward heterochromatin. Consequently, DAPI staining may not be effective in detecting any changes. Previous studies have reported that during meiosis, Ph1 affects centromere structure and behavior (Aragón-Alcaide et al., 1997; Vega and Feldman, 1998; Martinez-Perez et al., 2001), subtelomeric heterochromatin structure and behavior (Prieto et al., 2004), and chromosome conformation or condensation using 3D imaging approaches (Boden et al., 2009; Knight et al., 2010). All these observations are consistent with altered histone H1 phosphorylation in the presence and absence of Ph1.
Figure 4.
Histone Extraction and Relative Levels of Phosphorylation in Histone H1 Peptides.
(A) Coomassie blue-stained histone extraction separated by 4 to 13% Bis-Tris gel in 2-(N-morpholino)-ethane-sulfonic acid-SDS running buffer. The locations of the linker histone H1 and the core histone proteins are noted. M, molecular weight protein marker with masses in kD.
(B) Ratio of amount of phosphopeptide DAAVD(pT)PAAKPAK to amount of peptide DAAVDTPAAKPAK in seven biological replicates of wheat–rye Ph1+ samples and nine biological replicates of Ph1− samples.
(C) Ratio of amount of two regularly observed peptides from histone H1 (SGSTIAIGK and ILLTQIK) in three biological replicates of wheat–rye Ph1+ samples and three biological replicates of Ph1− samples.
(D) Ratio of amount of phosphopeptide DAAVD(pT)PAAKPAK to amount of peptide DAAVDTPAAKPAK in two biological replicates of wheat–rye Ph1+ samples untreated and treated with okadaic acid. Bars represent se from the mean. OA, okadaic acid.
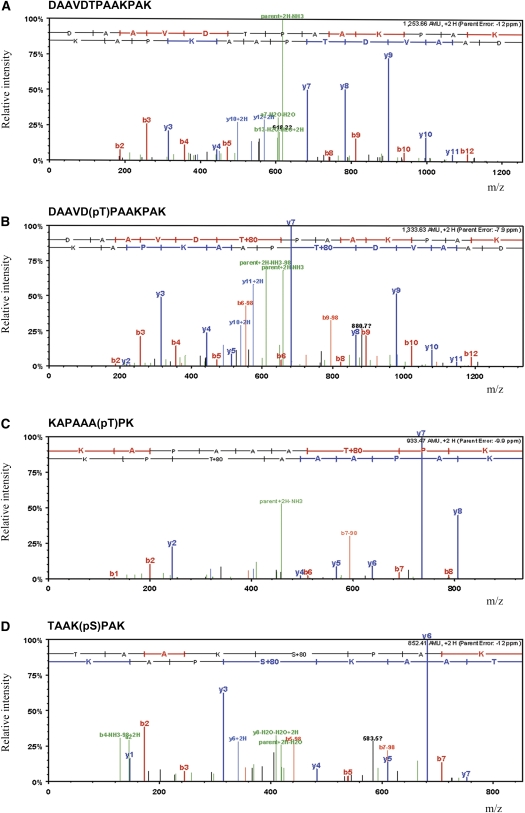
Figure 5.
Examples of Mass Spectra from Fragmented Phosphorylated and Nonphosphorylated Peptides.
The major ion peaks are annotated as the parent ion and a series of y and b ions depending on whether fragmentation has occurred from the N or C terminus of the peptide, respectively. Fragment ion scan spectrum annotated by Scaffold 3 (Proteome Software).
(A) Peptide DAAVDTPAAKPAK from histone H1 WH1A2 with Mascot score of 67.7.
(B) Phosphorylated form of peptide DAAVD(pT)PAAKPAK with Mascot score of 65.2. Evidence of a phosphopeptide is supported by the loss of 98 D (corresponding to the loss of neutral phosphate) from the parent ion and from fragment ions b6 and b9.
(C) Phosphorylated form of peptide KAPAAA(pT)PK with Mascot score of 43.4.
(D) Phosphorylated form of peptide TAAK(pS)PAK with Mascot score of 51.1.
A Model for Ph1 Regulation of Chromosome Pairing and Recombination
These results raise the question of how Ph1 affects chromosome pairing and recombination in the hybrids between wheat and its relatives. Recombination involves the formation and repair of DSBs. DSB repair exploiting homologs or homoeologs can lead to chromosome exchange, whereas repair using sister chromatids (sisters) leads to nonexchange. Chromosomes intimately align or synapse during early meiosis, and recombination can occur within this framework. Synapsis studies in wheat hybrids, where no homologs are present, have indicated that in both the presence and absence of Ph1, chromosomes can be fully synapsed (Gillies, 1987). However, this synapsis does not lead to recombination in the presence of Ph1. After dissolution of the SC, bivalents resolve as univalents, which do not fragment, implying that DSBs have been repaired using the sister chromatids. On the other hand, in the absence of Ph1, DSBs can be repaired with homoeologs, and chromosome exchange can occur.
Previous studies have revealed that Ph1 affects the structure of centromeric heterochromatin as well as sister chromatid separation during anaphase (Aragón-Alcaide et al., 1997; Vega and Feldman, 1998). Moreover, in wheat, the addition of B chromosomes can compensate for the effect of deleting Ph1 (Dover and Riley, 1972). It has recently been shown that the heterochromatin on B chromosomes also affects sister chromatid separation of both A and B chromosomes (Endo et al., 2008; Pereira et al., 2009), and alterations in sister chromatid cohesion would be expected to affect sister chromatid separation (Hoque and Ishikawa, 2002). We therefore propose that Ph1 affects sister chromatid cohesion through altering heterochromatin decondensation, and that whether DSBs are repaired with sister chromatids or homoeologs will be changed by alterations in sister chromatid cohesion.
At the onset of meiosis, heterochromatin decondenses just prior to the homologs pairing, leading to chromosome elongation and sister chromatid separation (Dawe et al., 1994; Martinez-Perez et al., 1999) before condensing later on in meiosis. It can be argued that the separation of sisters helps in the formation of a barrier to DSB repair with sisters. The extent of this heterochromatin decondensation depends on the presence of Ph1 (Prieto et al., 2004; Colas et al., 2008). Ph1 delays heterochromatin decondensation during the onset of meiosis in the wheat–rye hybrid, which may prevent the barrier to sister repair from being formed. In the absence of Ph1, decondensation is not delayed, and so the barrier to sister repair can be established. We propose that Ph1 regulates this process by altering the structural stability of heterochromatin. Recent work has shed some light on what provides the barrier to sister chromatid DSB repair. Asy1, an axial element protein, suppresses DSB repair via sister chromatids (Sanchez-Moran et al., 2007). Interestingly, reducing Asy1 in wheat mimics the phenotype of deleting Ph1 (Boden et al., 2009).
The homology that the defective Ph1-Cdk-like genes show to Cdk2 in mammals (Yousafzai et al., 2010a; Yousafzai et al., 2010b) and the phenotypic effects reported in this article are consistent with an enhancement of Cdk2-type activity in the absence of Ph1, which in mammals is known to affect chromatin structure, replication, and meiosis (Viera et al., 2009). Our hypothesis suggests that this increased activity alters the process of chromatin remodeling and recombination, leading to decreased specificity in the chromosome pairing process.
METHODS
Plant Material
The tillers used for this study came from crosses between rye (Secale cereale cv Petkus) and hexaploid wheat (Triticum aestivum cv Chinese Spring) either carrying or lacking the Ph1 locus (ph1b deletion) (Sears, 1977). Seeds from both genotypes were germinated on Petri dishes for 3 to 4 d. The seedlings were vernalized for 3 weeks at 4°C and then transferred to a controlled environmental room until meiosis with the following growth conditions: 16 h at 20°C and 8 h at 15°C, 85% humidity. Plants were collected after 6 to 7 weeks for meiosis studies.
Ph1 Cdk Sequences
The Ph1 locus contains seven kinases (Cdk)-like genes, and the related (homoeologous) loci on chromosomes 5A and 5D contain five and two Cdk-like genes, respectively. Of these genes, at least one copy (5B2, 5A3, and 5D2) on each genome has a full open reading frame. However, these Cdk-like genes are highly guanine-cytosine–rich, which creates sequencing problems (Yousafzai et al., 2010a). Standard sequencing suggested that 5B2, 5A3, and 5D2 possess a similar 351 amino acid protein sequence. However, ongoing transformation studies have revealed that whereas overexpression of both 5A3 and 5D2 have dramatic effects on plant viability and fertility, overexpression of 5B2 had no such effect, suggesting that it was defective compared with 5A3 and 5D2. Resequencing using different methodologies designed for guanine-cytosine–rich sequences revealed that whereas the 5B2, 5A3, and 5D2 protein sequences are similar for their first 314 amino acids, the final 31 amino acids of 5B2 and its stop codon are different, possibly providing an explanation for the phenotype observations. The GenBank accession numbers for the nucleotide and protein sequences of these three Cdks are FJ811975, FJ811976, and FJ883562.
Meiosis Staging
Tillers were harvested when the flag leaf was starting to emerge (around 1 cm) and kept in cold water to slow down meiosis while dissecting the anthers. For each dissected floret, one of the three synchronized anthers was squashed in acetocarmine stain and examined under the light microscope to identify the stage of meiosis. The two remaining anthers were kept on ice in a solution of 1× PBS containing protease inhibitors (Protease Inhibitor Cocktail Set 1; Calbiochem) and phosphatase inhibitors (Sigma Phosphatase Inhibitor Cocktails 1 and 2; Sigma-Aldrich). Anthers were then stored at −80°C until protein extraction.
Protein Extraction
Batches of 100 staged anthers were ground in liquid nitrogen and immediately washed with 400 μL urea buffer (25 mM triethylammonium bicarbonate, 8 M urea, 2% Triton X-100, 0.1% SDS) including the protease inhibitors and phosphatase inhibitors mentioned previously. The mixture was homogenized and pelleted by centrifugation (16,000g for 10 min). Histones were extracted from the pellet with 300 μL 0.2M HCl overnight at 4°C with shaking. After centrifugation (18,000g at 4°C for 10 min), the pellet containing the insoluble fraction was discarded. Soluble histones were precipitated by adding 4 volumes of ice-cold acetone overnight at −20°C. Acetone was removed after centrifugation (16,000g at 4°C for 10 min), and the pellet was washed twice with 70% ethanol. The pellet was left to air dry at 4°C, and the proteins were solubilized in double-distilled water. The histone quality and concentration were evaluated by PAGE.
In-Gel Digestion
Samples were run on NuPAGE Novex 4 to 12% Bis-Tris Gel 1.0 mm gel (Invitrogen) and stained with Coomassie brilliant blue R-250. Protein bands were excised from the gel and diced into cubes (~1 × 1 mm). Coomassie-stained gel pieces were destained with 50% acetonitrile incubated at 45°C. The samples were reduced in 55 mM ammonium bicarbonate containing 10 mM DTT and incubated for 30 min at 57°C. An equal volume of 50 mM iodoacetamide for alkylation was added to the sample and incubated in the dark at room temperature for 15 min. After discarding the solution, the gel pieces were rinsed with 50% acetonitrile/ammonium bicarbonate and washed in 100% acetonitrile to dehydrate the gel.
40 ng trypsin (Promega Gold, part V511A) in 20 μL of trypsin buffer and sufficient 50 mM ammonium carbonate buffer to cover the gel pieces were added to the dehydrated gel and then incubated overnight at 37°C. The reaction was stopped by heating at 95°C for 10 to 15 min. 80 ng of endoproteinase AspN (Roche Diagnostics) was added to the samples and incubated at 37°C for 8 h. The supernatant (containing the digested peptides) was transferred to a new tube. A total of 0.5% formic acid and 50% acetonitrile was added to the gel pieces, and the mix was sonicated for 5 min to extract the remaining peptides. The supernatants were pooled and lyophilized. Samples were resuspended in 0.5% formic acid prior to mass spectrometric analysis.
Liquid Chromatography–Tandem Mass Spectrometry Analysis
Liquid chromatography–tandem mass spectrometry (MS/MS) analysis was performed using an linear trap quadrupole–Orbitrap mass spectrometer (Thermo Scientific) and a nanoflow-HPLC system (nanoAcquity; Waters). Peptides were applied to a precolumn (Symmetry C18 5-μm beads, 180 μm × 20 mm column; Waters) connected to a 25-cm analytical column (BEH130 C18 1.7-μm beads, 75 μm × 250 mm column; Waters). Peptides were eluted by a gradient of 1 to 80% acetonitrile in 0.1% formic acid over 52 min at a flow rate of 250 nL min−1. Precursor ion scan was acquired in the Orbitrap at resolution 60,000, whereas up to five data-dependent acquisitions of MS/MS (selection of the five most abundant ions in each cycle) were acquired in the linear trap quadrupole:mass spectrometry mass-to-charge ratio (m/z) 300 to 2000, minimum signal 500, collision energy 35, 1 repeat hit, 60 s exclusion. The data-dependent acquisition used an inclusion list of m/z values listed below. Charge state screening using 1+, 2+, and 3+ ions was enabled from 16 to 24 min and charges 2+ and 3+ only for the remainder of the run. The mass spectrometer was operated in positive ion mode with a nanospray source (Proxeon) and a capillary temperature of 200°C, no sheath gas was used, and the source voltage and focusing voltages were optimized for the transmission of the peptide MRFA.
m/z inclusion list:
307.21 309.17 309.50 328.18 335.72 648.32 648.84 656.35 656.35 656.84
363.71 363.73 384.25 405.25 418.89 667.82 695.70 702.83 705.03 728.32
427.76 443.29 460.76 471.80 472.30 775.49 775.93 779.36 786.07 789.93
479.31 479.75 481.76 489.76 491.77 790.74 796.08 797.01 805.43 832.95
492.26 492.83 507.34 535.85 561.35 859.36 942.98 950.98 1036.03 1043.04
569.31 580.30 589.31 603.33 605.86 1051.04 1054.03 1067.03 1072.04 1075.03
627.84 627.84 627.84 638.83 640.85 1187.14 1215.65 1254.67 1845.87
Data Analysis
Peak list (dta) files were extracted by Quant.exe (MaxQuant, version 36) (Cox and Mann, 2009) and converted to Mascot generic format using an in-house Perl script. Peptides were assigned using Mascot version 2.3 (Matrix Science) against the Swiss-Prot and TrEMBL sequence database with taxonomy specified as Triticum (14,423,06 sequences), with the iodoacetamide derivative of Cys specified in Mascot as a fixed modification and oxidized Met and phosphorylation on Ser and Thr as variable modifications. Monoisotopic mass values were used with ±10 ppm on unrestricted protein masses. Fragment mass tolerance was ±0.5 D, and two missed cleavages were allowed with trypsin and AspN.
Protein Identification
Scaffold (version Scaffold_3_00_01; Proteome Software) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm (Keller, et al., 2002). Protein identifications were accepted if they could be established at greater than 95.0% probability and contained at least two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (Nesvizhskii et al., 2003). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Relative Quantification of Peptides by Liquid Chromatography-MS/MS
Peak areas were calculated by Progenesis (version 2.5; Non Linear Dynamics) using the following settings: Thermo RAW mass spectrometry files were aligned and overall intensity was normalized using the default settings, precursor ions features were filtered by charge states only (retaining 1+, 2+, and 3+ ions). Peptides were identified using a Progenesis-created Mascot generic file and searched with Mascot v 2.3 using the same database and parameters described in the mass spectrometry methods. Normalized peak areas were exported to a spreadsheet, and the peak area for each peptide was taken as the most intense peak measured at the specified m/z. To assess differences in phosphorylation levels of histone H1, normalized overall intensity ratios of the phosphorylated peptide DAAVD(p)TPAAKPAK to the unphosphorylated form were compared between seven biological replicates of wild-type (Ph1+) and nine biological replicates of mutant (Ph1−) samples using a two-tailed unpaired t test (Prism software), giving a P value of 0.003.
EdU Treatment and Detection
Tillers containing an immature premeiotic spike were cut immediately after an 8-h period in the dark and transferred immediately to a solution of 100 mM Suc and 1 mM EdU (Invitrogen; A10044). The cut tillers in individual tubes were left in the light for 4 h, after which the spike was dissected out and fixed in 4% formaldehyde solution, freshly made from paraformaldehyde (Prieto et al., 2007). The fixed samples were placed in biopsy cassettes and embedded in wax using a Tissue-tek vacuum infiltrator processor machine (Schwarzacher and Heslop-Harrison, 2000). They were then sectioned using a microtome (Armstrong et al., 2001).
EdU detection was performed using a Click-iT EdU Alexa Fluor 488 Imaging Kit, as per manufacturer’s instructions (Invitrogen; C10337). Fluorescence microscopy and image processing techniques used in the EdU detection and fluorescence in situ hybridization procedures have all been previously described (Prieto et al., 2007).
Fluorescence in Situ Hybridization
Protocols for making the telomere and wheat centromere (CCS1) probes have previously been described (Aragón-Alcaide et al., 1996; Colas et al., 2008). New primers were designed from the rye retrotransposon Bilby sequence to make the rye centromeric probe (F: gatagaacaccacaccgaactcgtacccgg, R: acttaaattgtgtaagaggtaagcacttgc). In situ hybridization was performed as previously described (Prieto et al., 2004). Wheat centromere probe was labeled with digoxigenin-11-deoxyuridine triphosphate and detected using antidigoxigenin-Cy5. Telomere and rye centromere probes were labeled with biotin-16-deoxyuridine triphosphate and detected using streptavidin-Cy3 conjugate. Chromosomes were counterstained with DAPI. Representative 3D data sets from this study are available at www.jic.ac.uk/staff/peter-shaw.
Accession Numbers
Sequence data for the Cdks mentioned in this article can be found at GenBank/EMBL databases under the following accession numbers: FJ811975, FJ811976, and FJ883562.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Graph Showing the Mean Number of Centromere Sites at the Different Stages of Replication.
Supplemental Table 1. Intensity Values of Phosphorylated (Pi) and Nonphosphorylated (Non-Pi) Peptide DAAVDTPAAK from Progenesis Software.
Supplemental Table 2. Intensity Values of Peptides SGSTIAIGK and ILLTQIK from Progenesis Software.
Supplemental Table 3. Intensity Values from Progenesis Software of Phosphorylated (Pi) and Nonphosphorylated (Non-Pi) Peptide DAAVDTPAAK Treated with Okadaic Acid.
Acknowledgments
We thank Gerhard Saalbach and Jan Sklenar for advice with proteomic analysis. We also thank Chris Burt for statistical analysis and advice. This work was supported by the UK Biological and Biotechnology Research Council (BBSRC) via a grant to the John Innes Centre, by the Gatsby Foundation (A.M.E.J.), and by a fellowship to A.C.M. from the Spanish Government (I-D+I 2008–2009) and the Institute of Sustainable Agriculture, Cordoba, Spain.
AUTHOR CONTRIBUTIONS
E.G. performed the EdU treatment and detection and the fluorescence in situ hybridization. A.C.M., A.P., I.C., and A.M.E.J. designed and performed the proteomic experiments and liquid chromatography-MS/MS analysis. G.M. and P.S. conceived the initial approach and designed the cell biology experiments.
References
- Able J.A., Langridge P. (2006). Wild sex in the grasses. Trends Plant Sci. 11: 261–263 [DOI] [PubMed] [Google Scholar]
- Ajiro K., Yoda K., Utsumi K., Nishikawa Y. (1996). Alteration of cell cycle-dependent histone phosphorylations by okadaic acid. Induction of mitosis-specific H3 phosphorylation and chromatin condensation in mammalian interphase cells. J. Biol. Chem. 271: 13197–13201 [DOI] [PubMed] [Google Scholar]
- Al-Kaff N., Knight E., Bertin I., Foote T., Hart N., Griffiths S., Moore G. (2008). Detailed dissection of the chromosomal region containing the Ph1 locus in wheat Triticum aestivum: With deletion mutants and expression profiling. Ann. Bot. 101: 863–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragón-Alcaide L., Miller T., Schwarzacher T., Reader S., Moore G. (1996). A cereal centromeric sequence. Chromosoma 105: 261–268 [DOI] [PubMed] [Google Scholar]
- Aragón-Alcaide L., Reader S., Miller T., Moore G. (1997). Centromeric behaviour in wheat with high and low homoeologous chromosomal pairing. Chromosoma 106: 327–333 [DOI] [PubMed] [Google Scholar]
- Armstrong S.J., Franklin F.C.H., Jones G.H. (2001). Nucleolus-associated telomere clustering and pairing precede meiotic chromosome synapsis in Arabidopsis thaliana. J. Cell Sci. 114: 4207–4217 [DOI] [PubMed] [Google Scholar]
- Boden S.A., Langridge P., Spangenberg G., Able J.A. (2009). TaASY1 promotes homologous chromosome interactions and is affected by deletion of Ph1. Plant J. 57: 487–497 [DOI] [PubMed] [Google Scholar]
- Colas I., Shaw P., Prieto P., Wanous M., Spielmeyer W., Mago R., Moore G. (2008). Effective chromosome pairing requires chromatin remodeling at the onset of meiosis. Proc. Natl. Acad. Sci. USA 105: 6075–6080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Mann M. (2009). MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Dawe R.K., Sedat J.W., Agard D.A., Cande W.Z. (1994). Meiotic chromosome pairing in maize is associated with a novel chromatin organization. Cell 76: 901–912 [DOI] [PubMed] [Google Scholar]
- Dhaliwal H.S., Gill B.S., Waines J.G. (1977). Analysis of induced homoeologous pairing in a ph mutant wheat x rye hybrid. J. Hered. 68: 206–209 [Google Scholar]
- Dover G.A., Riley R. (1972). Prevention of pairing of homoeologous meiotic chromosomes of wheat by an activity of supernumerary chromosomes of Aegilops. Nature 240: 159–161 [Google Scholar]
- Endo T.R., Nasuda S., Jones N., Dou Q., Akahori A., Wakimoto M., Tanaka H., Niwa K., Tsujimoto H. (2008). Dissection of rye B chromosomes, and nondisjunction properties of the dissected segments in a common wheat background. Genes Genet. Syst. 83: 23–30 [DOI] [PubMed] [Google Scholar]
- Garcia B.A., Busby S.A., Barber C.M., Shabanowitz J., Allis C.D., Hunt D.F. (2004). Characterization of phosphorylation sites on histone H1 isoforms by tandem mass spectrometry. J. Proteome Res. 3: 1219–1227 [DOI] [PubMed] [Google Scholar]
- Gillies C.B. (1987). The effect of the Ph gene alleles on synaptonemal complex formation in Triticum aestivum x T kotshyi hybrids. Theor. Appl. Genet. 74: 430–438 [DOI] [PubMed] [Google Scholar]
- Griffiths S., Sharp R., Foote T.N., Bertin I., Wanous M., Reader S., Colas I., Moore G. (2006). Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439: 749–752 [DOI] [PubMed] [Google Scholar]
- Hale T.K., Contreras A., Morrison A.J., Herrera R.E. (2006). Phosphorylation of the linker histone H1 by CDK regulates its binding to HP1alpha. Mol. Cell 22: 693–699 [DOI] [PubMed] [Google Scholar]
- Hoque M.T., Ishikawa F. (2002). Cohesin defects lead to premature sister chromatid separation, kinetochore dysfunction, and spindle-assembly checkpoint activation. J. Biol. Chem. 277: 42306–42314 [DOI] [PubMed] [Google Scholar]
- Keller A., Nesvizhskii A.I., Kolker E., Aebersold R. (2002). Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74: 5383–5392 [DOI] [PubMed] [Google Scholar]
- Knight E., Greer E., Draeger T., Thole V., Reader S., Shaw P., Moore G. (2010). Inducing chromosome pairing through premature condensation: Analysis of wheat interspecific hybrids. Funct. Integr. Genomics 10: 603–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasinska L., Besnard E., Cot E., Dohet C., Méchali M., Lemaitre J.-M., Fisher D. (2008). Cdk1 and Cdk2 activity levels determine the efficiency of replication origin firing in Xenopus. EMBO J. 27: 758–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez E., Shaw P., Aragón-Alcaide L., Moore G. (2003). Chromosomes form into seven groups in hexaploid and tetraploid wheat as a prelude to meiosis. Plant J. 36: 21–29 [DOI] [PubMed] [Google Scholar]
- Martinez-Perez E., Shaw P., Moore G. (2001). The Ph1 locus is needed to ensure specific somatic and meiotic centromere association. Nature 411: 204–207 [DOI] [PubMed] [Google Scholar]
- Martínez-Pérez E., Shaw P., Reader S., Aragón-Alcaide L., Miller T., Moore G. (1999). Homologous chromosome pairing in wheat. J. Cell Sci. 112: 1761–1769 [DOI] [PubMed] [Google Scholar]
- Nasmyth K. (1996). Viewpoint: Putting the cell cycle in order. Science 274: 1643–1645 [DOI] [PubMed] [Google Scholar]
- Nesvizhskii A.I., Keller A., Kolker E., Aebersold R. (2003). A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75: 4646–4658 [DOI] [PubMed] [Google Scholar]
- Okamoto M. (1957). A synaptic effect of chromosome V. Wheat Inf. Serv. 5: 6 [Google Scholar]
- Pereira H.S., Barão A., Caperta A., Rocha J., Viegas W., Delgado M. (2009). Rye Bs disclose ancestral sequences in cereal genomes with a potential role in gametophyte chromatid segregation. Mol. Biol. Evol. 26: 1683–1697 [DOI] [PubMed] [Google Scholar]
- Polit J.T., Kazmierczak A. (2007). Okadaic acid (1 microM) accelerates S phase and mitosis but inhibits heterochromatin replication and metaphase anaphase transition in Vicia faba meristem cells. J. Exp. Bot. 58: 2785–2797 [DOI] [PubMed] [Google Scholar]
- Prieto P., Moore G., Shaw P. (2007). Fluorescence in situ hybridization on vibratome sections of plant tissues. Nat. Protoc. 2: 1831–1838 [DOI] [PubMed] [Google Scholar]
- Prieto P., Shaw P., Moore G. (2004). Homologue recognition during meiosis is associated with a change in chromatin conformation. Nat. Cell Biol. 6: 906–908 [DOI] [PubMed] [Google Scholar]
- Riley R., Chapman V. (1958). Genetic control of the cytologically diploid behaviour of hexaploid wheat. Nature 182: 713–715 [Google Scholar]
- Roth S.Y., Allis C.D. (1992). Chromatin condensation: Does histone H1 dephosphorylation play a role? Trends Biochem. Sci. 17: 93–98 [DOI] [PubMed] [Google Scholar]
- Sánchez-Morán E., Benavente E., Orellana J. (2001). Analysis of karyotypic stability of homoeologous-pairing (ph) mutants in allopolyploid wheats. Chromosoma 110: 371–377 [DOI] [PubMed] [Google Scholar]
- Sanchez-Moran E., Santos J.-L., Jones G.H., Franklin F.C.H. (2007). ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 21: 2220–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzacher T., Heslop-Harrison J.S. (2000). Practical in Situ Hybridization. (Oxford, United Kingdom: BIOS Scientific Publishers; ). [Google Scholar]
- Sears E.R. (1977). An induced mutant with homoeologous pairing in common wheat. Can. J. Genet. Cytol. 19: 585–593 [Google Scholar]
- Sears E.R., Okamoto M. (1958). Intergenomic chromosome relationships in hexaploid wheat. Proc. Xth Intern. Congr. Genet. 2: 258–259 [Google Scholar]
- Thomson A.M., Gillespie P.J., Blow J.J. (2010). Replication factory activation can be decoupled from the replication timing program by modulating Cdk levels. J. Cell Biol. 188: 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega J.M., Feldman M. (1998). Effect of the pairing gene Ph1 on centromere misdivision in common wheat. Genetics 148: 1285–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viera A., Rufas J.S., Martínez I., Barbero J.L., Ortega S., Suja J.A. (2009). CDK2 is required for proper homologous pairing, recombination and sex-body formation during male mouse meiosis. J. Cell Sci. 122: 2149–2159 [DOI] [PubMed] [Google Scholar]
- Yamashita K., Yasuda H., Pines J., Yasumoto K., Nishitani H., Ohtsubo M., Hunter T., Sugimura T., Nishimoto T. (1990). Okadaic acid, a potent inhibitor of type 1 and type 2A protein phosphatases, activates cdc2/H1 kinase and transiently induces a premature mitosis-like state in BHK21 cell. EMBO J. 13: 4331–4338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousafzai F.K., Al-Kaff N., Moore G. (2010a). The molecular features of chromosome pairing at meiosis: The polyploid challenge using wheat as a reference. Funct. Integr. Genomics 10: 147–156 [DOI] [PubMed] [Google Scholar]
- Yousafzai F.K., Al-Kaff N., Moore G. (2010b). Structural and functional relationship between the Ph1 locus protein 5B2 in wheat and CDK2 in mammals. Funct. Integr. Genomics 10: 157–166 [DOI] [PubMed] [Google Scholar]
- Zickler D., Kleckner N. (1998). Meiotic chromosomes integrating structure and function. Annu. Rev. Genet. 32: 619–697 [DOI] [PubMed] [Google Scholar]