The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY (SPY) promotes cytokinin responses by unknown mechanisms. This work shows that SPY interacts with and is required for the activity of two closely related class I TCP transcription factors, TCP14 and TCP15, that promote cytokinin responses in leaves and flowers, affecting leaf shape and trichome development.
Abstract
O-linked N-acetylglucosamine (O-GlcNAc) modifications regulate the posttranslational fate of target proteins. The Arabidopsis thaliana O-GlcNAc transferase (OGT) SPINDLY (SPY) suppresses gibberellin signaling and promotes cytokinin (CK) responses by unknown mechanisms. Here, we present evidence that two closely related class I TCP transcription factors, TCP14 and TCP15, act with SPY to promote CK responses. TCP14 and TCP15 interacted with SPY in yeast two-hybrid and in vitro pull-down assays and were O-GlcNAc modified in Escherichia coli by the Arabidopsis OGT, SECRET AGENT. Overexpression of TCP14 severely affected plant development in a SPY-dependent manner and stimulated typical CK morphological responses, as well as the expression of the CK-regulated gene RESPONSE REGULATOR5. TCP14 also promoted the transcriptional activity of the CK-induced mitotic factor CYCLIN B1;2. Whereas TCP14-overexpressing plants were hypersensitive to CK, spy and tcp14 tcp15 double mutant leaves and flowers were hyposensitive to the hormone. Reducing CK levels by overexpressing CK OXIDASE/DEHYDROGENASE3 suppressed the TCP14 overexpression phenotypes, and this suppression was reversed when the plants were treated with exogenous CK. Taken together, we suggest that responses of leaves and flowers to CK are mediated by SPY-dependent TCP14 and TCP15 activities.
INTRODUCTION
Addition of O-linked N-acetylglucosamine (O-GlcNAc) to Ser and Thr residues by O-GlcNAc transferases (OGTs) regulates the posttranslational fate and function of target proteins (Hart et al., 2007; Butkinaree et al., 2010). O-GlcNAc modifications of animal cytosolic and nuclear proteins affect their localization, phosphorylation, interaction with other proteins, and/or stability (Roos and Hanover, 2000; Wells et al., 2001; Hanover et al., 2003; Butkinaree et al., 2010). In animals, this modification plays a key role in various cellular processes, such as signal transduction, transcription, and proteasomal degradation (Hanover, 2001; Wells et al., 2003; Zachara and Hart, 2004; Yang et al., 2008; Butkinaree et al., 2010; Hanover et al., 2010). Although a similar modification occurs in plant cells, only a few O-GlcNAc–modified plant proteins have been identified.
The Arabidopsis thaliana genome encodes two OGTs, SPINDLY (SPY) and SECRET AGENT (SEC). Whereas sec and spy embryos develop normally, double mutant embryos are not viable, suggesting that SEC and SPY have redundant functions during embryogenesis (Hartweck et al., 2002). Although sec plants have subtle growth defects (Hartweck and Olszewski, 2006), the pathways in which SEC functions have not been determined. By contrast, SPY functions in the gibberellin (GA), cytokinin (CK), red light response, and circadian pathways (Jacobsen and Olszewski, 1993; Tseng et al., 2004; Greenboim-Wainberg et al., 2005; Maymon et al., 2009). SPY is a negative regulator of GA signaling, and it has been hypothesized that SPY accomplishes this regulation by O-GlcNAc–modifying and activating DELLA proteins, which also negatively regulate GA responses (Shimada et al., 2006; Silverstone et al., 2007). In a previous study, we found that spy mutants are partially resistant to exogenously applied CK and proposed that SPY is a positive element in CK signal transduction (Greenboim-Wainberg et al., 2005; Maymon et al., 2009). Because GA also suppressed CK responses, we suggested that crosstalk between these hormones is partially mediated by SPY (Greenboim-Wainberg et al., 2005; Weiss and Ori, 2007). SPY promotes CK responses via a DELLA-independent pathway and thus appears to inhibit GA responses and enhance CK signaling through distinct mechanisms (Maymon et al., 2009).
The CK-signaling cascade in Arabidopsis starts with the binding of CK to the His kinase receptors and their autophosphorylation (Müller and Sheen, 2007). The phosphate group is then transferred by His phosphotransfer proteins (AHPs in Arabidopsis) to the nucleus to phosphorylate a set of transcriptional regulators known as type-B response regulators (RRs; or ARR in Arabidopsis). The phosphorylated type-B RRs induce the transcription of various CK-regulated genes, including type-A RRs (D’Agostino et al., 2000; Rashotte et al., 2006). Because GA and spy mutations affected numerous CK responses occurring at different stages of plant development, including induction of the CK primary response gene, type-A Arabidopsis RR5 (ARR5; Greenboim-Wainberg et al., 2005; Gan et al., 2007), it was suggested that SPY promotes and GA suppresses elements of the early CK-induced phosphorelay cascade.
To understand the mechanism by which SPY affects plant development, several groups have performed protein interaction assays. These revealed an interaction between SPY and GIGANTEA (GI), which may affect the activity of the circadian clock (Tseng et al., 2004). The Hordeum vulgare SPY homolog Hv-SPY was shown to interact with two transcriptional regulators from the MYB- and NAC-like families, which may affect GA responses in this plant (Robertson, 2004). Here, we show that SPY interacts with two class I TCP proteins: TCP14 and its closest homolog TCP15.
TCPs, named after the transcription factors TEOSINTE BRANCHED1 from Zea mays, CYCLOIDEA from Antirrhinum majus, and PROLIFERATING CELL NUCLEAR ANTIGEN FACTOR from Oryza sativa (Navaud et al., 2007), belong to the family of basic helix-loop-helix–type transcription factors. The TCP domain is a plant-specific DNA binding motif that binds specifically to cis-elements named site II elements. The Arabidopsis genome encodes 24 predicted TCP proteins, of which, based on the structure of the DNA binding domain, 13 are grouped as class I and 11 as class II (Cubas et al., 1999; Kosugi and Ohashi, 2002; Aguilar-Martínez et al., 2007; Navaud et al., 2007). Because the two classes appear to bind similar promoter elements, it was suggested that development is regulated by a balance in the activity of the two groups. Class I TCPs were suggested to promote and class II to restrict cell proliferation (Li et al., 2005; Li et al., 2011).
Class II TCPs affect floral organ asymmetry, axillary meristem development, branching, and leaf shape via the regulation of cell proliferation and/or growth (Martín-Trillo and Cubas, 2010). Some of these effects may be mediated by hormonal activities. TCP1 affects brassinosteroid biosynthesis by regulating the transcription of DWARF4, a key enzyme in brassinosteroid biosynthesis (Guo et al., 2010). TCP3 suppresses auxin responses probably by the transcriptional stimulation of the signaling repressor INDOLE-3-ACETIC ACID3/SHORT HYPOCOTYL2 (Koyama et al., 2010). TCP4 regulates the transcription of LIPOXYGENASE2, a key enzyme in jasmonate biosynthesis (Schommer et al., 2008), and the Solanum lycopersicum homolog of TCP4, LANCEOLATE, promotes GA biosynthesis (Yanai et al., 2011). TCP13 has been identified in yeast two-hybrid assays as an interactor with AHPs (Suzuki et al., 2001), which raises the possibility that TCP13 is regulated by the CK signal.
Much less is known about the role of class I TCPs in plant development. Most class I single mutants have mild or no phenotypes, probably as a result of genetic redundancy. However, overexpression of Arabidopsis TCP20 as a fusion with a transcriptional repressor domain caused phenotypes suggestive of roles in cell division, expansion, and differentiation (Hervé et al., 2009; Martín-Trillo and Cubas, 2010). TCP20 binds a specific motif in the site II elements (GCCC) found in the promoter of ribosomal protein genes (Tatematsu et al., 2005). TCP21 (CHE) is involved in the regulation of the circadian clock activity (Pruneda-Paz et al., 2009). This protein interacts with a core element of the clock oscillator, PSEUDO RESPONSE REGULATOR1 (PRR1)/TOC1, and together they suppress the transcription of CIRCADIAN AND CLOCK ASSOCIATED1. Similarly, TCP15 interacts with the core circadian clock component PRR5 (Giraud et al., 2010), and the diurnal oscillations in PPR5 transcript abundance were slightly affected in tcp15 plants. In addition, Li et al. (2011) have shown that TCP15 directly regulates cell cycle genes and inhibits endoreduplication. The Antirrhinum TCP14 homolog TIC interacts with a NAC domain transcription factor, CUPULIFORMIS, suggesting a role in the establishment of boundaries between lateral organs (Weir et al., 2004). Tatematsu et al. (2008) have shown that the Arabidopsis TCP14 has a role in GA regulation of embryo growth during seed germination and that the tcp14 mutant seeds are hypersensitive to exogenously applied abscisic acid and paclobutrazol. Recently, Kieffer et al. (2011) characterized the double mutant tcp14 tcp15 and demonstrated its redundant activities in the regulation of cell proliferation in bolting stem internodes, developing leaf blades, and floral tissues. Here, we show that TCP14 and TCP15 physically and genetically interact with SPY and provide evidence that they act together to promote CK responses in leaves and flowers. These CK responses affect leaf shape and trichome development.
RESULTS
SPY Interacts Physically with TCP14 and TCP15
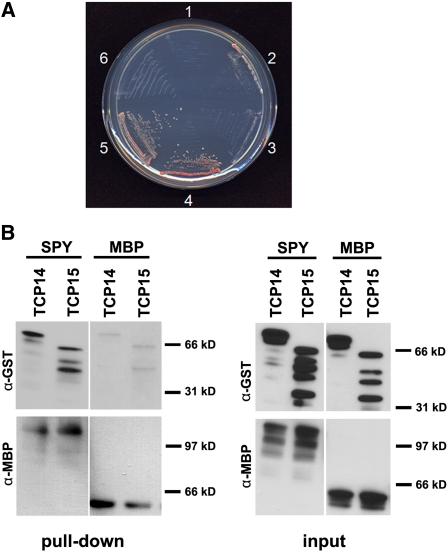
To understand the mechanism by which SPY affects plant development in general and hormone signaling in particular, we performed a yeast two-hybrid screen for SPY-interacting proteins (Tseng et al., 2004). One bait clone expressing a portion of TCP15 was recovered from this screen. Additional yeast two-hybrid assays demonstrated that full-length TCP15 and its closest homolog, TCP14, interacted in this yeast system with the SPY-TPR domain (Figure 1A). Similar to previous reports that TCPs autoactivate in a yeast-based assay (Kosugi and Ohashi, 2002; Giraud et al., 2010), TCP14 and TCP15 prey proteins weakly autoactivated reporter genes, but much stronger activation occurred in the presence of the SPY bait. To provide additional evidence for the physical interaction between SPY and TCP14 and TCP15, we performed in vitro pull-down assays using bacterially expressed maltose binding protein (MBP)-tagged full-length SPY and glutathione S-transferase (GST)-tagged TCP14 and TCP15. Whereas very low levels of nonspecific interaction between GST-TCP14/TCP15 and MBP were detected in the pull-down assay, the interaction between full-length GST-TCP14/TCP15 and MBP-SPY was much stronger (Figure 1 B). MBP-SPY interacted also with large truncated products of GST-TCP14/15. GST alone did not interact with SPY (see Supplemental Figure 1 online).
Figure 1.
SPY Interacts Physically with TCP14.
(A) The results from a yeast two-hybrid assay. Yeast cells containing the following vectors: 1, empty bait and empty prey; 2, empty bait and TCP14 prey; 3, empty bait and TCP15; 4, SPY-TPR bait and TCP14 prey; 5, SPY-TPR bait and TCP15 prey; and 6, SPY-TPR bait and empty prey, were plated on medium containing 10 mM 3-amino-1,2,4-triazole and lacking His. Although cells expressing the TCP14 or TCP15 prey protein alone (2 and 3) exhibited a small amount of growth, cells expressing the TCP bait and SPY-TPR prey proteins (4 and 5) grew much better.
(B) In vitro pull-down assay using E. coli–expressed MBP, MBP-SPY, GST-TCP14, and GST-TCP15. MBP or MBP-SPY (full-length SPY) were used as bait and incubated with equal amount of GST-TCP14 or GST-TCP15. Amylose resin was used to pull down the MBP or MBP-SPY and its interacting proteins. Left panels (pull-down): An anti-GST antibody was used to detect GST-TCP14 or GST-TCP15. The blot was striped and reprobed with anti-MBP antibody to detect MBP and MBP-SPY. Right panels (input): Amounts of proteins added to the assays. Full-length GST-TCP14 = 80.1 kD, GST-TCP15 = 62.5 kD, MBP-SPY = 145.7 kD, GST = 26.7 kD, and MBP = 50.8 kD.
[See online article for color version of this figure.]
TCP14 Activity Requires SPY
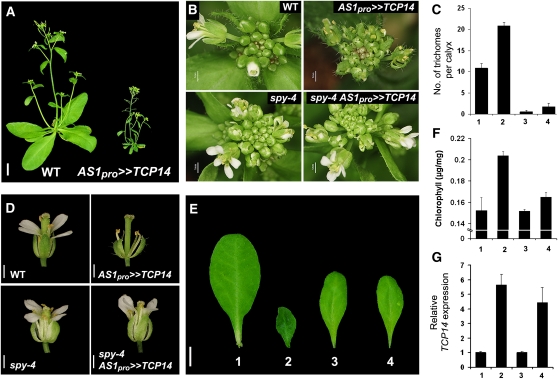
To assess the biological relevance of the physical interaction between SPY and TCP14, we examined whether the activity of TCP14 requires functional SPY. To this end, we first assayed TCP14 in vivo function by overexpression. Expressing TCP14 under the regulation of the 35S promoter severely altered plant development and occasionally caused lethality. We therefore expressed TCP14 under the control of promoters that are active in lateral-organ primordia but not in the shoot apical meristem proper. We evaluated the FILAMENTOUS FLOWER (FIL), CUP-SHAPED COTYLEDON2 (CUC2), and ASYMMETRIC LEAVES1 (AS1) promoters using a transactivation system (Moore et al., 1998). The phenotype found in plants expressing TCP14 under the regulation of the AS1 promoter (AS1pro>>TCP14) was stronger than that found in CUC2pro>>TCP14 or FILpro>>TCP14 plants (Figure 2A; see Supplemental Figure 2 online). Thus, AS1pro>>TCP14 plants were selected for this study. The AS1pro driver used for transactivation (AS1pro:LhG4) is active primarily in leaf primordia, young leaves, and young flowers (Eshed et al., 2001); this activity was confirmed by monitoring the expression of an operator (op):green fluorescent protein (GFP) transgene that was included in all crosses (AS1pro>>ER-GFP; see Supplemental Figures 3B and 3C online).
Figure 2.
Functional Interaction between SPY and TCP14.
(A) Four-week-old wild-type (WT) Ler and transactivated transgenic plants overexpressing TCP14 under the regulation of the AS1 promoter (AS1pro>>TCP14). The transgene suppressed leaf expansion and elongation growth.
(B) Inflorescences of the wild type, spy-4, and AS1pro>>TCP14 in the wild type or spy-4.
(C) Sepal trichome number of the sixth flower of wild-type Ler (1), AS1pro>>TCP14 (2), spy-4 (3), and AS1pro>>TCP14 in spy-4 (4). Values are means of 10 flowers ± se.
(D) Flowers (flower no. 6) of wild-type Ler, transgenic transactivated AS1pro>>TCP14, spy-4, and transactivated AS1pro>>TCP14 in spy-4.
(E) Fifth rosette leaf of wild-type Ler (1), AS1pro>>TCP14 (2), spy-4 (3), and AS1pro>>TCP14 in spy-4 (4).
(F) Chlorophyll content (μg chlorophyll per mg fresh weight) in the fifth rosette leaf of wild-type Ler (1), AS1pro>>TCP14 (2), spy-4 (3), and AS1pro>>TCP14 in spy-4 (4). Values are means of five leaves ± se.
(G) qRT-PCR analysis of TCP14 in seedlings of wild-type Ler (1), AS1pro>>TCP14 (2), spy-4 (3), and AS1pro>>TCP14 in spy-4 (4). Values are means of three biological replicates ± se.
Bars = 1 cm in (A) and (E) and 1 mm in (B) and (D).
Overexpression of TCP14 (AS1pro>>TCP14) resulted in inhibition of internode elongation, inhibition of petal growth, reduced fertility, promotion of trichome development on sepals, and formation of small dark-green leaves containing higher levels of chlorophyll (Figures 2A to 2F; see Supplemental Figure 3A online). Occasionally, elongated carpels were found in flowers of these lines (see Supplemental Figures 3A and 4 online). Similar phenotypic changes were observed when TCP15 was expressed under the control of the same promoter (see Supplemental Figure 4 online), suggesting that TCP15 and TCP14 have the potential to be engaged in similar functions.
We tested if TCP14 activity requires functional SPY by analyzing AS1pro>>TCP14 spy-4 plants. All TCP14 overexpression phenotypes (see above) were strongly suppressed by spy (Figures 2B to 2F; see Supplemental Figure 3A online). Importantly, the activity of the AS1 driver remained unchanged in the spy-4 background, as was evident by the pattern of GFP fluorescence in these lines (see Supplemental Figures 3B and 3C online). Quantitative RT-PCR (qRT-PCR) also confirmed that TCP14 expression was similar in AS1pro>>TCP14 wild type and AS1pro>>TCP14 spy-4 (Figure 2G). As expected, crossing AS1pro>>TCP14 spy-4 back to the wild type restored the overexpression phenotype (see Supplemental Figure 3A online). These results suggest that at least some TCP14 activities require functional SPY.
Several TCPs Are O-GlcNAc Modified by SEC
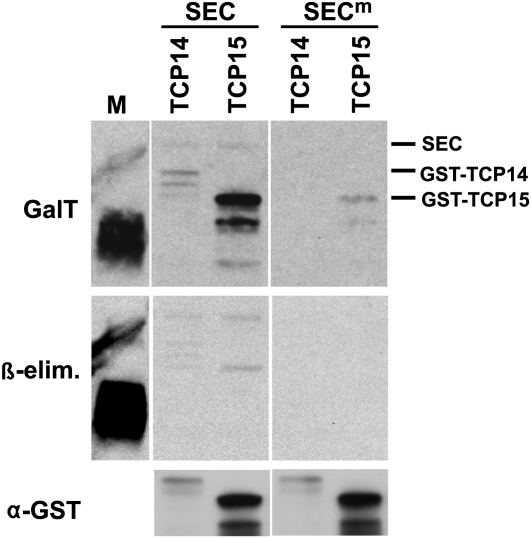
The interaction between SPY and TCP14/15 and its requirement for their activity raised the possibility that these proteins are O-GlcNAc modified by SPY. At present, there is no reliable plant-based system for in vivo analysis of O-GlcNAc modifications, but an Escherichia coli–based O-GlcNAc modification assay with SEC (the second Arabidopsis OGT) is available (Scott et al., 2006). We first tested if SEC can interact with TCP14/15 and found that they interact in an in vitro pull-down assay (see Supplemental Figure 5 online). We then tested if SEC modifies TCP14 and TCP15. When protein blots containing total proteins from E. coli expressing SEC and TCP14 or TCP15 were probed using an assay that labels terminal GlcNAc with 3H-Gal, both TCP14 and TCP15, but not E. coli proteins, were labeled (Figure 3). To determine if TCP14 and TCP15 undergo GlcNAc modification by SEC, they were coexpressed with a mutant form of SEC. This mutant form of SEC has a missense mutation (G676D) that, based on the crystal structure of the Xanthomonas campestris OGT (Martinez-Fleites et al., 2010), is predicted to affect the active site. Consistent with SEC-modifying TCP14 and TCP15, the mutation greatly reduced GlcNAc modification of these proteins (Figure 3). Since TCP14 and TCP15 were expressed as GST fusions, SEC could modify GST and not TCP14 or TCP15. However, this is unlikely since another protein (AAR5) fused to GST was not modified by SEC in this assay (see Supplemental Figures 6 and 7 online).
Figure 3.
TCP14 and TCP15 Are O-GlcNAc Modified by SEC.
GST-tagged TCP14 and TCP15 were coexpressed in E. coli with wild-type SEC (SEC) or mutant SEC (SECm). Two identical blots containing total E. coli proteins were prepared, and GlcNAc-modified proteins on one blot were labeled with 3H-Gal (GalT). This blot was then subjected to β-elimination to remove O-linked modifications and exposed to film for twice as long as the original exposure (β-elim.). The second blot was used to monitor TCP14 and TCP15 expression using an anti-GST antibody (αα-GST). Wild-type SEC modified both full-length TCPs and itself to a much greater extent than did SECm. β-Elimination removed most of label from the modified TCPs but not the ovalbumin marker (M), which has N-linked glycans, indicating that the modification is O-linked. The location of SEC, TCP14, and TCP15 on the blot is indicated. The experiment was repeated with similar results.
The labeling assay used to detect GlcNAc modifications labels both O- and N-linked GlcNAc modifications. O-linked modifications are removed by high pH (β-elimination), but N-linked modifications are stable under these conditions. The GlcNAc modifications on TCP14 and TCP15, but not a negative control bearing N-linked modifications, were removed when the blot was subjected to β-elimination, indicating that the GlcNAc modifications on TCP14 and TCP15 are O-linked (Figure 3). These results show that TCP14 and TCP15 have the potential to be O-GlcNAc modified. We tested other TCPs and found that SEC can also modify TCPs 2, 8, 19, 23, and 24 (see Supplemental Figure 7 online). Two other tested proteins were not modified in this assay (see Supplemental Figures 6 and 7 online).
The tcp14 tcp15 Double Mutant Plants Exhibit a Subset of the spy Phenotypes
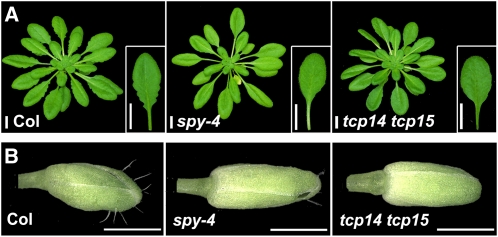
Plants with single and double mutations in TCP14 and TCP15 (tcp14-4 and tcp15-3) have been characterized before; in these mutants, a T-DNA lies within TCP14’s single exon in the tcp14-4 line, and no TCP15 transcripts could be detected in the tcp15-3 mutant line (Kieffer et al., 2011). We used these lines (all in the Columbia [Col] background) to examine the relationships between these class I TCPs and SPY. tcp14 and tcp15 single mutants have leaf margins slightly smoother than those of wild-type leaves (see Supplemental Figure 8A online). When grown under short-day conditions, the tcp14 tcp15 double mutant leaf margins were similar to spy-4 and much smoother than the wild type (Figure 4A). This effect was weaker under long-day conditions (data not shown). As shown before (Kieffer et al., 2011), stem internodes of the double mutant were slightly shorter than the wild type or either of the single mutants.
Figure 4.
Leaf Morphology and Sepal Trichome Development Are Similarly Affected in tcp14 tcp15 and spy Mutants.
(A) Serrated leaves of wild-type Col and smoother leaves of tcp14 tcp15 and spy-4. The seventh leaf of each plant is also shown.
(B) Trichomes on wild-type (Col) sepals and the glabrous sepals of tcp14 tcp15 and spy-4 (flower no. 4).
Plants were grown under short-day conditions (8 h of light). Bars = 1 cm in (A) and 1 mm in (B).
[See online article for color version of this figure.]
Mutation of TCP14 and TCP15 also affected flower morphology: The wild type and the tcp14 and tcp15 single mutants had several trichomes on each sepal (see Supplemental Figure 8B online), but similar to spy-4, tcp14 tcp15 sepals were glabrous (Figure 4B; see Supplemental Figure 8B online). Other typical spy-4 phenotypes, such as slender growth, early flowering, and insensitivity to the GA biosynthesis inhibitor paclobutrazol, were not observed in the double mutant (see Supplemental Figure 9 online). In fact, as reported previously for tcp14 (Tatematsu et al., 2008), tcp14 tcp15 seeds were slightly hypersensitive to paclobutrazol. It should be noted, however, that whereas SPY is expressed throughout the plant (Hartweck et al., 2006), TCP14 and TCP15 are expressed primarily in young developing leaves; TCP15 is expressed mainly in leaf margins, whereas TCP14 is expressed more widely in the leaf blade (Kieffer et al., 2011; Li et al., 2011). Both genes are also expressed in young flowers, mainly in sepals, and later in stamens and carpel.
We have found previously that leaf serration and trichome development on sepals are good markers for SPY-promoted CK responses (Greenboim-Wainberg et al., 2005; Maymon et al., 2009). Slender growth, early flowering, and resistance to paclobutrazol, on the other hand, are related to SPY-suppressed GA responses. Thus, it is possible that the interaction of SPY with TCP14/TCP15 has a significant role in CK but not GA responses.
TCP14 and TCP15 Promote Responses to CK
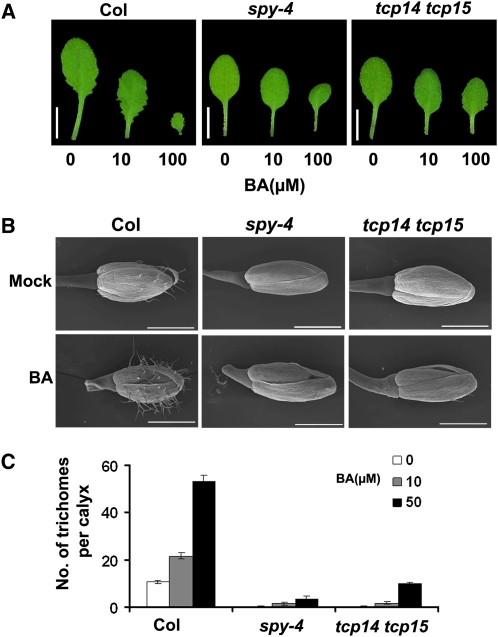
To test if TCP14 and TCP15 have a role in CK responses, we examined the sensitivity of the tcp14 tcp15 double mutant to the CK N6-benzyladenine (BA). We tested several responses to exogenous BA, including promotion of leaf serration, inhibition of inflorescence elongation, and trichome development on sepals (Greenboim-Wainberg et al., 2005; Gan et al., 2007; Maymon et al., 2009). Col, spy-4, and tcp14 tcp15 seedlings with two true leaves were sprayed twice a week with 0, 10, or 100 μM BA. BA-treated Col plants produced small and highly serrated leaves, but this effect was not apparent in spy-4 or the tcp14 tcp15 double mutant (Figure 5A). Furthermore, BA treatment strongly promoted trichome development on wild-type sepals but had a very mild effect on spy-4 and tcp14 tcp15 sepals (Figures 5B and 5C). BA treatment also inhibits inflorescence elongation. Whereas spy suppresses inhibition of inflorescence elongation by BA (Maymon et al., 2009), the inflorescence of tcp14 tcp15 plants responds normally to BA, demonstrating that there is incomplete spatial overlap in the functioning of SPY, TCP14, and TCP15.
Figure 5.
Leaves and Flowers of the tcp14 tcp15 Double Mutant and spy-4 Are Partially Resistant to CK.
(A) and (B) Wild-type Col, spy-4, and tcp14 tcp15 seedlings were treated with different concentrations of BA for 3 weeks. Representative rosette leaves (fifth leaf; [A]) and flowers (B) of treated and nontreated wild-type (Col), spy-4, and tcp14 tcp15 plants are presented.
(C) Number of trichomes on sepals (first flower) of the wild type (Col), spy-4, and tcp14 tcp15 treated with different BA concentrations. Values are means of 20 flowers ± se.
The experiment was performed five times with similar results. Bars = 1 cm in (A) and 1 mm in (B).
[See online article for color version of this figure.]
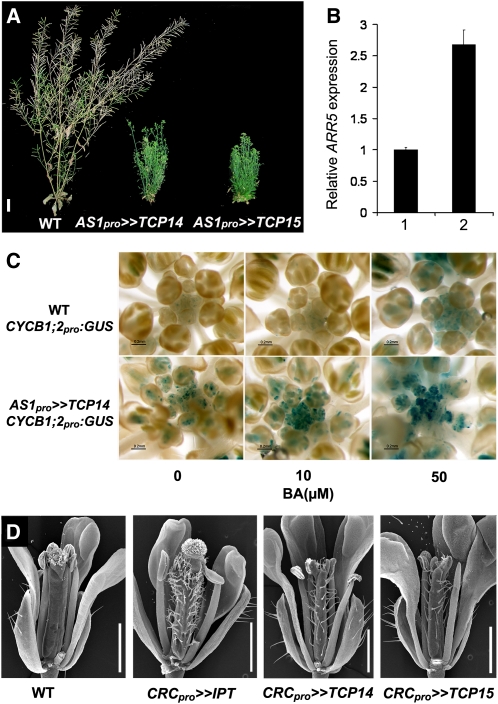
Some of the phenotypes observed in TCP14- and TCP15-overexpressing plants resemble the effect of CK treatment (e.g., promotion of trichome development on sepals). Furthermore, similar to CK application (Guo and Gan, 2011), TCP14 and TCP15 overexpression promoted shoot branching and delayed whole-plant senescence. Approximately 2 months after germination, when wild-type plants started to die, axillary buds in the transgenic AS1pro>>TCP14 and AS1pro>>TCP15 plants sprouted and produced lateral inflorescences. Three-month-old transgenic AS1pro>>TCP14 and AS1pro>>TCP15 plants were bushy and green (Figure 6A). Although TCP14 and TCP15 overexpression delayed whole-plant senescence, overexpression had no effect on leaf senescence. It should be noted, in this regard, that the AS1 promoter is not active in mature leaves or in mature inflorescence stems.
Figure 6.
Overexpression of TCP14 and TCP15 Caused Typical CK Responses.
(A) Three-month-old wild-type (WT) Ler and transactivated transgenic plants overexpressing TCP14 or TCP15, under the regulation of the AS1 promoter (AS1pro>>TCP14 and AS1pro>>TCP15). The transgenes delayed whole-plant senescence and promoted branching.
(B) Overexpression of TCP14 promoted ARR5 expression. RNA was extracted from 10-d-old wild-type Ler (1) and AS1pro>>TCP14 (2) seedlings and analyzed by qRT-PCR. Values are means of three biological replicates ± se.
(C) GUS activity in BA-treated inflorescences of wild-type (Ler) and AS1pro>>TCP14 transactivated plants, both expressing the GUS gene under the regulation of CYCB1;2 promoter.
(D) Flowers (flower no. 2) of control wild-type (Ler) and transactivated plants expressing IPT, TCP14, or TCP15 under the regulation of the CRC promoter (CRCpro>>IPT, CRCpro>>TCP14, and CRCpro>>TCP15).
Bars = 2 cm in (A), 0.2 mm in (C), and 1 mm in (D).
To test further if overexpression of TCP14 promotes CK responses, we analyzed the expression of the CK-induced gene ARR5 in the AS1pro>>TCP14 seedlings. qRT-PCR assays with RNA extracted from whole seedlings revealed an approximately twofold higher expression of ARR5 in the transgenic plants compared with the wild type (Figure 6B), suggesting increased CK responses in the transgenic plants.
CK promotes cell proliferation (Riou-Khamlichi et al., 1999), and a previous study has shown that several mitotic factor genes, including CYCLIN B1;2 (CYCB1;2), are downregulated in tcp14 tcp15 double mutant plants (Kieffer et al., 2011). Since our results suggest that TCP14 promotes CK responses, we tested if TCP14 and CK act in the same pathway to promote mitotic activity by analyzing their effect on CYCB1;2. To this end, we used transgenic wild-type and AS1pro>>TCP14 plants expressing the GUS reporter gene under the regulation of the CYCB1;2 promoter (Efroni et al., 2008). Inflorescences of CYCB1;2pro:GUS transgenic plants were treated with different BA concentrations and then GUS activity was detected. Transgenic CYCB1;2pro:GUS inflorescences exhibited very low GUS activity in young flower buds (Figure 6C). GUS activity in CYCB1;2pro:GUS/AS1pro>>TCP14 inflorescences was much higher and was maintained longer in developed flowers. CK application promoted CYCB1;2pro:GUS activity in both wild-type and AS1pro>>TCP14 inflorescences, but the effect of the hormone was much stronger in the TCP14 transactivated plants. These results support the previous suggestion that TCP14 promotes cell proliferation (Kieffer et al., 2011) and further suggest that it acts in the CK pathway to regulate this process. In addition, the results suggest that TCP14 enhances the sensitivity of the tissue to CK.
In a previous study, we showed that ectopic expression of the CK biosynthetic gene ISOPENTENYL TRANSFERASE (IPT) under the regulation of the carpel-specific CRABS CLAW (CRC) promoter induced trichome development on carpels (Greenboim-Wainberg et al., 2005; Figure 6B). Here, we found that expressing IPT, TCP14, or TCP15 under the regulation of the CRC promoter in transactivated lines (CRCpro>>IPT, CRCpro>>TCP14, and CRCpro>>TCP15) similarly induced trichome development on carpels (Figure 6D), suggesting increased CK responses in CRCpro>>TCP14 and CRCpro>>TCP15 carpels.
Reducing CK Levels Suppresses TCP14 Overexpression Effects
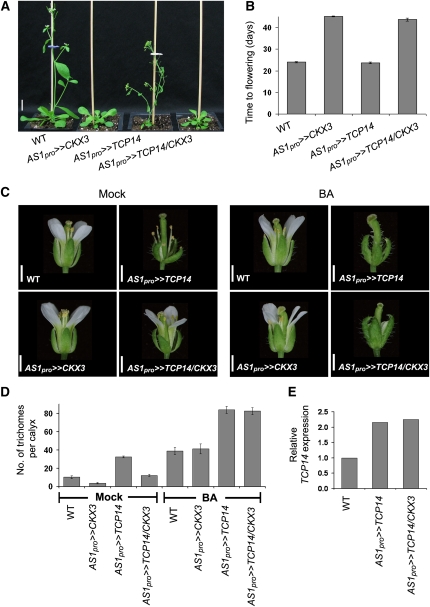
If TCP14 and TCP15 promote CK responses, they might, similar to other signaling components in this pathway (Müller and Sheen, 2007), be regulated by CK. We first tested if the expression of these genes is affected by CK by treating 10-d-old wild-type seedlings with 10 μM BA. qRT-PCR analyses showed that, while CK treatments promoted ARR5 expression, it had no effect on TCP14 and TCP15 mRNA levels (see Supplemental Figure 10 online). We then tested if CK affects TCP14 activity. To this end, we transactivated the Arabidopsis CK-degrading CK OXIDASE/DEHYDROGENASE3 (CKX3) gene (Shani et al., 2010) with the AS1pro driver line. The CKX3 genes have been used before for the generation of CK-deficient plants (Werner et al., 2003). AS1pro>>CKX3 plants were relatively normal but exhibited late flowering (Figures 7A and 7B). This effect of CKX3 overexpression was shown before (Werner et al., 2003). We then crossed AS1pro>>CKX3 to AS1pro>>TCP14 to generate AS1pro>>TCP14/CKX3 plants. Progenies containing all three transgenes were identified by PCR (see Supplemental Figure 11D online) and their phenotype was examined. While AS1pro>>TCP14/CKX3 plants showed the typical late-flowering phenotype of AS1pro>>CKX3 (Figures 7A and 7B), two AS1pro>>TCP14 phenotypes, inhibition of petal development and promotion of trichome development on sepals, were strongly suppressed (Figure 7C; see Supplemental Figures 11B and 12 online). Other effects of AS1pro>>TCP14 were only partially suppressed by CKX3 activity; leaves and siliques of AS1pro>>TCP14/CKX3 plants were larger than those of AS1pro>>TCP14 plants but smaller and darker green than those of wild-type and AS1pro>>CKX3 plants (see Supplemental Figures 11A to 11C online). qRT-PCR analysis showed that the TCP14 expression level was similar in AS1pro>>TCP14 and in AS1pro>>TCP14/CKX3 plants (Figure 7D). To confirm the presence of active TCP14 in these plants, AS1pro>>TCP14/CKX3 plants (heterozygous for both transgenes) were self-pollinated. Four phenotypic classes, the wild type, CKX3-, CKX3/TCP14-, and TCP14-like, were recovered, confirming the presence of active TCP14 in AS1pro>>TCP14/CKX3 parental plants.
Figure 7.
Reducing CK Levels Suppresses TCP14 Overexpression Effects.
(A) Four-week-old wild-type (WT) Ler, transactivated AS1pro>>CKX3, transactivated AS1pro>>TCP14, and transactivated AS1pro>>TCP14/CKX3 (expressing both CKX3 and TCP14) plants. Bar = 1 cm.
(B) Time to flowering of the wild type and the different transgenic lines. The number of days from germination to production of a 2-cm-long inflorescence was determined. Values are means of 10 plants ± se.
(C) Flowers (flower no. 4) of wild-type Ler, AS1pro>>TCP14, AS1pro>>CKX3, and AS1pro>>TCP14/CKX3 nontreated (left) and treated (right) with 50 μM BA. Bars = 1 mm.
(D) Sepal trichome number of the third flower of wild-type Ler, AS1pro>>CKX3, AS1pro>>TCP14, and AS1pro>>TCP14/CKX3 nontreated or treated with 50 μM BA. Values are means of 10 flowers ± se.
(E) qRT-PCR analysis of TCP14 in inflorescences of wild-type Ler, AS1pro>>TCP14, and AS1pro>>TCP14/CKX3 plants. The analysis was repeated three times with different plants and crosses, and similar results were obtained.
[See online article for color version of this figure.]
To examine if the TCP14 overexpression phenotypes in AS1pro>>TCP14/CKX3 plants were suppressed due to the low CK levels, we applied 50 μM BA directly to the young inflorescences of the different lines (the wild type, AS1pro>>TCP14, AS1pro>> CKX3, and AS1pro>>TCP14/CKX3). CK application to AS1pro>>TCP14/CKX3 restored the TCP14 overexpression phenotype, including the development of trichomes on sepals and inhibition of petal growth (Figure 7C; see Supplemental Figure 12 online). CK application to AS1pro>>TCP14 plants strongly promoted the development of numerous trichomes on sepals (see Supplemental Figure 12 online), in agreement with TCP14-overexpressing plants being hypersensitive to CK. These results suggest that CKX3 suppresses the effects of TCP14 overexpression by reducing the CK level. Thus, while the most logical explanation for these results is that CK promotes TCP14 activity, we cannot exclude the possibility that TCP14 upregulates CK levels.
DISCUSSION
The plant OGT SPY can affect developmental processes in Arabidopsis by suppressing GA signaling and/or promoting CK responses (Jacobsen and Olszewski, 1993; Greenboim-Wainberg et al., 2005). The mechanism by which SPY influences plant development and hormone responses is still unknown. Here, we report that SPY physically interacts with two class I TCPs: TCP14 and its closest homolog TCP15. We also show that functional SPY is required for TCP14 activity and provide evidence that TCP14 and TCP15 act together with SPY to promote CK responses. Whereas there is no direct evidence, it is possible that O-GlcNAc modification by SPY activates TCP14 and TCP15 to facilitate specific CK responses in leaves and flowers. The observation that E. coli–expressed SEC modifies TCP14 and TCP15 demonstrates that these proteins have the potential to be O-GlcNAc modified in planta. It should be noted that sec mutants do not exhibit phenotypes consistent with action in CK signaling or in TCP14/15 activity. Thus, it is possible that, whereas SEC can interact in vitro with TCP14 and TCP15 and modify them in E. coli, differences in expression pattern or cellular localization prevent it from modifying them in planta.
Several lines of evidence suggest that TCP14 and TCP15, following activation by SPY, promote specific CK responses: Loss of TCP14 and TCP15 function, similar to loss of SPY, suppressed marginal leaf serration and trichome development on sepals and attenuated the effect of exogenous CK on these processes. Furthermore, the phenotype of plants overexpressing TCP14 (and TCP15) was similar to that of plants treated with high doses of CK; both TCP14 overexpression and CK promoted branching and trichome formation on sepals and inhibited inflorescence elongation, leaf growth, and petal elongation (Greenboim-Wainberg et al., 2005; Maymon et al., 2009; Figures 2 and 5 to 7). These effects of TCP14 overexpression were strongly suppressed by the loss of SPY activity. Overexpression of TCP14 also promoted the expression of the CK response gene ARR5. Moreover, expressing TCP14 or TCP15 in the carpel had a similar effect to the expression of the CK-biosynthetic gene IPT; all promoted trichome development on the normally glabrous carpel. TCP14 overexpression also increased the activity of the promoter of the mitotic factor CYCB1;2 and enhanced the responsiveness of this promoter to CK. This result suggests that TCP14 overexpression not only promotes CK responses but also increases the sensitivity of the tissue to the hormone. The observations that TCP14 overexpression also enhanced the effects of CK on sepal trichome development further support this suggestion.
Whereas CK-promoted marginal leaf serration was suppressed in tcp14 tcp15, it was not affected by TCP14 overexpression. This discrepancy is probably caused by the different genetic backgrounds used in the different experiments. Whereas tcp14 tcp15 is in the Col background, TCP14 overexpression is in Landsberg erecta (Ler). Col leaves are slightly serrated, and CK strongly increases serration. Ler leaves, on the other hand, are smooth even after CK treatment.
Taken together, the results suggest that TCP14 and TCP15, similar to SPY, promote CK responses. However, while the loss of SPY affects CK responses of seedling and mature plants organs (Greenboim-Wainberg et al., 2005), the loss of TCP14/TCP15 suppressed CK activity only in leaves and sepals. Thus, TCP14 and TCP15 may mediate only a subset of SPY-regulated CK responses. This is expected, as SPY and TCP14/TCP15 expression have limited spatial overlap; SPY is expressed throughout the plant (Hartweck et al., 2006), while TCP14 and TCP15 are expressed mainly in emerging lateral roots, young leaves, and flowers (Kieffer et al., 2011; Li et al., 2011). It is possible, however, that additional TCPs are activated by SPY in other tissues/developmental stages to promote other CK responses. The fact that TCP8, TCP22, and TCP23, which are closely related to TCP14 and TCP15, could be O-GlcNAc modified by SEC supports this possibility.
Whereas very little is known about the developmental role of class I TCPs, it was proposed that they promote cell proliferation (Li et al., 2005; Martín-Trillo and Cubas, 2010; Kieffer et al., 2011; Li et al., 2011). Cell proliferation is also promoted by CK, and both class I TCPs (i.e., TCP20, TCP14, and TCP15) and CK enhance the expression of cyclin genes to regulate this process (Riou-Khamlichi et al., 1999; Li et al., 2005; Kieffer et al., 2011; Li et al., 2011; Figure 6). We show that TCP14 overexpression promotes the expression of the mitotic factor CYCB1;2 and enhances the induction of this gene by CK. This implies that TCP14 and TCP15 mediate the effect of CK on CYC genes to promote cell proliferation. Recently, it was shown that CK promotes cell proliferation at the leaf margin, leading to increased leaf serration and complexity (Shani et al., 2010). Both leaf margin serration and the promotion of it by CK are attenuated in spy and tcp14 tcp15 plants. Therefore, it is possible that TCP14 and TCP15, following activation by SPY, act in the CK pathway to promote cell proliferation at the leaf margin. This suggestion is supported by the expression pattern of these TCPs: TCP14 is expressed in leaf blades, and TCP15 is expressed mainly in leaf margins (Kieffer et al., 2011)
Our results show that reducing the endogenous levels of CK by overexpressing the CK-degrading enzyme CKX3 suppressed TCP14-induced trichome development on sepals and inhibition of petal development. These effects were restored when the plants (AS1>>TCP14/CKX3) were treated with exogenous CK. Several different models that are not mutually exclusive can explain these results; TCP14 could increase the CK level either by promoting its biosynthesis or inhibiting its degradation or CK signaling could promote TCP14 protein activity (Figure 8). Alternatively, TCP14 could perform a step in the CK signaling pathway that is rate limiting when CK levels are normal but not when CK levels are reduced by overexpression of CKX. The resistance of tcp14 tcp15 mutant to exogenously applied CK suggests that TCP14 and TCP15 are involved in CK signaling and not biosynthesis. Thus, we favor the possibility that CK signaling activates TCP14 and TCP15.
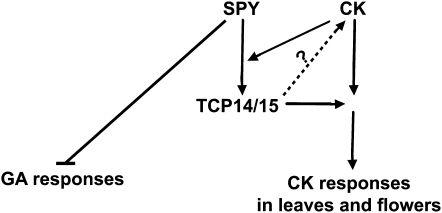
Figure 8.
A Model for the Interaction between SPY and TCP14/TCP15 and Its Effect on CK Responses.
Our results demonstrate that SPY interacts with TCP14 and TCP15 to facilitate CK responses in leaves and flowers. They also suggest that CK promotes TCP14 protein activity via SPY-dependent or -independent pathways. As the effects of TCP14 are CK dependent, it is possible that activated TCP14/15 can also regulate CK levels (dotted arrow with question mark). By contrast, SPY suppresses GA responses via a TCP14/TCP15-independent pathway.
Overexpression of TCP14 and TCP15 caused some phenotypic changes found in plants with suppressed GA responses (e.g., dwarfism and dark-green small leaves). These typical GA-regulated responses were suppressed by the loss of SPY, suggesting that TCP14 and TCP15 also mediate the suppression of GA responses by SPY. However, this is not supported by the phenotype of the double mutant tcp14 tcp15: Whereas a mutation in SPY affects typical GA responses, such as seed germination, cell elongation, and flowering time, the tcp14 tcp15 double mutant was not affected in these or other GA responses. Moreover, whereas spy is resistant to the GA biosynthesis inhibitor paclobutrazol, tcp14 and tcp14 tcp15 mutants are hypersensitive to the chemical (Tatematsu et al., 2008; see Supplemental Figure 9 online). Since different TCPs bind similar cis-regulatory elements (Martín-Trillo and Cubas, 2010), it is possible that some of the TCP14/15 overexpression phenotypes are caused by nonspecific activation/suppression of target genes, which are normally regulated by other TCPs. By that scenario, other TCPs may have a role in SPY’s suppression of GA responses, and, notably, SEC can modify several class I and class II TCPs.
A previous study has shown that SPY affects the activity of the circadian clock; mutations in SPY lengthen the free-running period of cotyledon movements, and overexpression of the gene shortens it (Tseng et al., 2004). It was proposed that SPY acts together with GI in the circadian clock pathway. Recently, several studies have shown interactions of clock elements such as TOC1 and PRR5 with different TCPs, including TCP15 and TCP21/CHE (Pruneda-Paz et al., 2009; Giraud et al., 2010). Since we show here that TCP15 interacts with SPY, it is possible that at least part of SPY’s effect on clock activity is mediated by TCP15.
In conclusion, the results of this study suggest that at least part of SPY’s activity in the regulation of plant development is mediated by class I TCP transcription factors. We propose that SPY interacts with TCP14 and TCP15 to promote CK effects in flowers and leaves (Figure 8). Such effects may be mediated by changes in CK signaling effectors or, alternatively, via increased CK levels in leaves and flowers, either by promoting its biosynthesis or inhibiting its degradation. As SPY also suppresses GA responses via a TCP14/TCP15-independent pathway, its pivotal role as a hub in hormone signaling integration is becoming evident.
METHODS
Plant Material, Growth Conditions, and Genetics
The Arabidopsis thaliana spy-4 mutant plants used in this study are in the Col-0 and in Ler backgrounds (Maymon et al., 2009). Single tcp14-4 and tcp15-3 and double tcp14 tcp15 mutants are in the Col-0 background (Kieffer et al., 2011). TCP14, TCP15, and CKX3 overexpression phenotypes and CYCB1;2pro:GUS are in Ler background. Arabidopsis seeds were sterilized, cold treated, and germinated on sterile Murashige and Skoog medium or in pots. Plants were grown in a growth room under controlled temperature (22°C) and long (16 h light/8 h dark) or short (8 h light/16 h dark) days.
Hormone Treatments
BA Treatment
Arabidopsis seedlings with two true leaves were sprayed with different concentrations of BA (Sigma-Aldrich) twice a week for ~3 weeks. In some cases, BA was applied directly to the inflorescence of mature plants.
Paclobutrazol Treatment
Arabidopsis seeds were germinated on plates containing Murashige and Skoog medium (Duchefa Biochemie), 0.8% (w/v) agar, and 1.5% (w/v) Suc with or without 2 mg L−1 paclobutrazol.
Scanning Electron Microscopy
Samples for scanning electron microscopy were fixed in 5% (w/v) gluteraldehyde in 0.1 M phosphate buffer, pH 7.2, transferred to ethanol (25% up to 100%), critical point dried with liquid carbon dioxide in a CPD 750 (Bio-Rad), sputter-coated with gold, and photographed with a Jeol scanning electron microscope (JSM-5410 LV).
Yeast Two-Hybrid Assays
Clones containing the full TCP14 and TCP15 coding regions were obtained from the ABRC. The TCP14 and TCP15 coding regions were PCR amplified using Platinum Taq DNA Polymerase High Fidelity (Invitrogen). The primers introduced NdeI and XhoI sites flanking the coding regions (primer sequences are detailed in Supplemental Table 1 online). After adding 3′ A-overhangs, the PCR products were cloned into pCR2.1-TOPO (Invitrogen). The coding regions were subcloned into the prey vector pGADT7 (Clontech) as NdeI to XhoI fragments. The bait plasmid expressing the SPY TPR region was described previously (Tseng et al., 2001). Bait and prey plasmids were transformed into the Saccharomyces cerevisiae strain HF7c, and the two hybrid screen was performed as described by Bai and Elledge (1997).
In Vitro Pull-Down Assays
TCP14 and TCP15 coding regions were cloned into pDEST15 (Invitrogen), and SEC and SPY coding regions were cloned into pRARE-MBP-DEST. The pRARE-MBP-DEST Gateway cloning vector expresses MBP-tagged proteins under the control of the isopropyl-β-d-thiogalactoside (IPTG)–inducible ptac promoter and also expresses several tRNAs that are rare in Escherichia coli. The vector was constructed by modifying the malE expression cassette of pMAL-c2 (New England Biolabs) for Gateway cloning. The region containing the lacIq gene and modified malE expression cassette was then subcloned into the pRARE plasmid (EMD Chemicals). The detailed sequences of primers used for cloning are presented in Supplemental Table 1 online. MBP and GST were expressed from pMALC1 (New England Biolabs) and pGEX (GE Healthcare), respectively. Plasmids for encoding MBP-SEC and MBP-SPY were expressed in the BL21-AI strain of E. coli (Invitrogen), and the other proteins were expressed in the Rosetta 2 strain (EMD Chemicals). For protein expression, cells were grown in Luria-Bertani medium containing 0.1% Glc and either 25 μg/mL chloramphenicol for BL21-AI cells or 25 μg/mL chloramphenicol and 75 μg/mL ampicillin for Rosetta 2 cells at 25°C. Protein expression was induced by the addition of IPTG to 1 mM to Rosetta 2 cultures or both IPTG and Ara to 0.2% to BL21-AI cultures when the OD600 was between 0.4 and 0.6. The cultures were grown for an additional 2 h, the cells were collected by centrifugation, and the cell pellet was stored at −20°C. Cell lysates were prepared as described previously (Tseng et al., 2004). We quantitated the prey (MBP, MBP-SEC, and MBP-SPY) and bait (GST, PST-TCP14, and GST-TCP15) proteins in the respective cell lysates by probing an immunoblot containing a dilution series for each lysate with anti-MBP or anti-GST, respectively. By comparing the signals from the different dilution series, it was possible to determine the relative amount of prey or bait in each lysate. Lysate mixtures for the different combinations of MBP-tagged and GST-tagged proteins were prepared. The MBP-tagged protein was pulled down, and immunoblots containing the pulled-down proteins were prepared as described previously (Tseng et al., 2004). GST-tagged proteins were detected using anti-GST horseradish peroxidase (HRP) conjugate (GE Healthcare Biosciences). The blot was then stripped and probed with anti-MBP HRP conjugate (New England Biolabs).
Generation of Transgenic Plants and the Transactivation System
The coding regions of both TCP14 and TCP15 were cloned in XhoI and HindIII sites under the control of an op array into pBJ36 (Moore et al., 1998), subcloned into the pMLBART binary vector (Goldshmidt et al., 2008), and transformed into Ler plants by floral dipping. At least 10 different T1 lines were examined, and representative lines with a single insertion were selected for further analysis. AS1pro:LhG4 (Sarojam et al., 2010), FILpro:LhG4, CRCpro:LhG4, op:GFP (Goldshmidt et al., 2008), op:CKX3 (Shani et al., 2010), CYCB1;2pro:GUS (Efroni et al., 2008), and CRCpro>>IPT (Greenboim-Wainberg et al., 2005) plants were previously described. CUC2pro:LhG4 was generated with PCR-based cloning of 3480 bp 5′ to the gene's ATG, and its expression largely matched the published CUC2 expression (primer sequences are detailed in Supplemental Table 1 online). Lines were crossed to generate the genotypes described in the article.
qRT-PCR and PCR Analyses
qRT-PCR analysis was performed using the Absolute Blue qPCR SYBR Green ROX Mix (AB-4162/B) kit (Thermo Fisher Scientific). Reactions were performed using a Rotor-Gene 6000 cycler (Corbett Research). A standard curve was obtained for each gene using dilutions of a cDNA sample. Each gene was quantified using Corbett Research Rotor-Gene software. At least three independent technical repeats were performed for each cDNA sample. Relative expression of each sample was calculated by dividing the expression level of the analyzed gene by that of TUBULIN. Gene-to-tubulin ratios were then averaged and presented as a proportion of the control treatment, set to a value of 1. All sequences of primers used for PCR and qRT-PCR analyses are presented in Supplemental Table 1 online.
O-GlcNAc Modification Assay
The SEC, TCP14, and TCP15 coding regions were amplified (primer sequences are detailed in Supplemental Table 1 online) using Pfx50 DNA polymerase (Invitrogen). The PCR products were Gateway cloned into pDONR221 (Invitrogen). The SEC coding region was transferred by the Gateway reaction from pDONR221 into the E. coli expression vector pCOLA-2-DEST (EMD Chemicals), and the TCP14 and TCP15 coding regions were transferred into the E. coli expression vector pDEST15 (Invitrogen). Rosetta 2 (DE3) E. coli cells were cotransformed with the pCOLA-2-DEST plasmid encoding SEC and the pDEST15 plasmid encoding one of the TCPs. The plasmids were selected using chloramphenicol (25 μg/mL) and ampicillin (75 μg/mL). Five-milliliter cultures were grown overnight with antibiotic selection at 37°C in Luria-Bertani medium and then 20-mL cultures were inoculated with ~1 mL of the saturated culture and grown at 22°C. At an OD600 of 0.4, SEC and TCP expression were induced by the addition of IPTG to a final concentration of 1.0 mM. Two hours after the induction of protein expression, cells from 1.5 mL of culture were harvested by centrifugation. The bacterial pellets were resuspended in 80 μL of SDS sample buffer, boiled for 5 min, and, if not used immediately, stored at −20°C. Fifteen microliters of each samples was resolved by SDS-PAGE and transferred to polyvinylidene fluoride membrane (Millipore). Terminal GlcNAc on membrane-bound proteins was detected by labeling with [3H] Gal, and the label was detected by fluorography (Scott et al., 2006). To determine if the modification was O- or N-linked, the blot was subjected to β-elimination (Duk et al., 1997). The blot was washed three times for 30 min in 95% ethanol to remove fluorography agent. It was then equilibrated in double distilled water for 5 min followed by equilibration in 55 mM NaOH for 5 min. The blot was then incubated in 55 mM NaOH for 16 h at 40°C and then washed four times in 10 mM HEPES, pH 7.9, for 5 min and air dried, and the residual radioactivity was detected by fluorography. Protein expression was assessed by probing duplicate blots with anti-GST HRP conjugate (GE Healthcare), which detects the TCP fusion proteins, or HisProbe-HRP (Thermo Scientific), which detects His-tagged SEC.
Detection of GUS Activity
Histochemical detection of GUS activity was performed using 5-bromo-4-chloro-3-indolyl-β-d-glucuronide as described before (Donnelly et al., 1999).
Chlorophyll Measurements
Chlorophyll was extracted and measured spectrophotometrically from fresh leaves (fifth leaf) and normalized to fresh weight (Arnon, 1949).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ARR5, At3g48100; CKX3, At5g56970; CUC2, At5g53950; CYCB1;2, At5g06150; SEC, At3g04240; SPY, At3g11540; TCP14, At3g47620; and TCP15, At3g69690.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. SPY Does Not Interact with Free GST.
Supplemental Figure 2. Overexpression of TCP14 under the Regulation of CUC2 or FIL Promoters Only Slightly Affected Plant Development.
Supplemental Figure 3. TCP14 Activity Requires Functional SPY.
Supplemental Figure 4. Overexpression of TCP14 and TCP15 Suppresses Petal Development.
Supplemental Figure 5. SEC Interacts with TCP14 and TCP15.
Supplemental Figure 6. SEC Does Not Modify ARR5 or GST.
Supplemental Figure 7. SEC Modifies TCP 2, 8, 19, 23, and 24.
Supplemental Figure 8. The Double Mutant tcp14 tcp15 and spy Share Similar Phenotypes.
Supplemental Figure 9. The loss of TCP14 and TCP15 Has No Effect on GA Responses.
Supplemental Figure 10. Expression of ARR5, but Not TCP14 or TCP15, Is Promoted by CK.
Supplemental Figure 11. Reducing CK Levels Suppresses TCP14 Overexpression Effects.
Supplemental Figure 12. Overexpressing CKX3 Suppresses TCP14 Activity and This Is Reversed by Exogenous CK.
Acknowledgments
This research was supported by research grants from the Israel Science Foundation (576-11) to D.W., from U.S.–Israel Binational Agricultural Research and Development Fund (US-3896-06) to D.W. and N.O., from the National Science Foundation (MCB-0820666) to N.O., and from Minerva and the Israel Science Foundation (1294-10) to Y.E. This work was also supported by the Pearlstein Fund for research in horticulture at Hebrew University; we thank the donors for their help. We thank Naomi Ori for her suggestions, comments, and helpful discussions.
AUTHOR CONTRIBUTIONS
D.W., Y.E., and N.O. designed the research. E.S., I.E., M.G., T.-S.T., and M.K. performed research. E.S., D.W., Y.E., K.S., and N.O. wrote the article.
References
- Aguilar-Martínez J.A., Poza-Carrión C., Cubas P. (2007). Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19: 458–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon D.I. (1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C., Elledge S.J. (1997). Gene identification using the yeast two-hybrid system. Methods Enzymol. 283: 141–156 [DOI] [PubMed] [Google Scholar]
- Butkinaree C., Park K., Hart G.W. (2010). O-linked beta-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim. Biophys. Acta 1800: 96–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P., Lauter N., Doebley J., Coen E. (1999). The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. 18: 215–222 [DOI] [PubMed] [Google Scholar]
- D’Agostino I.B., Deruère J., Kieber J.J. (2000). Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 124: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly P.M., Bonetta D., Tsukaya H., Dengler R.E., Dengler N.G. (1999). Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215: 407–419 [DOI] [PubMed] [Google Scholar]
- Duk M., Ugorski M., Lisowska E. (1997). Beta-elimination of O-glycans from glycoproteins transferred to immobilon P membranes: method and some applications. Anal. Biochem. 253: 98–102 [DOI] [PubMed] [Google Scholar]
- Eshed Y., Baum S.F., Perea J.V., Bowman J.L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol. 11: 1251–1260 [DOI] [PubMed] [Google Scholar]
- Efroni I., Blum E., Goldshmidt A., Eshed Y. (2008). A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Y.B., Liu C., Yu H., Broun P. (2007). Integration of cytokinin and gibberellin signalling by Arabidopsis transcription factors GIS, ZFP8 and GIS2 in the regulation of epidermal cell fate. Development 134: 2073–2081 [DOI] [PubMed] [Google Scholar]
- Giraud E., Ng S., Carrie C., Duncan O., Low J., Lee C.P., Van Aken O., Millar A.H., Murcha M., Whelan J. (2010). TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant Cell 22: 3921–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldshmidt A., Alvarez J.P., Bowman J.L., Eshed Y. (2008). Signals derived from YABBY gene activities in organ primordia regulate growth and partitioning of Arabidopsis shoot apical meristems. Plant Cell 20: 1217–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenboim-Wainberg Y., Maymon I., Borochov R., Alvarez J., Olszewski N., Ori N., Eshed Y., Weiss D. (2005). Cross talk between gibberellin and cytokinin: The Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell 17: 92–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Fujioka S., Blancaflor E.B., Miao S., Gou X., Li J. (2010). TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell 22: 1161–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Gan S. (2011). AtMYB2 regulates whole plant senescence by inhibiting cytokinin-mediated branching at late stages of development in Arabidopsis. Plant Physiol. 156: 1612–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanover J.A. (2001). Glycan-dependent signaling: O-linked N-acetylglucosamine. FASEB J. 15: 1865–1876 [DOI] [PubMed] [Google Scholar]
- Hanover J.A., Krause M.W., Love D.C. (2010). The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim. Biophys. Acta 1800: 80–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanover J.A., Yu S., Lubas W.B., Shin S.H., Ragano-Caracciola M., Kochran J., Love D.C. (2003). Mitochondrial and nucleocytoplasmic isoforms of O-linked GlcNAc transferase encoded by a single mammalian gene. Arch. Biochem. Biophys. 409: 287–297 [DOI] [PubMed] [Google Scholar]
- Hart G.W., Housley M.P., Slawson C. (2007). Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446: 1017–1022 [DOI] [PubMed] [Google Scholar]
- Hartweck L.M., Genger R.K., Grey W.M., Olszewski N.E. (2006). SECRET GENT and SPINDLY have overlapping roles in the development of Arabidopsis thaliana L. Heyn. J. Exp. Bot 57: 865–875 [DOI] [PubMed] [Google Scholar]
- Hartweck L.M., Olszewski N.E. (2006). Rice GIBBERELLIN INSENSITIVE DWARF1 is a gibberellin receptor that illuminates and raises questions about GA signaling. Plant Cell 18: 278–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartweck L.M., Scott C.L., Olszewski N.E. (2002). Two O-linked N-acetylglucosamine transferase genes of Arabidopsis thaliana L. Heynh. have overlapping functions necessary for gamete and seed development. Genetics 161: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervé C., Dabos P., Bardet C., Jauneau A., Auriac M.C., Ramboer A., Lacout F., Tremousaygue D. (2009). In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiol. 149: 1462–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen S.E., Olszewski N.E. (1993). Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5: 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer M., Master V., Waites R., Davies B. (2011). TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. 68: 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S., Ohashi Y. (2002). DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30: 337–348 [DOI] [PubMed] [Google Scholar]
- Koyama T., Mitsuda N., Seki M., Shinozaki K., Ohme-Takagi M. (2010). TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22: 3574–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Potuschak T., Colón-Carmona A., Gutiérrez R.A., Doerner P. (2005). Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl. Acad. Sci. USA 102: 12978–12983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.-Y., Li B., Dong A.-W. (October 12, 2011). The Arabidopsis transcription factor AtTCP15 regulates endoreduplication by modulating expression of key cell-cycle genes. Mol. Plant http://dx.doi.org/10.1093/mp/ssr086
- Martín-Trillo M., Cubas P. (2010). TCP genes: A family snapshot ten years later. Trends Plant Sci. 15: 31–39 [DOI] [PubMed] [Google Scholar]
- Martinez-Fleites C., He Y., Davies G.J. (2010). Structural analyses of enzymes involved in the O-GlcNAc modification. Biochim. Biophys. Acta 1800: 122–133 [DOI] [PubMed] [Google Scholar]
- Maymon I., Greenboim-Wainberg Y., Sagiv S., Kieber J.J., Moshelion M., Olszewski N., Weiss D. (2009). Cytosolic activity of SPINDLY implies the existence of a DELLA-independent gibberellin-response pathway. Plant J. 58: 979–988 [DOI] [PubMed] [Google Scholar]
- Moore I., Gälweiler L., Grosskopf D., Schell J., Palme K. (1998). A transcription activation system for regulated gene expression in transgenic plants. Proc. Natl. Acad. Sci. USA 95: 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B., Sheen J. (2007). Advances in cytokinin signaling. Science 318: 68–69 [DOI] [PubMed] [Google Scholar]
- Navaud O., Dabos P., Carnus E., Tremousaygue D., Hervé C. (2007). TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 65: 23–33 [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz J.L., Breton G., Para A., Kay S.A. (2009). A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte A.M., Mason M.G., Hutchison C.E., Ferreira F.J., Schaller G.E., Kieber J.J. (2006). A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc. Natl. Acad. Sci. USA 103: 11081–11085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou-Khamlichi C., Huntley R., Jacqmard A., Murray J.A.H. (1999). Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283: 1541–1544 [DOI] [PubMed] [Google Scholar]
- Robertson M. (2004). Two transcription factors are negative regulators of gibberellin response in the HvSPY-signaling pathway in barley aleurone. Plant Physiol. 136: 2747–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos M.D., Hanover J.A. (2000). Structure of O-linked GlcNAc transferase: Mediator of glycan-dependent signaling. Biochem. Biophys. Res. Commun. 271: 275–280 [DOI] [PubMed] [Google Scholar]
- Sarojam R., Sappl P.G., Goldshmidt A., Efroni I., Floyd S.K., Eshed Y., Bowman J.L. (2010). Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell 22: 2113–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer C., Palatnik J.F., Aggarwal P., Chételat A., Cubas P., Farmer E.E., Nath U., Weigel D. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott C.L., Hartweck L.M., de Jesús Pérez J., Chen D.H., García J.A., Olszewski N.E. (2006). SECRET AGENT, an Arabidopsis thaliana O-GlcNAc transferase, modifies the Plum pox virus capsid protein. FEBS Lett. 580: 5829–5835 [DOI] [PubMed] [Google Scholar]
- Shani E., Ben-Gera H., Shleizer-Burko S., Burko Y., Weiss D., Ori N. (2010). Cytokinin regulates compound leaf development in tomato. Plant Cell 22: 3206–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A., Ueguchi-Tanaka M., Sakamoto T., Fujioka S., Takatsuto S., Yoshida S., Sazuka T., Ashikari M., Matsuoka M. (2006). The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J. 48: 390–402 [DOI] [PubMed] [Google Scholar]
- Silverstone A.L., Tseng T.S., Swain S.M., Dill A., Jeong S.Y., Olszewski N.E., Sun T.P. (2007). Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol. 143: 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Sakurai K., Ueguchi C., Mizuno T. (2001). Two types of putative nuclear factors that physically interact with histidine-containing phosphotransfer (Hpt) domains, signaling mediators in His-to-Asp phosphorelay, in Arabidopsis thaliana. Plant Cell Physiol. 42: 37–45 [DOI] [PubMed] [Google Scholar]
- Tatematsu K., Nakabayashi K., Kamiya Y., Nambara E. (2008). Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J. 53: 42–52 [DOI] [PubMed] [Google Scholar]
- Tatematsu K., Ward S., Leyser O., Kamiya Y., Nambara E. (2005). Identification of cis-elements that regulate gene expression during initiation of axillary bud outgrowth in Arabidopsis. Plant Physiol. 138: 757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng T.-S., Salomé P.A., McClung C.R., Olszewski N.E. (2004). SPINDLY and GIGANTEA interact and act in Arabidopsis thaliana pathways involved in light responses, flowering, and rhythms in cotyledon movements. Plant Cell 16: 1550–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng T.-S., Swain S.M., Olszewski N.E. (2001). Ectopic expression of the tetratricopeptide repeat domain of SPINDLY causes defects in gibberellin response. Plant Physiol. 126: 1250–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir I., Lu J., Cook H., Causier B., Schwarz-Sommer Z., Davies B. (2004). CUPULIFORMIS establishes lateral organ boundaries in Antirrhinum. Development 131: 915–922 [DOI] [PubMed] [Google Scholar]
- Weiss D., Ori N. (2007). Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol. 144: 1240–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells L., Vosseller K., Hart G.W. (2001). Glycosylation of nucleocytoplasmic proteins: Signal transduction and O-GlcNAc. Science 291: 2376–2378 [DOI] [PubMed] [Google Scholar]
- Wells L., Whelan S.A., Hart G.W. (2003). O-GlcNAc: A regulatory post-translational modification. Biochem. Biophys. Res. Commun. 302: 435–441 [DOI] [PubMed] [Google Scholar]
- Werner T., Motyka V., Laucou V., Smets R., Van Onckelen H., Schmülling T. (2003). Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai O., Shani E., Russ D., Ori N. (2011). Gibberellin partly mediates LANCEOLATE activity in tomato. Plant J. 68: 571–582 [DOI] [PubMed] [Google Scholar]
- Yang X., Ongusaha P.P., Miles P.D., Havstad J.C., Zhang F., So W.V., Kudlow J.E., Michell R.H., Olefsky J.M., Field S.J., Evans R.M. (2008). Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature 451: 964–969 [DOI] [PubMed] [Google Scholar]
- Zachara N.E., Hart G.W. (2004). O-GlcNAc a mediator of cellular function: Characterizing a family of O-GlcNAc binding proteins. Glycobiology 14: 1063 [Google Scholar]