The NITRATE TRANSPORTER2.4 (NRT2.4) is a member of the NRT2 gene family in Arabidopsis thaliana and is preferentially expressed under nitrogen starvation. This study shows that NRT2.4 is a nitrate transporter involved both in root nitrate uptake at very low nitrate concentration and in delivering nitrate to the shoot phloem under nitrogen starvation.
Abstract
Plants have evolved a variety of mechanisms to adapt to N starvation. NITRATE TRANSPORTER2.4 (NRT2.4) is one of seven NRT2 family genes in Arabidopsis thaliana, and NRT2.4 expression is induced under N starvation. Green fluorescent protein and β-glucuronidase reporter analyses revealed that NRT2.4 is a plasma membrane transporter expressed in the epidermis of lateral roots and in or close to the shoot phloem. The spatiotemporal expression pattern of NRT2.4 in roots is complementary with that of the major high-affinity nitrate transporter NTR2.1. Functional analysis in Xenopus laevis oocytes and in planta showed that NRT2.4 is a nitrate transporter functioning in the high-affinity range. In N-starved nrt2.4 mutants, nitrate uptake under low external supply and nitrate content in shoot phloem exudates was decreased. In the absence of NRT2.1 and NRT2.2, loss of function of NRT2.4 (triple mutants) has an impact on biomass production under low nitrate supply. Together, our results demonstrate that NRT2.4 is a nitrate transporter that has a role in both roots and shoots under N starvation.
INTRODUCTION
Nitrate (NO3–) uptake from the soil and distribution through the plant can profoundly affect plant growth and productivity. Nitrogen (N) limitation decreases crop yield worldwide. To meet expanding food demands, the global use of N fertilizer in agricultural production is projected to increase threefold to reach 249 million tons annually by the year 2050 (Tilman et al., 2001). However, the recovery of N fertilizer by crops is low, with in some cases only 30 to 50% of the applied N being taken up by the crop (Peoples et al., 1995; Sylvester-Bradley and Kindred, 2009). The remainder is partly used by subsequent crops but can also be lost from the agro-ecosystem, and fertilizer runoff into aquatic systems leads to environmentally harmful eutrophication (Tilman, 1998). Therefore, improving N uptake efficiency is important to reduce the costs of crop production and pollution damage. Beside N uptake, N remobilization is another key step to improve N use efficiency in crops (Mickelson et al., 2003; Masclaux-Daubresse et al., 2008).
Plants have evolved versatile mechanisms to cope with N limitation and N starvation, and besides major adaptive changes of the root system architecture (Drew and Saker, 1975), root NO3– uptake characteristics are regulated in response to N availability (Clarkson et al., 1986; Lejay et al., 1999; Glass, 2003). Physiological studies have led to the conclusion that at least three NO3– uptake systems are responsible for the influx of NO3– into roots (reviewed in Crawford and Glass, 1998; Daniel-Vedele et al., 1998; Forde, 2000). Two high-affinity transport systems (HATS) operate to take up NO3– at low concentrations in the external medium, and both display saturable kinetics as a function of the external NO3– concentration, with saturation in the range of 0.2 to 0.5 mM. The first one, constitutive HATS, is active in plants that have not been supplied with NO3–whereas the second HATS is induced by NO3– supply. In addition to these systems, there is a low-affinity transport system with uptake activity that is linear as a function of external NO3– concentration.
At the molecular level, three types of NO3– transporters, NITRATE TRANSPORTER1 (NRT1), NRT2, and CLC (chloride channel family), have been identified in higher plants (reviewed in Dechorgnat et al., 2011). In Arabidopsis thaliana, CLCa and CLCb, both proton-NO3– exchangers located in the tonoplast, mediate NO3– accumulation in vacuoles (De Angeli et al., 2006; von der Fecht-Bartenbach et al., 2010). Among the other five Arabidopsis CLC proteins, CLCc is involved in chloride transport (Jossier et al., 2010) and CLCd and CLCg possess a selectivity filter in favor of chloride transport (Zifarelli and Pusch, 2009). In Arabidopsis, NRT1 and NRT2 are families of proton-coupled transporters with 53 and seven members, respectively. Four members of these families, Arabidopsis NRT1.1, NRT1.2, NRT2.1, and NRT2.2, have been shown to participate in root NO3– uptake (Tsay et al., 1993; Wang et al., 1998; Huang et al., 1999; Liu et al., 1999; Cerezo et al., 2001; Filleur et al., 2001; Orsel et al., 2004; Li et al., 2007). NRT2.1 and NRT2.2 act in the high-affinity range, and NRT2.1 transport activity depends on a second protein NAR2.1 (or NRT3.1) (Okamoto et al., 2006; Orsel et al., 2006), possibly functioning as a 2x2 tetramer through an unknown mechanism (Yong et al., 2010). NRT1.2 is active only in the low-affinity range, while NRT1.1 is a dual affinity transporter depending on its phosphorylation status (Liu and Tsay, 2003). In addition, NRT1.1 is also involved in the signaling of NO3– (Ho et al., 2009) and in auxin transport at low NO3– concentrations (Krouk et al., 2010).
More members of both families have been characterized, but none of them are involved in NO3– uptake from the soil. NO3– efflux activity has been demonstrated for two NRT1 proteins, NRT1.5, which is involved in xylem loading for root-to-shoot transport of NO3– (Lin et al., 2008), and NAXT (Segonzac et al., 2007). NRT1.8 (Li et al., 2010) and NRT1.9 (Wang and Tsay, 2011), which are expressed in xylem parenchyma and root phloem companion cells, respectively, seem to participate in retrieving NO3– from xylem sap. NRT1.6 and NRT2.7 are both expressed in seeds, and NRT1.6 is involved in the transport of NO3– from maternal tissue to developing embryos (Almagro et al., 2008), whereas NRT2.7 regulates seed NO3– content (Chopin et al., 2007). Several family members are expressed in shoots. NRT1.4 regulates leaf NO3– homeostasis (Chiu et al., 2004), and NRT1.7 is important for NO3– remobilization from source to sink tissues via phloem transport in response to N limitation (Fan et al., 2009). However, little is known about NO3– transporters involved in N starvation responses.
In this study, we show that NRT2.4 plays a role in roots and shoots in response to N starvation. It is involved in the uptake of NO3– by the root at very low external concentration and in shoot NO3– loading into the phloem.
RESULTS
Expression of NRT2.4 in Response to N Availability
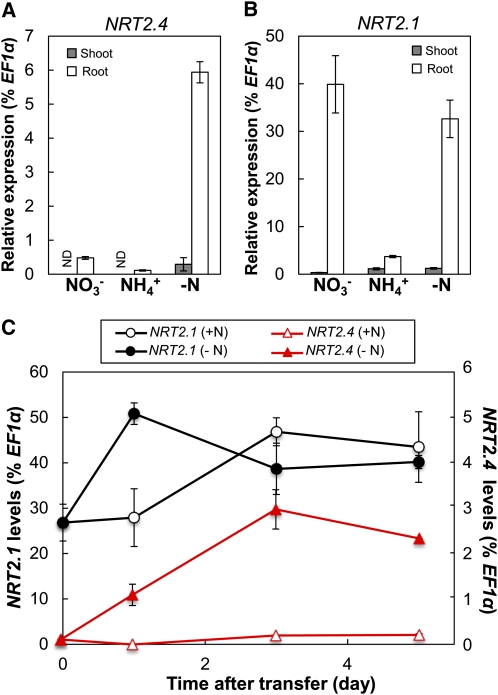
The expression of NRT2 genes in Arabidopsis is regulated by N availability (Lejay et al., 1999; Orsel et al., 2002). NRT2.4 and NRT2.1 expression patterns are different, with the NRT2.4 transcript level being highest under severe N starvation and repressed by resupply with a N source (Okamoto et al., 2003). NRT2.1 expression increases only transiently after the onset of N starvation, and a resupply of NO3– induces NRT2.1 expression (Lejay et al., 1999). We analyzed the expression of NRT2.4 in comparison to NRT2.1 in response to N supply in our experimental system using young seedlings grown in 10 mM NO3– (full N) for 7 d and then transferred to different N sources (10 mM NO3– or 10 mM ammonium) or to medium without N for 3 d. Compared with the NO3–-replete control where NRT2.4 was expressed at low levels only in roots, NRT2.4 expression in roots increased 12-fold under N starvation and became detectable in shoots (Figure 1A). No induction of NRT2.4 expression was observed under phosphate- or sulfate-starved conditions, indicating that the expression is not responding to general nutrient deficiency stress (see Supplemental Figure 1A online). After transfer to ammonium (NH4+), NRT2.4 expression was decreased (Figure 1A). This expression profile was different from that of NRT2.1 (Figure 1B). NRT2.1 was also mainly expressed in roots (Figure 1B), but NRT2.1 expression was high on NO3–91% lower on NH4+, and not significantly changed under starvation conditions compared with NO3–. NRT2.4 expression decreased steadily with increased NO3– concentration in the medium (decreased 98% between 0 and 10 mM NO3–see Supplemental Figure 1A online), whereas NRT2.1 expression decreased only 70% between 0 and 10 mM NO3–was unchanged between 0.1 and 1 mM NO3– supplies (see Supplemental Figure 1C online). However, NH4+ repression of NRT2.4 and NRT2.1 occurred at a similar concentration for both genes (see Supplemental Figures 1B and 1D online).
Figure 1.
Expression of NRT2.4 and NRT2.1 Is Differentially Regulated in Young Seedlings in Response to N Availability.
(A) and (B) NRT2.4 (A) and NRT2.1 (B) expression levels in the shoot and root of seedlings grown under different N conditions. Wild-type (Col-0) seedlings were germinated and grown on MGRL plates containing 10 mM NO3– (full N plate) for 7 d and then incubated for 3 d on MGRL plates containing 10 mM KNO3 (NO3–), 5 mM (NH4+)2 succinate (NH4+), or no N source (−N). Shoots and roots were sampled separately. ND, not detected.
(C) Expression levels of NRT2.4 and NRT2.1 in the root during N starvation. Wild-type seedlings grown on full N plates were transferred to full N (10 mM NO3–+N) or N-free (−N) plates and harvested at the indicated times. Relative expression levels are presented as a percentage of EF1α gene. Error bars in (A) to (C) represent sd of three biological replicates.
[See online article for color version of this figure.]
Given the increased expression of NRT2.4 in N-depleted roots, we followed the time course patterns of NRT2.4 and NRT2.1 expression during N starvation. Seven-day-old seedlings were transferred from full N medium to either N-free or full N medium, and samples were taken after 0, 1, 3, and 5 d of transfer for gene expression studies and NO3– measurements. NO3– content decreased rapidly after transfer to N free medium, attaining 15 and 53% in roots and shoots, respectively, after 24 h, and then dropped to barely detectable levels at 3 and 5 d, respectively (see Supplemental Figure 2 online). After transfer to N-free medium, NRT2.4 expression increased until day 3 and stayed high afterwards (Figure 1C). NRT2.1 expression increased at day 1 but decreased again and stayed at a slightly higher level than at day 0. The latter might be due to a general slight increase in NRT2.1 expression with developmental age as a similar time-dependent pattern was observed for expression of the same gene on full N medium. We also confirmed the induction of NRT2.4 expression by N starvation on older plants grown in hydroponic culture (see Supplemental Figure 3A online). Similar to the expression pattern in seedlings, the transfer of 35-d-old plants grown on 6 mM NO3– solution to a solution without N rapidly induced expression of NRT2.4, reaching a plateau after 2 d of starvation. In these hydroponically grown plants, we confirmed earlier data showing that NRT2.4 expression decreases when N was resupplied to N-starved plants (Okamoto et al., 2003, see Supplemental Figure 3B online), which was opposite to the expression response of NRT2.1, whose expression was induced specifically by NO3– after starvation (Okamoto et al., 2003; see Supplemental Figure 3B online).
NRT2.4 expression was lower than that for NRT2.1 in all the tested conditions (at least 99% lower under ample NO3– nutrition and 80% lower even in N starvation; Figure 1). This very specific NRT2.4 expression profile might be an indication of a distinct but complementary function for NRT2.4 in comparison to NRT2.1.
NRT2.4 Is Expressed in the Lateral Root Epidermis and the Shoot Vascular Tissue
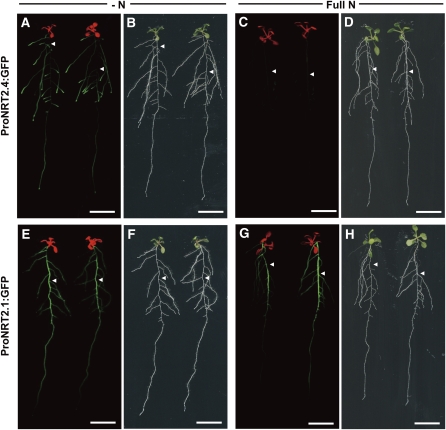
We further characterized the localization of NRT2.4 expression in comparison to NRT2.1 using fusions between either the NRT2.4 or the NRT2.1 promoter and the green fluorescent protein (GFP) reporter gene. These constructs were stably introduced into the Arabidopsis Columbia (Col-0) accession, and two individual transgenic lines were each studied. Figure 2 shows typical results of GFP fluorescence of one of these transformants growing either under N starvation or with full N supply. ProNRT2.4:GFP fluorescence was observed only under N starvation in laterals and in the younger parts of the primary root, but not in the older parts of the main root (Figures 2A to 2D). Under the same conditions ProNRT2.1:GFP fluorescence was observed in the older part of the main root of plants grown in full N and in the main root and at lower level in the laterals of N starved plants (Figures 2E to 2H). The preferential expression of NRT2.1 in older parts of the main root confirms previously reported results (Nazoa et al., 2003). We also confirmed the above-described NO3– dose-dependent expression of NRT2.4 (see Supplemental Figure 1 online) using the ProNRT2.4:GFP construct (see Supplemental Figure 4 online).
Figure 2.
GFP Fluorescence in ProNRT2.4:GFP and ProNRT2.1:GFP Transgenic Plants.
GFP fluorescence ([A], [C], [E], and [G]) and bright-field ([B], [D], [F], and [H]) images of ProNRT2.4:GFP ([A] to [D]) and ProNRT2.1:GFP ([E] to [H]) transgenic plants. Seven-day-old seedlings grown on full N plates were further incubated for 3 d either without a N source (−N; [A], [B], [E], and [F]) or on 10 mM NO3– (Full N; [C], [D], [G], and [H]) before detection. GFP fluorescence (green) was merged with chlorophyll autofluorescence (red) in (A), (C), (E), and (G). Arrowheads indicate the position of the main root. Bars = 1 cm.
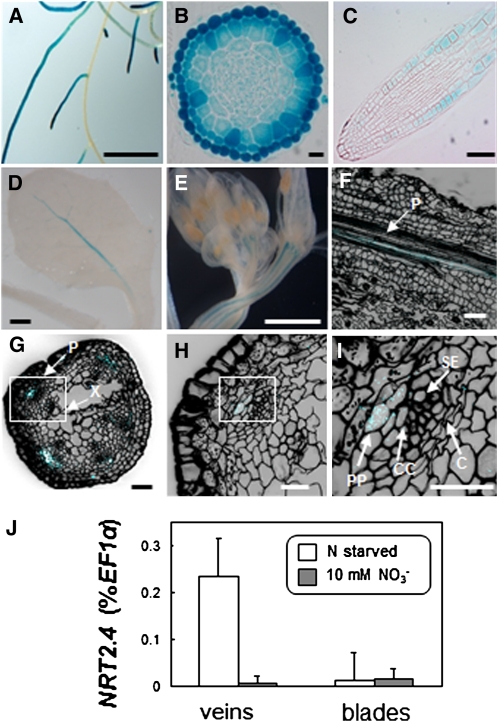
Expression studies using quantitative RT-PCR showed very low NRT2.4 expression in shoots (Figure 1), but no ProNRT2.4:GFP fluorescence was observed in shoots. Therefore, we further characterized the expression of NRT2.4 using a fusion between the NRT2.4 promoter and the β-glucuronidase (GUS) reporter gene, which is more sensitive than GFP (de Ruijter et al., 2003). This construct was again stably introduced into the Arabidopsis Col-0 plants, and two individual transgenic lines were studied. Figure 3 shows typical results from GUS staining of one of these transformants grown under N starvation. Confirming GFP results (Figure 2), GUS staining was observed chiefly in lateral roots (Figure 3A). In cross sections near the division zone (Figure 3B) and in longitudinal sections (Figure 3C), NRT2.4 promoter activity was observed mainly in the epidermis of lateral roots. In shoots, GUS staining was restricted to the vascular tissue. NRT2.4 promoter activity was observed mainly in the primary vein and sometimes in secondary veins (Figure 3D). The same localization was observed in flower stalks (Figure 3E). Staining cell walls with propidium iodide allowed the localization of GUS expression to or close to the phloem, probably to the phloem parenchyma (Figures 3F to 3I).
Figure 3.
NRT2.4 Expression in Roots and Shoots.
(A) to (C) Histochemical localization of GUS activity in ProNRT2.4:GUS plants grown on plates containing 0 mM NO3– for 16 d. Bars = 1 cm in (A), 10 μm in (B), and 50 μm in (C).
(A) Root system.
(B) Cross section near division zone of lateral root.
(C) Longitudinal section of lateral root. GUS activity is located in secondary roots, mainly in epidermal cells.
(D) to (I) Histochemical localization of GUS activity in ProNRT2.4:GUS plants grown on plates containing 0 mM NO3–for 19 d. Bar = 1 mM in (D), 1 mm in (E), 50 μm in (F) and (G), and 10 μm in (H) and (I).
(D) Young leaf
(E) Inflorescence
(F) Confocal image of longitudinal section of young leaf after propidium iodide staining. Blue color, GUS activity; black color, propidium iodide staining (cell walls); P, phloem
(G) to (I) Confocal image of cross section of young inflorescence stalk after propidium iodide staining. C, cambium; CC, companion cells; P, phloem; PP; phloem parenchyma; SE, sieve elements; X, xylem. Boxes in (G) and (H) are enlarged in (H) and (I), respectively. Blue color, GUS activity; black color, propidium iodide staining (cell walls).
(J) NRT2.4 expression levels in leaf veins and blades of 10-week-old plants grown on sand either under 10 mM NO3– (10 mM NO3–) or under N starvation (N starved) for the final 4 weeks. Data are means ± se (n = 6 biological replicates).
This expression pattern was confirmed by quantitative RT-PCR on RNA isolated from veins and leaf blades of adult plants grown either under full N or in N starvation. Despite very low expression levels, a significant increase of NRT2.4 expression was observed in veins of N-starved plants compared with plants grown on full N (Figure 3J).
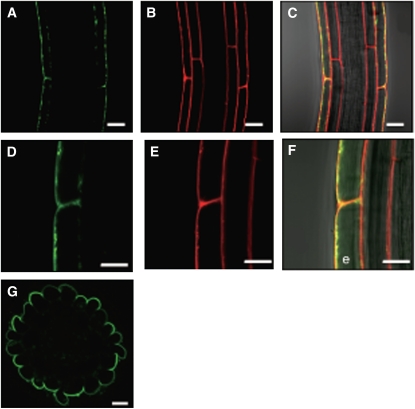
NRT2.4 Is Localized to the Plasma Membrane
The biological function of transporters depends highly on their subcellular localization. Therefore, a C-terminal translational fusion of GFP to NRT2.4 under the control of the NRT2.4 promoter (ProNRT2.4:NRT2.4-GFP) was introduced into Arabidopsis Col-0 plants, and confocal microscopy analyses were performed on homozygous lines. In the root epidermal cells under N deprivation, green fluorescence was detected in the plasma membrane (Figures 4A and 4D). This localization was confirmed by staining lipid membranes using the red fluorescent probe FM4-64 (Figures 4B and 4E) and by merging the two pictures (Figures 4C and 4F), indicating that NRT2.4 was located at the plasma membrane. The NRT2.4-GFP protein was predominantly found in the external (abaxial) membrane of the epidermal cells facing the nutrient solution. This polar localization was further visualized by a cross section (Figure 4G).
Figure 4.
Subcellular Localization of a Translational NRT2.4-GFP Fusion Protein in Root Epidermal Cells of the ProNRT2.4:NRT2.4-GFP Transgenic Plant.
(A) and (D) GFP fluorescence (green).
(B) and (E) FM4-64 (red).
(C) and (F) Merged images of GFP fluorescence, FM4-64, and bright field. Yellow color represents the superposition of green and red. e, epidermal cell.
(G) A cross section showing the GFP fluorescence image of ProNRT2.4:NRT2.4-GFP in the lateral root. Transgenic seedlings were grown on full N plates for 7 d and then incubated on MGRL plates without N source for 3 d.
Bars = 20 μm in (A) to (C) and (G) and 10 μm in (D) to (F).
NRT2.4 Transports NO3– in the High-Affinity Range
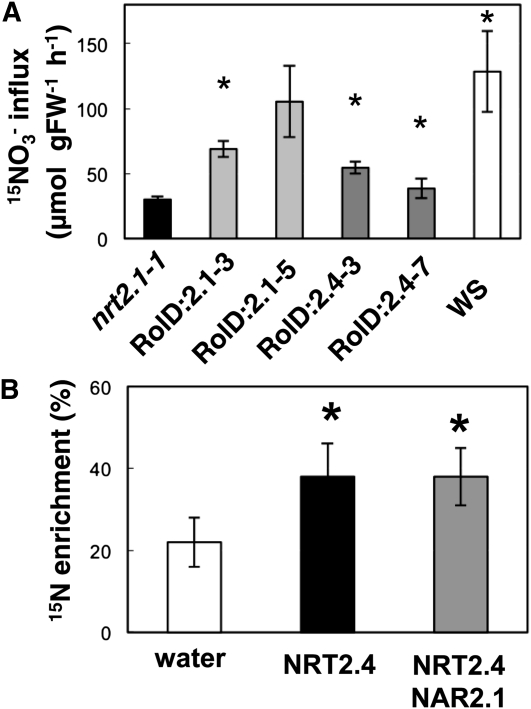
To evaluate the capacity of the NRT2.4 protein to transport NO3–we introduced the NRT2.4 cDNA driven by the root-specific RolD promoter (Fraisier et al., 2000) into the nrt2.1-1 mutant background. The nrt2.1-1 mutant is largely defective for high-affinity NO3– uptake as both NRT2.1 and NRT2.2 are not functional in this mutant (Filleur et al., 2001). In addition, we used RolD:NRT2.1-overexpressing mutant lines as a control. Homozygous transgenic plants (two independent lines each) were grown for 6 weeks in a hydroponic system on 0.2 mM NO3–. Root NO3– influx was measured at an external concentration of 0.2 mM using 15NO3– (atom% 15N: 99%). At this concentration, the difference in the HATS activity between the wild type and the nrt2.1-1 mutant was largest (Filleur et al., 2001). The overexpression of the two genes (NRT2.4 or NRT2.1) was correlated with an increase in the root 15NO3– influx (Figure 5A). Quantitative RT-PCR analyses confirmed that this increase in NO3– influx in the overexpressors was not due to compensation by altered expression of other genes of the NRT1 or NRT2 family (see Supplemental Table 1 online). Increased NO3– acquisition in the nrt2.1-1 mutant plants complemented by either NRT2.1 or NRT2.4 can also be deduced from the increased NO3– content in shoots and the increased shoot biomass in these transgenic lines compared with the nrt2.1-1 mutant (see Supplemental Figure 5 online).
Figure 5.
NRT2.4 Can Transport NO3–.
(A) Root 15NO3– influx in the nrt2.1-1 mutant and in ProRolD:NRT2.4 (RolD:2.4) and ProRolD:NRT2.1 (RolD:2.1) overexpressors in the mutant background. Plants were grown on 0.2 mM NO3– solution for 43 d. Root 15NO3– influx was measured in complete nutrient solution containing 0.2 mM 15NO3–. The values are means ± sd of five biological replicates. Asterisks indicate statistically significant differences between overexpressors and nrt2.1-1 mutant (P < 0.001). FW, fresh weight.
(B) Uptake of 15NO3– into Xenopus oocytes injected with either water or mRNA mixtures as indicated. Oocytes were incubated for 16 h in ND96 solution at pH 6.5 enriched with 0.5 mM Na 15NO3–. The percentage15N enrichment values are means ± sd for five oocytes and calculated as described previously (Tong et al., 2005). The asterisk indicates statistically significant differences between mRNA and water-injected oocytes (P < 0.05).
To confirm that NRT2.4 is able to transport NO3–we used a heterologous expression system. Xenopus laevis oocytes were injected with either NRT2.4 mRNA or nuclease-free water. After 3 d, oocytes were incubated in a solution enriched with 0.2 mM Na15NO3–and the 15N enrichment of individual oocytes was measured after 16 h. These measurements showed that NRT2.4 mRNA–injected oocytes took up significantly more NO3– than water-injected controls (Figure 5B). In addition, as NRT2.1 transport activity was dependent on coexpression with NAR2.1 (Orsel et al., 2006), we tested NO3– uptake in oocytes by not only injecting NRT2.4 mRNA alone, but also together with NAR2.1 mRNA. NRT2.4-driven NO3– enrichment was independent of the presence of NAR2.1 (Figure 5B).
nrt2.4 Mutants Show Decreased NO3– Uptake in the Very-High-Affinity Range
To investigate the in planta function of NRT2.4, two knockout mutants were characterized. nrt2.4-1 corresponds to a T-DNA insertion line in the Col-0 background (MDL-ArBrAr-125; Forsbach et al., 2003). The insertion of the T-DNA in the last exon of the NRT2.4 gene (position 2289 from the ATG) led to a deletion of 23 bp (cf. AJ506341 and AJ506342). nrt2.4-2 was obtained from the Syngenta Arabidopsis Insertion Library (SAIL) T-DNA insertion line collection (SAIL_205_F02, stock CS872100). T-DNA insertion occurred in the third exon. No expression of NRT2.4 could be detected by RT-PCR for either mutant (see Supplemental Figures 6A and 6B online). Neither mutant showed an obvious morphological or physiological phenotype in our growth conditions either under ample NO3– or under N starvation (see Supplemental Figures 6C and 6D online).
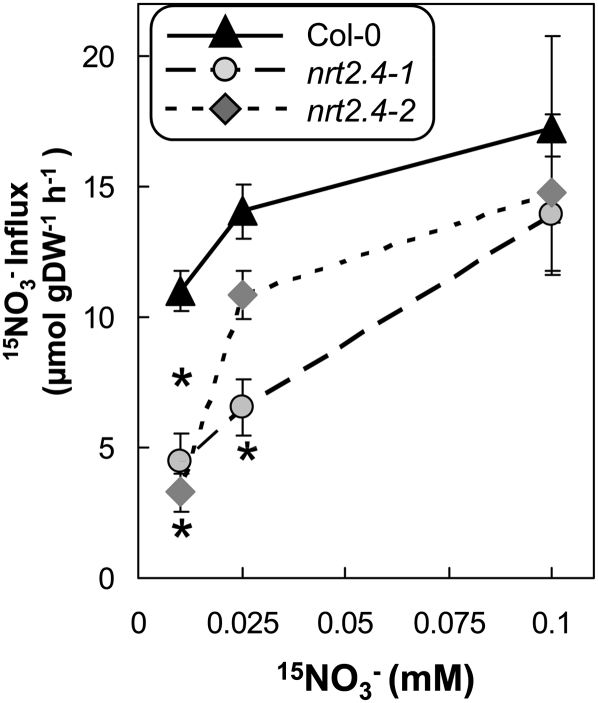
We measured NO3– influx on nrt2.4-1 and nrt2.4-2 mutant seedlings that had been grown first for 7 d on full N medium and were then transferred for 5 d on medium without N. Under these conditions, NRT2.4 expression was maximal in the wild type (Figure 1C). We used 15NO3– concentrations from 6 to 0.01 mM and found no differences in NO3– influx between the wild type and mutants for 6, 0.5, and 0.2 mM NO3– (see Supplemental Figure 7A online). However, when supplying very low NO3– concentrations, a significant decrease in NO3– influx (up to 30% less influx) was observed at both 0.025 and 0.01 mM external NO3– in both mutant lines (Figure 6). Similar data have been obtained measuring NO3– depletion from the medium when plants were incubated for 2 h in 0.2 and 0.01 mM NO3– solution (see Supplemental Figure 7B online).
Figure 6.
Decrease of NO3– Influx in the Very-High-Affinity Range in the nrt2.4 Mutants under N Starvation Condition.
Seedlings were grown on MGRL-based vertical plates for 8 d on 10 mM NO3–then for 5 d without N. 15NO3– influx was measured after 5 min of labeling in complete nutrient solution containing 0.01, 0.025, and 0.1 mM 15NO3–. The values are means ± se (n = 8) of biological replicates. Asterisks indicate statistically significant differences between the wild type (Col-0) and mutants (P < 0.01). DW, dry weight.
Our data show that N-starved nrt2.4 null mutants are impaired in NO3– uptake at very low NO3– concentrations, suggesting that NRT2.4 is a NO3– transporter operating in the very-high-affinity range.
nrt2.1 nrt2.2 nrt2.4 Triple Mutants Show Decreased Growth and NO3– Influx at Low External NO3–
It has been shown previously that the influx capacity of the nrt2.1 nrt2.2 double mutants (called nrt2.1-1 in Wassilewskija [Ws] and nrt2.1-2 in Col-0) at low NO3– concentration was decreased, but not zero (Cerezo et al., 2001; Filleur et al., 2001; Little et al., 2005; Li et al., 2007). We therefore generated triple null mutants that are deficient for the three root high-affinity NO3– transporters NRT2.1, NRT2.2, and NRT2.4.
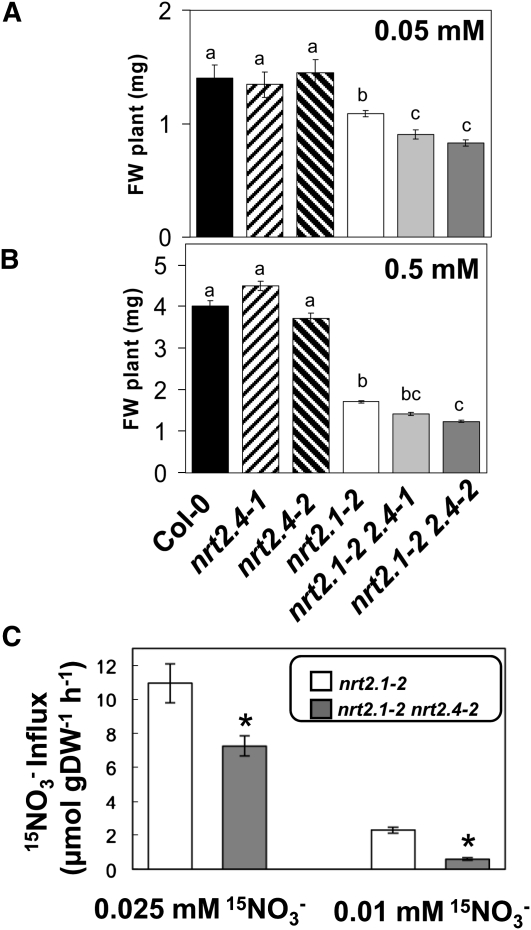
We analyzed the fresh weight of the triple mutants nrt2.1-2 2.4-1 and nrt2.1-2 2.4-2 in comparison to the double mutant nrt2.1-2, the single mutants nrt2.4-1 and nrt2.4-2, and the wild type when grown on agar plates containing different KNO3 concentrations (0.5 and 0.05 mM). No difference in fresh weight was observed for the single mutants compared with the wild type. As expected, fresh weight of the double mutant nrt2.1-2 was lower than that of the wild type under all conditions tested. Comparing triple and double mutants, fresh weight was similar on 0.5 mM NO3– but significantly decreased for the triple mutants grown on 0.05 mM KNO3 agar plates (Figures 7A to 7C).
Figure 7.
The nrt2.1 nrt2.2 nrt2.4 Triple Mutants Show Decreased Growth in Low NO3– Conditions.
(A) to (B) Fresh weight (FW) of Col-0, nrt2.4-1, nrt2.4-2, nrt2.1-2 nrt2.4-1, and nrt2.1-2 nrt2.4-2 mutant seedlings. Seedlings were grown for 12 d on MGRL-based vertical plates containing the indicated amount of N. Error bars represent sd of five biological replicates. Letters indicate statistically significant classes (P < 0.01).
(C) 15NO3– influx measurement in nrt2.1-2 and nrt2.1-2 nrt2.4 mutant seedlings. Seedlings were grown on MGRL-based plates 8 d on 10 mM NO3–then 5 d without N. 15NO3– influx was measured after 5 min of labeling in complete nutrient solution containing either 0.025 or 0.01 mM 15NO3–. Error bars represent se of eight biological replicates. Asterisks indicate statistically significant differences between nrt2.1-2 and nrt2.1-2 nrt2.4 (P < 0.01). DW, dry weight.
We then measured very-high-affinity NO3– influx on double and triple mutants that had been grown first for 8 d on full N medium and were then transferred for 5 d to medium without N. Indeed, NO3– influx was decreased in the triple mutants compared with the double mutant when measured in the presence of 0.025 or 0.01 mM external 15NO3– (Figure 7D), suggesting that the growth phenotype of triple mutant is attributed to reduced NO3– influx.
Although only the triple knockout of NRT2.1, NRT2.2, and NRT2.4, but not the single mutant nrt2.4 (Figures 7A to 7C), revealed a growth phenotype on low external NO3–this result demonstrates a role of NRT2.4 in sustaining plant growth at low N availability.
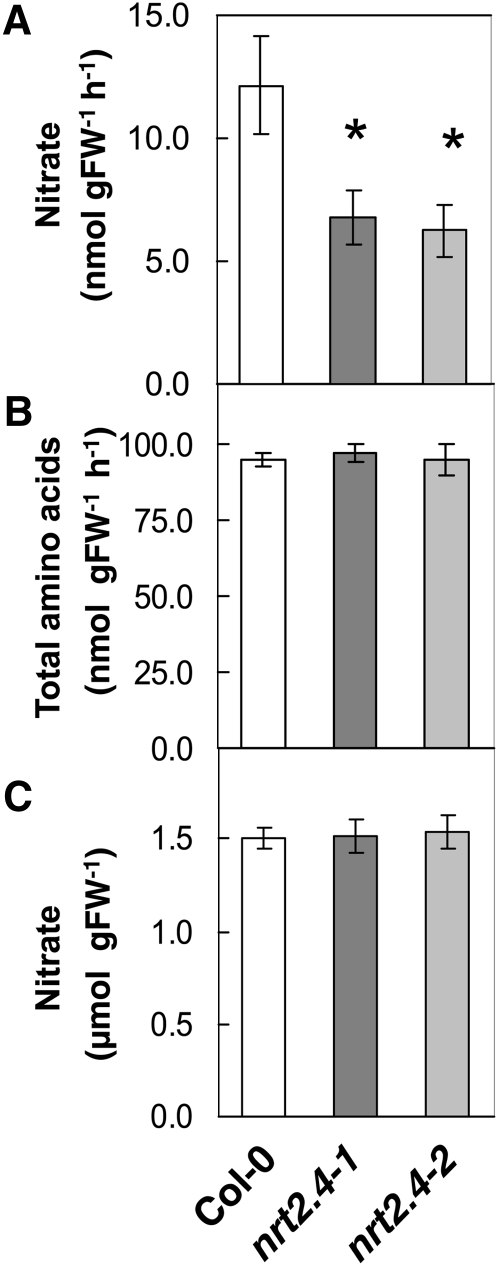
nrt2.4 Mutants Have Decreased NO3– Levels in Leaf Exudates
Since NRT2.4 is expressed in or close to the phloem of leaves, the amount of NO3– in the phloem sap in the wild type and the mutants was compared. Plants were grown on sand in short days for 6 weeks on full N and then N starved for a further 4 weeks. At that time, the plants had been severely starved but were not yet flowering. NO3– levels in the phloem exudates of mature leaves were decreased by 45 and 47% in the nrt2.4-1 and nrt2.4-2 mutants, respectively, compared with the wild type (Figure 8A). However, the levels of amino acids in the phloem exudates were similar between the wild type and mutants (Figure 8B), suggesting that the rate of exudation is comparable between the wild type and mutants and that the effect was specific to NO3–. Furthermore, whole-leaf NO3– levels were unchanged in the nrt2.4 mutants compared with the wild type (Figure 8C). The decrease in NO3– content in phloem exudate was only observed when plants were cultivated under N starvation conditions. Phloem exudate NO3– levels were similar for the wild type and mutants when plants were grown on ample NO3– (see Supplemental Figure 8 online). The expression of NRT1.7, the only other known NO3– transporter involved in shoot phloem NO3– content, was not changed between the wild type and mutants (see Supplemental Figure 9 online).
Figure 8.
nrt2.4 Mutants Have a Decreased Phloem NO3– Content.
Metabolite levels in plants grown in sand culture in the short-day condition, first in a 10 mM NO3– solution for 6 weeks and then N starved for 4 weeks.
(A) NO3– levels in leaf exudate per hour of exudation. FW, fresh weight.
(B) Total amino acid levels in leaf exudates per hour of exudation.
(C) NO3– levels in whole leaves. Data are means ± se of five biological replicates. Asterisks indicate statistically significant differences between the wild type (Col-0) and mutants (P < 0.0001).
This result is in agreement with the expression of NRT2.4 in veins (Figure 3D to 3I) and suggests a high-affinity scavenging role for NRT2.4 in NO3– remobilization during N starvation, in contrast with NRT1.7, which is involved in NO3– remobilization in the low-affinity range (Fan et al., 2009).
DISCUSSION
Arabidopsis NRT2.4 Is a Plasma MembraneNO3– Transporter Expressed in N-Starved Plants
In the Arabidopsis genome, seven closely related genes are designated as the NRT2 family (Arabidopsis Genome Initiative, 2000), and three members have been shown to encode high-affinity NO3– transporters. NRT2.4 is the NRT2 family member that shows the highest degree of homology to NRT2.1 and NRT2.2, which are the major high-affinity NO3– transporters in roots under N-replete conditions (Orsel et al., 2002; Chopin et al., 2007; Li et al., 2007). We first showed that overexpression of NRT2.4 in the high-affinity NO3– uptake-deficient nrt2.1-1 mutant partly restores NO3– uptake and plant growth (Figure 5A; see Supplemental Figure 5 online). This finding indicates that NRT2.4 is a NO3– transporter operating in the high-affinity range. This result was confirmed by functional studies in Xenopus oocytes and underpinned by the localization of NRT2.4 at the root plasma membrane. Interestingly, the functional studies in Xenopus oocytes indicate that NRT2.4, in contrast with NRT2.1 (Okamoto et al., 2006; Orsel et al., 2006), is able to transport NO3– in the absence of NAR2.1 (Figure 5B). NAR2.1 interacts directly with NRT2.1 on the plasma membrane (Yong et al., 2010), and in nar2.1 mutants, NRT2.1 is absent from the plasma membrane (Wirth et al., 2007). NO3– uptake in nar2.1 mutants is lower than in the nrt2.1 nrt2.2 double mutant (nrt2.1-1), which leads to the hypothesis that other NO3– transporter activities depend as well on NAR2.1 (Orsel et al., 2006). However, NRT2.4 is active in Xenopus oocytes without NAR2.1 (Figure 5B), which was also true for the activity of NRT2.7 (Chopin et al., 2007).
We showed that NRT2.4 takes part in root NO3– uptake at very low external concentrations, as uptake at concentrations below 0.025 mM NO3– was decreased in the knockout mutants (Figure 6). This result may be explained by the NRT2.4 localization patterns and/or a very high affinity of NRT2.4 for NO3–. Consistent with the former interpretation, NRT2.4 is predominantly expressed in the lateral root epidermis and localized to the outer face of the plasma membrane of these cells (Figures 3 and 4). A very high affinity of NRT2.4 for NO3– is implied because analyses of root uptake kinetics using the nrt2.1 nrt2.2 double mutants indicated the presence of residual root NO3– uptake at low micromolar NO3– concentrations (<0.025 mM; Cerezo et al., 2001; Li et al., 2007). In Chlamydomonas reinhardtii, very low Km (1.6 μM) has been reported for a NRT2-type NO3– transporter (Galván et al., 1996). However, further biochemical and in planta studies are required to calculate the exact Km for NRT2.4. Nevertheless, our results, together with the fact that NRT2.4 is overexpressed in the nrt2.1-1 plants (Orsel et al., 2004), provide strong evidence that NRT2.4 participates in very-high-affinity range NO3– uptake.
Historically, NO3– influx measured after long-term N starvation or when plants have not been exposed to NO3– was named constitutive HATS (cHATS; Jackson et al., 1973; Behl et al., 1988; Clarkson and Lüttge, 1991). Following this definition, NRT2.4 might be responsible for the so-called cHATS. However, as NRT2.4 expression is dramatically induced by N starvation and repressed in the presence of ammonium, the term cHATS seems misleading. The idea that distinct NO3– transporters are responsible for cHATS and inducible HATS (Okamoto et al., 2003) might need to be revised as already proposed after the finding that NAR2.1 and NRT2.1 are involved in both inducible HATS and cHATS (Okamoto et al., 2006; Orsel et al., 2006).
In the absence of NRT2.1, NRT2.2, and NRT2.4, NO3– influx at low NO3– concentrations is decreased but still detectable. More NO3– transporters, for example, other NRT2s, NRT1.1, or yet uncharacterized proteins, may be responsible for this uptake activity. NRT2.5 is a good candidate for such a role in high-affinity NO3– transport, as NRT2.5 expression increased greatly in the nrt2.1 nrt2.2 double mutant (nrt2.1-1; Orsel et al., 2004).
The Major High-Affinity NO3– Transporter NRT2.1 and the N Starvation–Inducible NO3– Transporter NRT2.4 Are Complementary in Roots
The expression of NRT2.4 is induced during N starvation and increased over time. By contrast, NRT2.1 transcript level is transiently derepressed during early starvation (Figure 1C; see Supplemental Figure 3A online). Furthermore, the tissue-specific expression of NRT2.1 and NRT2.4 differ, the first gene is expressed in older roots and to a lesser extent in lateral roots (Figure 2; Nazoa et al., 2003; Wirth et al., 2007), and the second one only in young lateral roots during N starvation (Figure 2). Lateral root growth is stimulated during N starvation, and the presence of an N starvation–induced high-affinity NO3– transporter in this organ should contribute to the adaptation of plants to N availability. Whereas NRT2.1 is expressed in the root cortex and under some conditions in the epidermis (Wirth et al., 2007), NRT2.4 is expressed mainly in the epidermis (Figures 3B and 3C), the outermost root cell layer, which is first in contact with external NO3–. Thus, NRT2.4 and NRT2.1 spatiotemporal expression pattern is complementary, with NRT2.1 being the main transporter and NRT2.4 only expressed under N starvation conditions with properties ideally suited for a NO3– transporter active under N-limiting conditions, being expressed in lateral roots and transporting NO3– with a very high affinity.
At the protein level, we showed a polar subcellular localization of NRT2.4-GFP to the outer periclinal side and the anticlinal faces of the plasma membrane (Figure 4). Located at the soil/root interface, NRT2.4 would be ideally placed to compete with soil microbes when available NO3– becomes scarce. Several recent studies have shown that the polar localization of mineral nutrient transporters in plant cells is involved in efficient uptake of the nutrient. In rice (Oryza sativa), the silicic acid channel Lsi1 (NIP2;1) and the silicon exporter Lsi2 localize to the outer (distal) and inner (proximal) plasma membrane domain of the exodermis and the endodermis, respectively, thus driving directional transcellular transport of silicon in rice roots (Ma et al., 2006, 2007). Similarly, the potato (Solanum tuberosum) high-affinity phosphate transporter PT2 localizes to the apical surface of the epidermal plasma membrane (Gordon-Weeks et al., 2003) and the boron transporters NIP5;1 and BOR1 localize to opposite plasma membrane domains, which illustrates the radial transport route of boron toward the stele (Takano et al., 2010). It is likely that the polar localization of NRT2.4-GFP to the outer plasma membrane domain in epidermal cells is important for efficient capture and uptake of NO3– from the soil solution into roots.
In nrt2.4 single mutants, NO3– transport in the very-high-affinity range was decreased, but a growth phenotype was not observed (Figures 7A to 7C; see Supplemental Figures 6C and 6D online). On the other hand, the loss of NRT2.4 activity in addition to loss of NRT2.1/2.2 function led to further decreases in NO3– uptake and to an impact on plant growth under low external NO3– (Figure 7). Considering that NRT2.1 and NRT2.2 also mediate NO3– uptake at concentrations below 0.025 mM (Cerezo et al., 2001; Filleur et al., 2001), it seems that the interplay between NRT2.1, NRT2.2, and NRT2.4 is required to ensure optimal adaptation to N limitation. Furthermore, these results are consistent with what was suggested for ammonium transport (Yuan et al., 2007), that the effective uptake is achieved by the proper spatial arrangement of transporters and the distribution of their transport capacities at different substrate affinities. Taken together, these results suggest that for all these nutrients, Si, B, NH4+, and NO3–when the resource is present at low concentrations (high-affinity ranges), the spatial distribution of root transporters becomes very important.
NRT2.4 Participates in Phloem NO3– Transport in Shoots
The phloem performs a variety of important roles in plants in addition to the basic function of transporting carbohydrates and reduced N to sink tissues. Genes specifically expressed in the phloem have been identified (Brady et al., 2007; Zhang et al., 2008), but the functions of relatively few phloem-expressed genes have been analyzed. NO3– has been shown to be transported not only in the xylem but also in the phloem. Phloem sap of plants growing in nonlimiting N availability contains 1.9 to 8.1 mM NO3– (Smith and Milburn, 1980; Hayashi and Chino, 1985, 1986; Allen and Smith, 1986). The first NO3– transporter implicated in phloem NO3– transport was NRT1.7, a plasma membrane low-affinity NO3– transporter, which is localized in the sieve element/companion cell complex. NRT1.7 is involved in moving excess NO3– from older leaves into developing tissues, indicating that apoplastic phloem loading is responsible for NO3– remobilization (Fan et al., 2009). A second plasma membrane NO3– transporter, NRT1.9, is expressed in phloem companion cells in roots. It seems that phloem-localized NRT1.9 can influence root-to-shoot xylem transport of NO3– when plants are grown on ample NO3– (Wang and Tsay, 2011). NRT2.4 is expressed in or close to the phloem in shoots, probably in phloem parenchyma (Figure 3), and phloem sap of nrt2.4 mutants cultivated under N starvation had decreased NO3– levels compared with the wild type (Figure 8A). These data suggest that NRT2.4 in source leaves is involved in delivering NO3– into the phloem for remobilization. The phloem parenchyma cells appear to be specialized in delivery of photosynthetic assimilate products from the bundle sheath into the sieve track (Haritatos et al., 2000). During N starvation, NO3– that was stored in the vacuoles of bundle sheath and mesophyll cells will be remobilized and might enter the phloem sap via the phloem parenchyma.
However, no consequences for plant growth or senescence have been detected for the nrt2.4 mutants, indicating that the decreased phloem NO3– levels in the mutants are not limiting for the adaptation to N starvation and that other transporters are involved in the same processes. For this reason, it might be interesting to study for example double mutants of NRT1.7 and NRT2.4.
In conclusion, we showed that N limitation induced NRT2.4 expression in the lateral root epidermis membrane facing the external medium and the shoot phloem. In planta and heterologous expression in addition to mutant analysis revealed a NO3– transport activity of the plasma membrane–located NRT2.4 in the high-affinity range. In the absence of the main high-affinity transporters NRT2.1 and NRT2.2, loss of function of NRT2.4 has an impact on plant fresh weight under low external NO3–. These data not only show that NRT2.4 is a NO3– transporter functioning in the high-affinity range and plays a role under N starvation but also illustrate the sophisticated interplay of multiple NO3– transporters in response to changes in N availability.
METHODS
Plant Material and Growth Conditions
The SAIL line CS872100 (nrt2.4-2) was obtained from the ABRC and was derived from a T-DNA–mutagenized population of the Col-0 accession (Alonso et al., 2003), while the nrt2.4-1 line was derived from a T-DNA–mutagenized population of the Col-0 accession (Forsbach et al., 2003). Homozygous mutant plants were isolated by PCR with the primers listed in Supplemental Table 2 online.
The nrt2.1 nrt2.2 double mutant (Col-0 accession, called nrt2.1-2) was obtained from the stock center (SALK_035429) and was characterized previously (Little et al., 2005; Li et al., 2007).
Another nrt2.1 nrt2.2 double mutant (Ws accession, called nrt2.1-1) was obtained initially from the Versailles T-DNA insertion line collection (FLAG lines) and was characterized previously (Filleur et al., 2001).
Triple null mutants were generated by crosses of the double mutant nrt2.1 nrt2.2 (nrt2.1-2) and the two nrt2.4 alleles (nrt2.4-1 and nrt2.4-2). The F1 seeds were grown and allowed to self-fertilize to produce a population of F2 plants. We determined the genotypes of the F2 plants by genomic PCR for NRT2.1, NRT2.2, and NRT2.4 (primers are given in Supplemental Table 2 online).
For studies on seedlings, wild-type Arabidopsis thaliana (accession Col-0 or Ws) as well as mutants or transgenic plants were grown on Molecular Genetics Research Laboratory (University of Tokyo) growth medium (MGRL)–based vertical agar plates (Naito et al., 1994) containing 1% Suc in 100 μmol m−2 s−1 fluorescent light in a long-day condition (16 h light/8 h dark). The full N MGRL plate (full N plate) contained 10 mM NO3–and MGRL plates with other N contents are indicated in the figures. For N-limiting conditions, the ion equilibrium of the medium was ensured by replacing KNO3 and CaNO3 by CaCl2 and KCl. For GUS expression, seedlings were grown in vitro on a medium containing either 0 or 10 mM NO3– (Estelle and Somerville, 1987). Adult plants were grown either under hydroponic conditions in a Sanyo growth chamber (SGC660/PL4) or on sand culture in a Seitha growth chamber with an 8-h-light/16-h-dark cycle at 21°C/17°C, respectively, 80% relative humidity, and 150 μmol m–2 s–1 irradiation. Seeds were stratified for 5 d at 4°C in the dark in a 0.15% agar solution in water and then sown and cultivated as already described (Orsel et al., 2002; Castaings et al., 2009). The nutrient solutions contained 6 mM NO3– as full N for hydroponic culture and 10 mM NO3– for full N for sand culture. For N starvation conditions, ion equilibrium of the medium was ensured by replacing KNO3 and Ca(NO3)2 by KCl and CaSO4.
Quantitative RT-PCR
RNA was extracted with the GenElute Mammalian Total RNA kit from Sigma-Aldrich, modified by adding a DNase step, which was performed with the Qiagen RNase-free DNase kit (according the protocols of the supplier), RNeasy Plant Mini kit (Qiagen), or Trizol method (Invitrogen). First-strand cDNAs were synthesized according to Daniel-Vedele and Caboche (1993) using Moloney murine leukemia virus reverse transcriptase and oligo(dT)15 primers (Promega) or using the SuperScript III Fast strand synthesis system (Invitrogen). PCR was performed on a LightCycler instrument (Roche), StepOnePlus real-time PCR system (Applied Biosystems), or on a Realplex Mastercycler (Eppendorf) with the LightCycler-FastStart DNA Master SYBR Green I kit (Roche) or SYBR Premix ExTaq II (Takara) according to the manufacturer's protocol. Each reaction was performed on a 1:20 dilution of the first cDNA strands, synthesized as described above, in a total reaction of 20 μL. With this dilution, the SYBR green signal was linear. Specific primer sets are given in Supplemental Table 2 online.
Complementation of the nrt2.1-1 Mutant with RolD:NRT2.1 or RolD:NRT2.4
The full-length NRT2.4 cDNA was amplified by RT-PCR from total RNAs extracted from Arabidopsis roots (accession Ws) using the start primer ATG-NRT2.4, the stop primer STOP-NRT2.4 (see Supplemental Table 2 online), and the high-fidelity Taq enzyme (Roche) and were cloned into pGEM-T Easy vector (Promega). The nucleotide sequence of the insert was checked before transferring the cDNA first into the pRT103 vector (Töpfer et al., 1987) downstream from the RolD promoter, followed by cloning of this whole chimeric gene into a pGREEN vector (Hellens et al., 2000) already containing the cauliflower mosaic virus 35S terminator. The full-length NRT2.1 cDNA was amplified from a cDNA clone (Filleur and Daniel-Vedele, 1999) using the start primer ATG-NRT2.1, the stop primer STOP-NRT2.1 (see Supplemental Table 2 online), and the subsequent procedures as for NRT2.4. Binary vectors were introduced into Agrobacterium tumefaciens strain C58C1 (pMP90). The nrt2.1-1 mutant (Filleur et al., 2001) was transformed by the in planta method using the surfactant Silwet L-77. Transgenic plants were selected on Estelle and Sommerville media (Estelle and Sommerville, 1987) containing 20 μg L–1 of hygromycin B.
Root 15N Influx
Influx of 15NO3– was assayed as previously described (Orsel et al., 2004). The plants were transferred first to 0.1 mM CaSO4 for 1 min and then to complete nutrient solution containing 15NO3– (atom% 15N: 99%) at the indicated concentrations for 5 min and finally to 0.1 mM CaSO4 for 1 min The roots were dried for 72 h at 80°C and analyzed using the ANCA-MS system (PDZ Europa). Influx of 15NO3– was calculated from the total N and 15N content of the roots.
NO3– Depletion Assay
NO3– depletion assays were conducted as by Liu et al. (1999) with some modifications. Wild-type (Col-0) and nrt2.4-2 seedlings were grown on MGRL plates containing 10 mM NH4NO3 for 7 d and then transferred to MGRL plates without a N source. On the fifth day, ~30 seedlings (0.2 g fresh weight) were washed twice with MGRL medium without N and suspended in 100 mL of MGRL solution with 0.2 mM NO3– or in 10 mL of MGRL solution with 0.01 mM NO3– in a flask. The flasks were rotated at 100 rpm under 100 μmol m–2 s–1 irradiation, and the assay media were collected at the indicated time points up to 2 h. The amount of NO3– left in the assay medium was determined by HPLC (Waters 2695 separation module and Waters 2996 photodiode array detector) using an anion exchange column (Waters Spherisorb S5 SAX). The mobile phase was pumped at a rate of 1 mL/min and consisted of 50 mM K2HPO4/KH2PO4, pH 3.7.
Xenopus laevis Oocyte Expression System
The pGEM-T Easy vector containing the full-length NRT2.4 cDNA was fully digested with NotI. The cDNA fragment was blunt-ended using the Klenow enzyme, dephosphorylated using calf intestinal alkaline phosphatase, and then subcloned into the EcoRV site of the pT7TS expression vector containing the 5′- and 3′-untranslated regions of the Xenopus β-globin gene (Cleaver et al., 1996). For in vitro synthesis of mRNA, the pT7TS clone was linearized by digestion with XbaI for NRT2.4 or with BamHI for NAR2.1. Synthesis of capped full-length mRNAs and Xenopus oocyte preparation were performed as previously described (Orsel et al., 2006). Healthy oocytes at stage V or VI were injected with 50 nL of water (nuclease free), NRT2.4 mRNAs at 1 μg·μL–1, or a mixture of NAR2.1 (Orsel et al., 2006) and NRT2.4 mRNA at 1 μg·μL–1 each. After 3 d incubation at 18°C, 5 to 10 oocytes were incubated in 3 mL of ND96 solution (96 mM NaCl, 2 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, and 5 mM HEPES), pH 6.5, enriched with 0.5 mM Na15NO3 (atom% 15N: 98%) during 16 h at 18°C. The oocytes were then thoroughly washed four times with ice-cooled 0.5 mM NaNO3 ND96 solution and dried at 60°C. The 15N/14N ratio of the single dried oocyte was measured as previously described (Orsel et al., 2006). The values are means ± sd of five replicates; results from a representative experiment are shown.
Construction of ProNRT2.4:GUS Fusions
Approximately 1.2 kb of DNA immediately 5′ of the ATG start codon of the NRT2.4 gene was amplified from Arabidopsis genomic DNA using Pfu polymerase (Stratagene Europe) and gene-specific primers (5′ProNRT2.4 and 3′ProNRT2.4; see Supplemental Table 2 online). PCR products were cloned into pZeroblunt (Stratagene), sequenced, and subcloned into pBI101 (Clontech Laboratories). The binary vector was transformed into Agrobacterium strain GV3101 pMP90 via electroporation (Koncz and Schell, 1986). Transformants were selected on yeast extract and beef plates containing rifampicin (100 μg L–1), gentamycin (20 μg L–1), and kanamycin (50 μg L–1) and confirmed positive via restriction digest of the recovered plasmid. Arabidopsis was transformed using the floral dip method as described by Clough and Bent (1998). Transgenic seedlings were recovered on Murashige and Skoog plates containing 1% Suc and 50 μg L–1 kanamycin.
GUS Staining
Histochemical GUS staining was performed according to the method described by Jefferson (1987) with some modifications. Plants were vacuum infiltrated for 5 min in a 50 mM potassium phosphate buffer, pH 7.0 (0.05% Triton, 2 mM ferro/ferricyanide, and 2 mM X-glucuronide). Subsequently, samples were incubated overnight in the dark at 37°C. Stained plants were cleared by incubation in an ethanol series (70 to 100% [v/v]) and then observed under a light microscope (Axioplan 2; Zeiss).
For sections, tissue was fixed in formaldehyde (4%). After dehydration in an ethanol series, tissue was embedded in Historesine (Kit Technovit 7100; Labonord). Eight-micrometer cuts were produced with a microtome (EICA JUNG RM 2055). Propidium iodide staining was performed as by Truernit et al. (2008). The fluorescence was recorded at 600 nm (excitation 488 nm) for the propidium iodide staining of cell walls and the reflection between 485 and 491 nm visualized the GUS staining (confocal microscope; Leica TCS-SP2-AOBS)
Construction of Transcriptional and Translational GFP Fusions
The NRT2.4 promoter (ProNRT2.4; 1886 bp upstream of the inferred initiation codon), NRT2.1 promoter (ProNRT2.1; 1974 bp upstream), and GFP fragments were amplified with KOD plus DNA polymerase (TOYOBO) and specific primers (pNRT2.4-F and pNRT2.4-R for ProNRT2.4 and pNRT2.1-F and pNRT2.1-R for ProNRT2.1; see Supplemental Table 2 online), and assembled in the pBA002a binary vector (Kiba et al., 2007) to generate ProNRT2.4:GFP and ProNRT2.1:GFP. A genomic fragment encompassing NRT2.4 promoter and NRT2.4 coding region (without a stop codon) was amplified using PrimeStar GXL DNA polymerase and specific primers (pNRT2.4-F and NRT2.4-R; see Supplemental Table 2 online). The fragment was cloned into pENTR/D-TOPO vector (Invitrogen) and sequenced, then integrated into the Gateway binary vector pBA002a-GFP, which is a derivative of pBA002a (Kiba et al., 2007), to generate C-terminal GFP fusion construct (ProNRT2.4:NRT2.4-GFP), using LR clonase (Invitrogen). These constructs were transferred into Agrobacterium strain GV3101 (pMP90), and positive colonies were confirmed by PCR. Wild-type (Col-0) plants were transformed with the floral dip method as described (Clough and Bent, 1998), and transformants were selected on Murashige and Skoog plates (1% Suc) containing 10 μg L–1 bialaphos sodium salt.
Whole-Plant GFP Imaging
GFP fluorescence and chlorophyll autofluorescence were visualized using the FluorImager 595 (Molecular Dynamics). GFP signal was obtained under 488-nm excitation using a 530DF30 filter. Chlorophyll autofluorescence of plants was detected under 488-nm excitation using a 610RG filter.
Confocal Microscopy Analysis of the GFP Signal
Transgenic seedlings grown on full N MGRL medium for 7 d and then transferred for 3 d on medium without N were observed with the Zeiss LSM510 META confocal imaging system. Cells in living roots were stained with 0.1 μg/mL of FM4-64 for 5 min before observation (Molecular Probes). The different fluorochromes were detected using laser lines 488 nm (Alexa 488, GFP, and FM4-64) and 543 nm (Alexa 568). The images were coded green (fluorescent isothiocyanate and GFP) and red (Alexa 568 and FM4-64), giving yellow colocalization in merged images. The samples were washed twice after staining before observation with the confocal microscope. Each image shown represents a single focal plane.
Phloem Sap Exudation
Mature leaves of 10-week-old plants were cut at the base with a sharp razor blade at the petiole with a drop of overlaying solution to avoid allowing air to enter the vascular tissue. The petiole was recut, covered by the solution, and then four leaves were placed with the petiole in a tube containing 800 μL solution (5 mM EDTA, pH8.5). After 16 h in a water-saturated atmosphere, the phloem exudate–enriched solution was collected and the leaf fresh weight was measured. Free Glc content in the phloem exudates was below the detection limit (30 nmol/g fresh weight), indicating that contamination by damaged cell extract in the exudates was low. Leaves from independent plants were used for measuring total leaf NO3– content.
NO3– Measurement in Plants
NO3– content in plants grown on agar plates was measured as by Chiu et al. (2004) with some modifications. Plants were separated into roots and shoots, weighed, and collected in tubes. One milliliter of milli-Q water was added to each tube, boiled for 20 min, and frozen at −80°C overnight. After centrifugation, the supernatant was subjected to NO3– quantification by HPLC system as described in the method of NO3– depletion assay. The NO3– content for plants grown on soil or in hydroponic culture was measured in ethanolic extracts as by Orsel et al. (2004) using Dionex HPLC measurement.
Metabolite Measurements in Phloem Exudates
N NO3– concentration in phloem exudates was measured using a modified Miranda method (Miranda et al., 2001; Chopin et al., 2007) using an appropriate EDTA concentration as blank. Glc and Suc content was measured using a standard NADPH-coupled assay for soluble sugar measurements (Matt et al., 2001). Total amino acid content was measured using Ninhydrine methods as by Matt et al. (2001).
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for genes mentioned in this article are as follows: At5g60770 (NRT2.4), At1g08090 (NRT2.1), At1g08100 (NRT2.2), At5g60390 (EF1α), At1g12940 (NRT2.5), At3g45060 (NRT2.6), At5g14570 (NRT2.7), At1g12110 (NRT1.1), At1g69870 (NRT1.7), At5g50200 (NAR2.1); RolD (Agrobacterium rhizogenes) GenBank accession number X64255.1; Germplasm, SAIL_205_F02 line CS872100 (nrt2.4-2), MDL-ArBrAr-125 (nrt2.4-1), SALK_035429 (nrt2.1 nrt2.2 double mutant in Col-0), and FLAG line EDP03 (nrt2.1 nrt2.2 double mutant in Ws).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Nitrogen Dose-Dependent Expression of NRT2.4 and NRT2.1.
Supplemental Figure 2. Nitrate Content in the Root and Shoot during N Starvation.
Supplemental Figure 3. Effect of N Starvation and N Resupply on the Expression of NRT2.4 in Adult Plants.
Supplemental Figure 4. Nitrogen Dose-Dependent Changes in GFP Fluorescence in proNRT2.4:GFP.
Supplemental Figure 5. NRT2.4 Can Partly Restore Nitrate Content and Shoot Biomass When Overexpressed in the nrt2.1-1 Mutant.
Supplemental Figure 6. nrt2.4 Single Mutants Showed No Growth Phenotype under N Starvation Condition.
Supplemental Figure 7. Nitrate Influx below 0.025 mM External Nitrate Is Affected in nrt2.4.
Supplemental Figure 8. Nitrate Content in Phloem Exudates Is Not Affected in nrt2.4 Mutant When Grown on High Nitrate.
Supplemental Figure 9. NRT1.7 Expression Is Not Affected in nrt2.4 Mutants under N Limitation.
Supplemental Table 1. Relative Gene Expression Levels of NRT2 and NRT1.1 Genes in Roots of the nrt2.1-1 Mutant Complemented by ProRolD:NRT2.1 (RolD:NRT2.1) or ProRolD:NRT2.4 (RolD:NRT2.4) Constructs.
Supplemental Table 2. Oligonucleotides Used for Quantitative RT-PCR, Mutant Screening, and Cloning.
Acknowledgments
We thank Georg Leggewie and Michael Udvardi for the gift of the ProNRT2.4:GUS T1 seeds and Renate Schmidt for the gift of the nrt2.4-1 insertion mutant. We thank Pascal Tillard and Yumiko Tsuchiya for 15N analyses, Jean-Christophe Palauqui for help and advice regarding propidium iodide staining, and Catherine Jeudy, Joël Talbotec, and Philippe Maréchal for taking care of the plants in the greenhouse. We also thank the Nottingham Arabidopsis Stock Centre and the ABRC for supplying T-DNA insertion lines. This work was supported in part by a Grant-in-Aid for Scientific Research on Innovative Areas (No. 21114005 to H.S.) from the Ministry of Education, Culture, Sports, Science and Technology, Japan; by a grant from the Agence Nationale de la Recherche (France, Nitrapool Project, ANR-08-BLAN-0008-02; F.D.-V. and A.K.); and by European Union Grant BIO4CT972231, Research Training Network “Plant use of nitrate” HPRN-CT-2002-00247 (A.K., F.D-V., A.M., and M.O.).
AUTHOR CONTRIBUTIONS
T.K., F.D.-V., H.S., and A.K. designed the research. T.K., A.-B.F.-B., F.L., L.L., S.B.-M., M.O., V.B., and A.M. performed research. T.K., A.-B.F.-B., F.L., L.L., M.O., and A.M. analyzed data. T.K and A.K. wrote the article.
References
- Allen S., Smith J.A.C. (1986). Ammonium nutrition in Ricinus communis: Its effect on plant growth and the chemical composition of the whole plant, xylem and phloem saps. J. Exp. Bot. 37: 1599–1610 [Google Scholar]
- Almagro A., Lin S.H., Tsay Y.F. (2008). Characterization of the Arabidopsis nitrate transporter NRT1.6 reveals a role of nitrate in early embryo development. Plant Cell 20: 3289–3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Behl R., Tischner R., Raschke K. (1988). Induction of a high-capacity nitrate-uptake mechanism in barley roots prompted by nitrate uptake through a constitutive lowcapacity mechanism. Planta 176: 235–240 [DOI] [PubMed] [Google Scholar]
- Brady S.M., Orlando D.A., Lee J.-Y., Wang J.Y., Koch J., Dinneny J.R., Mace D., Ohler U., Benfey P.N. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Castaings L., Camargo A., Pocholle D., Gaudon V., Texier Y., Boutet-Mercey S., Taconnat L., Renou J.P., Daniel-Vedele F., Fernandez E., Meyer C., Krapp A. (2009). The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 57: 426–435 [DOI] [PubMed] [Google Scholar]
- Cerezo M., Tillard P., Filleur S., Muños S., Daniel-Vedele F., Gojon A. (2001). Major alterations of the regulation of root NO(3)(-) uptake are associated with the mutation of Nrt2.1 and Nrt2.2 genes in Arabidopsis. Plant Physiol. 127: 262–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C.C., Lin C.S., Hsia A.P., Su R.C., Lin H.L., Tsay Y.F. (2004). Mutation of a nitrate transporter, AtNRT1:4, results in a reduced petiole nitrate content and altered leaf development. Plant Cell Physiol. 45: 1139–1148 [DOI] [PubMed] [Google Scholar]
- Chopin F., Orsel M., Dorbe M.-F., Chardon F., Truong H.N., Miller A.J., Krapp A., Daniel-Vedele F. (2007). The Arabidopsis ATNRT2.7 nitrate transporter controls nitrate content in seeds. Plant Cell 19: 1590–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson D.T., Hopper M.J., Jones L.H.P. (1986). The effect of root temperature on the uptake of nitrogen and the relative size of the root-system in Lolium perenne. 1. Solutions containing both NH4+ and NO3−. Plant Cell Environ. 9: 535–545 [Google Scholar]
- Clarkson D.T., Lüttge U. (1991). Inducible and repressible nutrient transport systems. Prog. Bot. 52: 61–83 [Google Scholar]
- Cleaver O.B., Patterson K.D., Krieg P.A. (1996). Overexpression of the tinman-related genes XNkx-2.5 and XNkx-2.3 in Xenopus embryos results in myocardial hyperplasia. Development 122: 3549–3556 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crawford N.M., Glass A.D.M. (1998). Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 3: 389–395 [Google Scholar]
- Daniel-Vedele F., Caboche M. (1993). A tobacco cDNA clone encoding a GATA-1 zinc finger protein homologous to regulators of nitrogen metabolism in fungi. Mol. Gen. Genet. 240: 365–373 [DOI] [PubMed] [Google Scholar]
- Daniel-Vedele F., Filleur S., Caboche M. (1998). Nitrate transport: A key step in nitrate assimilation. Curr. Opin. Plant Biol. 1: 235–239 [DOI] [PubMed] [Google Scholar]
- De Angeli A., Monachello D., Ephritikhine G., Frachisse J.M., Thomine S., Gambale F., Barbier-Brygoo H. (2006). The nitrate/proton antiporter AtCLCa mediates nitrate accumulation in plant vacuoles. Nature 442: 939–942 [DOI] [PubMed] [Google Scholar]
- Dechorgnat J., Nguyen C.T., Armengaud P., Jossier M., Diatloff E., Filleur S., Daniel-Vedele F. (2011). From the soil to the seeds: The long journey of nitrate in plants. J. Exp. Bot. 62: 1349–1359 [DOI] [PubMed] [Google Scholar]
- de Ruijter N.C.A., Verhees J., van Leeuwen W., van der Krol A.R. (2003). Evaluation and comparison of the GUS, LUC and GFP reporter system for gene expression studies in plants. Plant Biol. 5: 103–115 [Google Scholar]
- Drew M.C., Saker L.R. (1975). Nutrient supply and the growth of the seminal root system in barley. II. Localized, compensatory increases in lateral root growth and rates of nitrate uptake when nitrate supply is restricted to only one part of the root system. J. Exp. Bot. 26: 79–90 [Google Scholar]
- Estelle M.A., Somerville C. (1987). Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol. Gen. Genet. 206: 200–206 [Google Scholar]
- Fan S.-C., Lin C.-S., Hsu P.-K., Lin S.-H., Tsay Y.-F. (2009). The Arabidopsis nitrate transporter NRT1.7, expressed in phloem, is responsible for source-to-sink remobilization of nitrate. Plant Cell 21: 2750–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsbach A., Schubert D., Lechtenberg B., Gils M., Schmidt R. (2003). A comprehensive characterization of single-copy T-DNA insertions in the Arabidopsis thaliana genome. Plant Mol. Biol. 52: 161–176 [DOI] [PubMed] [Google Scholar]
- Fraisier V., Gojon A., Tillard P., Daniel-Vedele F. (2000). Constitutive expression of a putative high-affinity nitrate transporter in Nicotiana plumbaginifolia: Evidence for post-transcriptional regulation by a reduced nitrogen source. Plant J. 23: 489–496 [DOI] [PubMed] [Google Scholar]
- Filleur S., Daniel-Vedele F. (1999). Expression analysis of a high-affinity nitrate transporter isolated from Arabidopsis thaliana by differential display. Planta 207: 461–469 [DOI] [PubMed] [Google Scholar]
- Filleur S., Dorbe M.-F., Cerezo M., Orsel M., Granier F., Gojon A., Daniel-Vedele F. (2001). An Arabidopsis T-DNA mutant affected in Nrt2 genes is impaired in nitrate uptake. FEBS Lett. 489: 220–224 [DOI] [PubMed] [Google Scholar]
- Forde B.G. (2000). Nitrate transporters in plants: Structure, function and regulation. Biochim. Biophys. Acta 1465: 219–235 [DOI] [PubMed] [Google Scholar]
- Galván A., Quesada A., Fernández E. (1996). Nitrate and nitrate are transported by different specific transport systems and by a bispecific transporter in Chlamydomonas reinhardtii. J. Biol. Chem. 271: 2088–2092 [DOI] [PubMed] [Google Scholar]
- Glass A.D.M. (2003). Nitrogen use efficiency of crop plants: Physiological constraints upon nitrogen absorption. Crit. Rev. Plant Sci. 22: 453–470 [Google Scholar]
- Gordon-Weeks R., Tong Y., Davies T.G., Leggewie G. (2003). Restricted spatial expression of a high-affinity phosphate transporter in potato roots. J. Cell Sci. 116: 3135–3144 [DOI] [PubMed] [Google Scholar]
- Haritatos E., Medville R., Turgeon R. (2000). Minor vein structure and sugar transport in Arabidopsis thaliana. Planta 211: 105–111 [DOI] [PubMed] [Google Scholar]
- Hayashi H., Chino M. (1985). Nitrate and other anions in the rice phloem sap. Plant Cell Physiol. 26: 325–330 [DOI] [PubMed] [Google Scholar]
- Hayashi H., Chino M. (1986). Collection of pure phloem sap from wheat and its chemical composition. Plant Cell Physiol. 27: 1387–1393 [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Ho C.H., Lin S.H., Hu H.C., Tsay Y.F. (2009). CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194 [DOI] [PubMed] [Google Scholar]
- Huang N.C., Liu K.H., Lo H.J., Tsay Y.F. (1999). Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell 11: 1381–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson W.A., Flesher D., Hageman R.H. (1973). Nitrate uptake by dark-grown corn seedlings: some characteristics of apparent induction. Plant Physiol. 51: 120–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A. (1987). Assaying chimeric gene in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 5: 387–405 [Google Scholar]
- Jossier M., Kroniewicz L., Dalmas F., Le Thiec D., Ephritikhine G., Thomine S., Barbier-Brygoo H., Vavasseur A., Filleur S., Leonhardt N. (2010). The Arabidopsis vacuolar anion transporter, AtCLCc, is involved in the regulation of stomatal movements and contributes to salt tolerance. Plant J. 64: 563–576 [DOI] [PubMed] [Google Scholar]
- Kiba T., Henriques R., Sakakibara H., Chua N.H. (2007). Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 19: 2516–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C., Schell J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204: 383–396 [Google Scholar]
- Krouk G., et al. (2010). Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 18: 927–937 [DOI] [PubMed] [Google Scholar]
- Lejay L., Tillard P., Lepetit M., Olive F., Filleur S., Daniel-Vedele F., Gojon A. (1999). Molecular and functional regulation of two NO3- uptake systems by N- and C-status of Arabidopsis plants. Plant J. 18: 509–519 [DOI] [PubMed] [Google Scholar]
- Li J.Y., et al. (2010). The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 22: 1633–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Wang Y., Okamoto M., Crawford N.M., Siddiqi M.Y., Glass A.D.M. (2007). Dissection of the AtNRT2.1:AtNRT2.2 inducible high-affinity nitrate transporter gene cluster. Plant Physiol. 143: 425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.H., Kuo H.F., Canivenc G., Lin C.S., Lepetit M., Hsu P.K., Tillard P., Lin H.L., Wang Y.Y., Tsai C.B., Gojon A., Tsay Y.F. (2008). Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20: 2514–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little D.Y., Rao H., Oliva S., Daniel-Vedele F., Krapp A., Malamy J.E. (2005). The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc. Natl. Acad. Sci. USA 102: 13693–13698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K.H., Huang C.Y., Tsay Y.F. (1999). CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11: 865–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K.H., Tsay Y.F. (2003). Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J. 22: 1005–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.F., Tamai K., Yamaji N., Mitani N., Konishi S., Katsuhara M., Ishiguro M., Murata Y., Yano M. (2006). A silicon transporter in rice. Nature 440: 688–691 [DOI] [PubMed] [Google Scholar]
- Ma J.F., Yamaji N., Mitani N., Tamai K., Konishi S., Fujiwara T., Katsuhara M., Yano M. (2007). An efflux transporter of silicon in rice. Nature 448: 209–212 [DOI] [PubMed] [Google Scholar]
- Masclaux-Daubresse C., Reisdorf-Cren M., Orsel M. (2008). Leaf nitrogen remobilisation for plant development and grain filling. Plant Biol. (Stuttg.) 10(Suppl. 1): 23–36 [DOI] [PubMed] [Google Scholar]
- Matt P., Geiger M., Walch-Liu P., Engels C., Krapp A., Stitt M. (2001). The immediate cause of the diurnal changes of nitrogen metabolism in leaves of nitrate-replete tobacco: A major imbalance between the rate of nitrate reduction and the rates of nitrate uptake and ammonium metabolism during the first part of the light period. Plant Cell Environ. 24: 177–190 [Google Scholar]
- Mickelson S., See D., Meyer F.D., Garner J.P., Foster C.R., Blake T.K., Fischer A.M. (2003). Mapping of QTL associated with nitrogen storage and remobilization in barley (Hordeum vulgare L.) leaves. J. Exp. Bot. 54: 801–812 [DOI] [PubMed] [Google Scholar]
- Miranda K.M., Espey M.G., Wink D.A. (2001). A rapid, simple spectrophotometric method for simultaneous detection of nitrate and nitrite. Nitric Oxide 5: 62–71 [DOI] [PubMed] [Google Scholar]
- Naito S., Hirai M.Y., Chino M., Komeda Y. (1994). Expression of a soybean (Glycine max [L.] Merr.) seed storage protein gene in transgenic Arabidopsis thaliana and its response to nutritional stress and to abscisic acid mutations. Plant Physiol. 104: 497–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazoa P., Vidmar J.J., Tranbarger T.J., Mouline K., Damiani I., Tillard P., Zhuo D., Glass A.D.M., Touraine B. (2003). Regulation of the nitrate transporter gene AtNRT2.1 in Arabidopsis thaliana: Responses to nitrate, amino acids and developmental stage. Plant Mol. Biol. 52: 689–703 [DOI] [PubMed] [Google Scholar]
- Okamoto M., Kumar A., Li W., Wang Y., Siddiqi M.Y., Crawford N.M., Glass A.D. (2006). High-affinity nitrate transport in roots of Arabidopsis depends on expression of the NAR2-like gene AtNRT3.1. Plant Physiol. 140: 1036–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M., Vidmar J.J., Glass A.D.M. (2003). Regulation of NRT1 and NRT2 gene families of Arabidopsis thaliana: Responses to nitrate provision. Plant Cell Physiol. 44: 304–317 [DOI] [PubMed] [Google Scholar]
- Orsel M., Chopin F., Leleu O., Smith S.J., Krapp A., Daniel-Vedele F., Miller A.J. (2006). Characterization of a two-component high-affinity nitrate uptake system in Arabidopsis. Physiology and protein-protein interaction. Plant Physiol. 142: 1304–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsel M., Eulenburg K., Krapp A., Daniel-Vedele F. (2004). Disruption of the nitrate transporter genes AtNRT2.1 and AtNRT2.2 restricts growth at low external nitrate concentration. Planta 219: 714–721 [DOI] [PubMed] [Google Scholar]
- Orsel M., Krapp A., Daniel-Vedele F. (2002). Analysis of the NRT2 nitrate transporter family in Arabidopsis. Structure and gene expression. Plant Physiol. 129: 886–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peoples M.B., Herridge D.F., Ladha J.K. (1995). Biological nitrogen fixation: an efficient source of nitrogen for sustainable agricultural production? Plant Soil 174: 3–28 [Google Scholar]
- Segonzac C., Boyer J.C., Ipotesi E., Szponarski W., Tillard P., Touraine B., Sommerer N., Rossignol M., Gibrat R. (2007). Nitrate efflux at the root plasma membrane: Identification of an Arabidopsis excretion transporter. Plant Cell 19: 3760–3777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J., Milburn J. (1980). Osmoregulation and the control of phloem-sap composition in Ricinus communis L. Planta 148: 28–34 [DOI] [PubMed] [Google Scholar]
- Sylvester-Bradley R., Kindred D.R. (2009). Analysing nitrogen responses of cereals to prioritise routes to the improvement of nitrogen use efficiency. J. Exp. Bot. 116: 1–13 [DOI] [PubMed] [Google Scholar]
- Takano J., Tanaka M., Toyoda A., Miwa K., Kasai K., Fuji K., Onouchi H., Naito S., Fujiwara T. (2010). Polar localization and degradation of Arabidopsis boron transporters through distinct trafficking pathways. Proc. Natl. Acad. Sci. USA 107: 5220–5225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilman D. (1998). The greening of the green revolution. Nature 396: 211–212 [Google Scholar]
- Tilman D., Fargione J., Wolff B., D’Antonio C., Dobson A., Howarth R., Schindler D., Schlesinger W.H., Simberloff D., Swackhamer D. (2001). Forecasting agriculturally driven global environmental change. Science 292: 281–283 [DOI] [PubMed] [Google Scholar]
- Tong Y., Zhou J.J., Li Z., Miller A.J. (2005). A two-component high-affinity nitrate uptake system in barley. Plant J. 41: 442–450 [DOI] [PubMed] [Google Scholar]
- Töpfer R., Matzeit V., Gronenborn B., Schell J., Steinbiss H.-H. (1987). A set of plant expression vectors for transcriptional and translational fusions. Nucleic Acids Res. 15: 5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E., Bauby H., Dubreucq B., Grandjean O., Runions J., Barthélémy J., Palauqui J.-C. (2008). High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. Plant Cell 20: 1494–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay Y.F., Schroeder J.I., Feldmann K.A., Crawford N.M. (1993). The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72: 705–713 [DOI] [PubMed] [Google Scholar]
- von der Fecht-Bartenbach J., Bogner M., Dynowski M., Ludewig U. (2010). CLC-b-mediated NO-3/H+ exchange across the tonoplast of Arabidopsis vacuoles. Plant Cell Physiol. 51: 960–968 [DOI] [PubMed] [Google Scholar]
- Wang R., Liu D., Crawford N.M. (1998). The Arabidopsis CHL1 protein plays a major role in high-affinity nitrate uptake. Proc. Natl. Acad. Sci. USA 95: 15134–15139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-Y., Tsay Y.-F. (2011). Arabidopsis nitrate transporter NRT1.9 is important in phloem nitrate transport. Plant Cell 23: 1945–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth J., Chopin F., Santoni V., Viennois G., Tillard P., Krapp A., Lejay L., Daniel-Vedele F., Gojon A. (2007). Regulation of root nitrate uptake at the NRT2.1 protein level in Arabidopsis thaliana. J. Biol. Chem. 282: 23541–23552 [DOI] [PubMed] [Google Scholar]
- Yong Z., Kotur Z., Glass A.D. (2010). Characterization of an intact two-component high-affinity nitrate transporter from Arabidopsis roots. Plant J. 63: 739–748 [DOI] [PubMed] [Google Scholar]
- Yuan L., Loqué D., Kojima S., Rauch S., Ishiyama K., Inoue E., Takahashi H., von Wirén N. (2007). The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. Plant Cell 19: 2636–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Barthelson R.A., Lambert G.M., Galbraith D.W. (2008). Global characterization of cell-specific gene expression through fluorescence-activated sorting of nuclei. Plant Physiol. 147: 30–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zifarelli G., Pusch M. (2009). Conversion of the 2 Cl(-)/1 H+ antiporter ClC-5 in a NO3(-)/H+ antiporter by a single point mutation. EMBO J. 28: 175–182 [DOI] [PMC free article] [PubMed] [Google Scholar]