Abstract
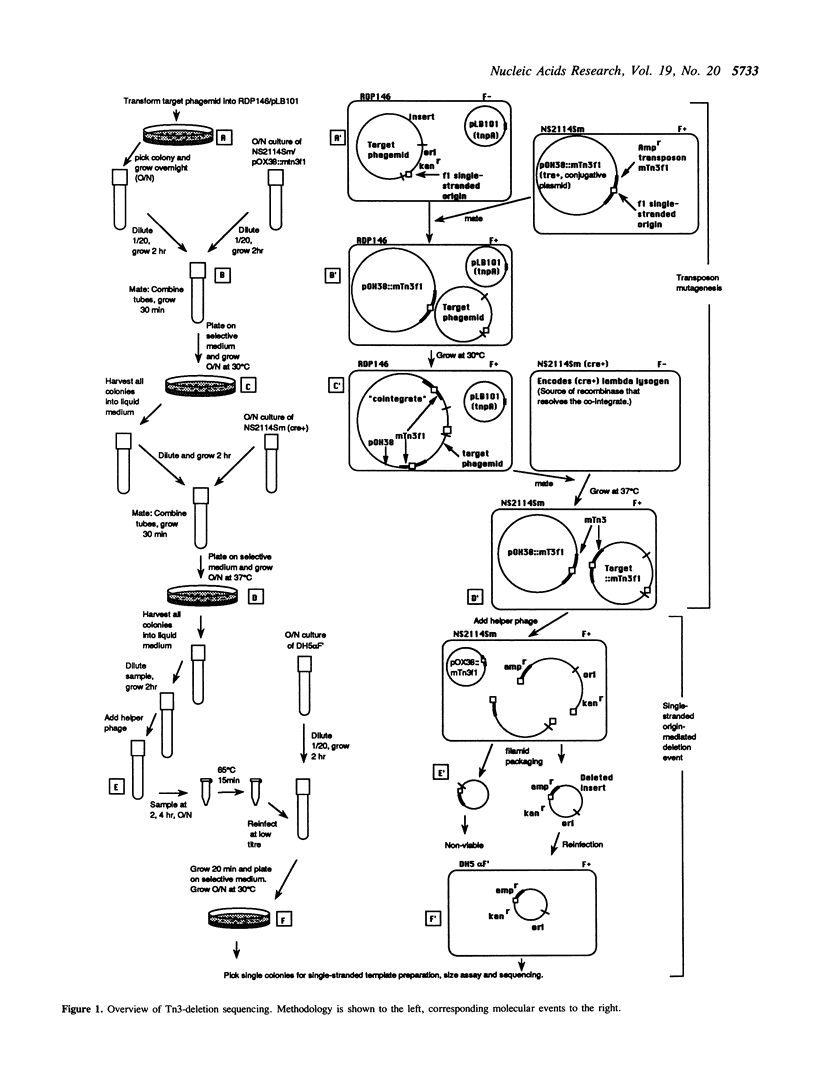
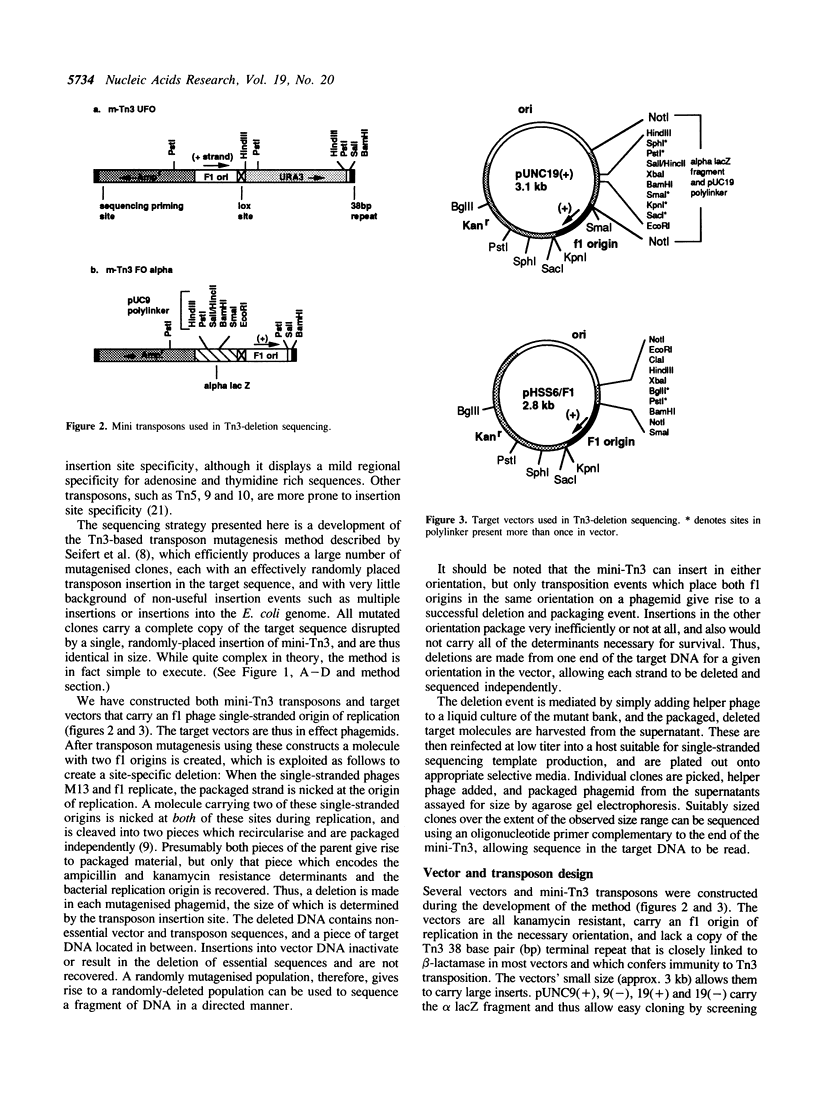
We have developed a directed DNA sequencing strategy based upon the Escherichia coli transposon Tn3. This transposon displays little sequence specificity for transposition and is thus well suited to this task. Both mini-Tn3 transposons and sequencing vectors bearing the phage f1 single stranded origin of replication have been constructed. Upon mutagenesis of a target sequence, a population is produced in which each clone has two f1 origins of replication, one of which is at a variable position depending upon the transposon insertion site. When helper phage is added to the mutagenised population, the two f1 origins present on each clone are nicked, dividing the packaged strand into two segments, each of which is packaged into a separate phage particle. One of these segments contains no resistance markers and is lost, whilst the other is recovered as a deleted clone with a single chimeric f1 origin. A unidirectionally, variably-deleted set of sequencing clones is produced, and appropriately sized clones are sequenced using a primer complimentary to the transposon end. In addition to being inexpensive, the method does not require the same degree of technical expertise needed for many in vitro, enzymatically based methods. The strategy has been used to determine 2.6 kilobases of nucleotide sequence in the Saccharomyces cerevisiae ADE 1 locus.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmed A. A rapid procedure for DNA sequencing using transposon-promoted deletions in Escherichia coli. Gene. 1985;39(2-3):305–310. doi: 10.1016/0378-1119(85)90328-2. [DOI] [PubMed] [Google Scholar]
- Crowley J. C., Kaback D. B. Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae: isolation of the ADE1 gene. J Bacteriol. 1984 Jul;159(1):413–417. doi: 10.1128/jb.159.1.413-417.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale R. M., McClure B. A., Houchins J. P. A rapid single-stranded cloning strategy for producing a sequential series of overlapping clones for use in DNA sequencing: application to sequencing the corn mitochondrial 18 S rDNA. Plasmid. 1985 Jan;13(1):31–40. doi: 10.1016/0147-619x(85)90053-8. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbole D. J., Zalkin H. Cloning and characterization of a 12-gene cluster from Bacillus subtilis encoding nine enzymes for de novo purine nucleotide synthesis. J Biol Chem. 1987 Jun 15;262(17):8274–8287. [PubMed] [Google Scholar]
- Eperon L. P., Graham I. R., Griffiths A. D., Eperon I. C. Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell. 1988 Jul 29;54(3):393–401. doi: 10.1016/0092-8674(88)90202-4. [DOI] [PubMed] [Google Scholar]
- Frischauf A. M., Garoff H., Lehrach H. A subcloning strategy for DNA sequence analysis. Nucleic Acids Res. 1980 Dec 11;8(23):5541–5549. doi: 10.1093/nar/8.23.5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hutchison C. A., 3rd, Swanstrom R., Loeb D. D. Complete mutagenesis of protein coding domains. Methods Enzymol. 1991;202:356–390. doi: 10.1016/0076-6879(91)02019-6. [DOI] [PubMed] [Google Scholar]
- Meyer T. F., Geider K., Kurz C., Schaller H. Cleavage site of bacteriophage fd gene II-protein in the origin of viral strand replication. Nature. 1979 Mar 22;278(5702):365–367. doi: 10.1038/278365a0. [DOI] [PubMed] [Google Scholar]
- Miasnikov A. N., Plavnik Iu A., Sasnauskas K. V., Gedminene G. K., Ianulaitis A. A. Nukleotidnaia posledovatel'nost' gena ADE 1 drozhzhei Saccharomyces cerevisiae. Bioorg Khim. 1986 Apr;12(4):555–558. [PubMed] [Google Scholar]
- Minton N. P., Atkinson T., Bruton C. J., Sherwood R. F. The complete nucleotide sequence of the Pseudomonas gene coding for carboxypeptidase G2. Gene. 1984 Nov;31(1-3):31–38. doi: 10.1016/0378-1119(84)90192-6. [DOI] [PubMed] [Google Scholar]
- Misra T. K. DNA sequencing: a new strategy to create ordered deletions, modified M13 vector, and improved reaction conditions for sequencing by dideoxy chain termination method. Methods Enzymol. 1987;155:119–139. doi: 10.1016/0076-6879(87)55012-1. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert H. S., Chen E. Y., So M., Heffron F. Shuttle mutagenesis: a method of transposon mutagenesis for Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1986 Feb;83(3):735–739. doi: 10.1073/pnas.83.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Automation of the computer handling of gel reading data produced by the shotgun method of DNA sequencing. Nucleic Acids Res. 1982 Aug 11;10(15):4731–4751. doi: 10.1093/nar/10.15.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathmann M., Hamilton B. A., Mayeda C. A., Simon M. I., Meyerowitz E. M., Palazzolo M. J. Transposon-facilitated DNA sequencing. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1247–1250. doi: 10.1073/pnas.88.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausbaugh L. D., Bourke M. T., Sommer M. T., Coon M. E., Berg C. M. Probe mapping to facilitate transposon-based DNA sequencing. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6213–6217. doi: 10.1073/pnas.87.16.6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedeman A. A., DeMarini D. J., Parker J., Smith J. M. DNA sequence of the purC gene encoding 5'-phosphoribosyl-5-aminoimidazole-4-N-succinocarboxamide synthetase and organization of the dapA-purC region of Escherichia coli K-12. J Bacteriol. 1990 Oct;172(10):6035–6041. doi: 10.1128/jb.172.10.6035-6041.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]




