Drug resistance in acute myeloid leukemia (AML) after treatment is a common problem, and the anti-apoptotic Bcl-2 family members Bcl-xL and Mcl-1 are believed to contribute to relapse. In this Perspective concerning a study by Strasser and coworkers (published in the January 15, 2012, issue), the authors discuss the finding that Mcl-1, but not other anti-apoptotic Bcl-2 family members, is essential for the development and survival of AML cells. How these findings can be used for therapeutic benefit is also discussed.
Keywords: acute myeloid leukemia, apoptosis, Mcl-1, Bcl-xL
Abstract
Bcl-2, Bcl-xL, Mcl-1, and A1 are the predominant anti-apoptotic members of the Bcl-2 family in somatic cells. Malignant B lymphocytes are critically dependent on Bcl-2 or Bcl-xL for survival. In contrast, a new study by Glaser and colleagues in the January 15, 2012, issue of Genes & Development (pp. 120–125) demonstrates that Mcl-1 is essential for development and survival of acute myelogenous leukemia cells. These results provide new impetus for the generation of selective Mcl-1 inhibitors.
Acute myelogenous leukemia (AML) is a hematological malignancy in which the bone marrow is replaced by clonal, immature cells of myeloid origin. Considered potentially curable with chemotherapy and/or stem cell transplantation, AML nonetheless accounts for >9000 deaths per year in the United States. Moreover, the fact that 70% of patients diagnosed with AML still die from this disorder highlights the urgent need for improved therapies. In the January 15, 2012, issue of Genes & Development, Glaser et al. (2012) reported that murine AML cells are critically dependent on the anti-apoptotic protein Mcl-1 for survival. In this perspective, we provide a context for this finding and discuss its potential implication for the therapy of AML as well as other neoplasms.
Mcl-1: family resemblances
Since its initial identification as a prosurvival factor by Vaux and coworkers in 1988 (Vaux et al. 1988), Bcl-2 has been the subject of extensive speculation and study. We now know that it is the founding member of a “family” of proteins that can be structurally and functionally subdivided into three groups (Strasser et al. 2011). Proteins in the first group, which consists of Bcl-2, Bcl-xL, Bcl-w, and A1, as well as Mcl-1, exhibit structural homology across multiple Bcl-2 homology (BH) domains and share the ability to inhibit apoptosis triggered by a variety of stimuli. Members of the second group, which consists of Bax, Bak, and Bok, also share extensive sequence homology with the anti-apoptotic family members but have acquired the ability to directly permeabilize the outer mitochondrial membrane, thereby releasing cytochrome c and other mitochondrial intermembrane components that contribute to activation of caspase-9 and subsequent apoptotic events. The third group, consisting of structurally dissimilar proteins such as Bad, Bid, Bik, Bim, Noxa, and Puma, share sequence homology only in an α-helical region known as the BH3 domain. These so-called BH3-only proteins are thought to be intracellular stress sensors that trigger apoptosis by either directly binding and activating Bax and Bak, as demonstrated for Bim, truncated Bid, and Noxa (Gavathiotis et al. 2010; Dai et al. 2011), or binding and neutralizing the anti-apoptotic family members, as exemplified by Bad. Conversely, the anti-apoptotic family members inhibit apoptosis by binding and sequestering activated BH3-only proteins as well as activated Bax and Bak (Cheng et al. 2001; Llambi et al. 2011).
The interactions between these various family members are becoming increasingly well understood. X-ray crystallography and nuclear magnetic resonance (NMR) have indicated that the anti-apoptotic family members, including Mcl-1, have the appearance of a catcher's mitt with an extended hydrophobic groove (Yan and Shi 2005). This groove accommodates the α-helical BH3 domain that is exposed on the surfaces of activated BH3-only proteins and is thought to be similarly, albeit transiently, exposed on Bax and Bak during their oligomerization (Yan and Shi 2005; Dewson et al. 2008; Llambi et al. 2011).
The turnstiles of Mcl-1 regulation
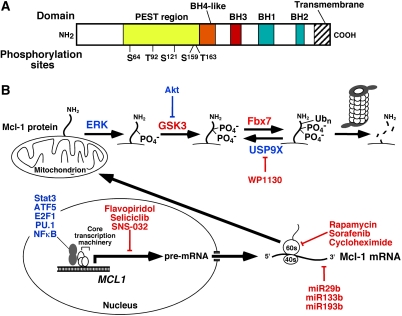
Despite the structural and functional similarities of the anti-apoptotic Bcl-2 family members, there are also increasingly well-recognized differences. Compared with Bcl-2 or Bcl-xL, Mcl-1 is particularly short-lived, with a half-life of only 2–4 h in most cells (Quinn et al. 2011). In addition, an exceptionally large number of pathways regulate Mcl-1 transcription, translation, and degradation (Fig. 1). Because of the rapid turnover of Mcl-1, these pathways can fine-tune Mcl-1 expression in response to a wide variety of cellular inputs on an hour-by-hour basis.
Figure 1.
Regulation of Mcl-1 levels. (A) Domain structure of Mcl-1. Shown are the four BH domains that Mcl-1 shares with Bcl-2 and other family members, as well as the localization of the proline/glutamic acid/serine/threonine-rich (PEST) region that contains many of the known Mcl-1 phosphorylation sites, including Ser 64 (Jun N-terminal kinase), Thr 92 (cyclin-dependent kinase 1), Ser 121 (extracellular signal-regulated kinase [ERK]), Ser 159 (glycogen synthase kinase-3 [GSK3] and PKCδ), and Thr 163 (ERK). (B) Transcriptional, translational, and post-translational regulation of Mcl-1 described in the text. Molecules depicted in blue increase Mcl-1 levels, whereas molecules rendered in red decrease Mcl-1 levels. In particular, ERK-mediated phosphorylation on Thr 163, which can stabilize Mcl-1 under certain circumstances, creates a recognition site for GSK3. GSK3-mediated phosphorylation of Mcl-1 on Ser 159 then creates the recognition site for the E3 ubiquitin ligase Fbx7, which generates Lys 48-linked ubiquitin chains that result in proteasome-mediated Mcl-1 degradation.
The short half-life of Mcl-1 is thought to reflect, in large part, unique features of its N terminus. Unlike other anti-apoptotic Bcl-2 family members, Mcl-1 contains at its N terminus two sequences rich in proline, glutamic acid, serine, and threonine (so-called PEST sequences) that target it for rapid proteasomal degradation (Warr and Shore 2008; Quinn et al. 2011). At least three E3 ubiquitin ligases are thought to contribute to this rapid turnover: MULE, β-TrCP, and SCFFBW7 (Zhong et al. 2005; Ding et al. 2007; Wertz et al. 2011). Of these E3s, FBW7 is particularly interesting in the context of neoplastic diseases because its mutational inactivation, which is known to occur in a subset of T-cell acute lymphocytic leukemias and cancers of the bile duct, stomach, pancreas, prostate, lung, and endometrium, can decrease Mcl-1 degradation, resulting in increased Mcl-1 protein levels and resistance to chemotherapeutic agents (Wertz et al. 2011). Mcl-1 can likewise be stabilized when the deubiquitinase USP9X, which is overexpressed in some malignancies, binds Mcl-1 and removes degradation-inducing Lys 48-linked polyubiquitin chains (Schwickart et al. 2010).
In addition to being modulated by expression of the various ligases and deubiquitinases, Mcl-1 ubiquitylation and deubiquitylation are regulated by Mcl-1 phosphorylation. In particular, the ability of E3 ligases to bind and modify Mcl-1 is dependent on phosphorylation of Mcl-1 at certain sites within its PEST sequences (De Biasio et al. 2007). For example, phosphorylation of Thr 163 by extracellular signal-regulated kinase (ERK) prolongs the Mcl-1 half-life. On the other hand, this Thr 163 phosphorylation can also serve as a priming phosphorylation for subsequent glycogen synthase kinase-3 (GSK3)-mediated phosphorylation at Ser 159, which promotes Mcl-1 ubiquitylation and degradation. Phosphorylation of Mcl-1 Thr 92 by the cyclin-dependent kinase 1 (cdk1)/cyclin B1 complex also enhances Mcl-1 proteasomal degradation. In addition, binding to the BH3-only protein Noxa can enhance Mcl-1 degradation, although the mechanism is incompletely understood.
Like Mcl-1 degradation, Mcl-1 expression is regulated on multiple levels. For example, several transcription factors, including ATF5, E2F1, STAT3, PU.1, and NFκB, bind and activate the MCL1 promoter (Warr and Shore 2008; Quinn et al. 2011). Like all transcriptional mechanisms, these are often cell type- and cellular context-specific.
Once synthesized, Mcl-1 transcripts can be influenced by microRNAs (miRs), which regulate both mRNA stability and translation. Three miRs have been demonstrated to regulate Mcl-1 to date: miR29b, miR133b, and miR193b, with miR29b being the most extensively studied (Mott et al. 2007; Garzon et al. 2009). This miR appears to reduce Mcl-1 cellular protein levels by reducing translation, as steady-state Mcl-1 mRNA levels are unchanged by miR29b overexpression (Mott et al. 2007). Conversely, loss of miR29b is a mechanism for enhanced Mcl-1 protein expression in cancers, especially AML, as described in greater detail below (Garzon et al. 2009).
Given the rapid cellular turnover of Mcl-1, it is not surprising that its cellular levels are also sensitive to other factors that decrease or increase translation. For example, endoplasmic stress-associated eIF2α phosphorylation inhibits translation and represses cellular Mcl-1 levels (Fritsch et al. 2007), whereas activation of the mammalian target of rapamycin complex 1 (mTORC1) enhances translation and promotes tumor cell survival by increasing Mcl-1 levels (Mills et al. 2008).
A unique role for Mcl-1 in hematopoietic cells
In contrast to BCL2, which was originally cloned because of its location at a common breakpoint in follicular lymphoma, MCL1 was originally identified because the message it encodes is up-regulated during phorbol ester-induced maturation of ML-1 AML cells (Kozopas et al. 1993). Subsequent studies demonstrated that Mcl-1 is expressed at particularly high levels during the commitment of cells to myeloid differentiation (Craig 2002).
In addition, Mcl-1 plays a critical role in the survival of hematopoietic stem cells and early myeloid precursors. Growth factors such as stem cell factor and IL-3, acting through the Jak/STAT and Akt pathways, up-regulate Mcl-1 in early myeloid progenitors (Craig 2002; Opferman 2007). Conversely, conditional knockout of Mcl1 results in the death of hematopoietic stem cells and complete marrow ablation within 12–21 d (Opferman et al. 2005). In addition to hematopoietic stem cells, the committed progenitors of granulocytes and macrophages appear to be particularly dependent on Mcl-1 (Opferman 2007).
Extending the results to AML
The term AML encompasses a genetically and phenotypically heterogeneous group of neoplasms in which the marrow is replaced to varying degrees with clonal, immature hematopoietic cells and production of normal marrow elements is generally impaired. A wide variety of pathogenic alterations occur in this group of disorders (Fröhling et al. 2005), including (but not limited to) (1) activating alterations in the c-Kit and FLT3 receptor tyrosine kinases, (2) activating mutations in the NRAS gene, (3) chromosomal translocations that impair the ability of retinoic acid receptorα to facilitate maturation of progranulocytes, (4) alterations in core-binding factor that result in recruitment of nuclear corepressor complexes and impaired expression of genes normally involved in hematopoietic differentiation, and (5) fusion of the N terminus of the MLL1 gene to a wide range of partners, creating long-lived products that activate expression of homeobox (HOX) genes and disrupt programs required for lineage commitment of hematopoietic cells (Liu et al. 2009). Despite the genetic heterogeneity of AML, recent studies indicate that a core group of pathways leading to proliferation and self-renewal are activated in this group of disorders (Kvinlaug et al. 2011). The work of Glaser et al. (2012) now raises the possibility of a common, critical role for one particular anti-apoptotic Bcl-2 family member, Mcl-1, in AML as well.
Using gene targeted murine marrow cells that express a tamoxifen-activatable Cre recombinase and contain loxP sites flanking one or both alleles of Mcl1 or Bclx as a starting point, Glaser et al. (2012) initially infected cells with retroviruses encoding one of five oncogenic constructs (MLL-ENL, MLL-AF9, AML1-ETO9a, MixL1, or Hoxa9) to generate AML lines of various genotypes in which levels of Mcl-1 or Bcl-xL could be experimentally manipulated. Strikingly, independent of the initiating leukemogenic lesion, activation of Cre recombinase led to almost complete loss of viability in AML cells containing two floxed Mcl1 alleles. In contrast, deletion of both Bclx alleles or one Mcl1 allele had little impact on leukemic cell viability over and above the (modest) anti-survival effect of Cre recombinase itself. Simultaneous inhibition of Bcl-2, Bcl-xL, and Bcl-w using the pharmacological agent ABT-737 likewise had limited impact on the survival of the AML cells. Remarkably, T cells transformed to acute lymphocytic leukemia by a retrovirus encoding Notch 1 also showed prolonged survival in vitro after deletion of both Mcl1 genes, suggesting that the Mcl-1 dependence might be somewhat unique to AML.
In light of the prior results of Opferman et al. (2005) showing ablation of normal marrow after conditional Mcl1 knockout, Glaser et al. (2012) next examined the impact of Cre recombinase on survival of normal murine hematopoietic stem cells and committed myeloid progenitors bearing the same floxed alleles. While 90% of committed myeloid progenitors died within 96 h of Mcl1 deletion, about one-third of normal myeloid stem cells survived, raising the possibility of a therapeutic window.
To explore the possibility of a therapeutic window in vivo, Glaser et al. (2012) reconstituted lethally irradiated mice with MLL–ENL retrovirus-infected bone marrow-derived hematopoietic progenitor cells. Activation of Cre recombinase as long as 21 d after reconstitution resulted in a twofold prolongation of survival (from 60 to >120 d) if the leukemia harbored two floxed Mcl1 alleles, but not if the leukemia harbored a single floxed Mcl1 allele or floxed Bclx alleles. Moreover, activation of Cre recombinase earlier after reconstitution resulted in a more prolonged survival (e.g., six of six mice were cured when tamoxifen was administered 5 d after reconstitution).
Targeting human AML—the good, the bad, and the unknown
Glaser et al. (2012) next attempted to determine whether human AML displays a similar requirement for the Mcl-1 protein. Because small-molecule inhibitors that selectively target Mcl-1 are not available, they used retroviruses encoding wild-type BimS (binds all anti-apoptotic family members), BimSL62A/F69A (“BimS2A,” which selectively binds Mcl-1), and BimS containing the Bad BH3 domain in place of the endogenous BH3 domain (selectively binds Bcl-2 and Bcl-xL). While wild-type BimS and BimSL62A/F69A were able to selectively diminish survival of cells in two samples of clinical AML, three other samples showed unimpaired survival.
In evaluating the claims of Glaser et al. (2012), it is important to ask whether anyone else has reported similar findings. Early studies comparing levels of Bcl-2 family members in the same acute leukemia at the time of diagnosis and at relapse indicated that Mcl-1 is often elevated at relapse, suggesting a possible role for this protein in survival of leukemia cells after chemotherapy (Kaufmann et al. 1998). A more recent report indicated that restoration of miR29b in AML cell lines and primary samples induces apoptosis and dramatically reduces tumorigenicity in a xenograft leukemia model (Garzon et al. 2009), in agreement with the suggestion of Glaser et al. (2012) that Mcl-1 plays a critical role in AML.
The results of Glaser et al. (2012), coupled with these earlier studies, highlight the potential importance of Mcl-1 in AML cell survival. Nonetheless, several critical issues need to be addressed before we will know whether Mcl-1 is an important future target for AML therapy or not. First, Glaser et al. (2012) examined the importance of Mcl-1 in only a small number of genetic settings (two involving MLL1 rearrangements, one involving a core-binding factor alteration, and one involving homeobox protein overexpression). Whether Mcl-1 will likewise be critical for survival of leukemias driven by mutations in NRAS or FLT3 or by the myriad other genetic abnormalities observed in AML (Fröhling et al. 2005) remains to be established, as Glaser et al. (2012) acknowledge.
Second, the observations in clinical AML specimens need to be expanded and understood. Sixty percent of the clinical AML specimens failed to respond in vitro to BIM transduction. This sobering fact might be explained by any of a variety of possibilities, including (1) alterations at the level of Bax and Bak activation, as suggested by diminished Bax expression and altered Bak mobility in one of the specimens examined by Glaser et al. (2012), and/or (2) blocks in caspase activation downstream from cytochrome c that have been previously described in AML (Schimmer et al. 2003). While the investigators attribute the limited induction of apoptosis to selection for resistant cells by prior therapy, it is noteworthy that three of the five samples (including one of the three resistant samples) came from patients without prior treatment. Clearly, effects of Mcl-1 inhibition or down-regulation in a much larger number of clinical AML specimens, including specimens with a wide range of pathogenic changes harvested at initial diagnosis and at relapse, are required to better define the subsets of AML that are most likely to benefit from agents that target Mcl-1.
Multitasking and Mcl-1 biology
Translation of the findings of Glaser et al. (2012) into improved AML therapy might also be accelerated by improved understanding of how Mcl1 deletion selectively kills AML cells. Glaser et al. (2012) report that Bim down-regulation protects AML cells from Mcl1 gene deletion. At first glance, this suggests that Mcl1 deletion is creating an imbalance between pro- and anti-apoptotic proteins. On the other hand, Bclx deletion does not have the same effect, which argues that the balance between pro- and anti-apoptotic family members might not be the full explanation. Does Mcl-1 perform other jobs promoting AML survival, and could these functions also be inhibited by binding BH3-only proteins? The answers to these questions remain ambiguous. Mcl-1 deficiency in mice results in peri-implantation embryonic lethality, an effect not clearly related to an anti-apoptotic role. Alternative roles for Bcl-2 proteins in mitochondrial biology are currently emerging. For example, Bcl-xL reportedly regulates ATP synthesis by interacting with the F1F0 ATP synthase (Alavian et al. 2011). A fast-mobility isoform of Mcl-1 resulting from protease cleavage of the N terminus, like a subfraction of Bcl-xL, is localized to the mitochondrial matrix, suggesting other mitochondrial functions for Mcl-1 (Huang and Yang-Yen 2010), although this finding remains controversial (Warr et al. 2011). Bax and Bak have also been implicated in the dynamics of mitochondrial fusion and fission (Martinou and Youle 2011), and binding of Bak by Mcl-1 might also alter mitochondrial function by altering these dynamics. Thus, in addition to neutralizing proapoptotic Bcl-2 family members, Mcl-1 might have other essential functions in AML cells that remain to be elucidated.
Linking leukemia and solid tumor biology: is Mcl-1 the gold chain?
A recent survey of anti-apoptotic Bcl-2 family member expression in breast, brain, colon, lung, ovarian, renal, and melanoma cell lines revealed that Mcl-1 mRNA is more abundant than Bcl-2 or Bcl-xL (Placzek et al. 2010). Furthermore, high-resolution analyses of somatic copy number alterations in 26 histological types of cancers identified frequent MCL1 gene amplification in lung, breast, neural, and gastrointestinal cancers (Beroukhim et al. 2010). These observations suggest that there might be some benefit in targeting Mcl-1 in solid tumors. However, unlike AML cells, solid tumors do not necessarily undergo spontaneous apoptosis following Mcl-1 knockdown, but often require an additional stimulus (Taniai et al. 2004), raising the possibility that findings in AML might not immediately translate into new therapies for solid tumors. Nonetheless, Mcl-1 appears to play a major role in cancer cell survival, making it an attractive therapeutic target.
Silver daggers for Mcl-1
If Mcl-1 is indeed a linchpin for survival of AML and various cancers, how can it be therapeutically targeted? A number of strategies for either reducing cellular Mcl-1 levels or blocking its hydrophobic BH3-binding groove are being actively explored.
Indirect approaches: pharmacological agents that cause Mcl-1 depletion
Over the past several years, several agents have been shown to diminish Mcl-1 expression by inhibiting Mcl-1 production or enhancing Mcl-1 degradation.
Sorafenib
This multikinase inhibitor is currently FDA-approved for use in renal cell and hepatocellular carcinomas. In tissue culture, high sorafenib concentrations diminish Mcl-1 protein levels. Multiple explanations have been suggested, including inhibition of protein translation due to activation of ER stress pathways, dephosphorylation of signal transducer and activator of transcription 3 (Stat3), inhibition of NFκB, and enhanced Mcl-1 degradation (Dai and Grant 2007). Although sorafenib by itself exhibits little anti-leukemic efficacy in the clinic (Pratz et al. 2010), promising evidence of activity has been observed in combination with standard cytotoxic chemotherapy (Ravandi et al. 2010). Whether the efficacy of this combination is due to the ability of sorafenib to down-regulate Mcl-1 or inhibit the oncogenic receptor tyrosine kinase FLT3 is currently unknown. Nonetheless, sorafenib-containing combinations certainly merit further investigation.
Cdk inhibitors
Flavopiridol, another agent already in clinical trials for AML, potently reduces Mcl-1 levels in a variety of cell types in vitro (for review, see Lee et al. 2006). This effect appears to reflect inhibition of Stat3 and/or inhibition of Cdk9, with the latter effect leading to diminished phosphorylation and activity of RNA polymerase II. Likewise, SNS-032, which inhibits several cdks including cdk9, also decreases Mcl-1 levels. Early clinical studies have shown activity of flavopiridol-containing combinations in AML (Karp et al. 2011), although it has been difficult to demonstrate consistent Mcl-1 down-regulation in leukemia cells in the clinical setting (Karp et al. 2005).
USP9X inhibitors: facilitating Mcl-1 degradation
In contrast to the agents described above, which inhibit Mcl-1 production, it might also be possible to facilitate Mcl-1 degradation. As indicated above, overexpression of the deubiquitinase USP9X by malignant cells causes deubiquitylation and stabilization of Mcl-1, resulting in apoptosis resistance. As a result, USP9X is a promising target for selectively reversing the Mcl-1 overexpression observed in certain malignancies. The small molecule WP1130 directly inhibits USP9X, lowers Mcl-1 levels in chronic myelogenous leukemia, and enhances sensitivity to apoptosis (Sun et al. 2011). Whether this agent or other USP9X inhibitors will be suitable for clinical development, either alone or in combination with conventional chemotherapy, remains to be determined.
BH3 mimetics: agents that neutralize anti-apoptotic Bcl-2 family members
Rather than altering Mcl-1 levels, it might also be possible to directly inhibit the action of Mcl-1. BH3 mimetics are small organic molecules that mimic the binding of BH3-only proteins to the hydrophobic grooves of anti-apoptotic Bcl-2 proteins, thereby neutralizing them. The most promising of these, navitoclax (ABT-263), is currently in phase II and III clinical trials (Walensky 2011). Unfortunately, this agent does not bind to the BH3-binding groove of Mcl-1, leaving Mcl-1 free to bind any BH3-only proteins displaced from Bcl-2 or Bcl-xL by navitoclax. As a consequence, Mcl-1 overexpression confers navitoclax resistance.
While navitoclax does not inhibit Mcl-1, several extended-spectrum BH3 mimetics do. Obatoclax, the first of these developed, binds Mcl-1 in addition to Bcl-2 and Bcl-xL (Nguyen et al. 2007), and the binding to Mcl-1 disrupts Mcl-1•Bak interactions. Given the neurotoxicity of obatoclax, however, as well as its yet unclear risk/benefit ratio, its potential for approval as an anti-cancer drug is uncertain. Likewise, the recently described apogossypol derivative sabutoclax and the terphenyl BH3-M6 bind and neutralize multiple Bcl-2 anti-apoptotic proteins, including Mcl-1.
The problem with all of the current small-molecule BH3 mimetics, however, is their lack of selectivity. None of them has been shown to bind and inhibit Mcl-1 preferentially, as one would need in order to pharmacologically recapitulate the selective Mcl1 gene deletion used so elegantly by Glaser et al. (2012) On the other hand, selective Mcl-1 inhibition does appear to be feasible. A derivative of the Mcl-1 BH3 peptide has been reported to selectively disrupt Mcl-1 binding to Bak, thereby sensitizing cancer cell lines to apoptosis (Stewart et al. 2010). Further work to convert this peptide into a peptidomimetic small molecule has a high likelihood of moving this area forward.
A challenge for the future
The study that prompted this perspective (Glaser et al. 2012) provides elegant genetic evidence that Mcl-1 plays a unique and critical role in the survival of AML cells in vitro and in vivo. The limited studies of Glaser et al. (2012) in clinical AML samples also suggest that there will be challenges in translating the murine findings into effective new clinical therapies. Hopefully, by highlighting the importance of Mcl-1, Glaser et al. (2012) will spur the development of new pharmacological agents that can be used in preclinical and early clinical studies to assess the impact of selectively inhibiting Mcl-1. Without these agents, it will be impossible to translate the elegant genetics into clinically meaningful results.
Acknowledgments
We gratefully acknowledge helpful discussions with members of our laboratories and the support of R01 DK59427 (to G.J.G) and CA69008 (to S.H.K.). We apologize to the many authors whose seminal contributions to this field could not be cited because of space limitations.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.186189.111.
References
- Alavian KN, Li H, Collis L, Bonanni L, Zeng L, Sacchetti S, Lazrove E, Nabili P, Flaherty B, Graham M, et al. 2011. Bcl-xL regulates metabolic efficiency of neurons through interaction with the mitochondrial F1FO ATP synthase. Nat Cell Biol 13: 1224–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. 2010. The landscape of somatic copy-number alteration across human cancers. Nature 463: 899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng EH, Wei MC, Weiler S, Flavell RA, Mak TW, Lindsten T, Korsmeyer SJ 2001. BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol Cell 8: 705–711 [DOI] [PubMed] [Google Scholar]
- Craig RW 2002. MCL1 provides a window on the role of the BCL2 family in cell proliferation, differentiation and tumorigenesis. Leukemia 16: 444–454 [DOI] [PubMed] [Google Scholar]
- Dai Y, Grant S 2007. Targeting multiple arms of the apoptotic regulatory machinery. Cancer Res 67: 2908–2911 [DOI] [PubMed] [Google Scholar]
- Dai H, Smith A, Meng XW, Schneider PA, Pang Y-P, Kaufmann SH 2011. Transient binding of an activator BH3 domain to the Bak BH3-binding groove initiates Bak oligomerization. J Cell Biol 194: 39–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Biasio A, Vrana JA, Zhou P, Qian L, Bieszczad CK, Braley KE, Domina AM, Weintraub SJ, Neveu JM, Lane WS, et al. 2007. N-terminal truncation of antiapoptotic MCL1, but not G2/M-induced phosphorylation, is associated with stabilization and abundant expression in tumor cells. J Biol Chem 282: 23919–23936 [DOI] [PubMed] [Google Scholar]
- Dewson G, Kratina T, Sim HW, Puthalakath H, Adams JM, Colman PM, Kluck RM 2008. To trigger apoptosis, Bak exposes its BH3 domain and homodimerizes via BH3:groove interactions. Mol Cell 30: 369–380 [DOI] [PubMed] [Google Scholar]
- Ding Q, He X, Hsu JM, Xia W, Chen CT, Li LY, Lee DF, Liu JC, Zhong Q, Wang X, et al. 2007. Degradation of Mcl-1 by β-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Mol Cell Biol 27: 4006–4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch RM, Schneider G, Saur D, Scheibel M, Schmid RM 2007. Translational repression of MCL-1 couples stress-induced eIF2α phosphorylation to mitochondrial apoptosis initiation. J Biol Chem 282: 22551–22562 [DOI] [PubMed] [Google Scholar]
- Fröhling S, Scholl C, Gilliland DG, Levine RL 2005. Genetics of myeloid malignancies: Pathogenetic and clinical implications. J Clin Oncol 23: 6285–6295 [DOI] [PubMed] [Google Scholar]
- Garzon R, Heaphy CE, Havelange V, Fabbri M, Volinia S, Tsao T, Zanesi N, Kornblau SM, Marcucci G, Calin GA, et al. 2009. MicroRNA 29b functions in acute myeloid leukemia. Blood 114: 5331–5341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavathiotis E, Reyna DE, Davis ML, Bird GH, Walensky LD 2010. BH3-triggered structural reorganization drives the activation of proapoptotic BAX. Mol Cell 40: 481–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser SP, Lee EF, Trounson E, Bouillet P, Wei A, Fairlie WD, Izon DJ, Zuber J, Rappaport AR, Herold MJ, et al. 2012. Anti-apoptotic Mcl-1 is essential for the development and sustained growth of acute myeloid leukemia. Genes Dev 26: 120–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CR, Yang-Yen HF 2010. The fast-mobility isoform of mouse Mcl-1 is a mitochondrial matrix-localized protein with attenuated anti-apoptotic activity. FEBS Lett 584: 3323–3330 [DOI] [PubMed] [Google Scholar]
- Karp JE, Bauer KS, Passaniti A, Goji I, Kaufmann SH, Bible K, Greer J, Briel J, Tidwell ML, Ross DD, et al. 2005. Phase I study of timed sequential therapy of relapsed and refractory adult acute leukemias with flavopiridol followed by Ara-C and mitoxantrone. Clin Cancer Res 11: 8403–8412 [DOI] [PubMed] [Google Scholar]
- Karp JE, Smith BD, Resar LS, Greer JM, Blackford A, Zhao M, Moton-Nelson D, Alino K, Levis MJ, Gore SD, et al. 2011. Phase 1 and pharmacokinetic study of bolus-infusion flavopiridol followed by cytosine arabinoside and mitoxantrone for acute leukemias. Blood 117: 3302–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann SH, Karp JE, Svingen PA, Krajewski S, Burke PJ, Gore SD 1998. Elevated expression of the apoptotic regulator Mcl-1 at the time of leukemic relapse. Blood 91: 991–1000 [PubMed] [Google Scholar]
- Kozopas KM, Yang T, Buchan HL, Zhou P, Craig RW 1993. MCL1, a gene expressed in programmed myeloid cell differentiation, has sequence similarity to BCL-2. Proc Natl Acad Sci 90: 3516–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvinlaug BT, Chan WI, Bullinger L, Ramaswami M, Sears C, Foster D, Lazic SE, Okabe R, Benner A, Lee BH, et al. 2011. Common and overlapping oncogenic pathways contribute to the evolution of acute myeloid leukemias. Cancer Res 71: 4117–4129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Isham CR, Kaufmann SH, Bible KC 2006. Flavopiridol disrupts STAT3/DNA interactions, attenuates STAT3-directed transcription and combines with the Jak kinase inhibitor AG490 to achieve cytotoxic synergy. Mol Cancer Ther 5: 138–148 [DOI] [PubMed] [Google Scholar]
- Liu H, Cheng EH, Hsieh JJ 2009. MLL fusions: Pathways to leukemia. Cancer Biol Ther 8: 1204–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llambi F, Moldoveanu T, Tait SW, Bouchier-Hayes L, Temirov J, McCormick LL, Dillon CP, Green DR 2011. A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol Cell 44: 517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinou JC, Youle RJ 2011. Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Dev Cell 21: 92–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills JR, Hippo Y, Robert F, Chen SM, Malina A, Lin CJ, Trojahn U, Wendel HG, Charest A, Bronson RT, et al. 2008. mTORC1 promotes survival through translational control of Mcl-1. Proc Natl Acad Sci 105: 10853–10858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott JL, Kobayashi S, Bronk SF, Gores GJ 2007. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene 26: 6133–6140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M, Marcellus RC, Roulston A, Watson M, Serfass L, Murthy Madiraju SR, Goulet D, Viallet J, Belec L, Billot X, et al. 2007. Small molecule obatoclax (GX15-070) antagonizes MCL-1 and overcomes MCL-1-mediated resistance to apoptosis. Proc Natl Acad Sci 104: 19512–19517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opferman JT 2007. Life and death during hematopoietic differentiation. Curr Opin Immunol 19: 497–502 [DOI] [PubMed] [Google Scholar]
- Opferman JT, Iwasaki H, Ong CC, Suh H, Mizuno S, Akashi K, Korsmeyer SJ 2005. Obligate role of anti-apoptotic MCL-1 in the survival of hematopoietic stem cells. Science 307: 1101–1104 [DOI] [PubMed] [Google Scholar]
- Placzek WJ, Wei J, Kitada S, Zhai D, Reed JC, Pellecchia M 2010. A survey of the anti-apoptotic Bcl-2 subfamily expression in cancer types provides a platform to predict the efficacy of Bcl-2 antagonists in cancer therapy. Cell Death Dis 1: e40 doi: 10.1038/cddis.2010.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratz KW, Cho E, Levis MJ, Karp JE, Gore SD, McDevitt M, Stine A, Zhao M, Baker SD, Carducci MA, et al. 2010. A pharmacodynamic study of sorafenib in patients with relapsed and refractory acute leukemias. Leukemia 24: 1437–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn BA, Dash R, Azab B, Sarkar S, Das SK, Kumar S, Oyesanya RA, Dasgupta S, Dent P, Grant S, et al. 2011. Targeting Mcl-1 for the therapy of cancer. Expert Opin Investig Drugs 20: 1397–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravandi F, Cortes JE, Jones D, Faderl S, Garcia-Manero G, Konopleva MY, O'Brien S, Estrov Z, Borthakur G, Thomas D, et al. 2010. Phase I/II study of combination therapy with sorafenib, idarubicin, and cytarabine in younger patients with acute myeloid leukemia. J Clin Oncol 28: 1856–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmer AD, Pedersen IM, Kitada S, Eksioglu-Demiralp E, Minden MD, Pinto R, Mah K, Andreeff M, Kim Y, Suh WS, et al. 2003. Functional blocks in caspase activation pathways are common in leukemia and predict patient response to induction chemotherapy. Cancer Res 63: 1242–1248 [PubMed] [Google Scholar]
- Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, Maecker H, O'Rourke K, Bazan F, Eastham-Anderson J, et al. 2010. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature 463: 103–107 [DOI] [PubMed] [Google Scholar]
- Stewart ML, Fire E, Keating AE, Walensky LD 2010. The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer. Nat Chem Biol 6: 595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser A, Cory S, Adams JM 2011. Deciphering the rules of programmed cell death to improve therapy of cancer and other diseases. EMBO J 30: 3667–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Kapuria V, Peterson LF, Fang D, Bornmann WG, Bartholomeusz G, Talpaz M, Donato NJ 2011. Bcr-Abl ubiquitination and Usp9x inhibition block kinase signaling and promote CML cell apoptosis. Blood 117: 3151–3162 [DOI] [PubMed] [Google Scholar]
- Taniai M, Grambihler A, Higuchi H, Werneburg N, Bronk SF, Farrugia DJ, Kaufmann SH, Gores GJ 2004. Mcl-1 mediates tumor necrosis factor-related apoptosis-inducing ligand resistance in human cholangiocarcinoma cells. Cancer Res 64: 3517–3524 [DOI] [PubMed] [Google Scholar]
- Vaux DL, Cory S, Adams JM 1988. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature 335: 440–442 [DOI] [PubMed] [Google Scholar]
- Walensky LD 2011. From mitochondrial biology to magic bullet: Navitoclax disarms BCL-2 in chronic lymphocytic leukemia. J Clin Oncol doi: 10.1200/JCO.2011.37.9339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warr MR, Shore GC 2008. Unique biology of Mcl-1: Therapeutic opportunities in cancer. Curr Mol Med 8: 138–147 [DOI] [PubMed] [Google Scholar]
- Warr MR, Mills JR, Nguyen M, Lemaire-Ewing S, Baardsnes J, Sun KL, Malina A, Young JC, Jeyaraju DV, O'Connor-McCourt M, et al. 2011. Mitochondrion-dependent N-terminal processing of outer membrane Mcl-1 protein removes an essential Mule/Lasu1 protein-binding site. J Biol Chem 286: 25098–25107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz IE, Kusam S, Lam C, Okamoto T, Sandoval W, Anderson DJ, Helgason E, Ernst JA, Eby M, Liu J, et al. 2011. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature 471: 110–114 [DOI] [PubMed] [Google Scholar]
- Yan N, Shi Y 2005. Mechanisms of apoptosis through structural biology. Annu Rev Cell Dev Biol 21: 35–56 [DOI] [PubMed] [Google Scholar]
- Zhong Q, Gao W, Du F, Wang X 2005. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell 121: 1085–1095 [DOI] [PubMed] [Google Scholar]