Abstract
Fetal chromosomal abnormalities are the most common reasons for invasive prenatal testing. Currently, G-band karyotyping and several molecular genetic methods have been established for diagnosis of chromosomal abnormalities. Although these testing methods are highly reliable, the major limitation remains restricted resolutions or can only achieve limited coverage on the human genome at one time. The massively parallel sequencing (MPS) technologies which can reach single base pair resolution allows detection of genome-wide intragenic deletions and duplication challenging karyotyping and microarrays as the tool for prenatal diagnosis. Here we reported a novel and robust MPS-based method to detect aneuploidy and imbalanced chromosomal arrangements in amniotic fluid (AF) samples. We sequenced 62 AF samples on Illumina GAIIx platform and with averagely 0.01× whole genome sequencing data we detected 13 samples with numerical chromosomal abnormalities by z-test. With up to 2× whole genome sequencing data we were able to detect microdeletion/microduplication (ranged from 1.4 Mb to 37.3 Mb of 5 samples from chorionic villus sampling (CVS) using SeqSeq algorithm. Our work demonstrated MPS is a robust and accurate approach to detect aneuploidy and imbalanced chromosomal arrangements in prenatal samples.
Introduction
Chromosomal abnormalities occur in 1 of 160 live births [1].The risk of giving birth to a child with chromosomal abnormalities, especially Down syndrome (OMIM# 190685), increases throughout a woman's reproductive years [2], [3]. Prenatal diagnosis of fetal chromosomal abnormalities is the most common indication for invasive prenatal testing. The prevalence of chromosomal abnormalities in fetuses with aneuploidy accounts for 6–11% of all stillbirths and neonatal deaths [4], [5]. Consequently, screening and diagnostic programs to detect the most common trisomies in live born infants are well established [6]. Currently, G-band karyotyping and several molecular genetic methods including multiplex ligation-dependent probe amplification (MLPA), fluorescence in situ hybridization (FISH), quantitative fluorescent PCR (QF-PCR) and microarray-based comparative genomic hybridization (arrayCGH) have been well established for prenatal diagnosis of chromosomal abnormalities in clinical labs [7], [8]. Although these testing methods have been proved to be highly reliable, the major limitation remains restricted resolution or can only achieve limited coverage on the human genome at one time [9], [10].
To overcome these limitations, in this study we have developed a new method based on massively parallel sequencing (MPS) platform to detect fetal chromosomal abnormalities, which is independent of particular genetic markers and cell culture in medical practice. We directly sequenced 62 DNA samples extracted from uncultured amniotic fluid (AF) of pregnant women. After statistical analysis, we can clearly detect numerical chromosomal abnormalities among 46 chromosomes. The whole process only takes 7 days and the results were validated by full karyotyping analysis. Subsequently, we investigated the presence of copy number variations among 5 prenatal samples previously identified by arrayCGH. We demonstrated that with different whole genome sequencing coverage MPS platform could be applied for identification of aneuploidy and imbalanced chromosomal arrangements, and those approaches are more sensitive and effective compared with conventional methods.
Results
Detection of Fetal Aneuploidy
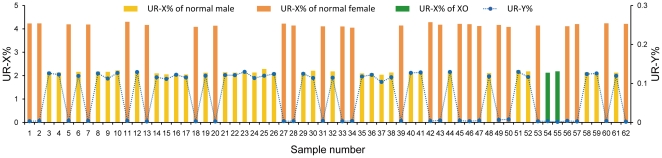
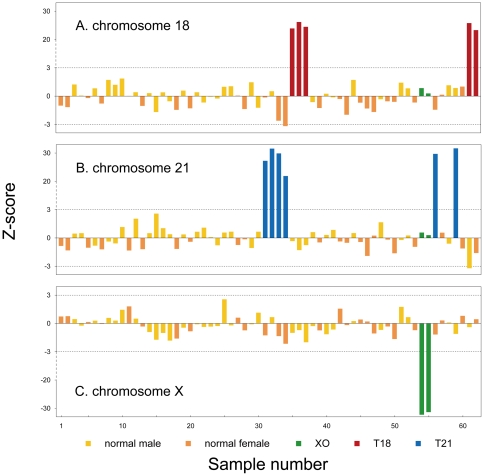
In this study, the 32 test cases were used for sequencing analysis under double-blind conditions. For all 62 AF samples (30 normal controls and 32 test cases), we obtained 0.55∼3.28 million 35-bp single-end reads (or 19.25 Mb∼114.80 Mb sequencing data) which corresponds to 0.006∼0.038× human genome depth. Consequently, 0.20∼1.15 million unique reads (UR) were obtained for each sample (Table 1). For each chromosome, the number of unique reads was counted and the UR% was calculated. The complete set of UR% for 62 samples is listed in Table S1. UR% for Chromosome X, Y also showed in Fig. 1. The gender of each sample can be determined from UR% of Chromosome X, Y. For 30 normal control samples, the mean and standard deviation (SD) of UR% for each chromosome were calculated. Then the z-scores of each of the chromosomes, except the Y chromosome, for each of the normal controls and test cases were calculated according to the formula described in Materials and methods (Table S2). From 32 test cases, 5 cases had a z-score>20 for chromosome 18 (indicating Trisomy 18), 6 cases had a z-score>20 for chromosome 21 (indicating Trisomy 21), and 2 cases had a z-score←20 for chromosome X (indicating XO). All the other chromosomes had z-scores within ±3 for all 32 test cases (indicating euploid chromosomes) (Fig. 2, Table. S1). The detection of fetal aneuploidy based on sequencing analysis agreed with karyotyping results.
Table 1. Raw data and UR% of chromosome 21, 18, 13, X, Y of 62 AF sample.
| Sample No. | Labrary No. | Fetal Karyotype | DNA Amount (ng) | Raw Data (reads) | Unique Data (reads) | UR-21% | UR-18% | UR-13% | UR-X% | UR-Y% |
| 1 | 92H13 | 46XX | 100 | 1 387 306 | 531 327 | 1.26909 | 2.91948 | 3.68963 | 4.22414 | 0.00320 |
| 2 | 08H9 | 46XX | 100 | 1 649 958 | 622 928 | 1.26638 | 2.90997 | 3.64248 | 4.22633 | 0.00385 |
| 3 | 09H10 | 46XY | 100 | 1 474 317 | 552 990 | 1.29496 | 3.05087 | 3.91725 | 2.16279 | 0.12586 |
| 4 | 10H11 | 46XY | 100 | 1 501 313 | 577 494 | 1.29664 | 2.98046 | 3.74255 | 2.12764 | 0.12295 |
| 5 | 11H12 | 46XX | 100 | 1 341 688 | 514 366 | 1.26525 | 2.96754 | 3.68026 | 4.18671 | 0.00447 |
| 6 | 12H14 | 46XY | 100 | 1 315 342 | 502 804 | 1.26968 | 3.02663 | 3.77026 | 2.15392 | 0.11913 |
| 7 | 13H15 | 46XX | 100 | 1 520 307 | 589 707 | 1.26164 | 2.93281 | 3.70743 | 4.17665 | 0.00407 |
| 8 | 15H18 | 46XY | 100 | 1 548 855 | 589 173 | 1.27874 | 3.07974 | 3.94434 | 2.16982 | 0.12560 |
| 9 | 17HCGY | 46XY | 100 | 749 354 | 309 033 | 1.27818 | 3.04466 | 3.87758 | 2.12275 | 0.12264 |
| 10 | 19H2 | 46XY | 100 | 1 929 900 | 744 124 | 1.30932 | 3.08860 | 3.90929 | 2.21683 | 0.12699 |
| 11 | 21H4 | 46XX | 100 | 3 285 106 | 1 244 513 | 1.26613 | 2.97819 | 3.82873 | 4.29276 | 0.00410 |
| 12 | 25H9 | 46XY | 100 | 1 575,322 | 667 903 | 1.32747 | 3.00182 | 3.86819 | 2.16540 | 0.12929 |
| 13 | 27H1 | 46XX | 100 | 947 196 | 377 647 | 1.27924 | 2.91754 | 3.67009 | 4.09139 | 0.02304 |
| 14 | 31H6 | 46XY | 100 | 1 248 950 | 476 205 | 1.29776 | 2.98863 | 3.57304 | 2.05458 | 0.13776 |
| 15 | 32H7 | 46XY | 100 | 693 356 | 268 692 | 1.34987 | 2.88137 | 3.58328 | 2.01383 | 0.12840 |
| 16 | 33H8 | 46XY | 100 | 929 156 | 371 080 | 1.31239 | 2.99262 | 3.70971 | 2.05805 | 0.13825 |
| 17 | 34H9 | 46XY | 100 | 1 647 104 | 646 180 | 1.30010 | 2.94190 | 3.62840 | 2.01244 | 0.13263 |
| 18 | 40H6 | 46XX | 100 | 728 707 | 278 944 | 1.26190 | 2.88158 | 3.41323 | 4.01586 | 0.01972 |
| 19 | 42H8 | 46XY | 100 | 2 021 819 | 829 302 | 1.29856 | 3.00795 | 3.82840 | 2.08272 | 0.13578 |
| 20 | 43H9 | 46XX | 100 | 685 181 | 270 538 | 1.27967 | 2.90236 | 3.52557 | 4.06486 | 0.02735 |
| 21 | 44H10 | 46XY | 100 | 993 450 | 400 726 | 1.30688 | 3.00180 | 3.72499 | 2.09894 | 0.13550 |
| 22 | 45H11 | 46XY | 100 | 636 527 | 250 931 | 1.30315 | 2.93308 | 3.64762 | 2.08464 | 0.13709 |
| 23 | 46H12 | 46XY | 100 | 1 010 133 | 399 849 | 1.30249 | 2.96512 | 3.72216 | 2.09129 | 0.14480 |
| 24 | 47H13 | 46XY | 100 | 764 726 | 301 493 | 1.28494 | 2.95828 | 3.58284 | 2.08827 | 0.13466 |
| 25 | 48H1 | 46XY | 100 | 3 003 505 | 1 149 789 | 1.29728 | 3.03621 | 3.86358 | 2.27746 | 0.11976 |
| 26 | 54H7 | 46XY | 100 | 1 561 427 | 589 196 | 1.29923 | 3.03974 | 3.85305 | 2.12900 | 0.12424 |
| 27 | 56H9 | 46XX | 100 | 2 322 635 | 885 707 | 1.27119 | 2.98022 | 3.78613 | 4.21618 | 0.00339 |
| 28 | 57H10 | 46XX | 100 | 1 278 926 | 512 310 | 1.29219 | 2.89961 | 3.57791 | 4.06414 | 0.02264 |
| 29 | 60H14 | 46XY | 100 | 1 618 924 | 616 422 | 1.26456 | 3.06495 | 3.95038 | 2.13555 | 0.12475 |
| 30 | 62H16 | 46XY | 100 | 661 358 | 263 458 | 1.30875 | 2.89420 | 3.73532 | 2.17530 | 0.12791 |
| 31 | 61HWMH | 47XX+21 | 100 | 833 041 | 340 693 | 1.85651 | 2.98157 | 3.71009 | 4.05644 | 0.01967 |
| 32 | 72HT21 | 47XY+21 | 100 | 766 103 | 311 852 | 1.93585 | 3.01072 | 3.82617 | 2.13627 | 0.12955 |
| 33 | 74H3 | 47XX+21 | 100 | 1 127 241 | 458 743 | 1.91087 | 2.81705 | 3.54033 | 4.03080 | 0.02180 |
| 34 | 81HWMH | 47XX+21 | 100 | 650 242 | 273 303 | 1.73434 | 2.77714 | 3.39257 | 3.96812 | 0.02378 |
| 35 | 02H2 | 47XY+18 | 90 | 1 091 649 | 429 544 | 1.28206 | 4.39862 | 3.64852 | 2.05544 | 0.12828 |
| 36 | 06H7 | 47XY+18 | 100 | 1 851 667 | 706 651 | 1.26689 | 4.54298 | 3.71881 | 2.10174 | 0.12198 |
| 37 | 82HCXP | 47XY+18 | 100 | 503 179 | 204 295 | 1.27707 | 4.42742 | 3.78815 | 1.99858 | 0.11601 |
| 38 | 04H4 | 46XY | 100 | 1 754 104 | 666 002 | 1.29819 | 2.94188 | 3.67957 | 2.12507 | 0.11662 |
| 39 | 05H6 | 46XX | 100 | 2 434 717 | 898 966 | 1.27680 | 2.90456 | 3.57889 | 4.13297 | 0.00389 |
| 40 | 63H17 | 46XY | 90.8 | 685 313 | 275 177 | 1.30970 | 2.98680 | 3.86915 | 2.04850 | 0.14064 |
| 41 | 65H4 | 46XY | 100 | 1 202 799 | 473 140 | 1.32244 | 2.97185 | 3.70208 | 2.07402 | 0.14879 |
| 42 | 66H9 | 46XX | 100 | 1 623 201 | 617 337 | 1.27888 | 2.96166 | 3.83680 | 4.27724 | 0.00340 |
| 43 | 67H10 | 46XX | 100 | 811 894 | 327 382 | 1.28443 | 2.85660 | 3.64566 | 4.10071 | 0.02474 |
| 44 | 68H11 | 46XY | 100 | 6 381 643 | 2 455 268 | 1.29619 | 3.07979 | 3.96616 | 2.15150 | 0.12931 |
| 45 | 69H12 | 46XX | 100 | 955 289 | 384 784 | 1.28384 | 2.93281 | 3.62723 | 4.14050 | 0.02339 |
| 46 | 70H13 | 46XX | 100 | 833 449 | 328 863 | 1.25554 | 2.91215 | 3.66147 | 4.13212 | 0.02037 |
| 47 | 71H14 | 46XX | 100 | 1 321 322 | 523 536 | 1.29943 | 2.87430 | 3.56537 | 4.04327 | 0.02636 |
| 48 | 73H2 | 46XY | 100 | 705 325 | 277 705 | 1.32443 | 2.94017 | 3.72230 | 2.07342 | 0.12639 |
| 49 | 77H8 | 46XX | 100 | 664 485 | 271 294 | 1.28422 | 2.95805 | 3.74686 | 4.10035 | 0.02101 |
| 50 | 83HSHD | 46XX | 100 | 551 560 | 223 988 | 1.26480 | 2.93632 | 3.66657 | 4.01227 | 0.02411 |
| 51 | 07H8 | 46XY | 100 | 1 383 844 | 525 106 | 1.28203 | 3.06243 | 3.92035 | 2.23364 | 0.12950 |
| 52 | 94HLCH | 46XY | 100 | 888 892 | 509 386 | 1.29097 | 3.02639 | 3.87113 | 2.17399 | 0.11661 |
| 53 | 95HSCJ | 46XX | 100 | 863 603 | 498 346 | 1.26719 | 2.93932 | 3.74920 | 4.13408 | 0.00301 |
| 54 | 96HLLR | 45XO | 100 | 880 178 | 508 298 | 1.29727 | 3.02775 | 3.67776 | 2.11766 | 0.00315 |
| 55 | 97HHH | 45XO | 100 | 984 935 | 565 960 | 1.29161 | 2.99420 | 3.87572 | 2.17719 | 0.00166 |
| 56 | 98HMCM | 47XX+21 | 100 | 525 892 | 303 307 | 1.89610 | 2.89344 | 3.47536 | 4.10475 | 0.00396 |
| 57 | 99HZQL | 46XX | 100 | 898 477 | 518 330 | 1.29705 | 2.98664 | 3.77655 | 4.19713 | 0.00251 |
| 58 | 118AXCR | 46XX | 99 | 1 132 088 | 440 245 | 1.27406 | 3.04603 | 3.82810 | 2.14335 | 0.12402 |
| 59 | 121ASSH | 47XY+21 | 100 | 1 045 028 | 394 985 | 1.93628 | 3.02847 | 3.72698 | 2.07983 | 0.12557 |
| 60 | 125A22 | 46XX | 100 | 1 217 046 | 469 095 | 1.26350 | 3.03755 | 3.79454 | 4.22857 | 0.00384 |
| 61 | 126AJYJ | 47XY+18 | 100 | 1 207 389 | 471 223 | 1.22044 | 4.51952 | 3.77189 | 2.11895 | 0.11948 |
| 62 | 129AGJ | 47XX+18 | 100 | 1 181 077 | 449 114 | 1.25358 | 4.37083 | 3.69461 | 4.20539 | 0.00178 |
Figure 1. UR-X% and UR-Y% of 62 amniotic fluid samples.
Figure 2. z-scores of 62 AF samples for chromosome 21,18 and X determined by MPS.
Broken lines indicate the z score cut-off value of ±3 for trisomy 21 and trisomy 18. All 11 trisomy cases have a z-score value greater than 3 and the 2 cases of Tuner syndrome have a z-score value less than minus 3.
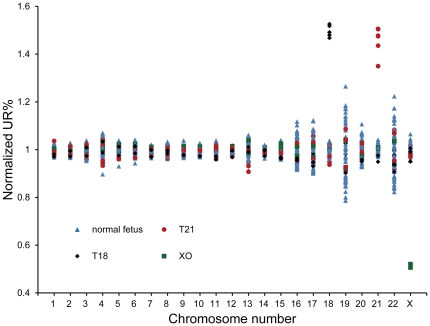
Normalized UR% values for all 22 autosomes and chromosome X were calculated (Fig. 3). 5 cases of trisomy 18, 6 cases of trisomy 21 and 2 cases of XO were successfully detected in the cohort of 32 test cases, which presented 100% of sensitivity and 100% of specificity.
Figure 3. The normalized UR% value per chromosome between different samples.
11 trisomy samples have a normalized UR% value of 1.5 and 2 cases of 45 XO have a normalized UR% value of 0.5.
Detection of imbalanced chromosomal arrangements
To further validated our developed and optimized analysis method among cases with imbalanced chromosomal arrangements, we obtained 44∼70 M sequence tags for the 5 “known” samples with 1.4 to 2.3× whole genome depth. To detect segmental copy number variations, a sequence-based Matlab CNV detection package, SegSeq, with YH reference genome as comparative genome were used. By applying the SegSeq analysis, we detected microdeletions and microduplications in all 5 samples (Table 2). All the 5 chromosomal copy number variations ranged from 1.4 Mb to 37.3 Mb were validated by arrayCGH and were mapped precisely to the correct location. Four pregnancies were terminated after genetic detection and only one pregnant gave a live birth (Table S3).
Table 2. Pathogenic chromosomal copy number changes and imbalance rearrangements identified by SeqSeq analysis which have been validated by arrayCGH.
| Array No. | Karyotype | arrayCGH | Segseq |
| A10071659 | 46,XY | deletion at 13q32.3q33.3(97091318∼106466788) | deletion at chr13: 97075576∼106516365(9.4 M) |
| A10021390 | 46,XY,der(5)t(5;18)(p13;q12.3) | deletion at 5p15.3∼p13.2(183931∼36816731);duplication at 18p12.3∼q23(39086755∼76067279) | deletion at chr5: 1∼36861739(36.9 M);duplication at chr18: 38768509∼76117152(37.3 M) |
| A10021383 | 46,XY,2q+ | duplication at 2q36∼q37.3(230369496∼242444380) | duplication at chr2: 230288772∼242427293(12.1 M) |
| A11091844 | 46,XX | deletion at 3q29(197216353∼198770242) | deletion at chr3: 197372027∼198831453(1.4 M) |
| A10071710 | 46,XY | deletion at 22q11.21(17289032∼19636115) | deletion at chr22: 17396815∼20248184(2.8 M) |
Two samples (A10021383, A10071659) were selected to further ascertain the results on the HumanOmni2.5 M chip (Illumina). CNV partition Algorithm plug-in, was used to detect copy number variations in these two samples. A duplication at 2q36∼q37.3 (230369496∼242444380) and a deletion at 13q32.3q33.3 (97091318∼106462788) were correctly detected, totally in concordance with the arrayCGH and sequencing analysis results.
Discussion
Massively parallel sequencing has been reported only to apply in noninvasive prenatal diagnosis of trisomy 21, 18 and 13 based on cell-free fetal DNA, due to it is limited amount of fragmented fetal DNA [11]. This also makes noninvasive MPS-based prenatal diagnosis difficult to detect all the chromosomal aneuploidies and the sexual chromosome abnormality accurately. In this study, we demonstrated for the first time that combined MPS with powerful bioinformatics analysis method can accurately diagnosis fetal aneuploidy and imbalanced chromosomal structural abnormalities. In fact, this study reports the first retrospective use of MPS (so called next generation sequencing) for prenatal diagnostics of chromosomal imbalance rearrangements to date and shows that it is practically feasible on a large-scale prenatal diagnosis of fetal chromosomal abnormalities.
By establishing a normal control sequencing tag data set, we have been able to demonstrate this new approach only requires a minimum among of DNA materials (100 ng) to achieve the identification of aneuploidies with a ultra low sequencing coverage (0.01×). Comparing to the golden standard (G-Band karyotyping) in clinical practice, MPS has no time limitation. Also as long as 100 ng genomic DNA can be extracted from tissues at any gestational weeks, MPS can be performed and report all fetal aneuploidies in 7 days. If necessary, more sequence reads can be performed to detect whether microdeletion or microduplication exists in the fetal genome which may result in severe developmental retardation. In our study, tissues obtained from fetus, such as amniotic fluid, CVS and placenta can be analyzed without cell culture since 100 ng genomic DNA is sufficient for library preparation and sequencing. So it can be used for the research into the molecular mechanism of miscarriage, stillbirth and fetal death when tissues are difficult to culture. Furthermore, our validation study on the 5 arrayCGH samples show that when more sophisticated sequencing protocols and bioinformatics algorithms are applied to the analysis, it is possible to detect smaller size chromosomal copy number variations as well as complex rearrangements across the whole genome of the fetus, such as balanced chromosomal arrangements,structure variations or even single-gene disorders. With the application of third-generation sequencing system in clinical laboratories, such as Miseq/Illumina and Ion Torrent PGM/Life Technologies, the whole process will take less time and acceptable price. Thus, it is likely that MPS will play an increasingly important role in the future development of prenatal screening and diagnosis.
A potential weakness of the study was that in the figure of describing the ratio of unique reads in each chromosome, chromosome19 and 22 have a differently huge coefficient of variation because of their extremely high GC content and made the detection of trisomy19 and trisomy22 difficult. Further study will be set up to deal with these problems, for example the computational correction of GC content among chromosomes. Other chromosomal abnormalities, such as balanced translocation or incomplete aneuploidy caused by mosaics or partial duplication or deletion of a chromosome should, in principle, also be detectable. Further studies are required to determine the effectiveness of massively parallel genomic sequencing in detecting these rare aberrations. Another weakness for the new methodology was the starting materials for library construction; with the conventional Illumina library construction approach100 ng genomic DNA of fetus was required. To further reduce the risk to pregnancies it would be important to reduce the amount for AF or CVS samples, other library construction methods such as using in vitro transposition may be an alternative solution.
In conclusion, we have demonstrated the usefulness of massively parallel sequencing to detect fetal aneuploidy and imbalanced chromosomal abnormalities of genomic DNA in prenatal samples. In principle, massively parallel sequencing can also reveal other features of the genomic material from amniotic fluid such as histone modifications as well as epigenetic DNA methylation. With the rapid reduction of sequencing cost, we expect that the strategy described in this article will become a powerful tool in the detection of all kinds of chromosomal abnormalities in clinical settings.
Materials and Methods
Subject Enrollment and Sample Recruitment
The study was approved by the Institutional Review Board of Beijing Obstetrics and Gynecology hospital of the Capital Medical University. Informed consent was obtained from each participant.
A total of 32 pregnant women at a high risk of Down's syndrome were recruited as test cases from the Beijing Obstetrics and Gynecology hospital during the period of January to May 2010. Amniocentesis was applied at 19∼22th gestational week and standard G-band karyotyping analysis was performed. Another 30 euploid AF samples (20 with male fetus and 10 with female fetus) were included as normal control at the same hospital. To validate the sensitivity of analysis method for detection of microdeletion/microduplication, 5 chorionic villus sampling (CVS) samples validated by karyotyping and arrayCGH were recruited from the Prenatal Genetic Diagnosis Centre (PGDC) at Department of Obstetrics & Gynecology, Chinese University of Hong Kong.
Sample Preparation and Sequencing
Genomic DNA was extracted from uncultured AF samples with Micro DNA Kit (Tiangen) and quantified with the Quant-iT dsDNA HS Assay Kit (Invitrogen). 100 ng genomic DNA from each sample was sheared into small fragments ranged from 100 to 400 bp with Bioruptor (Diagenode). After end-repair, “A”- overhanging and adapter-ligation, DNA fragments of 300 bp (±25 bp) in length were selected by 2% agarose gel electrophoresis and underwent 12 cycles of PCR with multiplex primers. PCR products were purified by Agencourt AMPure Kit (Beckman). Size distribution of the library was detected by Agilent Bioanalyzer DNA 1000 kit (Agilent Technologies) and the concentration was measured by quantitative PCR (qPCR). Libraries with different index tags were mixed in equal moles into a pool and sequenced with single-end 36 cycle multiplex sequencing on Illumina GAIIx platform.
For the 5 DNA samples from CVS tissues, sequencing libraries with the insert size of 500 bp (±25 bp) were prepared. Paired-end 100 cycle multiplex sequencing was performed on Illumina HiSeq 2000.
Bioinformatics analysis
z-score for detection of fetal aneuploidy
35-bp single-end reads from 62 AF samples (30 normal controls and 32 test cases) were aligned against repeat-masked human genome build 36 (hg18) by ELAND. Unique reads (UR), which can be mapped to reference genome sequence without any mismatches or alternative positions, were used for further analysis. The UR percentage (UR%) of chromosome N (UR-N%, N denotes chromosome number) can be used as an indicator of fetal trisomy for autosomes. To satisfy central-limit theorem, 30 euploid samples were used as normal controls. The z-score of chromosome N (N stands for chromosome numbers) is calculated as,
For autosomes, considering about the type I error rate (α) of 0.01, 3 was set as a cut-off value to determine the fetal trisomy.
SegSeq algorithm for detection of microdeletion/microduplication
We mapped the Illumina reads to the reference sequence of human genome (HG18, NCBI 36.3) by Short Oligonucleotide Analysis Package aligner (SOAP2) (http://soap.genomics.org.cn/) [12] with parameter about total allowed mismatches (-v 5), seed length (-s 40), minimal aligning length (-l 40) and insert DNA size enabled. Only unique reads were remained in following CNV analysis.
For the CNV detection, we employed a MATLAB packet, SegSeq (http://www.broadinstitute.org/cgi-bin/cancer/publications/pub_paper.cgi?mode=view&paper_id=182) [13], with YH Illumina reads [14] as reference control. Segments with copy ratio, calculated by SegSeq, less than 0.75 or greater than 1.25 were reported as variations. CNVs on critical regions of identified diseases would be an important signal for clinical screening/diagnose.
CNV Detection by arrayCGH and SNP typing array
Genomic chromosomal copy number variants (CNVs) were detected using a targeted high resolution 44 K oligonucleotide array specifically constructed for prenatal screening with the intention of targeting common trisomic aneuploidies and most known microdeletion and microduplication syndromes. This Fetal DNA chip included telomeric and pericentromeric regions, examining the genome to a resolution of 100 kb (http://www.fetalmedicine.hk/en/Fetal_DNA_Chip.asp) [15]. The Fetal DNA chip is specially (http://www.fetalmedicine.hk/en/Fetal_DNA_chip/Appendix_I.pdf) with most of the known common non-pathogenic CNVs regions removed. This chip provides a means to detect chromosomal aberrations with resolution of <100 Kb across the genome. The quality of the array was analysed using Agilent DNA analytics software and cases where the Derivative Log Ratio spread of the array was >0.25 were excluded from further data analysis. Data reporting variations in copy number were released after excluding known non-pathogenic chromosome copy number variants that have been listed at the Database of Genomic Variants.
To ascertain the accuracy of MSP-based CNV detection method, two CVS samples were analyzed with HumanOmni2.5-Quad Bead Chip according to Illumina manufacturer's protocol and CNV partition plug-in software was employed for DNA copy number analysis.
Supporting Information
The UR% of each chromosome for 62 AF samples.
(XLS)
The z-scores of each chromosome (except chr Y) for 62 AF samples.
(XLS)
Outcome and clinical data of CUHK cases.
(XLS)
Acknowledgments
We thank our colleagues at BGI for sequencing. We thank Dr. Lars Bolund and Dr. Qi Ming for revising the manuscript.
Footnotes
Competing Interests: The authors have read the journal's policy and have the following conflicts: Fuman Jiang, Fang Chen and Xiuqing Zhang have filed patent applications on the prenatal detection of aneuploidy by massively parallel sequencing of amniotic fluid. The patent number is 201010252849.X in China and the date is 2010-8-13. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials.
Funding: The study was supported by Beijing Nova Program (2006B08) and Beijing Municipal Science and Technology Commission (Z09090300830909). Choy KW and Lau TK were supported by Hong Kong General Research Fund (GRF) grant project number 463710. Leung TY was supported by Chinese University Direct grant project number 2041573. The study was also supported by Shenzhen City Key Laborabory Project (CXB200903110066A). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Driscoll DA, Gross S. Clinical practice. Prenatal screening for aneuploidy. N Engl J Med. 2009;360:2556–2562. doi: 10.1056/NEJMcp0900134. [DOI] [PubMed] [Google Scholar]
- 2.Hook EB. Rates of chromosome abnormalities at different maternal ages. Obstet Gynecol. 1981;58:282–285. [PubMed] [Google Scholar]
- 3.Morris JK, Wald NJ, Mutton DE, Alberman E. Comparison of models of maternal age-specific risk for Down syndrome live births. Prenat Diagn. 2003;23:252–258. doi: 10.1002/pd.568. [DOI] [PubMed] [Google Scholar]
- 4.Wapner RJ. Genetics of stillbirth. Clin Obstet Gynecol. 2010;53:628–634. doi: 10.1097/GRF.0b013e3181ee2793. [DOI] [PubMed] [Google Scholar]
- 5.Korteweg FJ, Bouman K, Erwich JJ, Timmer A, Veeger NJ, et al. Cytogenetic analysis after evaluation of 750 fetal deaths: proposal for diagnostic workup. Obstet Gynecol. 2008;111:865–874. doi: 10.1097/AOG.0b013e31816a4ee3. [DOI] [PubMed] [Google Scholar]
- 6.Irving C, Richmond S, Wren C, Longster C, Embleton ND. Changes in fetal prevalence and outcome for trisomies 13 and 18: a population-based study over 23 years. J Matern Fetal Neonatal Med. 2011;24:137–141. doi: 10.3109/14767051003758879. [DOI] [PubMed] [Google Scholar]
- 7.Williams ES, Cornforth MN, Goodwin EH, Bailey SM. CO-FISH, COD-FISH, ReD-FISH, SKY-FISH. Methods Mol Biol. 2011;735:113–124. doi: 10.1007/978-1-61779-092-8_11. [DOI] [PubMed] [Google Scholar]
- 8.Leung TY, Vogel I, Lau TK, Chong W, Hyett JA, et al. Identification of submicroscopic chromosomal aberrations in fetuses with increased nuchal translucency and an apparently normal karyotype. Ultrasound Obstet Gynecol. 2011 doi: 10.1002/uog.8988. [DOI] [PubMed] [Google Scholar]
- 9.Vermeesch JR, Fiegler H, de Leeuw N, Szuhai K, Schoumans J, et al. Guidelines for molecular karyotyping in constitutional genetic diagnosis. Eur J Hum Genet. 2007;15:1105–1114. doi: 10.1038/sj.ejhg.5201896. [DOI] [PubMed] [Google Scholar]
- 10.ACOG Practice Bulletin No. 88, December 2007. Invasive prenatal testing for aneuploidy. Obstet Gynecol. 2007;110:1459–1467. doi: 10.1097/01.AOG.0000291570.63450.44. [DOI] [PubMed] [Google Scholar]
- 11.Ehrich M, Deciu C, Zwiefelhofer T, Tynan JA, Cagasan L, et al. Noninvasive detection of fetal trisomy 21 by sequencing of DNA in maternal blood: a study in a clinical setting. Am J Obstet Gynecol. 2011;204(3):205.e1–11. doi: 10.1016/j.ajog.2010.12.060. [DOI] [PubMed] [Google Scholar]
- 12.Li R, Yu C, Li Y, Lam TW, Yiu SM, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25(15):1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 13.Chiang DY, Getz G, Jaffe DB, O'Kelly MJ, Zhao X, et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods. 2009;6:99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Wang W, Li R, Li Y, Tian G, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leung TY, Vogel I, Lau TK, Chong W, Hyett JA, et al. Identification of submicroscopic chromosomal aberrations in fetuses with increased nuchal translucency and an apparently normal karyotype. Ultrasound Obstet Gynecol. 2011;38:314–319. doi: 10.1002/uog.8988. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The UR% of each chromosome for 62 AF samples.
(XLS)
The z-scores of each chromosome (except chr Y) for 62 AF samples.
(XLS)
Outcome and clinical data of CUHK cases.
(XLS)