Abstract
The bacterial flagellar C-ring is composed of two essential proteins, FliM and FliN. The smaller protein FliN is similar to the C-terminus of the larger protein FliM, both being composed of SpoA domains. While bacterial type III secretion (T3S) systems encode many proteins in common with the flagellum, they mostly have a single protein in the place of FliM and FliN. This protein resembles FliM at its N-terminus and is as large as FliM, but is more like FliN at its C-terminal SpoA domain. We have discovered that a FliN-sized cognate indeed exists in the Yersinia T3S system to accompany the FliM-sized cognate. The FliN-sized cognate, YscQ-C, is the product of an internal translation initiation site within the locus encoding the FliM-sized cognate YscQ. Both intact YscQ and YscQ-C were found to be required for T3S, indicating that the internal translation initiation site, which is conserved in some but not all YscQ orthologs, is crucial for function. The crystal structure of YscQ-C revealed a SpoA domain that forms a highly intertwined, domain-swapped homodimer, similar to those observed in FliN and the YscQ ortholog HrcQB. A single YscQ-C homodimer associated reversibly with a single molecule of intact YscQ, indicating conformational differences between the SpoA domains of intact YscQ and YscQ-C. A ‘snap-back’ mechanism suggested by the structure can account for this. The 1:2 YscQ:YscQ-C complex is a close mimic of the 1:4 FliM:FliN complex and the likely building block of the putative Yersinia T3S system C-ring.
The type III secretion (T3S) system is essential to the pathogenesis of numerous Gram-negative bacteria (1, 2). This system transports select bacterial proteins, many of them virulence factors, into host cells. The transport is presumed to occur directly from the bacterial cytosol through a hollow needle (3), although an indirect route also appears to exist (4). The precise means by which proteins are recognized within the bacterium for transport is not understood. Among candidate recognition factors are T3S proteins related to proteins of the bacterial flagellar C-ring (5, 6), which is the most cytoplasmically disposed of the flagellar rings and which plays a role in both protein transport and flagellar rotation (7). The flagellar C-ring is composed of two proteins, FliM (38 kDa) and FliN (15 kDa), which are related. FliN is similar in sequence to the C-terminus of FliM, both being composed of SpoA domains that mediate the formation of a FliM/FliN complex (8–10). In Escherichia coli these complexes have been determined to have a 1:4 FliM:FliN stoichiometry (8), and in Salmonella ~32–36 FliM/FliN complexes have been estimated to form the C-ring (6).
While the T3S system version of the C-ring has not yet been visualized (11, 12), most T3S systems encode a single protein in the place of FliM and FliN that has been found to be essential for T3S (13–16). This protein resembles FliM at its N-terminus and is as large as FliM, but is more like FliN at its C-terminal SpoA domain. Most T3S systems appear to lack the smaller FliN-sized cognate to accompany the FliM-sized cognate. This is the case for Yersinia spp., which encode the FliM-sized cognate YscQ (34 kDa) but appear to lack the FliN-sized cognate. YscQ is essential for T3S and interacts with a number of T3S apparatus components (11, 13, 17, 18). A complex containing YscQ, YscN (the T3S ATPase), YscL (negative regulator of YscN), and YscK (undefined function) appears to co-assemble during formation of the T3S apparatus (11). YscQ is also reported to interact with YscP, a regulator of the needle length (19). Except for the association with the needle length regulator, these interactions are conserved in the YscQ orthologs Shigella Spa33 (12), Salmonella SpaO (14), and E. coli EscQ (SepQ) (15). In addition, the Chlamydia ortholog CdsQ has been reported to interact with the ortholog of YscL, CdsL, and the ortholog of the structural protein YscD, CdsD (20). Spa33 and SpaO along with CdsQ (21) have also been shown to associate with transported proteins, suggesting that these YscQ orthologs help in the recognition of T3S transported substrates (12). Both CdsQ and SpaO further interact with T3S chaperones (14, 21), which associate with certain transported proteins and are required for the transport of these proteins. In the case of SpaO, evidence exists that it acts as a sorting platform, such that the interactions with transported proteins are made in sequential order (14).
To understand the nature of the putative C-ring in the Yersinia T3S system, we isolated YscQ from Y. pseudotuberculosis and found that it exists as a complex composed of intact YscQ and a C-terminal fragment of YscQ that closely corresponds to FliN in size and sequence. We found that the C-terminal fragment, called YscQ-C, is a product of an internal translation initiation site, which is conserved in some but not all YscQ family members. This is similar to the recent discovery that in the Salmonella SPI-2 T3S system, the YscQ ortholog SsaQ is produced also as an intact protein and a C-terminal fragment due to an internal translation start site (16). However unlike the case for SsaQ in which the C-terminal fragment is dispensable for T3S, YscQ-C was found to be required for T3S. The crystal structure of YscQ-C was determined, showing that it consists of a SpoA domain that forms a highly intertwined, domain-swapped homodimer, resembling the structures of fragments of T. maritima FliN (8) and the Pseudomonas syringae ortholog of YscQ, HrcQB (22). We found that a single homodimer of YscQ-C associated with a single monomer of intact YscQ, indicating conformational differences between the SpoA domains of intact YscQ and YscQ-C. We propose a ‘snap-back’ mechanism suggested by the structure to account for this difference. The 1:2 YscQ:YscQ-C complex is a close mimic of the 1:4 FliM:FliN complex and the likely building block of the putative T3S C-ring in Yersinia.
EXPERIMENTAL PROCEDURES
DNA constructs and mutagenesis
The coding sequence for YscQ (residues 1–307, with an S2G substitution to accommodate a DNA restriction site) and YscQ-C (residues 218–307, with an S219G substitution to accommodate a DNA restriction site) was amplified by PCR from the pYV plasmid of Yersinia pseudotuberculosis 126 (23), and used for generation of pET28b(+) (EMD, San Diego, CA), and in the case of YscQ, also pBAD-A (Invitrogen, Carlsbad, CA) expression constructs. A PreScission protease cleavable C-terminal histidine tag was added to YscQ and YscQ-C in all pET28b(+) construct, and to YscQ in some pBAD constructs. YscQ(M218A) was expressed with a non-cleavable His-tag. Mutagenesis to generate YscQ(M218A) and other mutant proteins was performed using the QuikChange mutagenesis kit (Agilent). The integrity of all constructs was verified by DNA sequencing.
Protein expression in E. coli and purification
Wild-type YscQ and YscQ(M218A) were expressed using pET28b(+) in E. coli BL21 (DE3). Bacteria were grown at 37 °C in LB medium containing kanamycin (100 μg/ml) to an OD600 of 0.8–1.2, cooled to 18 °C for the induction of protein expression through the addition of 0.5 mM isopropyl-β-D-thio-galactoside, and then grown further at 18 °C overnight. Identical procedures were followed for expression of YscQ-C, except that the temperature was maintained at 37 °C and bacteria were grown for only 3 h following induction. Bacteria were pelleted by centrifugation (10 min, 8000 × g, 4 °C), and the pellet was resuspended in 150 mM NaCl, 50 mM Tris, pH 8.0 (TBS), and 1 mM phenylmethylsulfonyl fluoride (5 mL per g of wet cell paste). Bacteria were lysed using an EmulsiFlex-C5 (Avestin), cell debris was pelleted by centrifugation (30 min, 20,000 × g, 4 °C), and the supernatant was applied to a Ni2+-nitrilotriacetic acid (NTA) column (Sigma). Following extensive washing with TBS containing 5 mM imidazole, bound protein was eluted in TBS containing 250 mM imidazole. Eluted fractions were dialyzed or buffer exchanged by diafiltration (Amicon YM10 for YscQ and YscQ(M218A) or YM3 for YscQ-C) into TBS. For constructs with a cleavable His-tag, His-tagged PreScission protease was added at a 1:100 protease:substrate mass ratio and incubated overnight at 4°C. The sample was then re-applied to a Ni2+-NTA column, and cleaved YscQ was collected in the flow-through fractions. YscQ constructs were further purified on a Superdex 200 column (GE Healthcare), and concentrated to 6–15 mg/mL for storage at −80 °C. The concentration of YscQ constructs was determined using calculated molar absorption coefficients. For phasing purposes, the methionine substitution mutant YscQ-C(I248M) was constructed. Selenomethionine (SeMet) substituted YscQ-C(I248M) was prepared as previously described (24), and purified as described above, except that 5 mM 2-mercaptoethanol (2-ME) was included in all buffers.
N-terminal sequencing
N-terminal amino acid sequencing was carried out at the UCSD Division of Biological Sciences Protein Sequencing Facility.
Static Light Scattering
Absolute molecular masses were determined by multi-angle static light scattering (SLS). Protein samples in 150 mM NaCl, 50 mM Tris or HEPES, pH 7.4 were applied to a TSK-Gel G4000 WXL size-exclusion column (Tosoh, Bioscience) attached to a miniDAWN TREOS SLS detector and an Optilab T-rEX refractive index detector (Wyatt, Santa Barbara, CA). Data were processed using ASTRA 5 software (Wyatt, Santa Barbara, CA).
Protein crystallization
YscQ-C at 4–16 mg/mL in 10 mM Tris, pH 8.0 was crystallized by the vapor diffusion hanging drop method using 1.4–1.55 M (NH4)2SO4, 100 mM Tris, pH 7.8–8.8 as the precipitant at a 1:1 protein:precipitant ratio. SeMet-substituted YscQ-C(I248M) at 7.75 mg/mL in 10 mM Tris, pH 8.0, 5 mM 2-ME was crystallized by the sitting drop vapor diffusion method with an Oryx6 crystallization robot (Douglas Instruments, UK) using 100 mM Tris, pH 7.4–8.2, 40–120 mM MgCl2, 3–12% PEG 400 as the precipitant at 3:7 and 6:4 protein:precipitant ratios.
Structure Determination
Crystals of YscQ-C and YscQ-C(I248M) were cryoprotected using 1:2 (vol:vol) paratoneN:paraffin and cooled in liquid nitrogen. Diffraction data for crystals of YscQ-C were collected at APS beam line 23 ID-B (Argonne National Laboratory), as were single wavelength anomalous dispersion (SAD) data for crystals of SeMet-substituted YscQ-C(I248M). Diffraction data for YscQ-C and YscQ-C(I248M) were processed using XDS (25) and HKL2000 (26), respectively. Phases were calculated and refined using the programs Phaser and Resolve within Phenix (27). Two SeMet positions were identified in the asymmetric unit, which contains a dimer of YscQ-C. Automated model building using Phenix resulted in an initial model that contained 143 of the 172 residues in the final model. The initial model was manually modified using Coot through inspection of σA-weighted 2mFo-DFc and mFo-DFc maps (28), followed by cycles of refinement using Phenix. Default parameters and target weights defined by Phenix were used (3 macro cycles of bulk solvent correction and anisotropic scaling and up to 25 iterations of coordinate, atomic displacement parameters and occupancy refinement). The model was verified by inspection of composite omit maps calculated using CNS (5% of model omitted, σA-weighted 2mFo-DFc maps) (29). Waters were added in later stages of the refinement using Phenix with default parameters (3σ peak height in mFo-DFc maps), followed by inspection of maps. Continuous electron density for the main chain was evident for residues 229–313 of chain A and 228–313 of chain B, and all side chains were in electron density, except for Q291 of chain A and Q239 of chain B.
The structure of native, wild-type YscQ-C was determined by molecular replacement with Molrep using YscQ-C(Ile248SeMet) as the search model (30). The same refinement protocol described above was used, except that Cartesian simulated annealing was carried out in Phenix (using default parameters) in the first round of refinement. TLS refinement was performed in the final stages of structure refinement with 12 TLS groups (for chain A, 229–244, 245–250, 251–265, 266–271, 272–279, 280–290, 291–301, and 302–313; for chain B, 229–239, 240–271, 272–301, and 302–313) (31). Continuous electron density was observed for residues 229–313 of both chains. All side chains were in electron density with the exception of Q239 and Q291 of chain A and Q239 of chain B. The final map had correlation coefficients of 0.90 and 0.75 for the main and side chains, respectively, as calculated with OVERLAPMAN (30). The overall Molprobity score was 1.68, corresponding to the 97th percentile (32). Data collection and model statistics are presented in Table 1.
Table 1.
Data collection and refinement statistics
| YscQ-C | SeMet YscQ-C (I248M) | |
|---|---|---|
|
Data Collection
| ||
| λ(Å) | 0.96860 | 0.97948 |
|
| ||
| Resolution (Å)a | 34.42-2.25 (2.31-2.25) | 50-2.16 (2.20-2.16) |
|
| ||
| Space group | H32 | H32 |
| Cell dimensions (Å) | a=b=114.1, c=88.6 | a=b=114.2, c=91.5 |
|
| ||
| Unique reflections | 10,594 | 12,457 |
|
| ||
| Avg. multiplicitya | 10.8 (9.8) | 17.7 (3.6) |
|
| ||
| I/σIa | 32.1 (10.5) | 32.4 (2.1) |
|
| ||
| Completeness (%)a | 99.3 (94.2) | 97.4 (76.2) |
|
| ||
| Rmerge (%)a | 4.7 (23.2) | 8.3 (53.9) |
|
| ||
|
Refinement
|
||
| Resolution (Å)a | 34.42-2.25 (2.48-2.25) | |
|
| ||
| Number of reflections | 10,592 | |
|
| ||
| Number of atoms | ||
| Protein | 1,370 | |
| Solvent | 50 | |
|
| ||
| Rwork/Rfree (%)a | 20.2/23.3 (26.6/32.4) | |
|
| ||
| Rms deviation | ||
| Bond lengths (Å) | 0.009 | |
| Angles (°) | 1.244 | |
|
| ||
| Average B factor (Å2) | ||
| Protein | 47.6 | |
| Solvent | 48.5 | |
|
| ||
| Ramachandran (%)b | ||
| Favored | 98.8 | |
| Allowed | 1.2 | |
| Disallowed | 0 | |
Highest resolution shell in parenthesis.
Structure validation by Molprobity (32).
Molecular figures were made with PyMol (http://pymol.sourceforge.net). Structure-based sequence alignments were generated using Expresso (33) and displayed using ESPript (34). The crystal structure and structure factors have been deposited with the Protein Data Bank (accession code 3UEP).
Deletion of yscQ in Y. pseudotuberculosis
To generate Y. pseudotuberculosis (ΔyscQ), the entirety of yscQ, except for 22 bp at the 3′ end which contains a ribosome-binding site for yscR, was substituted in-frame with aph (kanamycin resistance) by homologous recombination using a PCR fragment, as described previously (35, 36). The PCR fragment (1799 bp) consisted of 504 bp of pYV sequence upstream of yscQ, followed by aph, the 22 bp from the 3′ end of yscQ, and 479 bp of pYV sequence downstream of yscQ. A minor modification was made to the published protocol in order to increase the induction time for expression of the λ red recombinase proteins. The proper substitution of yscQ by aph was verified by DNA sequencing.
Allelic replacement of yscQ
Replacement of aph by yscQ alleles in Y. pseudotuberculosis (ΔyscQ) through homologous recombination was carried out using plasmid pSB890 (37), as previously described with some modifications (38). The plasmid pWL204, which carries the λ red recombinase genes, was first transformed into Y. pseudotuberculosis (ΔyscQ) to facilitate recombination (35). This was followed by transformation of plasmid pSB890 carrying mutant yscQ alleles flanked by 504 and 457 bp of pYV sequence upstream and downstream, respectively, of yscQ. In addition, insertion of pSB890 into pYV was selected for by growth on agar plates containing tetracycline followed by growth in liquid media (BHI) containing tetracycline. The proper replacement of aph by yscQ alleles was verified by DNA sequencing.
Secretion Assay
Type III secretion was examined as previously published (38). Briefly, Y. pseudotuberculosis strains expressing yscQ from its native locus on the pYV plasmid were induced for type III secretion for 3 h in Ca2+-deficient media at 37 °C, whereupon the media was separated from bacteria by centrifugation (5 min, 14,000 × g, 23°C), and the media was precipitated with trichloroacetic acid (TCA) and resolubilized for analysis by Coomassie-stained SDS-PAGE. Prior to centrifugation of bacteria, the OD600 of samples was measured for normalization. An identical procedure was followed for Y. pseudotuberculosis (ΔyscQ) complemented with pBAD containing wild-type or variant YscQ, except that 0.1% L-arabinose was included in the growth media before the induction of secretion. Immediately prior to the induction of secretion, fresh L-arabinose was added to a final concentration of 0.2%.
Detection of YscQ in Y. pseudotuberculosis
Rabbit anti-YscQ polyclonal antibodies raised against YscQ-C as an antigen (Abgent, San Diego, CA) were purified using a YscQ-C affinity column (cross-linked to Aminolink resin, Pierce, Rockford, IL), and used as the primary antibody in Western blots, or cross-linked to Aminolink resin for isolation of YscQ from Y. pseudotuberculosis. For the latter, Y. pseudotuberculosis was grown in non-secreting conditions (BHI containing 2 mM CaCl2, 28°C) to OD600 0.64–0.66, at which point EGTA and MgCl2 were added to a final concentration of 10 mM each, and the culture was shifted to 37 °C to induce secretion and grown further for 3 h. Bacteria were harvested by centrifugation, resuspended in PBS, lysed by passing 3x through an EmulsiFlex-C5 (Avestin, Canada), and cell debris was removed by centrifugation (30 min, 20,000 × g, 4°C). The supernatant was then applied to the anti-YscQ-C antibody column, which was then washed extensively with PBS, and bound proteins were eluted with 0.1 M glycine, pH 2.9 and immediately neutralized with 1 M Tris, pH 9.0. The eluate was concentrated in a SpeedVac or precipitated with TCA prior to analysis by Western blot.
Western blot
Samples were separated on 12% SDS PAGE and transferred to a PVDF membrane using a Trans-Blot semi dry cell (Biorad, Hercules, CA) for 25–30 min at 15–30V. The membrane was blocked overnight at 4 °C in 5% nonfat dry milk in PBST (PBS including 0.1% Tween 80), washed once with PBST, and incubated for 1 h at 23 °C with primary antibody (1:2000 anti-His (Santa Cruz Biotechnology, Santa Cruz, CA) or 1:1000 purified anti-YscQ-C) in 5% nonfat dry milk in PBST, followed by 3 washes with PBST. An HRP-conjugated secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA) at 1:10000 was incubated with the membrane in 5% nonfat dry milk. Following extensive washes with PBST, the membrane was developed using ECL Western Blotting system (GE Healthcare, Piscataway, NJ) and visualized with Amersham Hyperfilm ECL (GE Healthcare, Piscataway, NJ).
Ni2+-NTA coprecipitation
Proteins were incubated overnight at 4 °C in 100 μL of PBS at 40 μM, except for the high concentration sample of YscQ-C-His6, which was at 280 μM. Samples were then centrifuged (1 min, 15,800 × g, 4 °C). Ten μL were reserved as the input sample. Eighty μL of the sample was then incubated for 30 min at 4 °C with end-over-end agitation with 50 μL of a ~50% HIS-select Ni2+ affinity gel (Sigma) slurry, which had been washed with PBS. The resin was washed 5x with 1 mL of 500 mM NaCl, 20 mM Tris, pH 8, 40 mM imidazole, and 0.1% Triton X-100, and then transferred to a new tube and washed once more. A centrifugation step (10 min, 720 × g, RT) separated unbound proteins from the resin after each wash. Bound proteins were eluted from the resin with 2x Tris-Tricine SDS-PAGE sample loading buffer and heated for 2 min at 55 °C. Samples were analyzed by 10% Tris-Tricine SDS-PAGE.
RESULTS
Two products from one gene
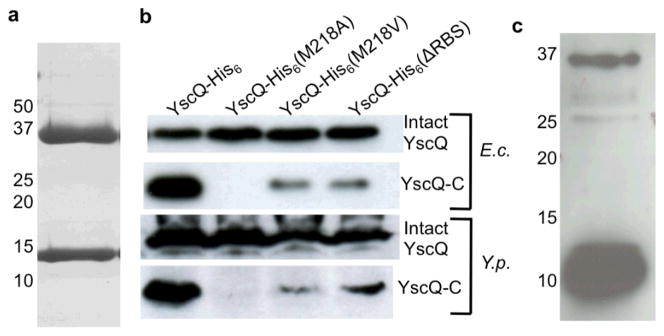
We expressed His-tagged YscQ recombinantly in E. coli and found that it co-purified as two protein products throughout Ni2+-chelation and size-exclusion chromatography (Fig. 1a). The first product migrated on SDS-PAGE at ~36 kDa, the expected size for intact YscQ (307 residues) containing a C-terminal His-tag, and the second migrated at ~11 kDa. The larger product was verified to be intact YscQ by N-terminal sequencing, which also revealed that the ~11 kDa product began at residue 218 and corresponded to the C-terminal portion of YscQ. While truncation fragments often arise due to inadvertent proteolysis, it was notable that residue 218 was a Met. This raised the possibility that translation initiation at codon 218 in yscQ had given rise to the C-terminal product, hereafter called YscQ-C (residues 218–307). Consistent with this hypothesis, a possible ribosome-binding site (RBS) upstream of codon 218 was found (Fig. S1).
Figure 1.
YscQ-C results from an internal translation initiation site. (a) YscQ expressed in E. coli and purified by Ni2+-chelation and size-exclusion chromatography, as visualized by Coomassie-stained SDS-PAGE. Molecular mass markers are indicated at left. Intact YscQ is the upper band at ~36 kDa and YscQ-C the lower band at ~11 kDa. (b) Wild-type and mutant YscQ expressed in E. coli (top two blots) and Y. pseudotuberculosis (bottom two), and detected by anti-His western blot from whole cell lysates. YscQ contains a C-terminal His-tag, and was detected as intact YscQ (upper panel in each set) and YscQ-C (lower panel in each set). (c) YscQ detected in Y. pseudotuberculosis by Western blot using anti-YscQ polyclonal antibodies. Prior to the Western blot, YscQ was captured from a whole cell Y. pseudotuberculosis lysate by affinity chromatography using the same anti-YscQ polyclonal antibodies. Molecular mass markers are indicated at left.
To test whether an internal translation initiation site existed, M218 was substituted with Ala. Expression of YscQ(M218A) in E. coli yielded the ~36 kDa intact YscQ product, but notably no YscQ-C (Fig. 1b), providing evidence for an internal translation initiation site. We next purified endogenous YscQ from Y. pseudotuberculosis, and detected both intact YscQ and YscQ-C (Fig. 1c). For this experiment, YscQ was captured and concentrated from a Y. pseudotuberculosis lysate using anti-YscQ polyclonal antibodies affixed to beads, and detected by Western blot using the same antibodies. These steps were necessary because the reactivity of the anti-YscQ polyclonal antibodies was poor. The imbalanced levels of intact YscQ and YscQ-C was an artifact, as the same imbalance was observed through this procedure for YscQ expressed in E. coli, in which equivalent levels of YscQ and YscQ-C were known to be produced (Fig. S2). The imbalance may be due to the fact that the antibodies were raised against YscQ-C, which appears to differ in conformation from intact YscQ (see below).
Internal translation initiation site
To overcome problems associated with the low sensitivity of our anti-YscQ polyclonal antibodies, we expressed His-tagged versions of YscQ in Y. pseudotuberculosis using the inducible araBAD promoter of the pBAD plasmid. As expected from the results with endogenous YscQ, His-tagged YscQ was expressed as two products in Y. pseudotuberculosis, intact YscQ and YscQ-C, as detected by an anti-His Western blot (Fig. 1b). Notably, expression of YscQ(M218A) in Y. pseudotuberculosis resulted in only one product, intact YscQ. No YscQ-C was observed, as had been the case in E. coli.
We next substituted the ATG codon at position 218 with GTG, which encodes valine and can function as an alternative initiation codon in bacteria. This substitution, YscQ(M218V), resulted in two protein products, intact YscQ and YscQ-C, in both Y. pseudotuberculosis and E. coli (Fig. 1b). GTG was less efficient than ATG as an initiation codon, and yielded less YscQ-C relative to intact YscQ. We also created a silent mutation in the putative ribosome-binding site upstream of codon 218 (GGAGTT→AGAATT). This mutation, YscQ(ΔRBS), substantially diminished the efficiency of the internal translation initiation site in Y. pseudotuberculosis and E. coli (Fig. 1b). In summary, these results provide strong evidence for the existence of an internal translation initiation site at codon 218, which results in two distinct proteins being produced from the single yscQ locus.
Structure of YscQ-C
YscQ-C was expressed in E. coli, purified, and crystallized. The 2.25 Å resolution limit structure of YscQ-C was determined by single wavelength anomalous dispersion (Table 1). Except for the first 11 residues, which were presumably flexible, the entirety of YscQ-C was visible and unambiguously traced. The structure revealed a highly intertwined dimer (Fig. 2a), characteristic of the SpoA fold in fragments of T. maritima FliN (8) (29% sequence identity, 3.2 Å rmsd, 75 Cα atoms, Z-score 7.4) and P. syringae HrcQB (22) (27% sequence identity, 2.3 Å rmsd, 70 Cα atoms, Z-score 7.4) (Figs. 2b and S3). As in FliN and HrcQB, each YscQ-C protomer consists of five β-strands (β1-β5) with a short helix (α1) between β1 and β2. The 10 anti-parallel β-strands in the YscQ-C dimer assemble to form a saddle-shaped structure. The YscQ-C protomers are related by approximate two-fold symmetry, but are not identical (rmsd 1.4 Å, 85 Cα atoms), which is most apparent at the β1 strand (Fig. 2a). In one of the protomers, this β-strand is disrupted (between β1a and β1b) and several residues within this disruption have orientations opposite to those in the other protomer (Fig. S3). Each protomer buries ~2140 Å2 of surface area at the largely apolar intermolecular interface (dominated by F242, W251, L262, V269, L294). The identity of these residues is not conserved in FliN and HrcQB, but their hydrophobic nature is (Fig. S4). The same is true for T3S orthologs of YscQ (Fig. S5). The hydrophobic interactions in YscQ-C are supplemented by a large number of β-sheet hydrogen bonds between the antiparallel β1 strands. The surface of YscQ-C contains extensive patches of hydrophobic residues (Fig. S6), consistent with it being involved in protein-protein interactions as part of the putative C-ring. One of these patches occurs at the dyad axis of the homodimer (Fig. S6b), at a location equivalent to one in FliN that has been shown to be functionally important in mediating interactions with the ATPase regulator FliH (39).
Figure 2.
Structure of YscQ-C. (a) The YscQ-C dimer in ribbon representation, with individual protomers in red and blue. (b) Superposition of YscQ-C (blue), T. maritima FliN (red), and P. syringae HrcQB (green) in coils representation. (c) The domain-swapped subdomain of YscQ-C is boxed. The subdomain could conceivably pivot at the disruption in the β1 strand and the loop between β2 and β3 to form intramolecular contacts and replace the equivalent blue domain. This would result in a monomeric form of the YscQ-C domain.
The highly intertwined nature of the YscQ-C homodimer is due to domain-swapping. A subdomain that consists of part of the β1 strand through the β2 strand from one protomer (Fig. 2c, boxed) reaches across and contacts the adjacent protomer. This same subdomain could conceivably reorient and ‘snap back’ to contact the same protomer from which it originated (Fig. 2c), thereby conferring a monomeric state to the SpoA domain, as has been suggested for FliM (9). A reasonable hinge for this ‘snap-back’ occurs at the disruption in the β1 strand in conjunction with the loop between the β2 and β3 strands.
Intact YscQ and YscQ-C form a 1:2 complex
The co-purification of intact YscQ with YscQ-C suggested that they formed a complex. To test this hypothesis, we separately expressed and purified intact YscQ, as produced by YscQ(M218A), and YscQ-C, and incubated the two proteins together. The resulting mixture ran on a gel filtration column at a position earlier than that of either YscQ(M218A) or YscQ-C (Fig. 3a). Significantly, this distinct and shifted position coincided precisely with the position of YscQ purified from E. coli, which consists of both intact YscQ and YscQ-C. These results confirm that YscQ and YscQ-C interact to form a complex, and show that the complex can be reconstituted from individual components. The YscQ/YscQ-C complex was determined to have a molecular mass of 52.8 kDa by static light scattering (Fig. 3b). This corresponds to a single molecule of intact YscQ (35.0 kDa calculated) bound to a homodimer of YscQ-C (21.5 kDa calculated). The homodimeric nature of YscQ-C, as observed in the crystal structure, was confirmed by static light scattering (19.8 kDa measured) (Fig. 3b). Such measurements were difficult with YscQ(M218A) because of high molecular mass impurities. However, we note that YscQ(M218A) ran between YscQ/YscQ-C and YscQ-C on the gel filtration column (Fig. 3a), indicative of a monomeric state for YscQ(M218A).
Figure 3.
Intact YscQ and YscQ-C form a 1:2 heterotrimer. (a) Gel filtration chromatographic profiles of YscQ-C, YscQ(M218A), YscQ(M218A) incubated with YscQ-C, and YscQ purified from E. coli (YscQ/YscQ-C). (b) Static light scattering of YscQ/YscQ-C and YscQ-C applied to a gel filtration column. The molar mass is indicated across the peaks. The proteins were run separately but the chromatograms were superimposed for display purposes. (c) Exchangeability of YscQ/YscQ-C and YscQ-C was evaluated by incubating YscQ-C-His6 separately with untagged YscQ/YscQ-C, YscQ(M218A), and YscQ-C. Association was detected by Ni2+-NTA coprecipitation and visualized by Coomassie-stained Tris-Tricine SDS-PAGE.
The 1:2 stoichiometry of the YscQ/YscQ-C complex leads to the conclusion that the sequence-identical SpoA domains of intact YscQ and YscQ-C take on different conformations. To test this unusual conclusion, we examined the exchangeability of components in the 1:2 YscQ:YscQ-C complex. We first confirmed that YscQ-C forms a non-exchangeable homodimer, as suggested by its extensive intermolecular interface. To do this, we incubated His-tagged YscQ-C with untagged YscQ-C, and observed no exchange of partners in a Ni2+-NTA coprecipitation assay, even with an excess of untagged YscQ-C (Fig. 3c). We next confirmed that intact YscQ, in the form of YscQ(M218A), associates with YscQ-C by incubating the two proteins together and observing the coprecipitation of YscQ(M218A) with His-tagged YscQ-C. We then asked whether free His-tagged YscQ-C would exchange with untagged YscQ-C from the YscQ/YscQ-C complex. We reasoned that if the SpoA domain in intact YscQ interacted with the SpoA domain in YscQ-C through the domain-swap observed for YscQ-C, then no exchange should occur. We instead found that a portion of the YscQ/YscQ-C complex had acquired His-tagged YscQ-C, providing evidence of exchange. These data indicate that the SpoA domain in intact YscQ does not engage in the irreversible, dimeric intertwining seen in YscQ-C, but instead takes on a different conformation.
Both intact YscQ and YscQ-C are required for type III secretion
We lastly examined the significance of the internal initiation site for type III secretion. We constructed an in-frame yscQ deletion mutant in Y. pseudotuberculosis, and showed that, as expected (13), the loss of YscQ abrogated type III secretion (Fig. 4, ΔyscQ). The deletion was nonpolar, as type III secretion was restored by complementation with wild-type, His-tagged YscQ expressed from pBAD (Fig. 4). Next, various alleles of yscQ were introduced by allelic exchange into their native locus on the pYV virulence plasmid of Y. pseudotuberculosis, and the resulting strains were tested for type III secretion. Notably, we found that Y. pseudotuberculosis expressing either the yscQ-M218A allele or the yscQ-C allele (encoding just residues 218–307) was deficient in type III secretion (Fig. 4). Type III secretion was restored in these strains by expressing YscQ-C or YscQ(M218), respectively, from a plasmid. These results indicate that the YscQ/YscQ-C complex is required for T3S, and that the internal translation initiation site is essential for function. It should be noted that the slight differences among samples in quantities of proteins secreted were not significant, as these were representative of typical variability in the procedure. No defect in type III secretion was seen for either the yscQ-M218V allele or the RBS mutated allele, and no effect was seen when additional YscQ-C was expressed from a plasmid in these strains. This means that an imbalanced ratio of intact YscQ relative to YscQ-C is not detrimental, and suggests that YscQ is produced in excess in Y. pseudotuberculosis.
Figure 4.
Both intact YscQ and YscQ-C are required for T3S. Proteins secreted via the T3S system by Y. pseudotuberculosis expressing wild-type (WT) or various mutant alleles of yscQ were detected by Coomassie-stained SDS-PAGE. Mutant alleles were incorporated into the native locus on the pYV plasmid by allelic exchange or expressed ectopically from the pBAD plasmid (designated by parentheses). The ‘M’ lane corresponds to molecular mass markers (kDa).
DISCUSSION
Type III secretion systems generally encode a single protein in the place of the two flagellar proteins FliM and FliN, which associate to form the flagellar C-ring. These T3S cognates are similar in their N-terminal domains to FliM and are as large as FliM, but more closely resemble FliN in their C-terminal SpoA domains. Most T3S systems appear to lack a smaller FliN-sized cognate to accompany the larger FliM-sized cognate. We have discovered that in the Yersinia T3S system, a FliN-sized cognate exists. The FliN-sized cognate, YscQ-C, is not encoded by a separate locus, but is instead produced by an internal translation initiation site within the locus encoding the FliM-sized cognate YscQ. This is similar to a recently described case in the Salmonella SPI-2 T3S system for the YscQ ortholog SsaQ (16). A C-terminal fragment of SsaQ, called SsaQs, arises from an internal translation start site, and this fragment associates with intact SsaQ. However, the mechanism of action of SsaQs and YscQ-C differ considerably. In contrast to the essential nature of YscQ-C for T3S in Yersinia, SsaQs was found to be dispensable for T3S in Salmonella (16). In the absence of SsaQs, proteins are still secreted by the T3S system, albeit at a somewhat diminished level (16). Notably, this diminution was observed to be complemented by overexpression of intact SsaQ. The dispensability of SsaQs is explained by the fact that SsaQs, rather than acting as a part of the T3S apparatus, is a chaperone for intact SsaQ, as evidenced by the fact that intact SsaQ has a shorter intrabacterial half-life in the absence of SsaQs (16). This chaperone mode of action for SsaQs is unusual and unprecedented for FliN family members. In contrast, our evidence suggests that YscQ-C mimics FliN in forming an essential part of the T3S.
The internal translation start site in SsaQ is at a slightly different location relative to the one in YscQ (Fig. S5). However, the N-terminus of YscQ-C is disordered and thus the exact location of the start site is unlikely to be consequential. An examination of other YscQ orthologs reveals that the M218 internal translation initiation site is precisely conserved in Pseudomonas aeruginosa PscQ, Aeromonas hydrophila AscQ, Photorhabdus luminescens LscQ, and Arsenophonus nasoniae SctQ (Fig. S5). In each of these cases, a potential ribosome binding site also occurs upstream of the Met codon (Fig. S1). This pattern of conservation indicates that these YscQ orthologs are also likely to produce FliN cognates through an internal translation initiation site. However, M218 is not conserved in all YscQ orthologs. For example, Shigella Spa33, Salmonella SpaO (from the SPI-1 T3S), and E. coli EscQ (SepQ) lack the equivalent of M218. They also do not have nearby ATG codons. Bacteria can also use TTG, GTG, and CTG as start codons, and while these exist near the equivalent of M218, it is impossible to say with confidence that these are used as internal translation initiation sites due to the absence of unambiguous ribosome binding sites upstream of these codons. It is possible that in these latter cases an alternative mechanism (e.g., proteolysis) exists to generate a FliN-sized cognate or that the putative T3S C-ring functions without one.
We determined that intact YscQ and YscQ-C associate to form a complex. Likewise, HrcQB interacts directly with the FliM-sized cognate HrcQA (22) and SsaQs interacts either directly or indirectly with the FliM-cognate SsaQ (16), but the stoichiometries of these associations are unknown. We determined the stoichiometry of the YscQ:YscQ-C complex to be 1:2, which is similar to the 1:4 complex reported for FliM:FliN in having a single large component bound to several smaller ones, but of course differs in the number of smaller ones (8). The FliN tetramer appears to consist of either an extended or donut-shaped dimer of dimers (22, 40, 41). The residues that mediate the dimer-of-dimer contacts, however, are not conserved in YscQ (Fig. S4), and no evidence was found in solution or in the crystal for tetramerization of YscQ-C. Interactions between FliM and FliN are mediated by their SpoA domains (10). While the SpoA domain in FliN confers homodimerization (8), the one in FliM does not. Instead, the SpoA domain in FliM forms an uncharacterized structure that supports association with FliN homodimers (9). Our observations with YscQ are consistent with these conclusions but extend them one step further, as the SpoA domains in FliM and FliN are only similar in sequence while those in intact YscQ and YscQ-C are exactly identical.
The SpoA domain of YscQ-C, as indicated by its crystal structure, exists as a domain-swapped homodimer. Consistent with its highly intertwined structure, YscQ-C was found to exist as a stable dimer and dissociate only upon denaturation. The structure of the SpoA domain in intact YscQ is unknown. However, the monomeric nature of intact YscQ and its exchangeability indicate that the SpoA domain in intact YscQ takes on a different conformation than the one in YscQ-C. We suggest that the N-terminal portions of YscQ disfavor the intermolecular, domain-swapped interactions of the SpoA domain seen in YscQ-C, and instead favor intramolecular, non-swapped interactions within the SpoA domain (Fig. 5). More specifically, we suggest that the N-terminal regions contact the SpoA domain, perhaps at the suggested ‘snap back’ hinge (i.e., disruption in the β1 strand along with the loop between the β2 and β3 strands), to favor these interactions. It is worth noting that the sequence at the disruption in the β1 strand is conserved among YscQ orthologs. In analogy with the FliM/FliN and the HrcQA/HrcQB complexes (10, 22), the non-swapped SpoA domain of intact YscQ is likely to mediate interactions with the domain-swapped SpoA domain of the YscQ-C homodimer.
Figure 5.
Schematic of the assembly of the 1:2 intact YscQ:YscQ-C complex. The SpoA domain of YscQ-C, in the absence of the N-terminal domain (‘N’) of YscQ, takes on a domain-swapped homodimeric conformation (red). In contrast, in the presence of the N-terminal domain, the SpoA domain is unable to form a domain-swapped homodimer, and instead forms intramolecular contacts (purple). Based on analogy with FliM/FliN, we also suggest that the monomeric, non-swapped SpoA domain in intact YscQ mediates association with the domain-swapped SpoA domains in the YscQ-C homodimer, resulting in the 1:2 intact YscQ:YscQ-C complex.
In summary, we have identified an essential internal translation initiation site in yscQ. This site is necessary for the synthesis of YscQ-C, a functionally required component of the 1:2 YscQ:YscQ-C complex, which is a close mimic of the 1:4 FliM:FliN complex and is the likely fundamental building block of the putative C-ring in Yersinia.
Supplementary Material
Acknowledgments
Funding Sources
This works was supported by NIH grant T32 CA009523 (BH) and R01 AI061452 (PG).
We thank the staff at beamline 23 ID-B for help in data collection, and Johanne Le Coq, Alicia Gamez, and other members of the Ghosh lab for helpful suggestions.
ABBREVIATIONS
- 2ME
2-mercaptoethanol
- RBS
ribosome-binding site
- SeMet
selenomethionine
- SLS
static light scattering
- T3S
type III secretion
Footnotes
Author Contributions
The manuscript was written through contributions of all authors.
ASSOCIATED CONTENT
Supporting Information. Figures S1–S6. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- 2.Cornelis GR. The type III secretion injectisome. Nat Rev Microbiol. 2006;4:811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- 3.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galan JE, Aizawa SI. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 4.Akopyan K, Edgren T, Wang-Edgren H, Rosqvist R, Fahlgren A, Wolf-Watz H, Fallman M. Translocation of surface-localized effectors in type III secretion. Proc Natl Acad Sci U S A. 2011;108:1639–1644. doi: 10.1073/pnas.1013888108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghosh P. Process of protein transport by the type III secretion system. Microbiol Mol Biol Rev. 2004;68:771–795. doi: 10.1128/MMBR.68.4.771-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas DR, Francis NR, Xu C, DeRosier DJ. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J Bacteriol. 2006;188:7039–7048. doi: 10.1128/JB.00552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erhardt M, Hughes KT. C-ring requirement in flagellar type III secretion is bypassed by FlhDC upregulation. Mol Microbiol. 2010;75:376–393. doi: 10.1111/j.1365-2958.2009.06973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown PN, Mathews MA, Joss LA, Hill CP, Blair DF. Crystal structure of the flagellar rotor protein FliN from Thermotoga maritima. J Bacteriol. 2005;187:2890–2902. doi: 10.1128/JB.187.8.2890-2902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarkar MK, Paul K, Blair DF. Subunit organization and reversal-associated movements in the flagellar switch of Escherichia coli. J Biol Chem. 2010;285:675–684. doi: 10.1074/jbc.M109.068676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathews MA, Tang HL, Blair DF. Domain analysis of the FliM protein of Escherichia coli. J Bacteriol. 1998;180:5580–5590. doi: 10.1128/jb.180.21.5580-5590.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diepold A, Amstutz M, Abel S, Sorg I, Jenal U, Cornelis GR. Deciphering the assembly of the Yersinia type III secretion injectisome. EMBO J. 2010;29:1928–1940. doi: 10.1038/emboj.2010.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morita-Ishihara T, Ogawa M, Sagara H, Yoshida M, Katayama E, Sasakawa C. Shigella Spa33 is an essential C-ring component of type III secretion machinery. J Biol Chem. 2006;281:599–607. doi: 10.1074/jbc.M509644200. [DOI] [PubMed] [Google Scholar]
- 13.Fields KA, Plano GV, Straley SC. A low-Ca2+ response (LCR) secretion (ysc) locus lies within the lcrB region of the LCR plasmid in Yersinia pestis. J Bacteriol. 1994;176:569–579. doi: 10.1128/jb.176.3.569-579.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lara-Tejero M, Kato J, Wagner S, Liu X, Galan JE. A sorting platform determines the order of protein secretion in bacterial type III systems. Science. 2011;331:1188–1191. doi: 10.1126/science.1201476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biemans-Oldehinkel E, Sal-Man N, Deng W, Foster LJ, Finlay BB. Quantitative Proteomic Analysis Reveals Formation of an EscL-EscQ-EscN Type III Complex in Enteropathogenic Escherichia coli. J Bacteriol. 2011;193:5514–5519. doi: 10.1128/JB.05235-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu XJ, Liu M, Matthews S, Holden DW. Tandem Translation Generates a Chaperone for the Salmonella Type III Secretion System Protein SsaQ. J Biol Chem. 2011;286:36098–36107. doi: 10.1074/jbc.M111.278663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson MW, Plano GV. Interactions between type III secretion apparatus components from Yersinia pestis detected using the yeast two-hybrid system. FEMS Microbiol Lett. 2000;186:85–90. doi: 10.1111/j.1574-6968.2000.tb09086.x. [DOI] [PubMed] [Google Scholar]
- 18.Blaylock B, Riordan KE, Missiakas DM, Schneewind O. Characterization of the Yersinia enterocolitica type III secretion ATPase YscN and its regulator, YscL. J Bacteriol. 2006;188:3525–3534. doi: 10.1128/JB.188.10.3525-3534.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riordan KE, Sorg JA, Berube BJ, Schneewind O. Impassable YscP substrates and their impact on the Yersinia enterocolitica type III secretion pathway. J Bacteriol. 2008;190:6204–6216. doi: 10.1128/JB.00467-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson DL, Stone CB, Mahony JB. Interactions between CdsD, CdsQ, and CdsL, three putative Chlamydophila pneumoniae type III secretion proteins. J Bacteriol. 2008;190:2972–2980. doi: 10.1128/JB.01997-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spaeth KE, Chen YS, Valdivia RH. The Chlamydia type III secretion system C-ring engages a chaperone-effector protein complex. PLoS Pathog. 2009;5:e1000579. doi: 10.1371/journal.ppat.1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fadouloglou VE, Tampakaki AP, Glykos NM, Bastaki MN, Hadden JM, Phillips SE, Panopoulos NJ, Kokkinidis M. Structure of HrcQB-C, a conserved component of the bacterial type III secretion systems. Proc Natl Acad Sci U S A. 2004;101:70–75. doi: 10.1073/pnas.0304579101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolin I, Norlander L, Wolf-Watz H. Temperature-inducible outer membrane protein of Yersinia pseudotuberculosis and Yersinia enterocolitica is associated with the virulence plasmid. Infect Immun. 1982;37:506–512. doi: 10.1128/iai.37.2.506-512.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Budisa N, Karnbrock W, Steinbacher S, Humm A, Prade L, Neuefeind T, Moroder L, Huber R. Bioincorporation of telluromethionine into proteins: a promising new approach for X-ray structure analysis of proteins. J Mol Biol. 1997;270:616–623. doi: 10.1006/jmbi.1997.1132. [DOI] [PubMed] [Google Scholar]
- 25.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Vol. 276. Academic Press; New York: 1997. [DOI] [PubMed] [Google Scholar]
- 27.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Brünger A. Crystallography & NMR system: a new software for macromolecular structure determination. Acta Crystallogr sect D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 32.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Tommaso P, Moretti S, Xenarios I, Orobitg M, Montanyola A, Chang JM, Taly JF, Notredame C. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011;39:W13–17. doi: 10.1093/nar/gkr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lathem WW, Price PA, Miller VL, Goldman WE. A plasminogen-activating protease specifically controls the development of primary pneumonic plague. Science. 2007;315:509–513. doi: 10.1126/science.1137195. [DOI] [PubMed] [Google Scholar]
- 36.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palmer LE, Hobbie S, Galan JE, Bliska JB. YopJ of Yersinia pseudotuberculosis is required for the inhibition of macrophage TNF-alpha production and downregulation of the MAP kinases p38 and JNK. Mol Microbiol. 1998;27:953–965. doi: 10.1046/j.1365-2958.1998.00740.x. [DOI] [PubMed] [Google Scholar]
- 38.Rodgers L, Mukerjea R, Birtalan S, Friedberg D, Ghosh P. A solvent-exposed patch in chaperone-bound YopE is required for translocation by the type III secretion system. J Bacteriol. 2010;192:3114–3122. doi: 10.1128/JB.00113-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paul K, Harmon JG, Blair DF. Mutational analysis of the flagellar rotor protein FliN: identification of surfaces important for flagellar assembly and switching. J Bacteriol. 2006;188:5240–5248. doi: 10.1128/JB.00110-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fadouloglou VE, Bastaki MN, Ashcroft AE, Phillips SE, Panopoulos NJ, Glykos NM, Kokkinidis M. On the quaternary association of the type III secretion system HrcQB-C protein: experimental evidence differentiates among the various oligomerization models. J Struct Biol. 2009;166:214–225. doi: 10.1016/j.jsb.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 41.Paul K, Blair DF. Organization of FliN subunits in the flagellar motor of Escherichia coli. J Bacteriol. 2006;188:2502–2511. doi: 10.1128/JB.188.7.2502-2511.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.