Abstract
The pathogenesis of type-1 diabetes is complicated, and a clear, single mechanism has yet to be identified. Reports have indicated that the activating receptor NKG2D plays an important role in the development of disease. Exploiting a natural phenomenon observed in tumors, plasmid DNA encoding for a soluble ligand to NKG2D (sRAE-1γ) was isolated and engineered into a plasmid expression system. A polymeric gene delivery system was developed to deliver the soluble RAE-1 plasmid locally to the pancreatic islets for the prevention of type-1 diabetes. The bioreducible cationic polymer poly(cystamine bisacrylamide – diamino hexane) (p(CBA-DAH)) was modified with poly(ethylene glycol) (PEG) and the targeting peptide CHVLWSTRC, known to target the EphA2 and EphA4 receptors. The PEG serves to improve stability and tissue selectivity, while the peptide will target EphA2 and A4, overexpressed in the pancreatic microvasculature. The targeting polymer Eph-PEG-p(CBA-DAH) shows selective uptake by the target cell line, indicative of the targeting properties that will be seen in systemic administration. Using the delivery system, the therapeutic plasmid can be delivered to the pancreas, reduce interactions between the beta-cells and infiltrating NKG2D positive lymphocytes, and effectively protect beta-cells from autoimmune destruction and prevent type 1 diabetes.
Keywords: Copolymer, Gene Therapy, Drug Delivery, Biodegradation
Introduction
Type 1 diabetes is an autoimmune disease in which the beta-cells in the pancreatic islets are destroyed by autoreactive immune cells [1, 2]. Specifically, CD4+ and CD8+ T-cells are considered essential in the development of type 1 diabetes [3–5]. Additionally, the interaction of co-stimulatory receptors on T-cells and their interaction with the ligands on beta-cells is thought to be critical for the autoimmune response. The activating receptor NKG2D is thought to be directly involved in the progression of type-1 diabetes [6–9]. Retinoic acid early inducible gene-1 (RAE-1) is a tightly regulated high affinity ligand for the NKG2D receptor in mice [10, 11]. The expression of RAE-1 on pancreatic islet beta-cells indicates a close relationship between this receptor and the pathogenesis of type-1 diabetes in the non obese diabetic (NOD) mouse model [12].
It has been shown that the systemic blockage of the NKG2D receptor with monoclonal antibodies can prevent the occurrence of type-1 diabetes in the NOD mouse [13]. However, systemic blockage of the NKG2D receptor is not clinically applicable, as NKG2D has important functions in immunosurveillence, including detection of cancerous and virally infected cells [14–17]. Similarly, by blocking the NKG2D receptor specifically around the pancreas, the beta cells can be protected from cell-mediated cytotoxicity. Specifically, delivering a gene coding for soluble RAE-1 locally to the pancreas will result in the binding and internalization of the NKG2D receptor of infiltrating lymphocytes. This will allow the pancreas to undergo “immune evasion,” and will halt the progression of type-1 diabetes.
The main purpose for designing a targeting gene carrier is to deliver therapeutic genes specifically to a target tissue or target cells, thereby minimizing side effects associated with systemic therapeutics [18]. By using a bioreducible polymer, pDNA will be released upon cell entry due to the difference in reduction potential [19, 20]. Additionally, the degradation of the polymer will reduce cytotoxicity and allow for clearance in vivo [21]. The targeting gene carrier consists of three basic components: a bioreducible cationic backbone, a PEG linker, and a targeting peptide. The cationic polymer is essential in complexing with DNA to form polyplex, providing protection from endonucleases, and supplying a mechanism for endonuclease escape [22]. By using a bioreducible polymer, pDNA will be released upon cell entry due to the difference in reduction potential [19, 20]. Additionally, the degradation of the polymer will reduce cytotoxicity and allow for clearance in vivo [21]. The addition of a PEG linker will not only act to connect the targeting ligand to the backbone polymer, but will also serve to stabilize polyplex, shield surface charge, and increase circulation time in vivo [23]. The targeting peptide NH2-GGGCHVLWSTRC (Eph) will be added to the distal end of the PEG chain. This peptide has been shown to target the receptors EphA2 and A4, which are overexpressed in the pancreatic islet microvasculature [24, 25].
Here, we report the design of an effective polymeric gene delivery system to deliver a plasmid coding for soluble RAE-1γ to the pancreatic islet microvasculature for the prevention of type-1 diabetes.
Materials and Methods
Materials
N-Boc-1,6-diaminohexane, N-Boc-DAH) was purchased from Alfa Aesar (Ward Hill, MA). Hyperbranched polyethlenimine (bPEI, Mw 25 kDa), trifluroacetic acid (TFA), triisobutylsilane (TIS), tithiothreitol (DTT), and 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium bromide (MTT) were purchased from Sigma-Aldrich (St. Louis, MO). N,N′-cystaminebisacrylamide (CBA) was purchased from PolySciences, Inc. (Warrington, PA). Heterobifunctional N-hydroxysuccinimide-poly(ethylene glycol)-maleimide (NHS-PEG-Mal, Mw 2000) was purchased from JenKem Technology (Allen, TX). The targeting peptide Eph (NH2-GGGCHVLWSTRC) was synthesized at the University of Utah Core Peptide Synthesis facility. SYBR Green Nucleic acid stain was purchased from Invitrogen (Carlsbad, CA). Luciferase assay system with reporter lysis buffer was purchased from Promega (Madison, WI). DMEM, phosphate buffered saline, penicillin/streptomycin, Trypsin-like enzyme (TrypLE), Trypan Blue, YOYO-1 nucleic acid dye, and AlexaFluor 647 carboxylic acid, succinimidyl ester were purchased from Invitrogen (Carlsbad, CA). Fetal bovine serum (FBS) was purchased from HyClone (Logan, UT). Luciferase Assay System and Reporter Lysis Buffer were purchased from Promega (Madison, WI). Superscript III First Strand Synthesis kit, Platinum Taq polymerase, pcDNA3.1/V5-His TOPO TA vector, Purelink PCR purification kit, NuPage Bis-Tris gels, nitrocellulose membranes, and anti-V5-AP antibody were purchased from Invitrogen (Carlsbad, CA). Alkaline Phosphatase development kit was purchased from BioRad (Hercules, CA). PCR primers were purchased from Integrated DNA Technologies (IDT, San Diego, CA). RNeasy Kit and Maxi-prep plasmid purification kit were purchased from Qiagen (Valencia, CA). KpnI and NotI restriction enzymes were obtained from New England Biolabs (Ipswich, MA). All materials and solvents were used as received without further purification.
Synthesis of p(CBA-DAH)
The polymer poly(CBA-DAH) was synthesized as previously reported [26].
Conjugation of NHS-PEG-Mal to p(CBA-DAH)
The conjugation of NHS-PEG-Mal to p(CBA-DAH) was adapted from manufacturer’s protocols. The NHS ester in heterobifunctional NHS-PEG-Mal was reacted with the primary amine in p(CBA-DAH. NHS-PEG-Mal (PEG Mw 2000) was dissolved in PBS (pH 7.4), and was added to the p(CBA-DAH) solution drop wise at a 1.5:1 molar ratio and reacted in the dark overnight while stirring. The polymer was collected and purified with dialysis (MWCO 3500) and lyophilized. Conjugation was confirmed with 1H-NMR (400 MHz, D2O).
Thiolation of Eph Peptide
The thiolation of the Eph peptide was carried out according to the manufacturers protocol. Eph peptide was dissolved in PBS, pH 8.0 + 2.0 mM EDTA. Traut’s reagent was dissolved and added with stirring to the peptide solution in a 2:1 molar ratio for one hour. The peptide was then purified with a D-Salt polyacrylamide desalting column (Pierce, Rockford, IL), MWCO 1.8 kDa. Elution was monitored with a nanoDrop ND-1000 spectrophotometer at 280 nm wavelength and characterized via MALDI-ToF.
Conjugation of Eph-SH to PEG-p(CBA-DAH)
The conjugation of Eph-SH to PEG-p(CBA-DAH) was completed in two steps. First, the conjugation of heterobifunctional NHS-PEG-Mal to p(CBA-DAH) was completed as described above. Prior to dialysis and lyophilization, thiolated peptide (Eph-SH) is added to the reaction. The thiolated peptide is dissolved in PBS, pH 7.4, and added drop wise with stirring. The peptide is added at a 1.5 fold molar excess over the amount of NHS-PEG-Mal to ensure adequate conjugation. Conjugation was confirmed with 1H-NMR (400 MHz, D2O).
Characterization of p(CBA-DAH)
The approximate molecular weight of p(CBA-DAH) was determined by size-exclusion chromatography (SEC). The AKTA FPLC system (Pharmacia) was used equipped with a Superose 12 column. Molecular weight was calculated using the Wyatt Optilab T-Rex and the Minidawn Treos, detecting refractive index and light scattering, respectively. The samples were run in a 0.3 M sodium acetate buffer (pH 5.0).
Agarose Gel Electrophoresis
Polyplex was formed in a HEPES buffer solution (20 mM HEPES, pH 7.4, 5% glucose) at weight:weight ratios ranging from 0.5 to 40. Samples were mixed via pipetting, vortexed at a low speed for 10 seconds, and incubated at room temperature for 30 minutes. Agarose gels (1% w/v) were prepared in Tris-acetate-EDTA (TAE) buffer with SYBR Green Nucleic Acid Gel stain. The samples were electrophoresed at 100V for 30 minutes. DNA was then visualized with a UV illuminator and analyzed using Gel Documentation Systems.
Polyplex Size Measurements
Dynamic light scattering (DLS) was used to determine the hydrodynamic radius of polyplexes [27]. Polyplexes were prepared by vortexing 60 μg pDNA in 500 μL HEPES buffer solution with an equal volume of polymer solution to yield the desired w:w ratios.
DNase Protection Assay
The ability of each polymer to protect plasmid DNA from enzymatic degradation was determined using a modified version of a previously reported method [28].
Cell lines
The cell lines MS1s, NIH 3T3s, and 293T that were used for experiments were obtained from ATCC (catalog numbers CRL-2279, CRL-1658, and CRL-11268, Manassas, VA). These cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin in a humidified incubator at 37°C with 5% CO2. All cell lines used for experiments were between passages 3–9.
Cellular Uptake Assay
Cellular uptake assay was performed with some modifications as previously described [29]. MS1 cells, NIH 3T3 cells, and 293 cells were seeded at a density of 2 × 105 cells/well in a 6-well plate in growth medium and grown to 70~80% confluence prior to transfection. Medium of each well was exchanged for 1 mL fresh serum-free medium, and cells were treated with polyplex solutions (2 μg of YOYO-1 labelled pDNA per well) at 10:1 w:w ratios for 4 h at 37 °C, control was given no treatment. For Eph competetion assay, 0.1 mM free Eph peptide was added to the media just prior to transfection. Following incubation, cells were washed two times with ice-cold D-PBS, incubated with 0.4 % trypan blue solution for 5 min, washed with DPBS, and collected via trypsinization. Uptake was examined with the BD FACScan analyzer at a minimum of 1 × 104 cells gated per sample. Analysis was performed by using Becton Dickinson CellQuest software.
Luciferase Transfection and Expression
Cells were plated at an initial density of 4.0 × 104 cells/well in 24 well plates. 24 hours following plating, luciferase (pCMV-Luc) was complexed with p(CBA-DAH), PEG-p(CBA-DAH), and Eph-PEG-p(CBA-DAH) at indicated w:w ratios in HEPES buffer and incubated for 30 minutes. Media in each well was replaced with 0.5 mL fresh, serum-free media. Polyplex was added to each well (0.5 μg DNA/well), and the cells were incubated for 4 hours at 37°C, 5% CO2. Medium was then replaced with 0.5 mL complete medium, and cells were incubated for an additional 44 hours. Cells were washed once with PBS, and 200 μL Reporter Lysis Buffer was added (Promega). Cells were then taken through one freeze-thaw cycle, scraped and cell lysate was collected. Experiments were performed in quadruplicate and analyzed via ANOVA.
Cytotoxicity Assay
MTT assay was performed. In parallel with the luciferase assay, cells were plated at 2 × 104 cells/well in 24-well plates. 24 hours following plating, cells were transfected as described for Luciferase transfection. After 20 hours, 20 μL MTT solution (5mg/mL) was added. After 4 hours, media was removed and 200 μL DMSO was added. 150 μL of the solution was transferred to a 96-well plate and absorbance was measured at 570 nm. Treated cells are relative to untreated control, were performed in quadruplicate, and analyzed via ANOVA.
Isolation of sRAE-1γ
Total RNA was collected from Min6 cells using a RNeasy mini kit according to manufacturers recommendation (Qiagen). A cDNA library was built via RT-PCR with the Superscript III first strand synthesis system (Invitrogen) and oligodT primers. To isolate the gene for soluble RAE-1γ, primers were designed with PrimerQuest (Integrated DNA Technologies, Inc) using a reported sequence for RAE-1γ (NCBI sequence ID: NM_009018) and screened on BLAST for cross-reactivity. The gene for soluble RAE-1γ was isolated using PCR amplification using gene specific primers with sequences as follows:
Forward: 5′ – ATGGCCAAGGCAGCAGTGA – 3′
Reverse: 5′ – AGAAGTAGAGTGGCTGGGA – 3′
The expected PCR product was 693 bp long.
Insertion of sRAE-1γ into Expression Cassette
The isolated gene was inserted into the multiple cloning site of pcDNA3.1/V5-His TOPO TA vector according to manufacturer’s instructions. Verification of gene insertion was confirmed by restriction enzyme digestion followed by gel electrophoresis. The orientation and open reading frame (ORF) integrity was confirmed via sequencing
Determine Expression Profile of pCMV-sRAE-1γ
The expression profile of pCMV-sRAE-1γ was characterized with verification of both transcription and translation. MS1 cells were transfected with pCMV-RAE-1γ/PEI polyplexes. The growth medium was collected at 24 and 48 hours following transfection, centrifuged at 14,000 × g for 2 minutes and stored at −20°C. Protein was separated via SDS-PAGE then transferred to a nitrocellulose membrane. Protein was detected with anti-V5-AP antibody and colorimetric detection.
Results
Synthesis, Conjugation, and Characterization of p(CBA-DAH), PEG-p(CBA-DAH), and Eph-PEG-p(CBA-DAH)
The bioreducible poly(disulfide amine) poly(cystaminebisacrylamide diaminohexane) p(CBA-DAH) was synthesized by a Michael-type addition reaction between the monomers N,N′-cystaminebisacrylamide (CBA) and N-Boc-1,6-diaminohexane (N-Boc-DAH). By using a monomer with one N-boc protected nitrogen, a linear, comblike polymer with a CBA backbone and DAH pendant chains result. This polymer contains one reducible disulfide bond, one tertiary amine, and one pendant primary amine per repeating unit. SEC data indicates the MW is 6kDa.
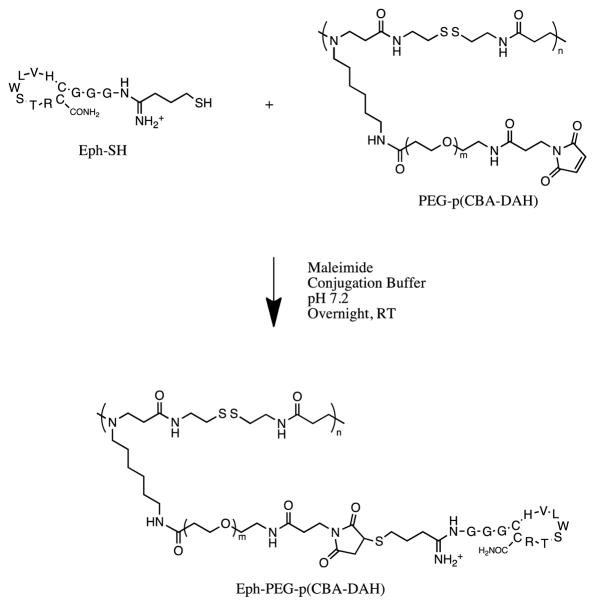
1H-NMR studies were used to verify the synthesis of p(CBA-DAH) and the conjugation of PEG-p(CBA-DAH) and Eph-PEG-p(CBA-DAH). Synthesis of p(CBA-DAH) was verified by comparing peaks to those previously published [26]. The appearance of a peak for PEG at δ 3.55 ppm indicates the successful conjugation of PEG. By calculating the ratio of the integral of the peak for PEG with one of the known peaks of p(CBA-DAH) (and normalizing to the relative number of protons in each peak per repeat unit and molecular weight), the ratio of PEG:p(CBA-DAH) was identified as 1.35:1 PEG:p(CBA-DAH).
For Eph-PEG-p(CBA-DAH), the thiolated targeting peptide Eph-SH was conjugated to PEGylated p(CBA-DAH). Again, by measuring the integral of the peaks produced by the methyl groups in the valine, leucine, and threonine (5 methyl groups, 15 protons), and comparing it to the integral of the peak produced for PEG, the relative number of PEG chains modified with the Eph peptide can be identified. After calculations, it was found that roughly 63% of the PEG chains had been modified with an Eph targeting peptide (Figure 1).
Figure 1.
Conjugation scheme for Eph-PEG-p(CBA-DAH)
Electrophoretic Mobility Shift Assay
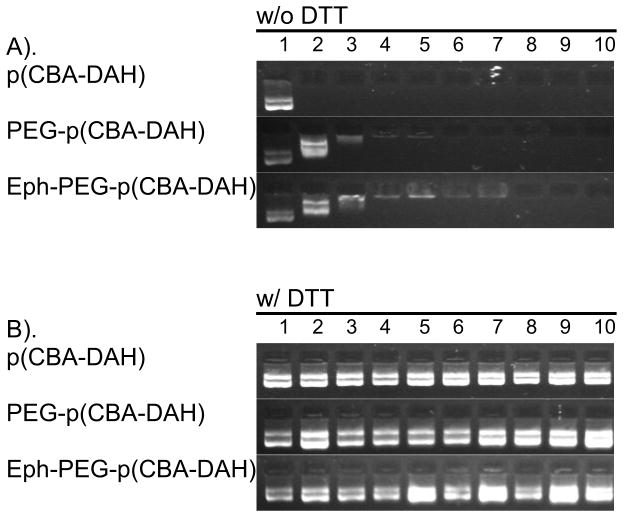
The ability of each of the three polymers to form polyplexes with plasmid DNA was investigated with an electrophoretic mobility shift assay. In Figure 2(A), the complexation ability of each polymer is examined. As can be seen, p(CBA-DAH) is capable of complexing pDNA at w:w ratios as low as 0.5:1. PEG-p(CBA-DAH) can form stable polyplex at a w:w ratio of 3:1, and Eph-PEG-pCBA-DAH can form polyplex at w:w ratios of 5:1. In Figure 2(B), the ability of the polyplexes to release their DNA cargo is shown by treating formed polyplex with 2.5 mM DTT for 30 minutes. Clearly, p(CBA-DAH), PEG-p(CBA-DAH), and Eph-PEG-p(CBA-DAH) are all able to release their plasmid DNA at w:w ratios up to 40:1. These results indicate that p(CBA-DAH), PEG-p(CBA-DAH), and Eph-PEG-p(CBA-DAH) will be able to unpackage the pDNA cargo upon entry into the reductive intracellular environment, enhancing gene expression [30, 31].
Figure 2.
Electrophoretic mobility shift assay of p(CBA-DAH), PEG-p(CBA-DAH), and Eph-PEG-p(CBA-DAH) with pDNA polyplex (A) without DTT and (B) with DTT (2.5 mM for 30m). Lane 1: naked pDNA; Lanes 2–10: polymer/pDNA polyplexes at N:P ratios of 0.5:1, 1:1, 3:1, 5:1, 10:1, 15:1, 20:1, 30:1, 40:1.
Polyplex Size Measurement
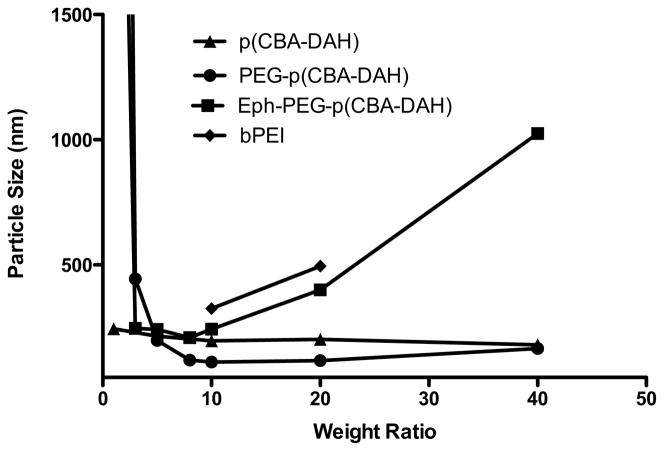
As can be seen in Figure 3, the particles have effective diameters of less than 250 nm at w:w ratios of 5:1 and above, reaching 200 nm around w:w ratios of 8:1. This is the ideal particle size for efficient cellular uptake via endocytosis and for circulation in vivo [32]. The PEG-p(CBA-DAH) particles remain small and well formed upto w:w ratios of 40:1, however Eph-PEG-p(CBA-DAH) shows an increased particle size above w:w ratio of 10:1. For this reason, w:w ratios up to 10:1 were used for experiments with Eph-PEG-pCBA-DAH.
Figure 3.
Average particle size of polymer/pDNA polyplexes at w:w ratios from 1:1 to 40:1. bPEI/pDNA polyplex sizes were determined at N:P ratios of 10:1 and 20:1. The large size of Eph-PEG-p(CBA-DAH) at w:w ratios above 10 is due to particle aggregation.
DNase Protection
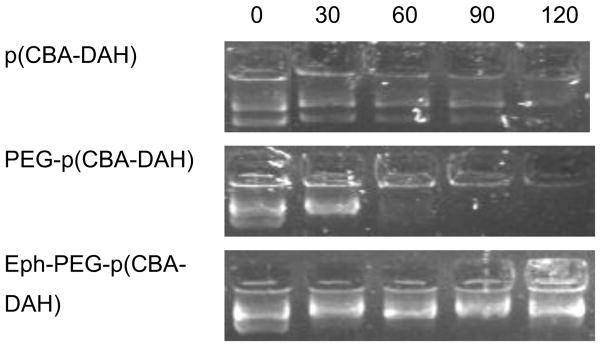
The ability of the three synthesized polymers to protect pDNA from enzymatic degradation was determined with a DNase protection assay. As can be seen in Figure 4, p(CBA-DAH) is capable of protecting plasmid DNA from enzymatic degradation up to two hours, while p(CBA-DAH) can protect up to roughly 90 min and PEG-p(CBA-DAH) is able to protect up to 30 min.
Figure 4.
DNase protecting ability of p(CBA-DAH), PEG-p(CBA-DAH), and Eph-PEG-p(CBA-DAH) at 0, 30, 60, 90, and 120 minutes. W:W ratios of 10:1 were used.
Cellular uptake of p(CBA-DAH), PEG-p(CBA-DAH), and Eph-PEG-p(CBA-DAH)
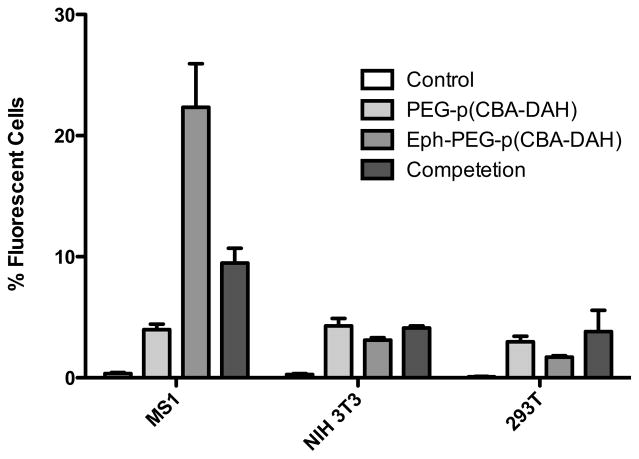
The uptake of PEG-p(CBA-DAH), and Eph-PEG-p(CBA-DAH) was analyzed in MS1s, NIH 3T3s, and 293Ts. MS1s, a pancreatic islet microvasculature cell line, is representative of the target cell and expresses EphA2 and EphA4 receptor. NIH 3T3 and 293T cells do not express EphA2 or EphA4, and are used as controls. As can be seen in Figure 5, the addition of PEG shields the polyplex and prevents non-specific uptake by all cell lines, as can be seen by the low fluorescence in cells treated with PEG-p(CBA-DAH). The recovery of uptake was observed in the MS1 cell line (the target cell line) by the addition of Eph targeting peptide, but not in NIH 3T3 or 293T cells. Furthermore, the functionality of Eph peptide targeting was verified by reducing the uptake in MS1 cells with a free competition assay. The addition of free peptide had no significant effect on the uptake of Eph-PEG-p(CBA-DAH) in NIH 3T3 cells and 293T cells. These results verify that the Eph-PEG-p(CBA-DAH) polymer can specifically target MS1 cells, the pancreatic islet microvasculature endothelial cells.
Figure 5.
Cellular uptake of polyplexes of pDNA with PEG-p(CBA-DAH) (PCD), and Eph-PEG-p(CBA-DAH) (EPCD). The effect of addition of free Eph peptide (0.1 mM) was examined in an uptake competition assay (EPCD Comp).
This result has implications for the ability of Eph-PEG-p(CBA-DAH) to deliver therapeutic genes specifically to the targeted tissue of the pancreatic islet microvasculature.
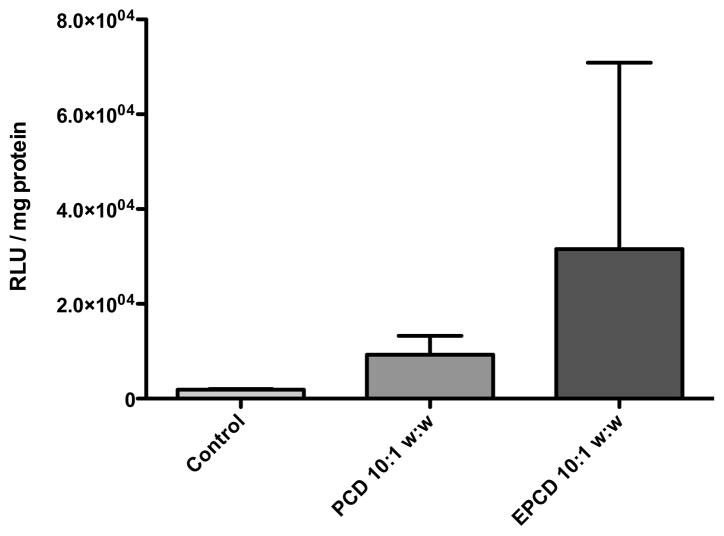
Luciferase Expression
Luciferase expression is indicative of uptake, endosomal escape, polyplex unpackaging, nuclear entry, transcription, and translation of a delivered gene. As can be seen in Figure 6, there is recovery of delivery with the addition of the Eph targeting peptide to PEG-p(CBA-DAH). However, there is variability in the Luciferase assay, and the results were not significant. The overall trend showed an increase in luciferase expression with delivery with the targeting peptide, which we assume was recovered through receptor-mediated uptake. As with the cellular uptake information, these results indicate that the targeting polymer Eph-PEG-p(CBA-DAH) is capable of delivering plasmid DNA specifically to target cells.
Figure 6.
Luciferase expression in MS1 cells transfected with pCMV-Luc delivered with PEG-p(CBA-DAH) (PCD), and Eph-PEG-p(CBA-DAH) (EPCD). Polyplexes were formed at a 10:1 w:w ratio.
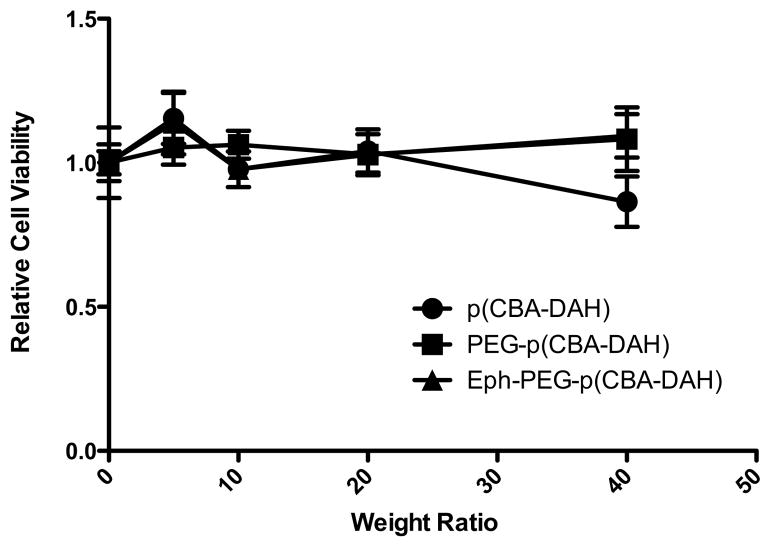
Cytotoxicity
The addition of PEG is well known to reduce toxicity of cationic polymers [33]. Additionally, the toxicity of bioreducible polymers such as p(CBA-DAH) has been shown to be markedly reduced compared to bPEI and PLL. Thus, it was expected that the polymers p(CBA-DAH), PEG-p(CBA-DAH), and Eph-PEG-p(CBA-DAH) would all show very little to no cytotoxicity. As can be seen in Figure 7, none of these three polymers show any statistically significant cytotoxicity up to a w:w ratio of 40:1.
Figure 7.
Cytotoxicity of p(CBA-DAH), PEG-p(CBA-DAH), and Eph-PEG-p(CBA-DAH) in MS1 cells at various w:w ratios.
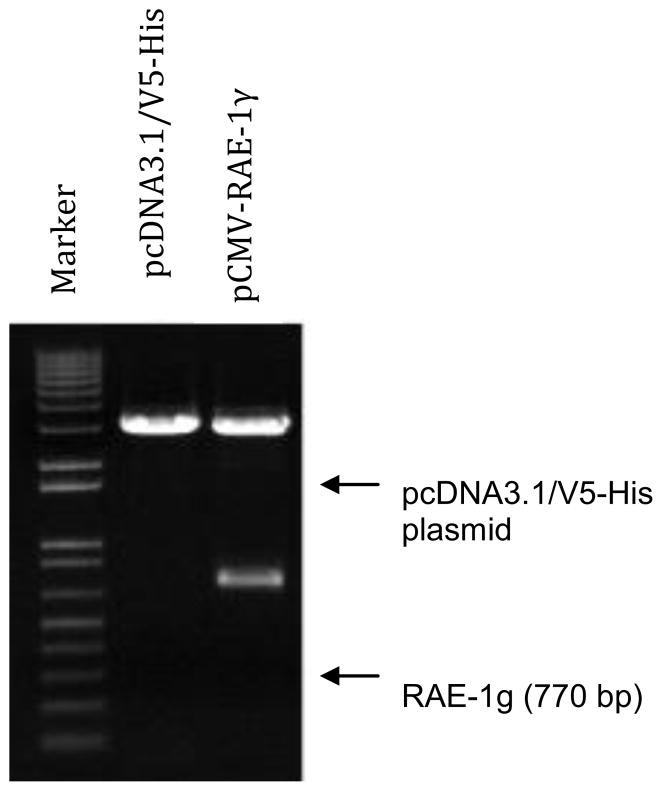
Construction of sRAE-1 Plasmid
Cloning of the sRAE-1 into the plasmid was also confirmed with restriction enzyme digestion. Digestion with NotI and KpnI produced a 770 bp fragment (sRAE-1), and a 5.4 kb fragment (vector), confirming the successful insertion of sRAE-1. The digestion of pCMV-RAE-1γ is shown in Figure 8. Lane 1 is a 1 Kb plus marker (Invitrogen), lane 2 shows the pcDNA3.1/v5-His vector alone, and lane 3 shows the complete constructed plasmid pCMV-RAE-1γ digested with KpnI and NotI.
Figure 8.
Digestion of pCMV-RAE-1γ with KpnI and NotI.
Characterization of pCMV-RAE-1γ
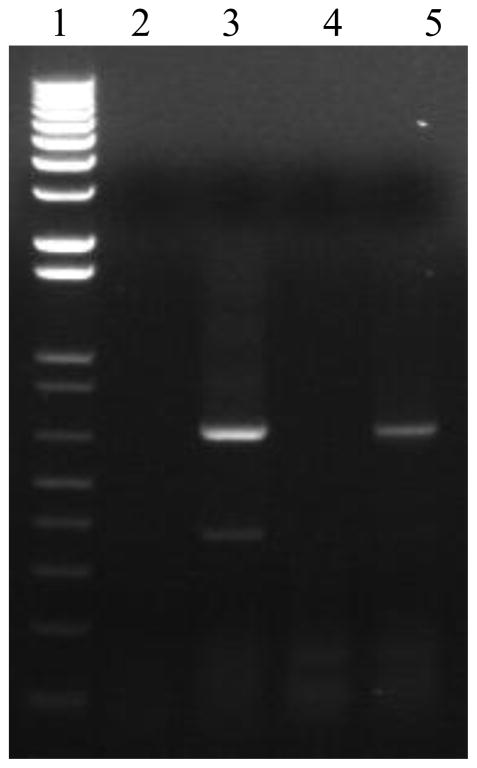
Expression and secretion of sRAE-1γ were evaluated following transfection of pCMV-RAE-1γ into pancreatic islet endothelial cells (MS1s). Transcription was verified using RT-PCR, and translation and secretion were verified using western blot. For RT-PCR, total RNA was collected from cells 24 hours following transfection. Reverse transcription was performed with both oligo dT primers and gene specific primers, and product was amplified using forward and reverse primers designed for initial isolation. As can be seen in Figure 9, sRAE-1γ is expressed in transfected cells (lanes 3 and 5), but not in untreated controls (lanes 2 and 4), verifying transcription of exogenous sRAE-1γ.
Figure 9.
RT-PCR verifies transcription of sRAE-1 γ. Lane 1: 1 kb+ size marker; lane 2: control nontransfected – oligo dT primer; lane 3: pCMV-RAE-1γ transfected – oligo dT primer; lane 4: control non-transfected – gene specific primers; lane 5: pCMV-RAE-1γ – gene specific primers.
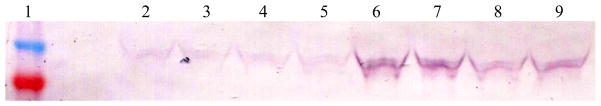
For western blot, cells were transfected with pCMV-RAE-1γ and samples of supernatant were collected at various time points. The total protein content was then assayed (BCA protein assay kit, Pierce) and equal amounts of protein were loaded and separated via SDS-PAGE. Following separation, protein was transferred to a nitrocellulose membrane, sRAE-1γ was stained with an anti-V5-AP antibody, and the blot was developed with an AP detection kit (BioRad). T there is a time dependent secretion of soluble RAE-1γ. Lane 1 shows the protein ladder, with bands at 40 and 50 kDa as shown in Figure 10. In lanes 2–5, samples collected at 24 hours can be seen, while in lanes 6–9, samples collected at 48 hours are seen.
Figure 10.
Western blot verifies translation and secretion of sRAE-1γ following transfection in MS1 cells. Lane 1: Novex Sharp prestained ladder (40 and 50 kDa bands shown). Lanes 2–5, 24h. Lanes 6–9, 48h.
Discussion
The use of p(CBA-DAH) as a polymeric gene carrier is attractive for several reasons. The inclusion of reducible disulfide bonds allows for the rapid triggered degradation upon entering the cytoplasmic compartment [34, 36]. The presence of primary and tertiary amines in the reducible polymer allow for the self-assembly of nucleic acid/polymer polyplexes [38]. Further, the presence of primary amines allows for straightforward modification of the polymer with PEG, and helps maintain solubility of the polymer [26]. Also, the polymer is synthesized from commercially available monomers, allowing for a straightforward synthesis.
The size of polyplexes plays a vital role in biodistribution for in vivo delivery [39–41]. Accumulation in the liver and spleen is prevalent with particle sizes that are too big (above 250 nm), so the ability to form polyplexes with diameters below that cutoff is important in designing a system for targeted in vivo delivery. All three polymers were able to reach sub-250 nm particle sizes at optimal w:w ratios, as can be seen in Figure 3.
The ability to condense DNA is essential for successful delivery to target tissues. The formation of dense polyplex between polymers and pDNA allows for the protection of the DNA from enzymatic degradation as well as packaging to facilitate cellular uptake [41, 42]. All three polymers were able to successfully condense pDNA at low w:w ratios, as was seen in the electrophoretic mobility shift assay in Figure 2 a). Furthermore, the ability to unpackage the pDNA following cellular uptake and endosomal escape enhances transfection efficiency [35–38]. The triggered release of plasmid DNA upon incubation in a reductive environment mimics the polyplex behavior in the cytoplasmic compartment. It was seen in Figure 2 b) that treatment of formed polyplexes with DTT can cause the triggered release of plasmid DNA, indicating that these polymers have an environment-responsive element for the triggered release following endosomal escape [29–45].
The ability to protect from enzymatic degradation is also essential for these polymers to be used for systemic delivery [46]. As was seen in Figure 5, the targeting polymer Eph-PEG-p(CBA-DAH) was able to protect plasmid DNA from DNase degradation up to 120 minutes (Figure 4). For p(CBA-DAH), some DNA degradation could be seen at 120 minutes. PEG-p(CBA-DAH) was only able to protect from Dnase degradation for roughly 30 minutes, however this result was not unexpected as the addition of PEG is known to interfere with condensation ability of cationic polymers [23].
The targeting ability of Eph-PEG-p(CBA-DAH) to the islet microvasculature endothelial cells was examined with transfection experiments with EphA2-expressing MS1 cells and EphA2-negative NIH 3T3 and 293T cells. It was expected that the addition of PEG to the backbone polymer would reduce cellular uptake of polyplex, as PEG has been previously reported to shield the cationic charge of polyplex and prevent non-specific interactions with cells [47–49]. This reduction in uptake was then expected to be recovered in the target cells, specifically MS1, the pancreatic islet microvasculature endothelial cell that expresses EphA2, with the addition of the targeting peptide, as has been shown with antibody fragments and other targeting peptides [50–52]. The uptake ability was not expected to recover in non-targeted cell lines, namely NIH 3T3 and 293T. The Eph-PEG-p(CBA-DAH) polymer was able to deliver plasmid DNA to MS1 cells with about 5-fold higher uptake efficiency than PEG-p(CBA-DAH) control in MS1 cells (Figure 5). In NIH 3T3 cells and 293T cells, however, there was no significant difference in uptake efficiency between Eph-PEG-p(CBA-DAH) and PEG-p(CBA-DAH). These results suggest that the Eph-PEG-p(CBA-DAH) polymer has specificity toward EphA2 expressing cells. The enhanced cellular uptake was verified to be due to specific interaction between the targeting peptide and the EphA2 receptor with a competition experiment carried out in the presence of free targeting peptide (0.1 mM) during transfection with Eph-PEG-p(CBA-DAH). The uptake efficiency of Eph-PEG-p(CBA-DAH) was significantly reduced to levels similar to that of PEG-p(CBA-DAH) complexes (Figure 5). In transfection experiments expressing luciferase, although there is a trend of increase in expression in the cells treated with Eph-PEG-p(CBA-DAH) compared to PEG-p(CBA-DAH), the results were not significant due to the variability in the experiment (Figure 6).
It is thought that this targeting polymer will have significant advantages when delivered in vivo. It is expected to have lower transfection efficiency with the addition of PEG to a backbone polymer, as most of the cellular uptake in unmodified cationic polymers is due to non-specific electrostatic interactions between polyplex and the anionic cell membrane [53].
The construction of an expression plasmid for a constitutively expressed soluble ligand to the NKG2D receptor, once successfully delivered locally to the pancreatic islets, will allow for the pancreatic islet beta cells to undergo immune evasion and prevent NKG2D mediated cytotoxicity to the beta cells. The therapeutic potential of the blockade of the NKG2D receptor for the prevention of type 1 diabetes remains largely unexplored, mainly due to the potential for side effects including the reduction in tumor surveillance, DNA damage detection, and viral infection [15, 54]. The systemic blockade of the NKG2D receptor is not an ideal therapy, yet remains the only in vivo tested therapeutic to prevent type 1 diabetes by receptor blockade in the form of systemic anti-NKG2D IgG delivery [13]. The construction of a therapeutic plasmid to deliver the gene that will encode for a protein to be synthesized locally around the pancreatic islets capable of blocking the NKG2D receptor of pancreatic infiltrating immunocytes will allow for the benefit of the blocking therapy while reducing the side effects expected from a systemic blockade.
The RAE-1γ expressing plasmid was designed for high constitutive expression with elements to aid in detection and purification [55]. The CMV promoter, in addition to being a strong, constitutively active promoter in many tissues, also shows relatively low transcription in hepatocytes [56]. The inclusion of the V5 epitope will enhance detection ability, both in vitro with western blot and in vivo with immunohistochemistry [57].
Conclusions
The synthesis of p(CBA-DAH) and modification to PEG-p(CBA-DAH) and Eph-PEG-p(CBA-DAH) were successfully completed. The polymers are all able to form polyplexes with pDNA and release that pDNA rapidly in a reductive environment due to the reduction of disulfide bonds in the backbone. All three polymers are capable of forming nano-sized particles when complexed with pDNA. Both p(CBA-DAH) and Eph-PEG-p(CBA-DAH) show the ability to protect pDNA from enzymatic degradation. It was shown that the addition of PEG to the backbone polymer reduced non-specific cellular uptake and transfection, and that the addition of a targeting peptide Eph could recover the cellular uptake and transfection efficiency selectively in a specific cell line. Additionally, the construction and characterization of expression of pCMV-RAE-1γ was successfully completed. The therapeutic gene was inserted in the expression plasmid, as was verified by sequencing and restriction enzyme digestion. The expression capabilities of the pCMV-RAE-1γ plasmid were verified with RT-PCR and western blot. The therapeutic protein srRAE-1γ was secreted, as was also verified by western blot. Therefore, the combination therapy of the polymer Eph-PEG-p(CBA-DAH) and the therapeutic pCMV-RAE-1γ is a viable therapeutic strategy for the prevention of type 1 diabetes.
Supplementary Material
Acknowledgments
Funding by NIH Grants DK085075.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Atkinson MA, Eisenbarth GS. Type 1 diabetes: new perspectives on disease pathogenesis and treatment. Lancet. 2001;358:221–229. doi: 10.1016/S0140-6736(01)05415-0. [DOI] [PubMed] [Google Scholar]
- 2.Atkinson MA, Maclaren NK. The pathogenesis of insulin-dependent diabetes mellitus. N Engl J Med. 1994;331:1428–1436. doi: 10.1056/NEJM199411243312107. [DOI] [PubMed] [Google Scholar]
- 3.Amrani A, Verdaguer J, Serra P, Tafuro S, Tan R, Santamaria P. Progression of autoimmune diabetes driven by avidity maturation of a T-cell population. Nature. 2000;406:739–742. doi: 10.1038/35021081. [DOI] [PubMed] [Google Scholar]
- 4.Harrison LC, Chu SX, DeAizpurua HJ, Graham M, Honeyman MC, Colman PG. Islet-reactive T cells are a marker of preclinical insulin-dependent diabetes. J Clin Invest. 1992;89:1161–1165. doi: 10.1172/JCI115698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagata M, Santamaria P, Kawamura T, Utsugi T, Yoon JW. Evidence for the role of CD8+ cytotoxic T cells in the destruction of pancreatic beta-cells in nonobese diabetic mice. J Immunol. 1994;152:2042–2050. [PubMed] [Google Scholar]
- 6.Maasho K, Opoku-Anane J, Marusina AI, Coligan JE, Borrego F. NKG2D is a costimulatory receptor for human naive CD8+ T cells. J Immunol. 2005;174:4480–4484. doi: 10.4049/jimmunol.174.8.4480. [DOI] [PubMed] [Google Scholar]
- 7.Caillat-Zucman S. How NKG2D ligands trigger autoimmunity? Hum Immunol. 2006;67:204–207. doi: 10.1016/j.humimm.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Ogasawara K, Lanier LL. NKG2D in NK and T cell-mediated immunity. J Clin Immunol. 2005;25:534–540. doi: 10.1007/s10875-005-8786-4. [DOI] [PubMed] [Google Scholar]
- 9.Van Belle TL, von Herrath MG. The role of the activating receptor NKG2D in autoimmunity. Mol Immunol. 2009 doi: 10.1016/j.molimm.2009.02.023. [DOI] [PubMed] [Google Scholar]
- 10.Cerwenka A, Bakker AB, McClanahan T, Wagner J, Wu J, Phillips JH, Lanier LL. Retinoic acid early inducible genes define a ligand family for the activating NKG2D receptor in mice. Immunity. 2000;12:721–727. doi: 10.1016/s1074-7613(00)80222-8. [DOI] [PubMed] [Google Scholar]
- 11.Steinle A, Li P, Morris DL, Groh V, Lanier LL, Strong RK, Spies T. Interactions of human NKG2D with its ligands MICA, MICB, and homologs of the mouse RAE-1 protein family. Immunogenetics. 2001;53:279–287. doi: 10.1007/s002510100325. [DOI] [PubMed] [Google Scholar]
- 12.Ogasawara K, Hamerman JA, Hsin H, Chikuma S, Bour-Jordan H, Chen T, Pertel T, Carnaud C, Bluestone JA, Lanier LL. Impairment of NK cell function by NKG2D modulation in NOD mice. Immunity. 2003;18:41–51. doi: 10.1016/s1074-7613(02)00505-8. [DOI] [PubMed] [Google Scholar]
- 13.Ogasawara K, Hamerman JA, Ehrlich LR, Bour-Jordan H, Santamaria P, Bluestone JA, Lanier LL. NKG2D blockade prevents autoimmune diabetes in NOD mice. Immunity. 2004;20:757–767. doi: 10.1016/j.immuni.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Mistry AR, O’Callaghan CA. Regulation of ligands for the activating receptor NKG2D. Immunology. 2007;121:439–447. doi: 10.1111/j.1365-2567.2007.02652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burgess SJ, Maasho K, Masilamani M, Narayanan S, Borrego F, Coligan JE. The NKG2D receptor: immunobiology and clinical implications. Immunol Res. 2008;40:18–34. doi: 10.1007/s12026-007-0060-9. [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Larrea C, Suarez-Alvarez B, Lopez-Soto A, Lopez-Vazquez A, Gonzalez S. The NKG2D receptor: sensing stressed cells. Trends Mol Med. 2008;14:179–189. doi: 10.1016/j.molmed.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 17.Raulet DH. Roles of the NKG2D immunoreceptor and its ligands. Nat Rev Immunol. 2003;3:781–790. doi: 10.1038/nri1199. [DOI] [PubMed] [Google Scholar]
- 18.Hughes JA, Rao GA. Targeted polymers for gene delivery. Exp Opin Drug Del. 2005;2:145–157. doi: 10.1517/17425247.2.1.145. [DOI] [PubMed] [Google Scholar]
- 19.Saito G, Swanson JA, Lee KD. Drug delivery strategy utilizing conjugation via reversible disulfide linkages: role and site of cellular reducing activities. Adv Drug Del Rev. 2003;55:199–215. doi: 10.1016/s0169-409x(02)00179-5. [DOI] [PubMed] [Google Scholar]
- 20.Wu G, Fang YZ, Yang S, Lupton JR, Turner ND. Glutathione metabolism and its implications for health. J Nutr. 2004;134:489–492. doi: 10.1093/jn/134.3.489. [DOI] [PubMed] [Google Scholar]
- 21.Hill IR, Garnett MC, Bignotti F, Davis SS. In vitro cytotoxicity of poly(amidoamine)s: relevance to DNA delivery. Biochim Biophys Acta. 1999;1427:161–174. doi: 10.1016/s0304-4165(99)00021-5. [DOI] [PubMed] [Google Scholar]
- 22.Eliyahu H, Barenholz Y, Domb AJ. Polymers for DNA delivery. Molecules. 2005;10:34–64. doi: 10.3390/10010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Vlerken LE, Vyas TK, Amiji MM. Poly(ethylene glycol)-modified nanocarriers for tumor-targeted and intracellular delivery. Pharm Res. 2007;24:1405–1414. doi: 10.1007/s11095-007-9284-6. [DOI] [PubMed] [Google Scholar]
- 24.Aspord C, Rome S, Thivolet C. Early events in islets and pancreatic lymph nodes in autoimmune diabetes. J Autoimmun. 2004;23:27–35. doi: 10.1016/j.jaut.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 25.Yao VJ, Ozawa MG, Trepel M, Arap W, McDonald DM, Pasqualini R. Targeting pancreatic islets with phage display assisted by laser pressure catapult microdissection. Am J Pathol. 2005;166:625–636. doi: 10.1016/S0002-9440(10)62283-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ou M, Wang XL, Xu R, Chang CW, Bull DA, Kim SW. Novel biodegradable poly(disulfide amine)s for gene delivery with high efficiency and low cytotoxicity. Bioconjug Chem. 2008;19:626–633. doi: 10.1021/bc700397x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ou M, Xu R, Kim SH, Bull DA, Kim SW. A family of bioreducible poly(disulfide amine)s for gene delivery. Biomaterials. 2009;30:5804–5814. doi: 10.1016/j.biomaterials.2009.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ou M, Kim T-i, Yockman JW, Borden BA, Bull DA, Kim SW. Polymer Transfected Primary Myoblasts Mediated Efficient Gene Expression and Angiogenic Proliferation. Journal of Controlled Release. 2010;142:61–69. doi: 10.1016/j.jconrel.2009.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christensen LV, Chang CW, Kim WJ, Kim SW, Zhong Z, Lin C, Engbersen JF, Feijen J. Reducible poly(amido ethylenimine)s designed for triggered intracellular gene delivery. Bioconjug Chem. 2006;17:1233–1240. doi: 10.1021/bc0602026. [DOI] [PubMed] [Google Scholar]
- 30.Kim TI, Ou M, Lee M, Kim SW. Arginine-grafted bioreducible poly(disulfide amine) for gene delivery systems. Biomaterials. 2009;30:658–664. doi: 10.1016/j.biomaterials.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaffer DV, Fidelman NA, Dan N, Lauffenburger DA. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol Bioeng. 2000;67:598–606. doi: 10.1002/(sici)1097-0290(20000305)67:5<598::aid-bit10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 32.Forrest ML, Koerber JT, Pack DW. A degradable polyethylenimine derivative with low toxicity for highly efficient gene delivery. Bioconjug Chem. 2003;14:934–940. doi: 10.1021/bc034014g. [DOI] [PubMed] [Google Scholar]
- 33.Kircheis R, Wightman L, Wagner E. Design and gene delivery activity of modified polyethylenimines. Adv Drug Del Rev. 2001;53:341–358. doi: 10.1016/s0169-409x(01)00202-2. [DOI] [PubMed] [Google Scholar]
- 34.Veronese FM, Pasut G. PEGylation, successful approach to drug delivery. Drug Discovery Today. 2005;10:1451–1458. doi: 10.1016/S1359-6446(05)03575-0. [DOI] [PubMed] [Google Scholar]
- 35.Read ML, Bremner KH, Oupicky D, Green NK, Searle PF, Seymour LW. Vectors based on reducible polycations facilitate intracellular release of nucleic acids. J Gene Med. 2003;5:232–245. doi: 10.1002/jgm.331. [DOI] [PubMed] [Google Scholar]
- 36.Kim SH, Ou M, Bull DA, Kim SW. Reductive Degradation Behavior of Bioreducible Poly(disulfide amine) for Enhancing SiRNA Efficiency. Macromolecular Bioscience. 2010 doi: 10.1002/mabi.200900482. [DOI] [PubMed] [Google Scholar]
- 37.Kim T-i, Rothmund T, Kissel T, Kim SW. Bioreducible Polymers with Cell Penetrating and Endosome Buffering Functionality for Gene Delivery Systems. Journal of Controlled Release. 2011;152:110–119. doi: 10.1016/j.jconrel.2011.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shim MS, Wang X, Ragan R, Kwon YJ. Dynamics of nucleic acid/cationic polymer complexation and disassembly under biologically simulated conditions using in situ atomic force microscopy. Microscope Research and Technique. 2010;73:845–856. doi: 10.1002/jemt.20856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y, Mounkes LC, Liggitt HD, Brown CS, Solodin I, Heath TD, Debs RJ. Factors influencing the efficiency of cationic liposome-mediated intravenous gene delivery. Nat Biotechnol. 1997;15:167–173. doi: 10.1038/nbt0297-167. [DOI] [PubMed] [Google Scholar]
- 40.Nishikawa M, Takakura Y, Hashida M. Pharmacokinetic evaluation of polymeric carriers. Adv Drug Del Rev. 1996;21:135–155. [Google Scholar]
- 41.Owens DE, III, Peppas NA. Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int J Pharm. 2006;307:93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 42.Takakura Y, Hashida M. Macromolecular carrier systems for targeted drug delivery: Pharmacokinetic considerations on biodistribution. Pharm Res. 1996;13:820–831. doi: 10.1023/a:1016084508097. [DOI] [PubMed] [Google Scholar]
- 43.Bloomfield VA. DNA condensation by multivalent cations. Biopolymers. 1997;44:269–282. doi: 10.1002/(SICI)1097-0282(1997)44:3<269::AID-BIP6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 44.Hansma HG, Golan R, Hsieh W, Lollo CP, Mullen-Ley P, Kwoh D. DNA condensation for gene therapy as monitored by atomic force microscopy. Nucleic Acids Res. 1998;26:2481–2487. doi: 10.1093/nar/26.10.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin C, Zhong Z, Lok MC, Jiang X, Hennink WE, Feijen J, Engbersen JFJ. Novel bioreducible poly(amido amine)s for highly efficient gene delivery. Bioconjug Chem. 2007;18:138–145. doi: 10.1021/bc060200l. [DOI] [PubMed] [Google Scholar]
- 46.Brus C, Petersen H, Aigner A, Czubayko F, Kissel T. Efficiency of polyethylenimines and polyethylenimine-graft-poly (ethylene glycol) block copolymers to protect oligonucleotides against enzymatic degradation. Eur J Pharm Biopharm. 2004;57:427–430. doi: 10.1016/j.ejpb.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 47.Hamidi M, Azadi A, Rafiei P. Pharmacokinetic consequences of pegylation. Drug Delivery. 2006;13:399–409. doi: 10.1080/10717540600814402. [DOI] [PubMed] [Google Scholar]
- 48.Lee M, Kim SW. Polyethylene glycol-conjugated copolymers for plasmid DNA delivery. Pharm Res. 2005;22:1–10. doi: 10.1007/s11095-004-9003-5. [DOI] [PubMed] [Google Scholar]
- 49.Ogris M, Steinlein P, Carotta S, Brunner S, Wagner E. DNA/polyethylenimine transfection particles: Influence of ligands, polymer size, and PEGylation on internalization and gene expression. AAPS PharmSci. 2001;3 doi: 10.1208/ps030321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jeong JH, Lee M, Kim WJ, Yockman JW, Park TG, Kim YH, Kim SW. Anti-GAD antibody targeted non-viral gene delivery to islet beta cells. J Controlled Release. 2005;107:562–570. doi: 10.1016/j.jconrel.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 51.Suh W, Han SO, Yu L, Kim SW. An angiogenic, endothelial-cell-targeted polymeric gene carrier. Molecular Therapy. 2002;6:664–672. [PubMed] [Google Scholar]
- 52.Lee M, Oh S, Ahn CH, Kim SW, Rhee BD, Ko KS. An Efficient GLP-1 Expression System Using Two-step Transcription Amplification. J Controlled Release. 2006;115:316–321. doi: 10.1016/j.jconrel.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 53.Khalil IA, Kogure K, Akita H, Harashima H. Uptake pathways and subsequent intracellular trafficking in nonviral gene delivery. Pharmacol Rev. 2006;58:32–45. doi: 10.1124/pr.58.1.8. [DOI] [PubMed] [Google Scholar]
- 54.Smyth MJ, Swann J, Cretney E, Zerafa N, Yokoyama WM, Hayakawa Y. NKG2D function protects the host from tumor initiation. J Exper Med. 2005;202:583–588. doi: 10.1084/jem.20050994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Makrides SC. Components of vectors for gene transfer and expression in mammalian cells. Protein Expr Purif. 1999;17:183–202. doi: 10.1006/prep.1999.1137. [DOI] [PubMed] [Google Scholar]
- 56.Najjar SM, Lewis RE. Persistent expression of foreign genes in cultured hepatocytes: expression vectors. Gene. 1999;230:41–45. doi: 10.1016/s0378-1119(99)00052-9. [DOI] [PubMed] [Google Scholar]
- 57.Lobbestael E, Reumers V, Ibrahimi A, Paesen K, Thiry I, Gijsbers R, Van den Haute C, Debyser Z, Baekelandt V, Taymans JM. Immunohistochemical detection of transgene expression in the brain using small epitope tags. BMC Biotechnology. 2010;10:16. doi: 10.1186/1472-6750-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.