In this comprehensive review, Reinberg and colleagues discuss the histone methyltransferase PR-Set7, which is emerging as an important player in multiple cellular processes. PR-Set7 and its H4K20me1 modification were recently identified in multiple pathways that regulate its cell cycle expression and activity. This review focuses on what is known about its function and regulation.
Keywords: PR-Set7, H4K20me1, chromatin, cell cycle, DNA damage
Abstract
Histone post-translational modifications impact many aspects of chromatin and nuclear function. Histone H4 Lys 20 methylation (H4K20me) has been implicated in regulating diverse processes ranging from the DNA damage response, mitotic condensation, and DNA replication to gene regulation. PR-Set7/Set8/KMT5a is the sole enzyme that catalyzes monomethylation of H4K20 (H4K20me1). It is required for maintenance of all levels of H4K20me, and, importantly, loss of PR-Set7 is catastrophic for the earliest stages of mouse embryonic development. These findings have placed PR-Set7, H4K20me, and proteins that recognize this modification as central nodes of many important pathways. In this review, we discuss the mechanisms required for regulation of PR-Set7 and H4K20me1 levels and attempt to unravel the many functions attributed to these proteins.
Post-translational modifications of histones have emerged as an important means by which the cell can demarcate specific regions of the genome for differential and dynamic regulation (Campos and Reinberg 2009). These modifications, such as phosphorylation, methylation, acetylation, and ubiquitination, occur extensively on the N-terminal tails of histones and have diverse functions and modes of action. A substantial amount of work has sought to determine how these “marks” regulate gene expression, but histone modifications have also been linked to other cellular processes, such as mitosis (Hirota et al. 2005; Kelly et al. 2010; Oda et al. 2010; Wang et al. 2010; Yamagishi et al. 2010), meiosis (Govin et al. 2010), and the DNA damage response (Burma et al. 2001; Polo and Jackson 2011). Furthermore, despite a lack of clear evidence, histone modifications are leading candidates for the means to propagate gene expression states epigenetically.
Histone H4 Lys 20 (H4K20) was one of the earliest modified histone residues to be identified, found in both pea and calf extracts in 1969 (DeLange et al. 1969). By analyzing metabolically labeled histone tails, H4K20 was subsequently identified as a major site of in vivo methylation, and it is the only identified and characterized methylated lysine residue on the H4 N-terminal tail (Zhang and Reinberg 2001). In metazoans, H4K20 can be modified to three separate degrees: mono-, di-, and trimethylation. These different methylation states are controlled by distinct methyltransferases in vivo, although a number of other enzymes have similar activity in vitro (Beisel et al. 2002; Marango et al. 2008; Schotta et al. 2008; Oda et al. 2009). Separate enzymatic activity specific for only monomethylation of H4K20 (H4K20me1) is present in Caenorhabditis elegans, whereas lower eukaryotes such as Schizosaccharomyces pombe contain a single methyltransferase for all H4K20 methylation (H4K20me) states, while Saccharomyces cerevisae contains little to no H4K20me at all (Wang and Jia 2009; Balakrishnan and Milavetz 2010; Edwards et al. 2011). Thus, the more recent appearance of H4K20me1 suggests an expansion of the chromatin modification canon in higher eukaryotes.
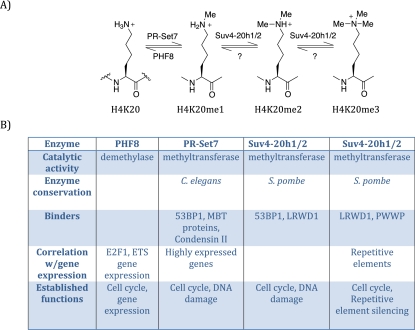
In mammals, at least three methyltransferases (PR-Set7, Suv4-20h1, and Suv4-20h2) and one demethylase (PHF8) directly regulate H4K20 methylation (Fig. 1; Liu et al. 2010; Qi et al. 2010). PR-Set7/Set8/KMT5a is the sole monomethyltransferase (Oda et al. 2009), whereas Suv4-20h1/h2 catalyzes the transition from H4K20me1 to H4K20me2/3 (Schotta et al. 2008). PHF8 demethylates H4K20me1 to H4K20me0, while a demethylase for H4K20me2/3 remains to be identified (Liu et al. 2010; Qi et al. 2010). Although separate enzymes control each level of methylation, H4K20me2/3 are dependent on H4K20me1. Furthermore, each of the H4K20me states exhibits unique genomic distributions and is recognized by specific binding proteins, underscoring its distinct roles. Regulation of H4K20me is essential, since knockout studies have shown that PR-Set7 and at least one of Suv4-20h1/2 are required for development in mice and that loss of these enzymes causes increased DNA damage and cell cycle defects (Schotta et al. 2008; Oda et al. 2010). Not surprisingly, loss of the monomethyltransferase PR-Set7 causes a more severe and complex phenotype, as this precludes the catalysis of all levels of H4K20 methylation.
Figure 1.
Overview of H4K20me. (A) Schematic diagram of the different methylation states of histone H4K20; from left to right: unmethylated (H4K20), monomethylated (H4K20me1), dimethylated (H4K20me2), and trimethylated (H4K20me3), with the various mammalian enzymes that perform these post-translational modification. (B) Properties of each of the enzymes that affect H4K20 methylation.
PR-Set7 was initially identified as a H4K20 monomethyltransferase by unbiased conventional biochemical purification (Nishioka et al. 2002). PR-Set7 displays robust enzymatic activity toward nucleosomal substrates, but not histone octamers or histone H4 alone. PR-Set7 was shown to be strictly a monomethyltransferase through a combination of nuclear magnetic resonance (NMR), mass spectroscopy, and in vitro experiments (Couture et al. 2005; Xiao et al. 2005). Providing further support and general insight as to how methyltransferases can be monomethyl-specific, crystallographic studies demonstrated that the substrate recognition channel of the enzyme is too narrow to accommodate a trimethylated species. This structure also helped determine that PR-Set7 acts specifically at H4K20 due to an array of required contacts with the surrounding residues on H4.
PR-Set7 catalytic activity is essential for viability. In Drosophila, loss of PR-Set7 causes lethality at the third instar larval stage, potentially through defects in embryonic patterning (Karachentsev et al. 2005). In mice, PR-Set7 knockout has a much more severe phenotype and results in embryonic lethality and arrest between the four- and eight-cell stage (Oda et al. 2009). Rescue experiments in mouse embryos elegantly showed that the lack of catalytic activity of PR-Set7 causes the lethality of PR-Set7−/− embryos. Injection of wild-type PR-Set7 mRNA into one of the PR-Set7−/− blastomeres at the two-cell stage led to blastocoel cavity formation from the progeny of injected cells, which was not observed for the progeny of cells that were injected with mRNA encoding a catalytically inactive mutant of PR-Set7. Further analysis showed that the lack of PR-Set7 catalytic activity causes a growth arrest due to increased DNA damage and defects in cell cycle progression in embryonic stem (ES) cells (Oda et al. 2009). The later lethality in the case of fly embryos may be explained by increased maternal contribution of PR-Set7 transcript, possibly delaying depletion of H4K20me1.
We hypothesize that the primary function of PR-Set7 and the requirement for enzymatic activity is regulation of H4K20me levels. Both PR-Set7 and H4K20me1 oscillate during the cell cycle, and maintenance of H4K20me levels is critical for proper cell cycle progression through its role in protecting genome stability, DNA replication, mitosis, and transcription. The emergence of PR-Set7 during evolution suggests a role specific to higher eukaryotes, which may be related to the increased complexities of mitosis, DNA replication initiation, or gene regulation. However, why PR-Set7 methyltransferase activity is essential for cell viability in higher eukaryotes is still only partially understood. Despite many thorough investigations into the function of PR-Set7, we believe that we are only now on the cusp of fully understanding the role of H4K20me1. In the following sections, we first discuss the substrates and regulation of PR-Set7, followed by its role in proper cell cycle progression, the DNA damage response, gene regulation, and higher-order chromatin structure.
PR-Set7 substrates and effectors
Although other yet-to-be-identified substrates may exist, methylation of H4 is likely to be the major effector of PR-Set7 function and a crucial mediator of its impact on viability. As is potentially the case for other histone modifications, H4K20me could act directly through altering nucleosome structure or indirectly through specific binders that mediate downstream functions. These two possibilities are further complicated in the case of PR-Set7, since the loss of H4K20me1 also causes a decrease in H4K20me2/3. However, the phenotype of Suv4-20h1/2−/− mice (Suv4-20 double knockout) is much less severe than PR-Set7−/− mice, suggesting a unique function for H4K20me1 independent of H4K20me2/3 (Schotta et al. 2008).
A wealth of evidence has emerged that supports both direct and indirect functions for H4K20me1. In vitro studies have shown that H4K20 forms internucleosomal contacts with the H2A C terminus on adjacent nucleosomes, possibly to promote compaction, and H4K20 methylation may also directly affect higher-order chromatin structure due to its position at the junction of DNA and the nucleosome (Lu et al. 2008). Furthermore, acetylation of a neighboring residue, H4K16, has been demonstrated to regulate higher-order chromatin dynamics and may be affected by H4K20me (Nishioka et al. 2002; Shogren-Knaak et al. 2006; Pesavento et al. 2008; Kapoor-Vazirani et al. 2011). This role of H4K20me in higher-order chromatin structure was also demonstrated by in vivo experiments in which loss of PR-Set7 resulted in large-scale chromatin decondensation in the interphase nucleus and expanded chromosome territory, signifying less compact chromatin (Oda et al. 2009). Moreover, further analysis of the loss of PR-Set7 showed a large change in overall subnuclear morphology (H Oda, D Beck, and D Reinberg, unpubl.). Therefore, PR-Set7 alone, presumably due to its activity toward H4K20, may contribute directly to chromatin structure without other factors.
In addition, H4K20me1 likely conveys its functions through the proteins that bind to it. There are three extensively investigated H4K20me1 binders that each carry a unique binding domain and contribute to distinct cellular functions: L3MBTL1 (Li et al. 2007; Trojer et al. 2007), 53BP1 (Sanders et al. 2004; Oda et al. 2009; Spektor and Rice 2009), and Condensin II (Liu et al. 2010). L3MBTL1 binds to both mono- and dimethylated H4K20 residues through its MBT domains. It works as a transcriptional repressor and can cause direct compaction of chromatin (see also PR-Set7 and Transcription). 53BP1 recognizes methylated lysine residues through its tandem tudor domain and functions within the DNA damage response pathway (see below, Function in DNA Damage). Condensin II recognizes H4K20me1 through its N-CAPD3 and N-CAPG2 subunits via two HEAT domains and is involved in mitotic progression (see below, Role in Cell Cycle Progression). In addition, the LRWD1/ORCA-associated factor of the origin of replication complex (ORC) binds to H4K20me3 and may link regulation of H4K20me to initiation of replication (Shen et al. 2010; Vermeulen et al. 2010).
It is difficult to attribute the essential function of PR-Set7 to any single signaling pathway, as deletion of any one of the specific binders of H4K20me1 does not result in a phenotype similar to PR-Set7 deletion (Ward et al. 2003; Qin et al. 2010). Moreover, loss of H4K20me2/3 may also contribute to the PR-Set7−/−phenotype. These findings suggest that PR-Set7 is functioning through multiple different pathways, thereby causing a compound phenotype. Thus, PR-Set7 and H4K20me1 operate at the convergence of important regulatory processes.
PR-Set7 interacts with a number of nuclear proteins, most notably PCNA, a processivity factor involved in DNA replication (Jorgensen et al. 2007; Huen et al. 2008). This interaction recruits PR-Set7 to sites of DNA damage and replication and is also essential for the CRL4Cdt2-dependent degradation of PR-Set7 (Abbas et al. 2010; Centore et al. 2010; Oda et al. 2010). PCNA-dependent recruitment of PR-Set7 to sites of DNA damage is required for recruitment of 53BP1 (Oda et al. 2010; see below, Regulation of PR-Set7 Expression Level, Activity, and Function; and Function in DNA Damage). Besides its ability to bind to monomethylated lysine residues such as H4K20me1, L3MBTL1 also interacts directly with PR-Set7 (Kalakonda et al. 2008). One attractive possibility is that this latter interaction could serve to propagate the H4K20me1 mark. Furthermore, PR-Set7 has been shown to interact with LEF3 (Z Li et al. 2011), RNA polymerase II (Y Li et al. 2011), ER (Y Li et al. 2011), and TWIST (Yang et al. 2011), and these interactions may direct PR-Set7 activity to specific genes during transcription, although there is limited evidence for the potential functions of these associations.
Besides H4K20, PR-Set7 methylates p53 at lysine residue 382, preventing p53 promoter binding and thereby suppressing transcriptional activation of p21 and PUMA (Shi et al. 2007). This event results in an inhibition of apoptosis (Shi et al. 2007). Thus, PR-Set7 may also regulate the DNA damage response and cell cycle progression via methylation of p53, a key checkpoint protein (Sherr 2004). However, the DNA damage and cell cycle defects caused by PR-Set7 knockdown are not entirely p53-dependent, and p53−/− cells do not show massive DNA damage, suggesting that the major impact of PR-Set7 is independent of p53 (Donehower et al. 1992; Tardat et al. 2007; Houston et al. 2008). Moreover, in stark contrast to PR-Set7−/− cells, p53−/− cells are viable (Donehower et al. 1992). For these reasons, we focus our attention here on H4K20me when discussing PR-Set7 function.
Regulation of PR-Set7 expression level, activity, and function
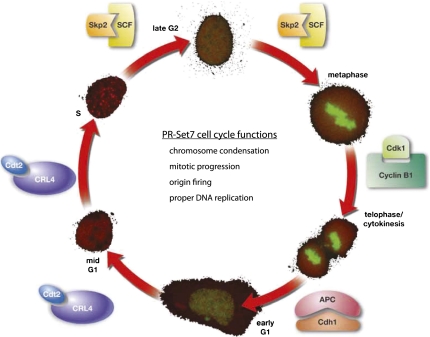
PR-Set7 protein expression is tightly regulated during the cell cycle; it is highest during G2/M and early G1 and is absent during S phase. (Fig. 2; Oda et al. 2009). This regulation is also reflected by a similar oscillation in the levels of H4K20mel during the different phases of the cell cycle. PR-Set7 levels are controlled primarily through post-translational modification by multiple enzymes, including kinases, SUMO, and ubiquitin ligases (Abbas et al. 2010; Centore et al. 2010; Oda et al. 2010; Tardat et al. 2010; Spektor et al. 2011). Yet recent evidence supports that PR-Set7 are also regulated at the transcriptional level (Wakabayashi et al. 2009). We speculate that there are additional, as-yet-unidentified factors that further modulate PR-Set7 activity and recruitment to chromosomes and specific loci and that these proteins may provide key insights into the regulation and function of H4K20me1.
Figure 2.
PR-Set7 is highly regulated during, and is important for multiple aspects of, the cell cycle. PR-Set7 levels oscillate—as shown by YFP-PR-Set7 (green) expression in the outer circle—during the different stages of the cell cycle (mCherry-PCNA). PR-Set7 is post-translationally regulated by multiple different enzymes during the cell cycle, including CRL4-Cdt2, SCF-Skp2, APC-Cdh1, and Cyclin B1/Cdk1. The various functions of PR-Set7 and H4K20me1 are listed within the cell cycle circle.
Post-translational regulation of PR-Set7
To date, we know of multiple mechanisms of post-translational regulation of PR-Set7—most prominently, ubiquitination and phosphorylation. Recently, multiple laboratories simultaneously demonstrated that PR-Set7 is a direct substrate of the E3 ubiquitin ligase complex CRL4Cdt2 (Abbas et al. 2010; Centore et al. 2010; Oda et al. 2010). CRL4Cdt2 causes the ubiquitin-dependent degradation of its substrates in response to UV damage and during S phase (Abbas and Dutta 2011). The complex is thought to be active only when PCNA is bound to chromatin, which is restricted normally to S phase (Arias and Walter 2006). CRL4Cdt2 targets various cellular proteins, such as p21 (Abbas et al. 2008; Kim et al. 2008) and Cdt1 (Arias and Walter 2007; Abbas and Dutta 2011), both of which contain a specialized type of PCNA interaction motif (PIP [PCNA-interacting peptide] domain) termed a “PIP degron” (Havens and Walter 2009). PR-Set7 has two PIP domains, yet only one qualifies as a PIP degron (Havens and Walter 2009; Abbas et al. 2010; Oda et al. 2010). In the absence of this PIP degron, PCNA, or CRL4Cdt2, PR-Set7 protein levels are stabilized during S phase and after UV damage, when they would normally be degraded (Abbas et al. 2010; Centore et al. 2010; Oda et al. 2010). These results confirm that CRL4Cdt2 is the major regulator of PR-Set7 protein levels during the cell cycle. Furthermore, all identified CRL4Cdt2 substrates play pivotal roles in regulating the onset of DNA replication (see below, Role in Cell Cycle Progression). What remains unclear is how the function of CRL4Cdt2 during S phase is linked to its function after UV damage and whether PR-Set7 recruitment by PCNA and degradation within these two contexts actually reflects a similar role for PR-Set7.
In addition, PR-Set7 is regulated by the E3 ubiquitin ligase SCF/Skp2 (Yin et al. 2008). Skp2 degrades substrates during S and G2 phase and could contribute partially to the steep decrease in PR-Set7 levels during S phase (Frescas and Pagano 2008). Silencing of Skp2 causes increased PR-Set7 expression levels. Furthermore, Skp2 was shown to be responsible for PR-Set7 degradation after UV damage, outside the chromatin context (Oda et al. 2010). BBAP, another E3 ligase, also regulates PR-Set7 protein levels (Yan et al. 2009). However, neither Skp2 nor BBAP have been shown to directly ubiquitinate PR-Set7.
Phosphorylation has also been linked to regulation of PR-Set7 levels. Multiple phosphorylation sites on PR-Set7 were identified by phospho-proteome mass spectroscopy studies (Dephoure et al. 2008; Huttlin et al. 2010; Olsen et al. 2010). One such site, Ser 29, is phosphorylated in vivo by Cyclin B/cdk1 during mitosis (Wu et al. 2010). Ser 29 phosphorylation causes PR-Set7 to dissociate from mitotic chromosomes at anaphase and relocate to the extrachromosome space, where it is dephosphorylated by cdc14 phosphatase and subsequently subjected to ubiquitination by APC/Cdh1 (Wu et al. 2010). This destruction probably does not contribute to the prominent cell cycle oscillation of its level, since PR-Set7 expression is highest during mitosis. Although this evidence suggests a role for mitotic phosphorylation in regulating PR-Set7 function, other phospho-proteome studies have failed to identify Ser 29, and further examination will be required to assess the abundance and biological importance of this modification (Olsen et al. 2006; Daub et al. 2008; Huttlin et al. 2010). Thus, distinct ubiquitin ligases and kinases control the dynamic changes of PR-Set7 protein during the cell cycle, and although it is not yet fully understood, each seems to play a unique role in regulating PR-Set7 and thereby H4K20me1 production. The coordination of these multiple post-translational processes suggests that it is necessary to maintain precise levels of H4K20me1 and PR-Set7 for proper cell cycle progression.
Transcriptional regulation of PR-Set7
Although PR-Set7 mRNA transcript levels change during the cell cycle, these changes do not appear to correlate well with the dynamic fluctuation of PR-Set7 protein levels (Rice et al. 2002). Along these lines, expression of a PR-Set7 transgene under the control of a constitutive promoter leads to oscillations in protein abundance similar to the endogenous PR-Set7 during the cell cycle (Oda et al. 2010). Yet, unexpectedly, transcriptional control of PR-Set7 was demonstrated to be important for adipocyte development. In this system, PPARγ was shown to directly regulate the mRNA expression of multiple SET domain-containing genes, including PR-Set7, although it is not clear whether these changes in transcript levels are accompanied by changes in protein levels (Wakabayashi et al. 2009). In addition, recent work implicated an important role for PR-Set7 in skin development as a transcriptional target of c-Myc (Driskell et al. 2011). These studies suggest a role for PR-Set7 in adipocyte and skin differentiation, indicating that regulation of PR-Set7 gene expression levels may also be important for lineage development and maintenance.
Role in cell cycle progression
PR-Set7 is unique among histone methyltransferases in that its protein levels are regulated during the cell cycle. This oscillation reflects the specific roles of PR-Set7 and H4K20me1 during different phases of the cell cycle that are essential for proper cell cycle progression. Loss-of-function studies established a role for PR-Set7 in proper mitotic condensation, S-phase progression, and genome integrity. Further insights have emerged from determining that PR-Set7 is degraded through PCNA-coupled CRL4Cdt2-mediated ubiquitination prior to S phase. These studies have implicated a role for PR-Set7 and H4K20me1 in DNA replication. Moreover, these efforts have defined PR-Set7 as an important controller of both S and G2/M phase and potentially link these two processes together.
Role in mitosis, S phase, or both?
In Drosophila, complete loss of PR-Set7 is lethal at the third instar larval stage. Neuroblasts from these flies show delays in mitosis and a decreased S-phase population (Sakaguchi and Steward 2007). This cell cycle arrest was rescued upon loss of the fly homolog of ATR (ataxia telangiectasia and Rad3-related protein), a critical checkpoint protein regulating DNA damage and proper DNA replication. These results suggest that cell cycle defects that occur upon loss of PR-Set7 are mediated by activation of a checkpoint. The vast majority of mitotic figures were abnormal in PR-Set7−/− flies, and further analysis showed that mitotic progression was disrupted because chromosome condensation occurred before spindle formation (Sakaguchi and Steward 2007). These findings implicated PR-Set7 in regulating mitotic chromosome condensation and entry into S phase.
Our group was able to expand on these studies by generating knockout and conditional knockout mice as a means to better understand PR-Set7 function (Oda et al. 2009). In both PR-Set7−/− embryos and PR-Set7 conditional knockout cells, complete elimination of PR-Set7 caused the cells to accumulate within the G2/M phase of the cell cycle. Similarly, transient depletion of PR-Set7 in HEK293 cells caused an increase in G2/M populations, correlating with a striking increase in abnormal mitotic figures and an overall decrease in chromosome compaction (Houston et al. 2008). These defects might be linked to either of the H4K20me1-binding proteins L3MBTL1 or condensin II, both of which are involved in chromatin compaction (Trojer et al. 2007; Liu et al. 2010).
Deletion or depletion of PR-Set7 in a variety of synchronized cell lines demonstrated that the protein is also required for proper S-phase progression (Jorgensen et al. 2007; Tardat et al. 2007; Houston et al. 2008; Oda et al. 2009). Similarly, the loss of PR-Set7 causes an accumulation of U2OS cells in S phase, although these cells are nonreplicating (Jorgensen et al. 2007; Tardat et al. 2007). This accumulation supports the hypothesis that PR-Set7 functions prior to S phase during G1 or the previous G2/M phases, yet is required for proper DNA replication during S phase.
In addition, loss of PR-Set7 correlates with increased DNA damage, which only occurs after entry into S phase (Jorgensen et al. 2007). This damage, however, is not observed when DNA replication is inhibited by aphidicolin or siRNA-mediated knockdown of CDC45. Upon loss of PR-Set7, this damage could potentially cause the activation of the intra-S-phase checkpoint, leading to a cell cycle arrest. Indeed, along these lines, inhibition of ATR or chk1, critical intra-S-phase checkpoint proteins, rescues the cell cycle delays, although DNA damage still accumulates (Jorgensen et al. 2007). These results cumulatively suggest that upon loss of PR-Set7, cells acquire DNA damage in S phase, causing activation of ATR and the intra-S-phase checkpoint, leading to a subsequent cell cycle arrest. This function could partially be explained by a role for H4K20me in replication fork stability and, potentially, homologous recombination, although this has not been clearly elucidated (Jorgensen et al. 2007).
A major indication of the fundamental role of H4K20 methylation in cell cycle progression comes from analyses of the various enzymes that regulate the overall methylation status. Loss of Suv4-20h1/2 causes defects in S-phase entry along with abnormal mitotic figures, although loss of PR-Set7 causes far more substantial defects (Schotta et al. 2008; Oda et al. 2009). In addition, disruption of the H4K20me1 demethylase PHF8 leads to increased H4K20me1, a delay in the G1/S phase transition, and an increase in the G2/M population (Liu et al. 2010). It is thus clear that regulation of H4K20me states is critical for proper cell cycle progression. Further studies will shed light on the relative contributions of each level of methylation and their specific binders.
In summary, in addition to the initial observation that PR-Set7 and H4K20me1 oscillate during the cell cycle, early studies have demonstrated that PR-Set7 methyltransferase activity is essential for cell cycle progression. Although PR-Set7 methyltransferase activity is undoubtedly essential for cell cycle progression, it is not entirely clear why and how the various cell cycle defects and DNA damage observed upon PR-Set7 loss are functionally related. Addressing these questions will help determine exactly how mechanistically H4K20me1 contributes to the rhythm of the cell cycle.
The role of PR-Set7 and H4K20me1 in DNA replication
In addition to its role in mitotic condensation, PR-Set7 was also shown to be important for maintaining proper replication fork speed, active replication sites, and the length of replicated tracks. These findings, however, were puzzling, since PR-Set7 is undetectable during DNA replication (Oda et al. 2009). Of note, after inhibition of proteasome-mediated degradation, PR-Set7 localizes to sites of active DNA replication (Tardat et al. 2007). Further insights into PR-Set7 function during DNA replication emerged through the study of its individual domains, specifically its PIP domain (Jorgensen et al. 2007; Huen et al. 2008). Stabilization of PR-Set7 during S phase by mutation of its PIP domain causes a cell cycle arrest in the G2 phase, with a concomitant decrease in the S-phase population (Abbas et al. 2010; Centore et al. 2010).
Previously identified CRL4Cdt2 substrates are critical for proper origin of DNA replication licensing (Abbas and Dutta 2011). The proper timing of origin licensing is critical to prevent rereplication or multiple uncoordinated rounds of DNA synthesis per cell cycle (Arias and Walter 2007). Inhibition of CRL4Cdt2-mediated degradation of PR-Set7 causes rereplication exemplified by increased nuclear volume, a cell population of >4n DNA content, and altered DNA sedimentation (Abbas et al. 2010; Tardat et al. 2010). Although this rereplication phenotype may partially be due to a decrease in degradation of other CRL4Cdt2 substrates, experimental evidence from our group and others has demonstrated a more direct role for PR-Set7 in DNA replication. First, a catalytically inactive PR-Set7 that is also mutant in its PIP domain does not cause rereplication, substantiating that appropriate regulation of PR-Set7 catalytic activity is required for proper S phase (Abbas et al. 2010; Tardat et al. 2010). Second, defects in cell cycle progression observed upon knockdown of CRL4Cdt2 can be rescued by simultaneous knockdown of PR-Set7 (Abbas and Dutta 2011; D Beck, H Oda, and D Reinberg, unpubl.). Finally, as mentioned previously, loss of PR-Set7 catalytic activity results in S-phase delays accompanied by origin of replication firing defects (Jorgensen et al. 2007; Tardat et al. 2007).
The mechanism by which PR-Set7 regulates DNA replication is still unclear. PR-Set7 knockdown decreases the loading of the ORC complex, and artificial recruitment of PR-Set7 to a genomic locus results in the recruitment of ORC1/MCM5 to that locus (Tardat et al. 2010). Unbiased biochemical analysis identified LRWD1 as an H4K20me3-binding protein, which also associates with ORC (Shen et al. 2010; Vermeulen et al. 2010), suggesting that PR-Set7 may control recruitment of ORC to chromatin through regulating methylation of H4K20. Another possible role for H4K20me1 in S-phase progression is through transcriptional regulation of E2F-1 target genes that are important in the G1–S-phase transition (Abbas et al. 2010; Liu et al. 2010). Alternatively, H4K20me1 may affect the status of H4 acetylation, which can also modulate origin of replication licensing (Nishioka et al. 2002; Iizuka et al. 2006; Miotto and Struhl 2010). Finally, L3MBTL1 was also shown to directly bind components of the replication machinery, potentially linking H4K20me1 with DNA replication (Gurvich et al. 2010). Further investigation will be required to ascertain which functions associated with PR-Set7 that occur outside of S phase contribute to proper DNA replication. We predict that PR-Set7 primarily functions in origin licensing through direct methylation of H4K20 at replication origins. This role in origin licensing and initiation of replication would satisfy the requirement of a process unique to higher eukaryotes that might necessitate independent enzymatic control of H4K20me1.
Attempts at unraveling the function of PR-Set7 during the cell cycle have been hampered by the phenotypic differences exhibited by deletion versus transient depletion of PR-Set7, differences exhibited by primary and transformed cell lines, and discrepancies arising from overexpression versus depletion studies. A major predicament in this cell cycle analysis is exactly how PR-Set7 and H4K20me1 affect DNA replication, given that PR-Set7 is undetectable and H4K20me1 levels are low during S phase. Future work will help clarify the mechanism by which PR-Set7 and H4K20me1 modulate such effects, the contribution of modification binders, the processing of H4K20me1 levels, and the potential effect of transcriptional regulation and mitosis. These insights will establish how chromatin modification can coordinate multiple phases of the cell cycle.
Function in DNA damage
Spontaneous DNA damage in the absence of PR-Set7
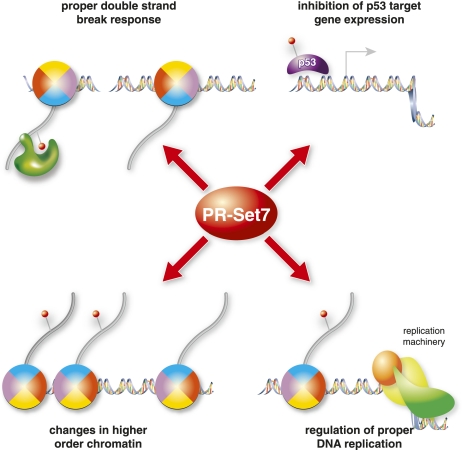
As described earlier, deletion of PR-Set7 in ES cells results in massive DNA damage, as shown by immunofluorescence staining with γ-H2AX antibodies (Oda et al. 2009). Moreover, in an unbiased RNAi screen, PR-Set7 was identified as a factor whose depletion leads to an increase in γ-H2AX foci in HeLa cells (Paulsen et al. 2009). It is unlikely that this increase in spontaneous DNA damage upon loss of PR-Set7 is the result solely of an impaired DNA damage pathway, since there is normally very little spontaneous damage in the cell (Vilenchik and Knudson 2003). Instead, the massive damage seen after loss of PR-Set7 is more likely attributable to increased genomic instability due to defects during DNA replication or mitosis (Fig. 3). In the absence of PR-Set7, DNA damage accumulates only if the cells are allowed to enter S phase, which suggests that the defect occurs during DNA replication (Jorgensen et al. 2007). Furthermore, this damage can be suppressed by depletion of RAD51, a key regulator of homologous recombination, placing PR-Set7 as a potential effector of this type of repair (Jorgensen et al. 2007). Interestingly, the increase of γ-H2AX foci was not observed in PR-Set7 knockout embryos even though they exhibited the same G2/M arrest as their ES cell counterparts. This finding suggests that checkpoint and DNA damage signaling in the early embryonic stages is different from that in later stage ES cells or differentiated cells.
Figure 3.
Role of PR-Set7 in genome integrity. PR-Set7 has multiple functions that are important for proper genome stability and DNA damage response. (Top left) PR-Set7 methylates H4K20me1, which is important for recruiting 53BP1 recruitment to sites of DNA damage. (Top right) PR-Set7 alters p53-dependent gene expression, potentially regulating checkpoint and repair processes. (Bottom left) H4K20me alters chromatin compaction and mitotic condensation, thereby affecting higher-order chromatin structure. Finally, PR-Set7 and H4K20me are required for proper DNA replication, possibly through regulation of fork stability.
H4K20me1–53BP1 pathway
Histone modifications are an attractive means to recruit DNA damage response proteins to sites of genotoxic stress because of their ability to dynamically mark the genome. In fact, a large number of chromatin-modifying enzymes have been implicated in the DNA damage response (Ciccia and Elledge 2010; Polo and Jackson 2011). Following ionizing radiation (IR), a cascade of proteins is recruited to sites of DNA damage for recognition and repair, including 53BP1, an important marker for proper signaling. 53BP1 functions as a scaffolding protein for efficient DNA repair and checkpoint activation (FitzGerald et al. 2009). The mechanism of 53BP1 recruitment after IR is complex, involving multiple post-translational modifications of different proteins, including phosphorylation and ubiquitination of histones, which serve as binding platforms for proper repair (Kolas et al. 2007; Doil et al. 2009; FitzGerald et al. 2009; Stewart et al. 2009).
The importance of H4K20 methylation in the DNA repair pathway was originally demonstrated in S. pombe. In this organism, Crb2, the homolog of mammalian 53BP1, has indispensable roles during the cell cycle and in DNA damage repair signaling (Saka et al. 1997). Crb2 binds H4K20me, which is catalyzed by Set9 in S. Pombe, through its tandem tudor domain (Sanders et al. 2004; Greeson et al. 2008). This domain is conserved in 53BP1 and is required for the proper recruitment of each protein to DNA damage foci. The tandem tudor domain of 53BP1 binds directly to H4K20me1/2 and H3K79me2 both in vitro and in vivo, although more substantial evidence has emerged for a physiological role in the case of H4K20me1 binding by 53BP1 (Fig. 3; Huyen et al. 2004; Houston et al. 2008; Oda et al. 2010). Transient reduction of PR-Set7 abrogates 53BP1 recruitment to sites of DNA double-strand breaks (Oda et al. 2010). Complete loss of Suv4-20h1/2 slightly decreased 53BP1 recruitment to DNA damage sites, suggesting that both H4K20me1 and H4K20me2 contribute to recruitment of 53BP1 (Schotta et al. 2008).
These findings led to investigations of how H4K20me is regulated after DNA damage, whether pre-existing H4K20me is dynamically exposed, or whether methylation of H4K20 is deposited de novo. Early mass spectroscopy evidence suggested that 53BP1 recruitment is accomplished in large part through exposure of pre-existing H4K20me. These studies showed that the majority of cellular histone H4K20 is dimethylated, leaving only a small pool of available substrate to modify after damage (Schotta et al. 2008). Thus, it is hypothesized that 53BP1 recruitment after DNA damage is facilitated by changes in the chromatin structure that expose the already present H4K20me, making it accessible for binding. Various chromatin remodeling factors, KAP-1 and RNF168, have been implicated in this dynamic chromatin relaxation process, yet very little experimental evidence has been provided to support this model (Goodarzi et al. 2008; Doil et al. 2009; Stewart et al. 2009).
On the other hand, there is supportive evidence for H4K20me being deposited de novo at sites of DNA damage. We recently showed that PR-Set7 is rapidly recruited to sites of DNA double-strand breaks through its binding to PCNA. 53BP1 recruitment is disrupted by siRNA-mediated knockdown of endogenous PR-Set7 and by transfection of an siRNA-resistant catalytically inactive mutant (Oda et al. 2010). Chromatin immunoprecipitation (ChIP) experiments revealed that H4K20me1 increased rapidly at sites of DNA double-strand breaks induced by site-specific endonucleases (Pei et al. 2011). In addition, more recent mass spectroscopy results showed that different cell lineages have widely variable amounts of each degree of H4K20 methylation, which may be indicative of a highly dynamic modification (P Voigt and D Reinberg, unpubl.). These findings suggest that PR-Set7-mediated de novo monomethylation is necessary to recruit 53BP1 to the sites of DNA damage. Quite surprisingly, despite all of the evidence for a role of H4K20me in 53BP1 recruitment, none of the H4K20me-regulating enzymes have been shown to affect 53BP1 foci in unbiased RNAi screens (Fig. 3; Doil et al. 2009; Stewart et al. 2009).
53BP1 recruitment is also affected by histone-modifying enzymes that indirectly modulate H4K20me levels or accessibility. One such enzyme is the ubiquitin ligase BBAP that targets H4K91 (Yan et al. 2009). In addition, enzymes other than PR-Set7 and Suv4-20 have been reported to methylate H4K20, although these enzymes most likely affect H4K20me indirectly (Pei et al. 2011). For example, MMSET, a histone methyltransferase, was recently demonstrated to dynamically dimethylate H4K20 at sites of DNA damage and regulate recruitment of 53BP1. Silencing of MMSET leads to decreased H4K20me2/3 and impaired 53BP1 recruitment at sites of DNA damage. However, other more thorough studies have shown that, both in vitro and in vivo, the major targets of MMSET are H3K36 and H4K44 (Li et al. 2009; Nimura et al. 2009). Therefore, although methylation of H4K20 is critical for a proper damage response, MMSET is unlikely to be the enzyme responsible for this modification and most likely works indirectly by modifying other histone residues to regulate H4K20me-dependent 53BP1 recruitment.
In addition to its role in DNA double-strand break repair, 53BP1 has been implicated in the regulation of the G2/M checkpoint (Harrigan et al. 2011; Lukas et al. 2011). Recently, time-lapse microscopy analysis of GFP-tagged 53BP1 revealed that 53BP1 forms spontaneous foci without any exogenously mediated damage. These foci are present in the G1 phase of the cell cycle at sites of inappropriately underreplicated genomic loci, which have already passed through mitosis. Loss of these 53BP1 foci leads to destabilization and improper repair of the associated DNA lesions (Lukas et al. 2011). Thus, PR-Set7 may regulate 53BP1 foci in G1 phase to protect the integrity of chromosomes exposed to replication stress during the previous S phase. Defective 53BP1 recruitment may partially explain both the spontaneous damage and cell cycle defects observed upon the loss of PR-Set7.
PR-Set7 and transcription
Despite numerous efforts, the role of PR-Set7 and H4K20me1 in gene transcription has remained elusive. Removal of PR-Set7 causes various cellular changes that can indirectly affect gene expression, making it difficult to determine its direct functions. For this reason, we are currently dependent on correlative studies, which have been inconclusive and imply complex roles for these factors in transcription. This is apparent when comparing the conclusions reached from initial subcellular localization studies suggesting a role for H4K20me1 in transcriptional repression with those from more recent genome-wide ChIP analysis implicating H4K20me1 in transcriptional activation.
Supporting a role in repression, H4K20me1 could operate directly by promoting chromatin compaction (Lu et al. 2008; Oda et al. 2009) or indirectly through its progression toward H4K20me2/3 and/or recruitment of modification-specific binders such as L3MBTL1, a transcriptional repressor (Trojer et al. 2007). Moreover, insights into a potential role for PR-Set7 in repression of transcription were initially made in Drosophila, wherein H4K20me1 localizes exclusively to regions on polytene chromosomes that are devoid of active transcription (Nishioka et al. 2002). Furthermore, deletion of PR-Set7 causes a suppression of the variegation phenotype in Drosophila, suggesting that PR-Set7 acts to silence gene expression (Karachentsev et al. 2005). However, this result is complicated in that Suv4-20 is also a suppressor of variegation, and the suppression of variegation phenotype identified after loss of PR-Set7me may be secondary to that of decreased H4K20me2/3 levels. For example, H4K20me3 has a well-established role in gene repression, specifically at repetitive elements (Schotta et al. 2004, 2008; Fodor et al. 2010). In addition to these results from Drosophila, localization studies in mouse cells showed an enrichment of H4K20me1 on the inactive X chromosome, a classic example of facultative heterochromatin (Kohlmaier et al. 2004).
Several additional studies support a role for PR-Set7 in gene repression. In-depth ChIP studies on a large subset of the genome in HeLa cells demonstrated that H4K20me1 localized primarily within gene bodies. Loss of PR-Set7 causes increased expression of H4K20me1-enriched genes, implying a role for PR-Set7 in gene repression (Congdon et al. 2010). In addition, genomic regions enriched for H4K20me1 can repress luciferase expression in an artificial promoter system (Congdon et al. 2010). Still other studies showed that H4K20me1 localized to repressed genes regulated by E2F (Abbas et al. 2010). Finally, knockdown of PHF8, which demethylates H4K20me1, causes increased repression of E2F-responsive genes due to increased H4K20me1 and L3MBTL1 binding (Liu et al. 2010).
On the other hand, results from genome-wide ChIP analyses argue against a role in repression. In T lymphocytes, H4K20me1 peaks were similarly found to localize downstream from the transcription start site (TSS), yet in contrast to the above-mentioned HeLa cell study, H4K20me1 correlates with highly transcribed genes (Barski et al. 2007; Cui et al. 2009). Further analysis revealed that H4K20me1 is one of the histone modifications most highly correlated with active transcription (Wang et al. 2008). This correlation between H4K20me1 and active transcription had previously been shown at a limited set of loci in murine erythroleukemia cells, adipocytes, and HeLa cells (Talasz et al. 2005; Vakoc et al. 2006; Wakabayashi et al. 2009). These findings suggest that PR-Set7 and H4K20me1 may function during active transcription as well as transcriptional repression.
Further evidence from gene-specific studies showed that H4K20me1 levels increased at estrogen-responsive genes after their activation (Y Li et al. 2011). In another focused analysis, H4K20me1 and PR-Set7 were enriched specifically at wnt3a-responsive gene promoters after wnt3a stimulation (Z Li et al. 2011). Upon PR-Set7 knockdown, wnt3a-dependent activation of wnt response genes was abolished. This regulation was shown to be physiologically relevant, since PR-Set7 participates in the wnt developmental pathway in zebrafish (Z Li et al. 2011). These results are slightly complicated by the fact that PR-Set7 was found to both stimulate and repress the wnt3a response genes potentially by cooperating with Lef3, a known transcriptional activator and repressor. A similar type of dual transcriptional regulation was demonstrated for PR-Set7 cooperation with TWIST, an important regulator of epithelial-to-mesenchymal transition (Yang et al. 2011).
Thus, despite numerous studies examining the role of H4K20me1, this modification still has an inconclusive role in gene expression, with evidence supporting a correlation with both gene expression and repression. Some gene-specific analyses, along with all genome-wide ChIP studies to date, have correlated H4K20me1 with active transcription, while a subset of non-genome-wide ChIP and biochemical studies have correlated H4K20me1 with gene repression.
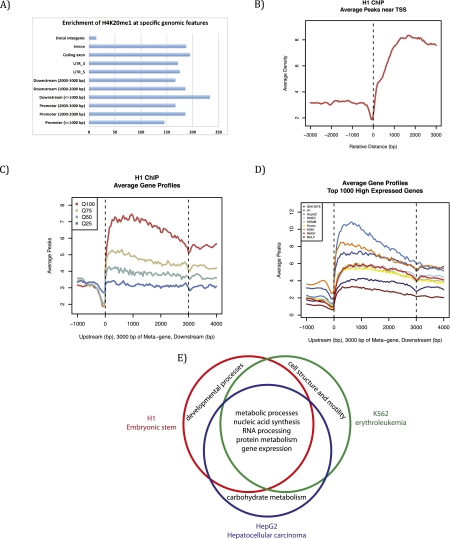
To better decipher the role of H4K20me1 in regulation of transcription, we embarked on a comparative analysis of recently released ENCODE (Encyclopedia of DNA Elements) data sets, established as a means to identify important functional genomic elements (The ENCODE Project Consortium 2004; Ernst et al. 2011). This analysis allows for the first cross-comparison of data sets between different cell lineages that, importantly, were generated using similar experimental procedures and data analysis. After comparing H4K20me1 ChIP sequencing data from the nine human cell lines examined by the ENCODE project, we found that H4K20me1 is specifically enriched in intragenic regions in all data sets (Fig. 4A). Supporting earlier observations, H4K20me1 also correlates with highly expressed genes and is found predominantly downstream from the TSS (Fig. 4B), although the magnitude of these peaks varies between cell lines (Fig. 4C,D). We further analyzed the gene ontologies enriched with H4K20me1 that are unique or conserved between three ENCODE cell types: H1 ES, HepG2 hepatocellular carcinoma, and K562 erythroleukemia cells. Genes with H4K20me1 peaks that were conserved across all three cell lines were primarily highly expressed metabolic and cellular housekeeping genes. In contrast, genes with H4K20me1 peaks unique to one specific cell line tended to be highly expressed tissue-specific genes functionally inherent to that lineage; i.e., development for H1, carbohydrate metabolism for HepG2, and cellular structure for K562 (Fig. 4). These combined results suggest that H4K20me1 correlates with highly expressed genes, independent of the lineage, and does not seem to be very specific to any functional subset of genes. Although these findings do not directly demonstrate a function of PR-Set7 and H4K20me1 in transcription, they suggest that these factors may participate in active transcription.
Figure 4.
Genome-wide distribution and features of H4K20me1 from the ENCODE project. Bioinformatic analysis (MACS) was used to analyze H4K20me1 genome-wide distribution from ENCODE data sets from nine different human cell lines: H1 (human ES cells), K562 (erythrocytic leukemia), GM12878 (B-lymphoblastoid), HepG2 (hepatocellular carcinoma), NHEK (normal human epidermal keratinocytes), HSMM (primary human skeletal muscle myoblasts), NHLF (normal human lung fibroblasts), Huvec (primary human umbilical vein endothelial cells), and HMEC (primary human mammary epithelial cells). (A) H4K20me1 is enriched on intragenic features in H1 cells, representative of other cell lines. (B) H4K20me1 is enriched downstream from the TSS in H1 cells, representative of other cell lines. (C) H4K20me1 is found within the gene body and is correlated best with the highly expressed genes (Q100) and less so with the lower expressed genes in H1 cells, representative of other cell lines. (D) Comparison across all nine cell lines; Q100 genes show a similar distribution, with differences in the level of enrichment. (E) Venn diagram comparing gene ontology term analysis of genes within 3 kb of an H4K20me1 peaks. Conserved genes with H4K20me1 peaks across all three cell lines are housekeeping genes required in all cell types, whereas cell-specific genes enriched with H4K20me1 are genes that are only highly expressed in that specific lineage.
These apparently conflicting experimental results concerning PR-Set7 function in gene expression may be explained by multiple, potentially confounding biological and technical variables. One major variable may be the proliferation rates of different cell lines, given the cell cycle-dependent variation in PR-Set7 and H4K20me1 levels. The role of H4K20me1 may also be context-dependent. For example, neighboring modifications on the H4 tail may affect H4K20me1 function. The presence of H4K20me1 at highly expressed genes may potentially be affected by the co-occurrence of neighboring H4K16 acetylation (H4K16ac), which has an established role in activating gene expression. H4K16ac could potentially override a subset of H4K20me1 function, creating a different role for H4K20me1 in gene expression in this context. Other subsets of H4K20me1, specifically at E2F target genes, may be important for gene repression and are demethylated by PHF8 during the cell cycle (Liu et al. 2010). Still other H4K20me1 may be destined to progress to H4K20me2/3, which repress repetitive DNA elements (Schotta et al. 2004, 2008). Finally, the H4K20me1 that is not sterically masked may be regulating transcription through L3MBTL1 recruitment (Trojer et al. 2007). Detailed investigation of H4K20me1 and specific binders of H4K20me1 and H4K16Ac in synchronized cells will be necessary to parse out the meaning of the differential localization patterns of H4K20me1 and determine the exact correlation between gene expression and cell cycle progression. These combined possibilities reveal a plethora of potential functions for H4K20me1. An outstanding resolution, although unlikely and difficult to prove, is that the modification may not have a specific role in gene expression even though potential correlations abound.
Final thoughts
Over the last 10 years, we have made great strides toward understanding the function of PR-Set7, yet we still have far to go. A major question remains as to why PR-Set7 and independent regulation of H4K20me1 emerged during evolution. To date, the least-characterized function of PR-Set7 and H4K20me1 is in transcription and gene expression. Clarification of how this modification is regulated genome-wide and the relevance of its distribution may better explain the major functions of PR-Set7 in cell cycle progression and in the DNA damage response. Recently, PR-Set7 was shown to potentially have physiological importance in cancer progression, making understanding the function of H4K20me1 even more important (Yang et al. 2011).
Although our understanding of the function of H4K20me1 has become more comprehensive, we still have not elucidated the mechanism by which one modification can have such a vast array of functions. The answer to these questions will help characterize a unique and essential modification and provide key insights into how a single modification can regulate so many cellular functions. Hopefully, further analyses, including knockout studies, will lead to a detailed understanding of the role of each modification binder in regulating gene expression and cell cycle progression. This key information will help decipher the contributions of PR-Set7 and H4K20me1 that are determinant to cell viability.
Acknowledgments
We thank Lynne Vales, Philipp Voigt, Maria Elena Torres-Padilla, Luca Colnaghi, and Erin Savner for critical reading of the manuscript. This work was supported by grants from the National Institutes of Health (GM64844, GM037120 to D.R.) and the Howard Hughes Medical Institute (to D.R.) and the Ministry of Education, Science, Sports, and Culture Grant-in-aid for scientific research (23616009 to H.O.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.177444.111.
References
- Abbas T, Dutta A 2011. CRL4Cdt2: Master coordinator of cell cycle progression and genome stability. Cell Cycle 10: 241–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, Dutta A 2008. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes & Dev 22: 2496–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas T, Shibata E, Park J, Jha S, Karnani N, Dutta A 2010. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol Cell 40: 9–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias EE, Walter JC 2006. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat Cell Biol 8: 84–90 [DOI] [PubMed] [Google Scholar]
- Arias EE, Walter JC 2007. Strength in numbers: Preventing rereplication via multiple mechanisms in eukaryotic cells. Genes & Dev 21: 497–518 [DOI] [PubMed] [Google Scholar]
- Balakrishnan L, Milavetz B 2010. Decoding the histone H4 lysine 20 methylation mark. Crit Rev Biochem Mol Biol 45: 440–452 [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Beisel C, Imhof A, Greene J, Kremmer E, Sauer F 2002. Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature 419: 857–862 [DOI] [PubMed] [Google Scholar]
- Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ 2001. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem 276: 42462–42467 [DOI] [PubMed] [Google Scholar]
- Campos EI, Reinberg D 2009. Histones: Annotating chromatin. Annu Rev Genet 43: 559–599 [DOI] [PubMed] [Google Scholar]
- Centore RC, Havens CG, Manning AL, Li JM, Flynn RL, Tse A, Jin J, Dyson NJ, Walter JC, Zou L 2010. CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol Cell 40: 22–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ 2010. The DNA damage response: Making it safe to play with knives. Mol Cell 40: 179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congdon LM, Houston SI, Veerappan CS, Spektor TM, Rice JC 2010. PR-Set7-mediated monomethylation of histone H4 lysine 20 at specific genomic regions induces transcriptional repression. J Cell Biochem 110: 609–619 [DOI] [PubMed] [Google Scholar]
- Couture JF, Collazo E, Brunzelle JS, Trievel RC 2005. Structural and functional analysis of SET8, a histone H4 Lys-20 methyltransferase. Genes & Dev 19: 1455–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K 2009. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell 4: 80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Korner R, Greff Z, Keri G, Stemmann O, Mann M 2008. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol Cell 31: 438–448 [DOI] [PubMed] [Google Scholar]
- DeLange RJ, Fambrough DM, Smith EL, Bonner J 1969. Calf and pea histone IV. II. The complete amino acid sequence of calf thymus histone IV; presence of ϵ-N-acetyllysine. J Biol Chem 244: 319–334 [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP 2008. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci 105: 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al. 2009. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136: 435–446 [DOI] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA Jr, Butel JS, Bradley A 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356: 215–221 [DOI] [PubMed] [Google Scholar]
- Driskell I, Oda H, Blanco S, Nascimento E, Humphreys P, Frye M 2011. The histone methyltransferase Setd8 acts in concert with c-Myc and is required to maintain skin. EMBO J doi: 10.1038/emboj.2011.421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards CR, Dang W, Berger SL 2011. Histone H4 lysine 20 of Saccharomyces cerevisiae is monomethylated and functions in subtelomeric silencing. Biochemistry 50: 10473–10483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2004. The ENCODE (Encyclopedia Of DNA Elements) Project. Science 306: 636–640 [DOI] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzGerald JE, Grenon M, Lowndes NF 2009. 53BP1: Function and mechanisms of focal recruitment. Biochem Soc Trans 37: 897–904 [DOI] [PubMed] [Google Scholar]
- Fodor BD, Shukeir N, Reuter G, Jenuwein T 2010. Mammalian Su(var) genes in chromatin control. Annu Rev Cell Dev Biol 26: 471–501 [DOI] [PubMed] [Google Scholar]
- Frescas D, Pagano M 2008. Deregulated proteolysis by the F-box proteins SKP2 and β-TrCP: Tipping the scales of cancer. Nat Rev Cancer 8: 438–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA 2008. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell 31: 167–177 [DOI] [PubMed] [Google Scholar]
- Govin J, Dorsey J, Gaucher J, Rousseaux S, Khochbin S, Berger SL 2010. Systematic screen reveals new functional dynamics of histones H3 and H4 during gametogenesis. Genes & Dev 24: 1772–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greeson NT, Sengupta R, Arida AR, Jenuwein T, Sanders SL 2008. Di-methyl H4 lysine 20 targets the checkpoint protein Crb2 to sites of DNA damage. J Biol Chem 283: 33168–33174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurvich N, Perna F, Farina A, Voza F, Menendez S, Hurwitz J, Nimer SD 2010. L3MBTL1 polycomb protein, a candidate tumor suppressor in del(20q12) myeloid disorders, is essential for genome stability. Proc Natl Acad Sci 107: 22552–22557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrigan JA, Belotserkovskaya R, Coates J, Dimitrova DS, Polo SE, Bradshaw CR, Fraser P, Jackson SP 2011. Replication stress induces 53BP1-containing OPT domains in G1 cells. J Cell Biol 193: 97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havens CG, Walter JC 2009. Docking of a specialized PIP Box onto chromatin-bound PCNA creates a degron for the ubiquitin ligase CRL4Cdt2. Mol Cell 35: 93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Lipp JJ, Toh BH, Peters JM 2005. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 438: 1176–1180 [DOI] [PubMed] [Google Scholar]
- Houston SI, McManus KJ, Adams MM, Sims JK, Carpenter PB, Hendzel MJ, Rice JC 2008. Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J Biol Chem 283: 19478–19488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Sy SM, van Deursen JM, Chen J 2008. Direct interaction between SET8 and proliferating cell nuclear antigen couples H4-K20 methylation with DNA replication. J Biol Chem 283: 11073–11077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP 2010. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143: 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyen Y, Zgheib O, Ditullio RA Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD 2004. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432: 406–411 [DOI] [PubMed] [Google Scholar]
- Iizuka M, Matsui T, Takisawa H, Smith MM 2006. Regulation of replication licensing by acetyltransferase Hbo1. Mol Cell Biol 26: 1098–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen S, Elvers I, Trelle MB, Menzel T, Eskildsen M, Jensen ON, Helleday T, Helin K, Sorensen CS 2007. The histone methyltransferase SET8 is required for S-phase progression. J Cell Biol 179: 1337–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalakonda N, Fischle W, Boccuni P, Gurvich N, Hoya-Arias R, Zhao X, Miyata Y, Macgrogan D, Zhang J, Sims JK, et al. 2008. Histone H4 lysine 20 monomethylation promotes transcriptional repression by L3MBTL1. Oncogene 27: 4293–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor-Vazirani P, Kagey JD, Vertino PM 2011. SUV420H2-mediated H4K20 trimethylation enforces RNA polymerase II promoter-proximal pausing by blocking hMOF-dependent H4K16 acetylation. Mol Cell Biol 31: 1594–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karachentsev D, Sarma K, Reinberg D, Steward R 2005. PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes & Dev 19: 431–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AE, Ghenoiu C, Xue JZ, Zierhut C, Kimura H, Funabiki H 2010. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science 330: 235–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Starostina NG, Kipreos ET 2008. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes & Dev 22: 2507–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, Wutz A 2004. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol 2: e171 doi: 10.1371/journal.pbio.0020171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318: 1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Fischle W, Wang W, Duncan EM, Liang L, Murakami-Ishibe S, Allis CD, Patel DJ 2007. Structural basis for lower lysine methylation state-specific readout by MBT repeats of L3MBTL1 and an engineered PHD finger. Mol Cell 28: 677–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Trojer P, Xu CF, Cheung P, Kuo A, Drury WJ III, Qiao Q, Neubert TA, Xu RM, Gozani O, et al. 2009. The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. J Biol Chem 284: 34283–34295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sun L, Zhang Y, Wang D, Wang F, Liang J, Gui B, Shang Y 2011. The histone modifications governing TFF1 transcription mediated by estrogen receptor. J Biol Chem 286: 13925–13936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Nie F, Wang S, Li L 2011. Histone H4 Lys 20 monomethylation by histone methylase SET8 mediates Wnt target gene activation. Proc Natl Acad Sci 108: 3116–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, Ohgi KA, Benner C, Garcia-Bassets I, Aggarwal AK, et al. 2010. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature 466: 508–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Simon MD, Chodaparambil JV, Hansen JC, Shokat KM, Luger K 2008. The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure. Nat Struct Mol Biol 15: 1122–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS, Grofte M, Chan KL, Hickson ID, Bartek J, et al. 2011. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol 13: 243–253 [DOI] [PubMed] [Google Scholar]
- Marango J, Shimoyama M, Nishio H, Meyer JA, Min DJ, Sirulnik A, Martinez-Martinez Y, Chesi M, Bergsagel PL, Zhou MM, et al. 2008. The MMSET protein is a histone methyltransferase with characteristics of a transcriptional corepressor. Blood 111: 3145–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto B, Struhl K 2010. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol Cell 37: 57–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, Schwartz RJ, Kaneda Y 2009. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome. Nature 460: 287–291 [DOI] [PubMed] [Google Scholar]
- Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P, Tempst P, Steward R, et al. 2002. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell 9: 1201–1213 [DOI] [PubMed] [Google Scholar]
- Oda H, Okamoto I, Murphy N, Chu J, Price SM, Shen MM, Torres-Padilla ME, Heard E, Reinberg D 2009. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol 29: 2278–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Hubner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL, Reinberg D 2010. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol Cell 40: 364–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M 2006. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127: 635–648 [DOI] [PubMed] [Google Scholar]
- Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, et al. 2010. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal 3: ra3 doi: 10.1126/scisignal.2000475 [DOI] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, et al. 2009. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell 35: 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei H, Zhang L, Luo K, Qin Y, Chesi M, Fei F, Bergsagel PL, Wang L, You Z, Lou Z 2011. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 470: 124–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesavento JJ, Yang H, Kelleher NL, Mizzen CA 2008. Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. Mol Cell Biol 28: 468–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Jackson SP 2011. Dynamics of DNA damage response proteins at DNA breaks: A focus on protein modifications. Genes & Dev 25: 409–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi HH, Sarkissian M, Hu GQ, Wang Z, Bhattacharjee A, Gordon DB, Gonzales M, Lan F, Ongusaha PP, Huarte M, et al. 2010. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature 466: 503–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Van Buren D, Huang HS, Zhong L, Mostoslavsky R, Akbarian S, Hock H 2010. Chromatin protein L3mbtl1 is dispensable for development and tumor suppression in mice. J Biol Chem 285: 27767–27775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JC, Nishioka K, Sarma K, Steward R, Reinberg D, Allis CD 2002. Mitotic-specific methylation of histone H4 Lys 20 follows increased PR-Set7 expression and its localization to mitotic chromosomes. Genes & Dev 16: 2225–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y, Esashi F, Matsusaka T, Mochida S, Yanagida M 1997. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes & Dev 11: 3387–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi A, Steward R 2007. Aberrant monomethylation of histone H4 lysine 20 activates the DNA damage checkpoint in Drosophila melanogaster. J Cell Biol 176: 155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T 2004. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119: 603–614 [DOI] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T 2004. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes & Dev 18: 1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Sengupta R, Kubicek S, Malin S, Kauer M, Callen E, Celeste A, Pagani M, Opravil S, De La Rosa-Velazquez IA, et al. 2008. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes & Dev 22: 2048–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z, Sathyan KM, Geng Y, Zheng R, Chakraborty A, Freeman B, Wang F, Prasanth KV, Prasanth SG 2010. A WD-repeat protein stabilizes ORC binding to chromatin. Mol Cell 40: 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ 2004. Principles of tumor suppression. Cell 116: 235–246 [DOI] [PubMed] [Google Scholar]
- Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW, Dutta S, Appella E, Gozani O 2007. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell 27: 636–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL 2006. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311: 844–847 [DOI] [PubMed] [Google Scholar]
- Spektor TM, Rice JC 2009. Identification and characterization of posttranslational modification-specific binding proteins in vivo by mammalian tethered catalysis. Proc Natl Acad Sci 106: 14808–14813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spektor TM, Congdon LM, Veerappan CS, Rice JC 2011. The UBC9 E2 SUMO conjugating enzyme binds the PR-Set7 histone methyltransferase to facilitate target gene repression. PLoS ONE 6: e22785 doi: 10.1371/journal.pone.0022785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, et al. 2009. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136: 420–434 [DOI] [PubMed] [Google Scholar]
- Talasz H, Lindner HH, Sarg B, Helliger W 2005. Histone H4-lysine 20 monomethylation is increased in promoter and coding regions of active genes and correlates with hyperacetylation. J Biol Chem 280: 38814–38822 [DOI] [PubMed] [Google Scholar]
- Tardat M, Murr R, Herceg Z, Sardet C, Julien E 2007. PR-Set7-dependent lysine methylation ensures genome replication and stability through S phase. J Cell Biol 179: 1413–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardat M, Brustel J, Kirsh O, Lefevbre C, Callanan M, Sardet C, Julien E 2010. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat Cell Biol 12: 1086–1093 [DOI] [PubMed] [Google Scholar]
- Trojer P, Li G, Sims RJ III, Vaquero A, Kalakonda N, Boccuni P, Lee D, Erdjument-Bromage H, Tempst P, Nimer SD, et al. 2007. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell 129: 915–928 [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Sachdeva MM, Wang H, Blobel GA 2006. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol 26: 9185–9195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, et al. 2010. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142: 967–980 [DOI] [PubMed] [Google Scholar]
- Vilenchik MM, Knudson AG 2003. Endogenous DNA double-strand breaks: Production, fidelity of repair, and induction of cancer. Proc Natl Acad Sci 100: 12871–12876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi K, Okamura M, Tsutsumi S, Nishikawa NS, Tanaka T, Sakakibara I, Kitakami J, Ihara S, Hashimoto Y, Hamakubo T, et al. 2009. The peroxisome proliferator-activated receptor γ/retinoid X receptor α heterodimer targets the histone modification enzyme PR-Set7/Setd8 gene and regulates adipogenesis through a positive feedback loop. Mol Cell Biol 29: 3544–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jia S 2009. Degrees make all the difference: The multifunctionality of histone H4 lysine 20 methylation. Epigenetics 4: 273–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Dai J, Daum JR, Niedzialkowska E, Banerjee B, Stukenberg PT, Gorbsky GJ, Higgins JM 2010. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science 330: 231–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward IM, Minn K, van Deursen J, Chen J 2003. p53 binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol Cell Biol 23: 2556–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Wang W, Kong X, Congdon LM, Yokomori K, Kirschner MW, Rice JC 2010. Dynamic regulation of the PR-Set7 histone methyltransferase is required for normal cell cycle progression. Genes & Dev 24: 2531–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Jing C, Kelly G, Walker PA, Muskett FW, Frenkiel TA, Martin SR, Sarma K, Reinberg D, Gamblin SJ, et al. 2005. Specificity and mechanism of the histone methyltransferase Pr-Set7. Genes & Dev 19: 1444–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y, Honda T, Tanno Y, Watanabe Y 2010. Two histone marks establish the inner centromere and chromosome bi-orientation. Science 330: 239–243 [DOI] [PubMed] [Google Scholar]
- Yan Q, Dutt S, Xu R, Graves K, Juszczynski P, Manis JP, Shipp MA 2009. BBAP monoubiquitylates histone H4 at lysine 91 and selectively modulates the DNA damage response. Mol Cell 36: 110–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Sun L, Li Q, Han X, Lei L, Zhang H, Shang Y 2011. SET8 promotes epithelial-mesenchymal transition and confers TWIST dual transcriptional activities. EMBO J 31: 110–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Yu VC, Zhu G, Chang DC 2008. SET8 plays a role in controlling G1/S transition by blocking lysine acetylation in histone through binding to H4 N-terminal tail. Cell Cycle 7: 1423–1432 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Reinberg D 2001. Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes & Dev 15: 2343–2360 [DOI] [PubMed] [Google Scholar]