Abstract
The transcription factor D-Pax2 is required for the correct differentiation of several cell types in Drosophila sensory systems. While the regulation of its expression in the developing eye has been well studied, little is known about the mechanisms by which the dynamic pattern of D-Pax2 expression in the external sensory organs is achieved. Here we demonstrate that early activation of D-Pax2 in the sensory organ lineage and its maintenance in the trichogen and thecogen cells are governed by separate enhancers. Furthermore, the initial activation is controlled in part by proneural proteins whereas the later maintenance expression is regulated by a positive feedback loop.
Keywords: D-Pax2, proneural genes, Drosophila, external sensory organs, transcriptional regulation, enhancers
INTRODUCTION
The development of external sensory (es) organs in Drosophila melanogaster is a sequential process beginning with the selection of a sensory organ precursor (SOP) from a proneural cluster (PNC) of cells, each of which is originally competent to adopt a proneural fate (Cubas et al., 1991; Skeath and Carroll, 1991). All but one cell in each PNC, however, are prevented from becoming SOPs via lateral inhibition due to Notch signaling (Heitzler and Simpson, 1991; Lai, 2004). The SOP then proceeds through a series of divisions which produces five cells, four of which eventually form the mature es organ (Hartenstein and Posakony, 1989; Gho et al., 1999; Reddy and Rodrigues, 1999). The division of the SOP generates a pIIa and pIIb cell. Next, the pIIb cell divides to produce a pIIIb cell and an extraneous glial cell which undergoes apoptosis (Gho et al., 1999; Reddy and Rodrigues, 1999). This cell is believed to be a vestigial component remaining from an ancestral sense organ lineage (Lai and Orgogozo, 2004). The pIIa cell divides, giving rise to the tormogen (socket) cell and trichogen (shaft) cell and the pIIIb cell divides to produce the thecogen (sheath) cell and the neuron. Cell fates among the SOP progeny are specified via a combination of intrinsic and extrinsic mechanisms, such as the asymmetric localization of Numb protein and Notch signaling between the members of the lineage, respectively (Posakony, 1994). Once the appropriate cell fates have been allocated, each cell executes its specific differentiative program resulting in a functional es organ.
One of the key players in the differentiation of es organ cell types is the Drosophila homolog of the mammalian Pax2 transcription factor (D-Pax2). Two classes of D-Pax2 mutants have been identified; mutants in the shaven (sv) class affect primarily es organs whereas those of the sparkling (spa) class affect eyes (Fu et al., 1998). Study of these mutants has demonstrated that D-Pax2 plays multiple roles during the development of both es organs and the eye (Fu and Noll, 1997; Kavaler et al., 1999; Dziedzic et al., 2009). During the development of the es organs, D-Pax2 protein is present in the SOP a few hours after it has been selected from the PNC. Shortly after it is first detectable, the SOP divides. D-Pax2 is present in all the cells of the es organ lineage throughout the divisions. The function of D-Pax2 during this early specification phase is unclear. The strongest sv mutants lead to trichogen-to-tormogen (double socket) cell fate transformations among the macrochaetes, albeit at a very low frequency (Lees and Waddington, 1942; Kavaler et al., 1999). During the differentiation of es organ cells, D-Pax2 expression is lost from the tormogen cell and neuron, but remains at high levels in the trichogen and thecogen cells. At this late stage, D-Pax2 functions to guide the appropriate differentiation of these cell types (Kavaler et al., 1999). The elements that regulate D-Pax2 expression in the eye are separable from the elements that regulate its expression in the es organs. Whereas a 350 bp region in the fourth intron of the gene is responsible for D-Pax2 expression in the eye, a less studied region upstream of the gene (originally termed the bristle enhancer) controls expression in the es organ lineage (Fu et al., 1998; Flores et al., 2000; Swanson et al., 2010).
The bristle enhancer was defined by rescue experiments in which the upstream region was cloned in front of a D-Pax2 cDNA and tested for its ability to rescue the defects found in either sv or spa mutants (Fu et al., 1998). A 2.1 KB upstream fragment was capable of partially rescuing sv defects while a larger 6.7 KB fragment produced a complete rescue. In this report, we have examined the ability of comparable upstream regions to drive gene expression in es organ cells in vivo by generating GFP reporter constructs. We demonstrate that early D-Pax2 expression and its later refined expression are controlled by separate enhancers. Furthermore, the early expression of D-Pax2 in the SOP lineage is initiated by the activity of proneural proteins while the late expression in the trichogen and thecogen cells is dependent on D-Pax2 protein itself via a positive feedback loop.
RESULTS
A 3.1 kb enhancer region is sufficient to drive gene expression in all D-Pax2 + es organ cells
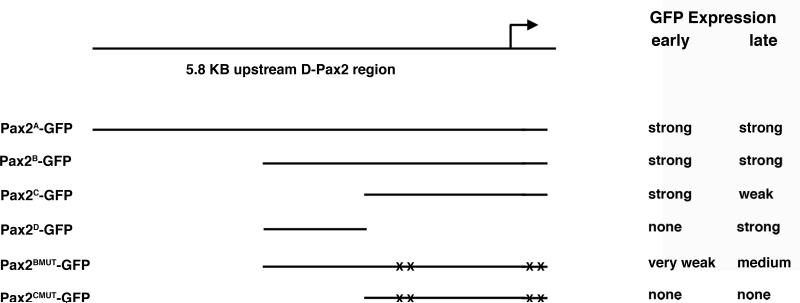
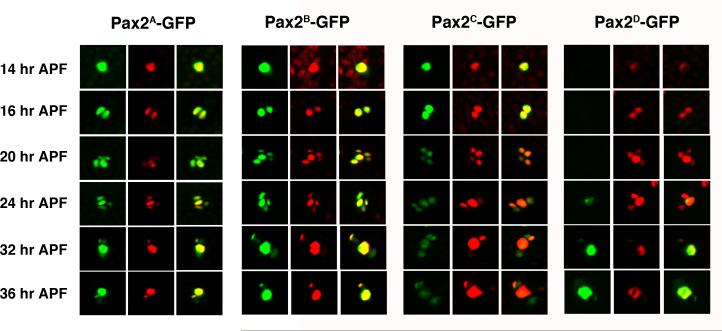
Previous work has exquisitely dissected the regulation of D-Pax2 expression in the eye (Fu et al., 1998; Flores et al., 2000; Swanson et al., 2010). In contrast, little is known about the mechanisms that regulate D-Pax2 in the es organ lineage. The information required to produce a complete D-Pax2 expression pattern in the es organs is likely present in a 6.7 KB upstream region including a small portion of the 5' UTR (Fu et al., 1998). To examine this enhancer region in more detail, we generated nuclear GFP reporter constructs bearing upstream fragments of 5.8 kb, 3.1 kb and 2.2 kb plus 100 bp of 5' UTR (Figure 1). These constructs were used to generate transgenic fly lines (Pax2A-GFP, Pax2B-GFP, Pax2C-GFP respectively) and GFP expression in the microchaete lineage in pupal nota was examined. All reporter constructs were generated using the GFP vector pH-Stinger (Barolo et al., 2000) which includes insulator elements that flank the multiple cloning site and GFP. These elements were originally introduced into the vector to minimize position effects on expression. We have observed no qualitative differences (and little to no quantitative differences) between lines generated from the same construct. Three independent lines were generated for each transgene construct described in this report. The Pax2A-GFP, Pax2B-GFP and Pax2C-GFP transgenes all showed expression in microchaete SOPs just prior to their division (Figure 2). GFP expression was also observable at the two cell stage and in all cells of the SOP lineage at 20 and 24 hours APF. The expression seen at each time point corresponded well to the actual presence of D-Pax2 protein in the cells, as detected by an anti-D-Pax2 antiserum. After the es organ lineage is established, the pattern of D-Pax2 expression changes. By 32 and 36 hours APF, D-Pax2 is restricted to the differentiating trichogen and thecogen cells. Both the 5.8 kb and 3.1 kb reporter trangenes exhibited strong GFP expression in these two cell types at this late time point and weak expression in the remaining two cells. In contrast, the 2.2 kb reporter transgene showed relatively weak expression in all es organ cells at this time.
Figure 1. List of D-Pax2 GFP reporter constructs.
Top diagram shows the upstream region of the D-Pax2 gene examined in this study. The name of each resulting construct generated is shown on the left. The corresponding position of the cloned DNA relative to the upstream region is marked by a straight line. Mutations of putative proneural protein binding sites are denoted by X's. The activity of each construct in both early (14-20 hours APF) and late (32-32 hours APF) es organ development is listed on the right.
Figure 2. GFP reporter expression driven by different es organ enhancer regions.
Pupal nota from Pax2A-GFP, Pax2B-GFP, Pax2C-GFP and Pax2D-GFP reporter lines were dissected at 14, 16, 20, 24, 32 and 36 hours APF and stained with an anti-D-Pax2 antiserum. For each line and time point, the left hand panel shows GFP (green), the center panel shows D-Pax2 (red) and right hand panel is the merged image. Each panel shows a single es organ position in the developing microchaete field.
Early and late D-Pax2 expression are dependent upon separate elements in the 3.1 kb enhancer region
The difference in the abilities of the 3.1 kb and 2.2 kb reporter transgenes to drive GFP expression in the trichogen and thecogen during the late stages of es organ development suggested that important late acting enhancer elements might exist in the 1 kb region directly upstream from the 2.2 kb fragment. Therefore, we created a reporter construct bearing only this region and used it to generate transgenic fly lines (Pax2D-GFP). These lines exhibited no GFP expression in the SOP or the es organ lineage cells during the early stages. At 24 hours APF, faint expression can be seen in a single cell, which is most likely the trichogen cell. However, by 36 hours APF, GFP expression in these transgenic lines was strong in both the trichogen and thecogen, the two D-Pax2+ cells (Figure 2). Therefore, the 3.1 kb upstream region provides all the information necessary to recapitulate D-Pax2 expression in the es organ lineage and this region can be subdivided into an approximately 2 kb proximal fragment, which drives strong expression early in the lineage (and weaker expression late), and a 1 kb distal fragment, responsible for strong expression late in the lineage, specifically in the differentiating trichogen and thecogen cells.
Control of early D-Pax2 expression by the proneural proteins
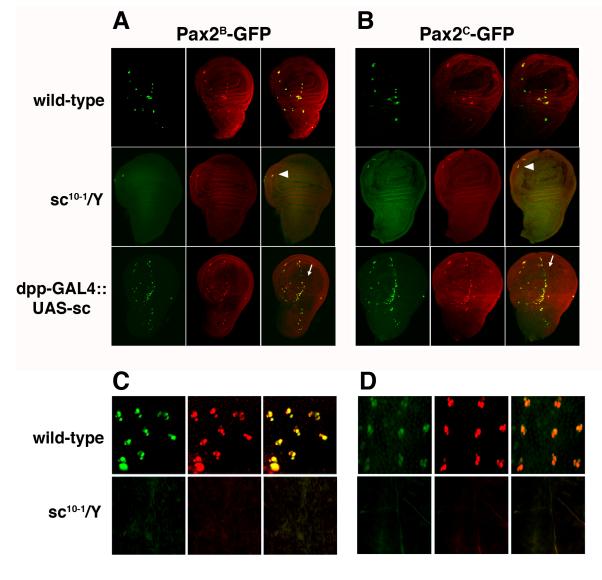
D-Pax2 expression follows the high level expression of the proneural genes in the SOP. The binding specificity of the proneural proteins has been well described (Singson et al., 1994; Powell et al., 2004; Powell et al., 2008) and a search of the 2.2 kb proximal enhancer region led to the identification of four canonical proneural binding sites, each containing the proneural E-box core CAGGTG (Table 1). Two of the E-boxes are upstream of the transcription start site at positions -1678 and -1593 while the other two reside just downstream in the 5' untranslated region of the first exon at positions +18 and +56. Although the exact position and number varies, the appearance of proneural E-boxes near the D-Pax2 gene is conserved among other Drosophila species (Table 2). To assess the importance of the proneural proteins to the activity of the D-Pax2 early enhancer, we first crossed both the 2.2 kb and 3.1 kb reporter transgenes into the sc10-1 background. The sc10-1 mutation removes the functions of both the achaete (ac) and scute (sc) proneural genes, which are required for the specification of the es organ SOP (Garcia-Bellido, 1979; Villares and Cabrera, 1987). Neither enhancer region was able to function in the absence of proneural gene function. In the wild-type background, D-Pax2 protein is detectable just before SOP division and can be seen along with GFP in the earliest developing SOPs in the late third instar wing imaginal discs from both Pax2B-GFP and Pax2C-GFP larvae. In contrast, wing discs from both sc10-1; Pax2B-GFP and sc10-1; Pax2C-GFP larvae showed neither D-Pax2 nor GFP expression where es organ SOPs are normally seen (Figure 3A), although the wing disc's chordotonal organ, which is specified by atonal (Jarman and Ahmed, 1998) and therefore unaffected in the sc10-1 mutant, shows both D-Pax2 and GFP expression (arrowheads). Additionally, no GFP expression was seen in the microchaete SOP lineage at pupal stages (Figure 3B). Secondly, we examined the ability of ectopic proneural gene expression to drive the reporter transgenes. We used a dpp-GAL4 driver (Staehling-Hampton et al., 1994) to ectopically express sc in the larval wing imaginal disc. This GAL4 line drives expression along the anterior/posterior boundary of the wing disc (Figure 3A, arrows). The misexpression of sc via dpp-GAL4 leads to the generation of supernumerary SOPs along the A/P boundary, as can be seen by the ectopic expression of D-Pax2 protein. Both the Pax2B-GFP and Pax2C-GFP reporter transgenes are active in these extra SOPs as well. The responsiveness of the transgenes to both loss and gain of proneural gene expression is consistent with a role for the proneurals in the regulation of D-Pax2 but does not address the question of whether proneural regulation of DPax2 is direct.
Table 1.
The position of proneural E-boxes in the D-Pax2 es organ enhancer.
| site | position | strand | wild-type | mutant |
|---|---|---|---|---|
| E-Box1 | -1678 | + | CAGGTG | CgtGcG |
| E-Box2 | -1593 | - | CAGGTG | CgtGcG |
| E-Box3 | +18 | - | CAGGTG | tAGtcG |
| E-Box4 | +56 | + | CAGGTG | CgtGcG |
Table 2.
Conservation of proneural E-boxes near D-Pax2 among Drosophila species.
| Species | CAGGTG E-Boxes position |
|---|---|
| D. melanogaster | -1678, -1593, +18, +56 |
| D. simulans | +18, +56 |
| D. sechellia | -2100,-1971,+18, +56 |
| D. yakuba | -1573,-1489,+18, +56 |
| D. erecta | -1638, -1554, +18, +56 |
| D. ananassae | -1984, -1857,-982 |
| D. pseudoobscura | -1621, -1129, -875, -683, -242, -30, +1305 |
| D. persimilis | -1621, -1129, -875, -683, -242, -30, +1306 |
Figure 3. Proneural gene control of es organ enhancers.
A. Wing imaginal discs from Pax2B-GFP, sc10-1/Y;Pax2B-GFP, and dpp-GAL4::UAS-sc,Pax2B-GFP lines. B. Wing imaginal discs from Pax2C-GFP, sc10-1/Y;Pax2C-GFP (E), and dpp-GAL4::UAS-sc;Pax2C-GFP lines. C. 24 hour APF pupal nota from Pax2B-GFP and sc10-1/Y;Pax2B-GFP lines. D. 24 hour APF pupal nota from Pax2C-GFP and sc10-1/Y;Pax2C-GFP lines. Discs and nota were stained with an anti-D-Pax2 antiserum. For each disc or notum, the left hand panel shows GFP (green), the center panel shows D-Pax2 (red) and right hand panel is the merged image. Arrowheads in the merged images in A and B point to remaining chordotonal organ in sc10-1 mutants. Arrows in the merged images in A and B indicate supernumerary SOPs forming along anterior/posterior boundary in the discs from larvae with ectopic sc expression.
The proneural E-boxes are required for early D-Pax2 enhancer activity
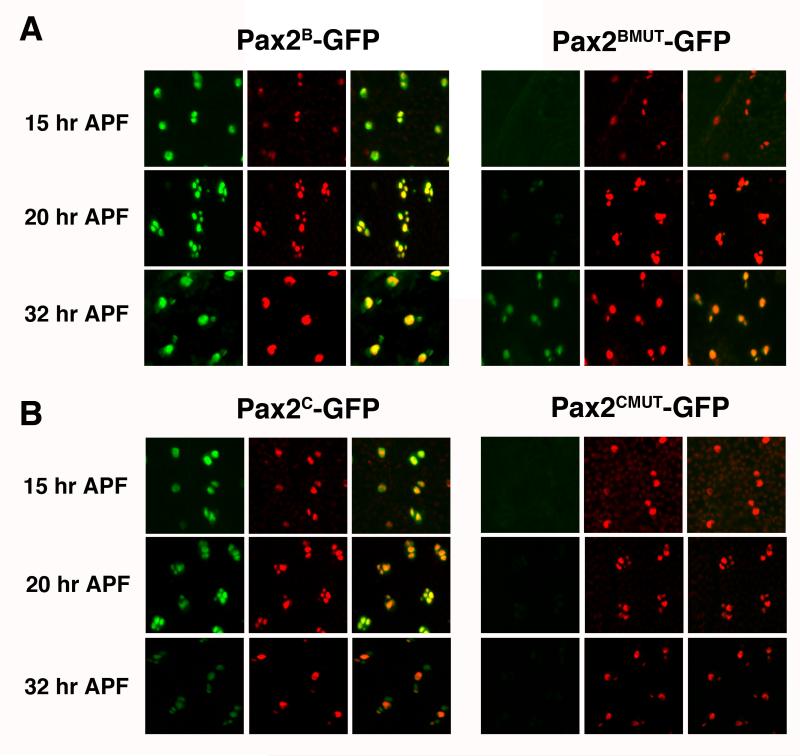
If proneural proteins are directly regulating D-Pax2, it is likely that they operate through any or all of the four identified proneural E-boxes present in the 2.2 kb enhancer region. To test the importance of the E-boxes to this enhancer region, we introduced mutations into all four E-boxes in both the Pax2B-GFP and Pax2C-GFP reporter constructs (Figure 1, Table 1). These mutated constructs were used to generate transgenic lines (Pax2BMUT-GFP, Pax2CMUT-GFP, respectively). In both cases, expression of the GFP reporter was reduced during the early stages of es organ development (Figure 4). Early expression of GFP in the SOP lineage was greatly diminished in the both the Pax2BMUT-GFP and Pax2CMUT-GFP lines. The Pax2BMUT-GFP lines did show faint GFP expression in one or two cells around 20 hours APF. Additionally, the proneural site mutations in the Pax2BMUT-GFP lines did not strongly affect GFP expression during the late stages of es organ development as GFP was clearly evident in the differentiating trichogen and thecogen cells at 32 hours APF.
Figure 4. Early and full es organ enhancers require E-boxes for early but not late reporter expression.
Pupal nota from Pax2B-GFP and Pax2BMUT-GFP (A) and Pax2C-GFP and Pax2CMUT-GFP (B) lines were dissected at 15 hours APF, 20 hours APF and 32 hours APF. Nota were stained with an anti-D-Pax2 antiserum. For each line and time point, the left hand panel shows GFP (green), the center panel shows D-Pax2 (red) and right hand panel is the merged image. Each panel shows a region of the developing microchaete field containing about 5-8 es organs.
Late D-Pax2 enhancer activity is dependent upon D-Pax2 function
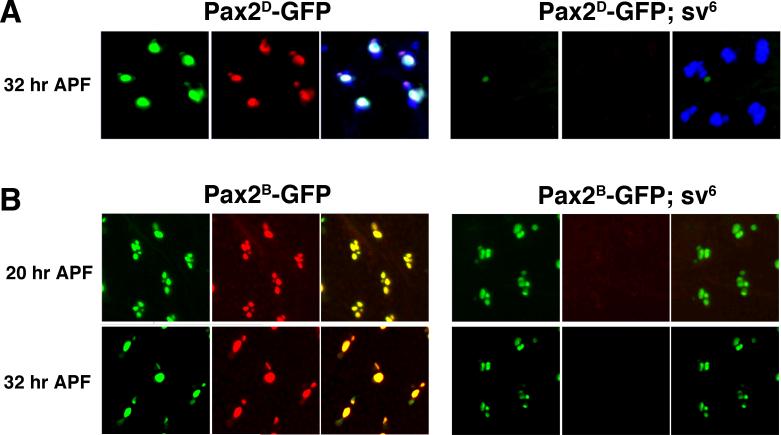
Previous work on vertebrate Pax2 genes have suggested or shown that expression can be maintained via an autoregulatory loop (Pfeffer et al., 2002). To examine the role of D-Pax2 on the late enhancer specifically, we introduced Pax2D-GFP reporter transgene into a strong D-Pax2 mutant (sv6). The sv6 mutant exhibits strongly reduced levels of D-Pax2 protein during early es organ development and D-Pax2 is virtually undetectable during the late stages of differentiation. The lack of D-Pax2 in sv6 flies does not lead to es organ cell death nor does it alter the cell fate decisions in the microchaete lineage. However, the sv6 mutants do exhibit differentiative defects in the trichogen and thecogen cells (Kavaler et al., 1999). Although GFP expression is clearly visible in the trichogen and thecogen cells in Pax2D-GFP nota, this expression is lost when the Pax2D-GFP transgene is placed in a sv6 background (Figure 5A). The progeny of the SOP are still present, as can be demonstrated via their expression of Cut protein (Blochlinger et al., 1993). This result suggested that the high levels of late D-Pax2 expression in the trichogen and thecogen cells might result from a positive feedback loop and that the 1 kb upstream enhancer is sensitive to D-Pax2 protein levels. We also introduced the Pax2B-GFP reporter transgene into the sv6 background and tested the ability of this reporter to function in the absence of D-Pax2 itself (Figure 5B). When sv6 pupal nota were examined at 20 hours APF, GFP expression could be seen in all four es organ cells at levels similar to those seen in the wild-type background, suggesting that early expression does not require a positive feedback loop. Surprisingly, 32 hour APF sv6 nota also exhibited GFP expression in four cells in contrast to wild-type nota, in which GFP expression was restricted to the trichogen and thecogen cells. Also, the sizes of the four nuclei in the sv6 nota were similar and relatively small. In wild-type es organ development, the trichogen and tormogen cells at this stage have undergone several rounds of endoreplication and can be distinguished by the large size of their nuclei.
Figure 5. D-Pax2 helps maintain its late expression in the trichogen and thecogen cells.
A. Nota from wild-type and sv6 pupae bearing the Pax2D-GFP reporter dissected at 32 hours APF. Nota were stained with an anti-D-Pax2 antiserum and an anti-Cut monoclonal antibody to identify all es organ cells. In each case, the left hand panel shows GFP (green), the center panel shows D-Pax2 (red) and right hand panel is the merged image including Cut (blue). B. Nota from wild-type and sv6 pupae bearing the Pax2B-GFP reporter dissected at 20 hours APF and 32 hours APF. Nota were stained with an anti-D-Pax2 antiserum. The left hand panels show GFP (green), center panels show D-Pax2 (red) and right hand panels are merged images. All panels show a developing microchaete field with approximately 5 es organs.
DISCUSSION
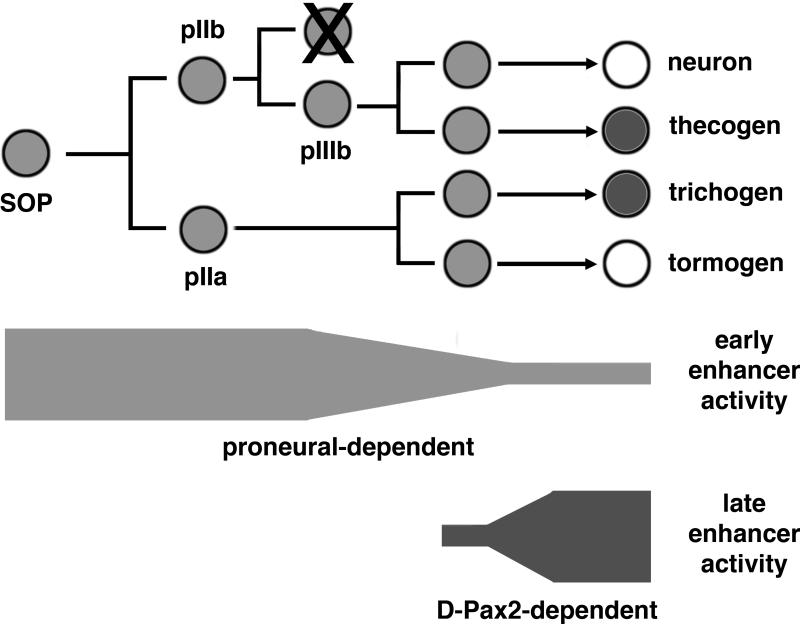
The development of adult es organs in Drosophila relies on the correct specification of cell types among the SOP progeny cells followed by their differentiation . Specification is controlled by Notch signaling through the divisions, which ultimately results in the expression of unique combinations of differentiation factors in each cell type (Lai and Orgogozo, 2004). The mechanisms by which these differentiation factors are regulated are not well understood. The D-Pax2 gene encodes a critical differentiation factor for proper es development. Its expression pattern is dynamic and complex and can be divided into an early stage, during which it is expressed in the SOP and all its progeny cells, and a late stage, during which it is restricted to the trichogen and thecogen cells (Kavaler et al., 1999). The function of the D-Pax2 transcription factor during early es organ development has not been established, but it is most likely involved as an antagonist of Notch signaling. Loss of function D-Pax2 mutants show occasional cell fate transformations leading to double socket phenotypes (Lees and Waddington, 1942; Kavaler et al., 1999) and loss of one functional copy of D-Pax2 greatly enhances the dominant double socket phenotype of Hairless mutations (J.K. unpublished observations). D-Pax2 functions during late es organ development to promote the differentiation of the trichogen and thecogen cells. In this paper, we identify two key enhancer regions that control early and late D-Pax2 expression and demonstrate that proneural proteins are in part responsible for driving early D-Pax2 expression whereas late D-Pax2 expression relies upon a positive feedback loop (Figure 6).
Figure 6. A working model for the transcriptional regulation of D-Pax2 in es organs.
The es organ lineage is shown from left to right starting with the SOP and ending with the differentiation of the four cell types. The light shading of the cells represents the early expression of D-Pax2 in all cells of the lineage and the dark shading represents the late expression in the trichogen and thecogen cells. Below the lineage are bars representing the relative activity of early and late enhancers. The activity of the early enhancer is dependent upon proneural proteins is responsible for the intial expression in the lineage but decreases in strength by the time the es organ cells are differentiating. The activity of the late enhancer is dependent upon D-Pax2 itself, starts soon after the divisions and increases with strength as the es organ cells differentiate.
Rescue experiments by (Fu et al., 1998) demonstrated that the es organ enhancer for D-Pax2 expression was located upstream from the D-Pax2 transcription start site. A region encompassing a small amount of leader sequence and approximately 6.7 kb of upstream sequence driving the D-Pax2 gene was capable of providing a complete rescue of two D-Pax2 bristle mutants, svn and svde. These experiments demonstrated a functional requirement for a large regulatory region but did not give a precise boundary for regulatory elements and did not provide information on the spatial and temporal control of D-Pax2 expression. To examine the relationship between this regulatory region and the expression of D-Pax2 during es organ development, we decided to dissect the upstream region using GFP reporter constructs. Our initial reporter experiments showed that a slightly smaller 5.8 kb region of DNA (Pax2A-GFP) was able to drive GFP expression in a complete D-Pax2 pattern, including all of the cells of the SOP lineage early and the trichogen and thecogen cells late. This large segment of DNA spans not only the region upstream of D-Pax2 but encroaches upon a neighboring gene, activin-β, which is oriented in the opposite direction. When we shortened our initial D-Pax2 es organ enhancer down to 3.1 kb (Pax2B-GFP), we obtained similar results, suggesting that all the information required to drive all aspects of D-Pax2 expression in the es organ is located within approximately 3 kb upstream of the transcription start site.
When we further shortened the 3.1 kb enhancer down to a 2.2 kb region (Pax2C-GFP), the intense late expression of GFP in the differentiating trichogen and thecogen cells was lost. For this reporter construct, early expression of GFP in the SOP and its progeny during the cell divisions remained and weak expression was visible in all four cells at least as late as 36 hours APF. A similar 2.1 kb enhancer region driving D-Pax2 expression in sv mutants has been shown to provide only partial rescue, compared to the greater efficacy of the 6.7 kb region discussed above (Fu et al., 1998). The failure of this enhancer to effect a complete rescue may therefore result from its inability to maintain a high level of D-Pax2 expression in the trichogen and thecogen cells during their differentiation. In contrast, a 1 kb fragment (Pax2D-GFP) representing the remaining part of the 3.1 kb enhancer was unable to drive GFP expression in the SOP and its progeny during the divisions. However, by 32 hours APF, GFP was evident in two cells, one large and one small, that showed coincident expression of D-Pax2 protein and so can be identified as the trichogen and thecogen cells. The activities of the 2.2 kb early enhancer and the 1 kb late enhancer are to some extent complementary and these results suggest that the initial expression of D-Pax2 in the lineage is controlled in a separate manner from its later expression during the differentiation of the cells of the es organ. The 2.2 kb early enhancer does generate a clearly visible GFP signal in the four es cells as late as 36 hours APF and the 3.1 kb complete enhancer also shows comparable expression in the tormogen cell and neuron at this time point. Conceivably, the early enhancer region retains some ability to activate D-Pax2 expression at later time points and there may be an unidentified repressor element required to completely extinguish D-Pax2 expression in the tormogen and neuron. Alternatively, the perdurance of GFP protein masks a sharper delineation of the transcriptional regulation.
There are several transcription factors expressed in the SOP that might potentially regulate the early expression of D-Pax2. Of special note are the proneural genes of the ac-sc complex. Of the four members of the complex, three are involved adult es organ development - ac, sc and ase (Bertrand et al., 2002). Both ac and sc are expressed in PNC and in the SOP and define the SOP fate. Loss of both ac and sc leads to virtually complete loss of SOPs and therefore es organs from the surface of the adult fly. (Garcia-Bellido, 1979; Cubas et al., 1991; Skeath and Carroll, 1991). The third member, ase, is not expressed in the PNC and whereas ac and sc are found exclusively in the SOP , ase is expressed in other members of the lineage and is regulated directly by ac and sc (Jarman et al., 1993). However, the functions of all three genes show some redundancy and ectopic expression of each leads to the appearance of supernumerary es organs (Rodriguez et al., 1990). Mutants in ase, however, show no obvious defects in notum microchaete development, although es organs in other regions do exhibit phenotypes suggestive of lineage defects (Rodriguez et al., 1990; Dominguez and Campuzano, 1993). The function of the D-Pax2 early enhancer requires ac and sc, as pupal nota from sc10-1 flies bearing the early enhancer reporter showed no GFP expression. Furthermore, ectopic expression of sc also leads to ectopic activation of the early enhancer reporter. Given the requirement for the proneural proteins for the establishment and maintenance of the SOP fate, these results are not surprising. The proneural proteins are therefore required for D-Pax2 expression but the question of whether proneural proteins directly regulate D-Pax2 is not addressed by this experiment.
If one or more proneural proteins regulates D-Pax2 directly, one would expect to find proneural binding sites located in the D-Pax2 enhancer. All of the proneural proteins are basic helix-loop-helix transcription factors and recognize a core E box sequence of CAGG/CTG. Upon examination of the 2.2 kb early enhancer, we identified four CAGGTG E box sequences. Two appear approximately 1.6 kb upstream of the transcription start site and the other two are located just 3' of the transcription start site in the 5' UTR. The presence of these sites is conserved in all the Drosophila strains for which we could identify the D-Pax2 ortholog, although the number and position of the sites varies. To address the function of the Drosophila melanogaster sites, all four E boxes were mutated in both the 3.1 KB full and 2.2 KB early enhancers. Neither mutated construct elicited GFP expression during the early stages of D-Pax2 expression. Therefore, the four proneural E boxes are necessary for the function of the early enhancer. Because the early enhancer does not drive GFP expression in the absence of the proneural proteins Ac and Sc and because it does not function when the proneural binding sites are mutated, we conclude that one or more of the proneural proteins are involved in the direct regulation of D-Pax2 in the bristle lineage. Mutation of the proneural binding sites in the 3.1 KB full enhancer did not disrupt its ability to drive late expression of GFP. Faint expression of GFP was seen at 20 hours APF and strong expression in the trichogen and tormogen cells was observable by 36 hours APF. This result indicates the involvement of the late enhancer in the maintenance of D-Pax2 expression in these two cells is independent of earlier proneural protein function.
We have not determined which proneural proteins are directly responsible for D-Pax2 expression. Ac and Sc are expressed only in the SOP and so D-Pax2 expression in the cells of the lineage after SOP division is unlikely to be driven by them. Ase can be found in the lineage but loss of Ase function does not lead to any notum microchaete defects and even mild sv mutants show misshapen shafts (Fu et al., 1998; Kavaler et al., 1999). Furthermore, the early enhancer provides weak expression of GFP at late time points, well after all the known proneurals are expressed. Conceivably, the proneural proteins are required to initiate D-Pax2 expression in the SOP and this initial event is required for continued expression which may be controlled by other unidentified factors.
The late enhancer is sufficient to drive gene expression in the differentiating trichogen and thecogen cells well after the fates of the progeny of the SOP have been specified. Interestingly, the late enhancer is dependent upon D-Pax2 protein itself. In a strong sv mutant background, GFP expression driven by the late enhancer alone disappears, indicating that the maintenance of D-Pax2 expression in the trichogen and thecogen cells is governed by a positive feedback loop. The utilization of a positive feedback loop to stabilize gene expression in particular cell types is common. Indeed, the activation and maintenance of Pax2 expression along the midbrainhindbrain boundary in mice has been shown to be controlled by separate enhancers and the maintenance enhancer is regulated directly by Pax2 itself (Pfeffer et al., 2002). In this case, we have no evidence that the regulation is direct; there is no full canonical D-Pax2 binding site (Dziedzic et al., 2009) in the 1 KB enhancer region. Possibly, sites that do not exactly match the full binding site sequence can function in this enhancer. Alternatively, D-Pax2 protein may control other transcription factors that feed back to keep D-Pax2 upregulated. A positive feedback loop appears to play little if any role in the early expression in the lineage. The 3.1 KB reporter was unaffected by loss of D-Pax2 function at 20 hours APF. Surprisingly, the 3.1 KB reporter also exhibits expression at 32 hours APF in four cells. We note that this reporter does show weak expression in the tormogen and neuron normally at this time point. The loss of DPax2 prevents the differentiation of the trichogen and thecogen cells and those cells could conceivably be arrested in an “early” state and more responsive to the early enhancer elements. Alternatively, the complete 3.1 KB enhancer may operate in a qualitatively different manner than the separated early and late enhancers do.
The work presented here demonstrates separable regulatory regions responsible for the initiation of D-Pax2 expression and its maintenance during the differentiation of the trichogen and thecogen cells. We also identify a partial complement of the factors that control this early and late expression. Factors aside from the proneural proteins are almost certainly involved in the early expression and still need to be uncovered. The maintenance of late expression via a positive feedback loop implicates D-Pax2 in its own regulation but the mechanism by which it does so remains unknown.
EXPERIMENTAL PROCEDURES
Drosophila stocks and crosses
Flies were cultured on standard yeast-cornmeal-molasses-agar medium at 25°C in a humidified chamber. Six new transgenic GFP reporter lines were generated: w;Pax2A-GFP, w;Pax2B-GFP, w;Pax2BMUT2-GFP, w;Pax2C-GFP, w;Pax2CMUT2-GFP, w;Pax2D-GFP. Transgenic fly strains were generated by embryo injections of plasmid constructs along with the helper plasmid pUChsΔ2-3 according to standard methods (Cripps and Bernstein, 2000). w;Pax2B-GFP and w;Pax2C-GFP transgenes were crossed to sc10-1 w/FM4 (Villares and Cabrera, 1987) and w;UAS-sc (Parras et al., 1996) strains, producing the following lines: sc10-1 w/FM4;Pax2B-GFP, sc10-1 w/FM4;Pax2B-GFP, w;UAS-sc,Pax2B-GFP, w;UAS-sc;Pax2C-GFP. w;Pax2C-GFP and w;Pax2D-GFP transgenes were crossed to w;sv6/Act-GFP to produce the following lines: w;Pax2C-GFP; sv6/Act-GFP and w;Pax2D-GFP; sv6/Act-GFP. The sv6 allele was generated previously (Kavaler et al., 1999) and was placed over a GFP marked fourth chromosome (Flybase ID FBti0076463) to allow identification of mutant larvae and pupae.
Plasmid constructs
Pax2-GFP constructs
Four regions primarily upstream of the D-Pax2 gene were amplified by PCR from w1118 genomic DNA using Phusion DNA polymerase (New England Biolabs), cloned into the BamHI site of pBluescript KS+ (Stratagene) and subcloned into pH-Stinger (Barolo et al., 2004). Three of the amplicons were generated with the same 3' end primer situated in the gene's 5' untranslated region, 5'-CTGAGTCAAGGATCCTAAGCC (introduced Bam HI site at +101). The 5' primers and the positions of the BamHI cloning site relative to the D-Pax2 transcription start site are as follows.
Pax2A-GFP: 5'- GCATTCAGGATCCAGCTATCTATG (-5712)
Pax2B-GFP: 5'- AAGCCAGGATCCACGATTGAAAT (-2109)
Pax2C-GFP: 5'- ACACACGGATCCACGGACATAC (-3027)
The fourth amplicon spanned the region from -3027 to -2028
Pax2D-GFP: 5' primer - 5'- ACACACGGATCCACGGACATAC
3' primer - 5'- TTTGTAGGATCCGCCTGGTAGC
Pax2MUT-GFP constructs
The four proneural binding sites were mutated in the 2.2 and 3.1 KB pBluescript KS+ via the Phusion site-directed in vitro mutagenesis kit (New England Biolabs) to introduce the changes shown in Table 1. The mutated enhancers were then subcloned into pH-Stinger to generate Pax2BMUT2-GFP and Pax2CMUT2-GFP.
All plasmid construct inserts were confirmed by DNA sequence analysis.
Immunohistochemistry
Imaginal discs from late third instar larvae or notum tissue from pupae were isolated by dissection in PBS, fixed for 30 minutes in 4% paraformaldehyde, and washed extensively in PBS with 0.1% Triton-X 100. The tissue was then incubated with the primary antibody for 3 hours at room temperature or overnight at 4°C, washed, and incubated with the appropriate secondary reagent in the same manner. Samples were then washed, mounted on slides in Biomedia Gel/Mount (Electron Microscopy Sciences) and images were take using an Axiovision D1 fluorescence microscope (Zeiss). The primary anti-D-Pax2 rabbit antiserum, generated and affinity purified for us by SDIX, was used at a 1:5000 dilution. The secondary goat anti rabbit Ig-Alexafluor 568 (Invitrogen), was used at a 1:1000 dilution. All antibody dilutions were made with PBS with 0.1% Triton-X 100 and 2% bovine serum albumin.
Comparative sequence analysis of D-Pax2 enhancer
D-Pax2 loci from other Drosophila species were identified using Flybase (Crosby et al., 2007) and UCSC Genome Browser (Kent et al., 2002) BLAST search functions with the D-Pax2 coding region or protein sequence serving as the query sequence. Sequence spans of 3 KB in each direction from the hypothetical transcription start site of each ortholog were examined for the proneural E-box sequence CAGGTG.
ACKNOWLEDGMENTS
We are grateful to Emily Hilton and Emily Bradford for critical reading of this manuscript. This project was supported by Award Number R15-HD04369-01A1 from the National Institutes of Health. The project and S. J., K. H., and F. S were supported by Award Number P20-RR016463 from the National Center for Research Resources. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health.
Funded by:
National Institutes of Health: R15-HD04369-01A1, P20 RR-016463
REFERENCES
- Barolo S, Carver LA, Posakony JW. GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques. 2000;29:726, 728, 730, 732. doi: 10.2144/00294bm10. [DOI] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Blochlinger K, Jan LY, Jan YN. Postembryonic patterns of expression of cut, a locus regulating sensory organ identity in Drosophila. Development. 1993;117:441–450. doi: 10.1242/dev.117.2.441. [DOI] [PubMed] [Google Scholar]
- Cripps RM, Bernstein SI. Generation of transgenic Drosophila melanogaster by P element-mediated germline transformation. In: Norton PA, Steel LF, editors. Gene Transfer Methods. Eaton Publishing; Natick, MA: 2000. pp. 93–125. [Google Scholar]
- Crosby MA, Goodman JL, Strelets VB, Zhang P, Gelbart WM. FlyBase: genomes by the dozen. Nucleic Acids Res. 2007;35:D486–491. doi: 10.1093/nar/gkl827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P, de Celis JF, Campuzano S, Modolell J. Proneural clusters of achaete-scute expression and the generation of sensory organs in the Drosophila imaginal wing disc. Genes Dev. 1991;5:996–1008. doi: 10.1101/gad.5.6.996. [DOI] [PubMed] [Google Scholar]
- Dominguez M, Campuzano S. asense, a member of the Drosophila achaete-scute complex, is a proneural and neural differentiation gene. Embo J. 1993;12:2049–2060. doi: 10.1002/j.1460-2075.1993.tb05854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziedzic K, Heaphy J, Prescott H, Kavaler J. The transcription factor D-Pax2 regulates crystallin production during eye development in Drosophila melanogaster. Dev Dyn. 2009;238:2530–2539. doi: 10.1002/dvdy.22082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores GV, Duan H, Yan H, Nagaraj R, Fu W, Zou Y, Noll M, Banerjee U. Combinatorial signaling in the specification of unique cell fates. Cell. 2000;103:75–85. doi: 10.1016/s0092-8674(00)00106-9. [DOI] [PubMed] [Google Scholar]
- Fu W, Duan H, Frei E, Noll M. shaven and sparkling are mutations in separate enhancers of the Drosophila Pax2 homolog. Development. 1998;125:2943–2950. doi: 10.1242/dev.125.15.2943. [DOI] [PubMed] [Google Scholar]
- Fu W, Noll M. The Pax2 homolog sparkling is required for development of cone and pigment cells in the Drosophila eye. Genes Dev. 1997;11:2066–2078. doi: 10.1101/gad.11.16.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bellido A. Genetic Analysis of the Achaete-Scute System of DROSOPHILA MELANOGASTER. Genetics. 1979;91:491–520. doi: 10.1093/genetics/91.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gho M, Bellaiche Y, Schweisguth F. Revisiting the Drosophila microchaete lineage: a novel intrinsically asymmetric cell division generates a glial cell. Development. 1999;126:3573–3584. doi: 10.1242/dev.126.16.3573. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Posakony JW. Development of adult sensilla on the wing and notum of Drosophila melanogaster. Development. 1989;107:389–405. doi: 10.1242/dev.107.2.389. [DOI] [PubMed] [Google Scholar]
- Heitzler P, Simpson P. The choice of cell fate in the epidermis of Drosophila. Cell. 1991;64:1083–1092. doi: 10.1016/0092-8674(91)90263-x. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Ahmed I. The specificity of proneural genes in determining Drosophila sense organ identity. Mech Dev. 1998;76:117–125. doi: 10.1016/s0925-4773(98)00116-6. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Brand M, Jan LY, Jan YN. The regulation and function of the helix-loop-helix gene, asense, in Drosophila neural precursors. Development. 1993;119:19–29. doi: 10.1242/dev.119.Supplement.19. [DOI] [PubMed] [Google Scholar]
- Kavaler J, Fu W, Duan H, Noll M, Posakony JW. An essential role for the Drosophila Pax2 homolog in the differentiation of adult sensory organs. Development. 1999;126:2261–2272. doi: 10.1242/dev.126.10.2261. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- Lai EC, Orgogozo V. A hidden program in Drosophila peripheral neurogenesis revealed: fundamental principles underlying sensory organ diversity. Dev Biol. 2004;269:1–17. doi: 10.1016/j.ydbio.2004.01.032. [DOI] [PubMed] [Google Scholar]
- Lees AD, Waddington CH. The development of the bristles in normal and some mutant types of Drosophila melanogaster. Proc. R. Soc. B. 1942;131:87–110. [Google Scholar]
- Parras C, Garcia-Alonso LA, Rodriguez I, Jimenez F. Control of neural precursor specification by proneural proteins in the CNS of Drosophila. Embo J. 1996;15:6394–6399. [PMC free article] [PubMed] [Google Scholar]
- Pfeffer PL, Payer B, Reim G, di Magliano MP, Busslinger M. The activation and maintenance of Pax2 expression at the mid-hindbrain boundary is controlled by separate enhancers. Development. 2002;129:307–318. doi: 10.1242/dev.129.2.307. [DOI] [PubMed] [Google Scholar]
- Posakony JW. Nature versus nurture: asymmetric cell divisions in Drosophila bristle development. Cell. 1994;76:415–418. doi: 10.1016/0092-8674(94)90105-8. [DOI] [PubMed] [Google Scholar]
- Powell LM, Deaton AM, Wear MA, Jarman AP. Specificity of Atonal and Scute bHLH factors: analysis of cognate E box binding sites and the influence of Senseless. Genes Cells. 2008;13:915–929. doi: 10.1111/j.1365-2443.2008.01217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell LM, Zur Lage PI, Prentice DR, Senthinathan B, Jarman AP. The proneural proteins Atonal and Scute regulate neural target genes through different E-box binding sites. Mol Cell Biol. 2004;24:9517–9526. doi: 10.1128/MCB.24.21.9517-9526.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy GV, Rodrigues V. A glial cell arises from an additional division within the mechanosensory lineage during development of the microchaete on the Drosophila notum. Development. 1999;126:4617–4622. doi: 10.1242/dev.126.20.4617. [DOI] [PubMed] [Google Scholar]
- Rodriguez I, Hernandez R, Modolell J, Ruiz-Gomez M. Competence to develop sensory organs is temporally and spatially regulated in Drosophila epidermal primordia. Embo J. 1990;9:3583–3592. doi: 10.1002/j.1460-2075.1990.tb07569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singson A, Leviten MW, Bang AG, Hua XH, Posakony JW. Direct downstream targets of proneural activators in the imaginal disc include genes involved in lateral inhibitory signaling. Genes Dev. 1994;8:2058–2071. doi: 10.1101/gad.8.17.2058. [DOI] [PubMed] [Google Scholar]
- Skeath JB, Carroll SB. Regulation of achaete-scute gene expression and sensory organ pattern formation in the Drosophila wing. Genes Dev. 1991;5:984–995. doi: 10.1101/gad.5.6.984. [DOI] [PubMed] [Google Scholar]
- Staehling-Hampton K, Jackson PD, Clark MJ, Brand AH, Hoffmann FM. Specificity of bone morphogenetic protein-related factors: cell fate and gene expression changes in Drosophila embryos induced by decapentaplegic but not 60A. Cell Growth Differ. 1994;5:585–593. [PubMed] [Google Scholar]
- Swanson CI, Evans NC, Barolo S. Structural rules and complex regulatory circuitry constrain expression of a Notch- and EGFR-regulated eye enhancer. Dev Cell. 2010;18:359–370. doi: 10.1016/j.devcel.2009.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villares R, Cabrera CV. The achaete-scute gene complex of D. melanogaster: conserved domains in a subset of genes required for neurogenesis and their homology to myc. Cell. 1987;50:415–424. doi: 10.1016/0092-8674(87)90495-8. [DOI] [PubMed] [Google Scholar]