A balance between progenitor cells and terminally differentiated layers is required in the epidermis. How this balance is achieved in mature somatic tissues is not completely understood. Here, Khavari and colleagues used high-throughput sequencing to characterize the transcriptome during somatic differentiation and identified a novel long noncoding RNA (ANCR) with a role in suppressing progenitor differentiation. The authors demonstrated that depletion of ANCR led to differentiation of progenitor cells and thus identified a novel lncRNA needed to maintain the undifferentiated state in the epidermis.
Keywords: lncRNA, RNA-Seq, epidermis, skin, differentiation, noncoding RNA, gene regulation
Abstract
Long noncoding RNAs (lncRNAs) regulate diverse processes, yet a potential role for lncRNAs in maintaining the undifferentiated state in somatic tissue progenitor cells remains uncharacterized. We used transcriptome sequencing and tiling arrays to compare lncRNA expression in epidermal progenitor populations versus differentiating cells. We identified ANCR (anti-differentiation ncRNA) as an 855-base-pair lncRNA down-regulated during differentiation. Depleting ANCR in progenitor-containing populations, without any other stimuli, led to rapid differentiation gene induction. In epidermis, ANCR loss abolished the normal exclusion of differentiation from the progenitor-containing compartment. The ANCR lncRNA is thus required to enforce the undifferentiated cell state within epidermis.
The human genome encodes several thousand long nonprotein coding transcripts >200 base pairs (bp) in length (The ENCODE Project Consortium 2007; Kapranov et al. 2007; Guttman et al. 2009; Cabili et al. 2011). A small subset of those long noncoding RNAs (lncRNAs) have been shown to be essential for a variety of biological processes, such as epigenetic control of chromatin (Rinn et al. 2007; Wang et al. 2011), promoter-specific gene regulation (Feng et al. 2006; Martianov et al. 2007), X-chromosome inactivation (Lee 2009; Tian et al. 2010), imprinting (Bartolomei et al. 1991; Lyle et al. 2000), nuclear import (Willingham et al. 2005), and maintenance of nuclear body structure (Mao et al. 2011). More recently, loss-of-function studies with a large number of lncRNAs expressed in mouse embryonic stem cells showed that lncRNAs regulate gene expression patterns that control pluripotent state or lineage commitment programs in stem cells (Guttman et al. 2011). However, the majority of lncRNAs functionally characterized to date regulate developmental processes, and their potential role in controlling mature tissue homeostasis and differentiation remains largely uncharacterized.
Epidermis is a stratified surface epithelium that provides a barrier to the external environment. Human epidermis self-renews approximately every 4 wk, undergoing continual regeneration from progenitor cells located in the basal layer. During the course of epidermal differentiation, a subset of basal keratinocytes withdraw from the cell cycle, detach from the basement membrane, and migrate outward while engaging a large differentiation gene expression program responsible for formation of the spinous, granular, and cornified layers (Blanpain and Fuchs 2006). A precise balance between the progenitor compartment and terminally differentiated layers is needed to maintain a functional epidermis. The mechanism by which this balance is maintained is not completely understood, and involvement of lncRNAs in somatic progenitor maintenance in this setting has not, to our knowledge, been firmly estab-lished. Here we used high-throughput transcriptome sequencing (RNA-Seq) along with high-resolution human lncRNA tiling arrays to identify ANCR (anti-differentiation ncRNA) as a lncRNA required to enforce the undifferentiated state in somatic progenitor populations of epidermis.
Results and Discussion
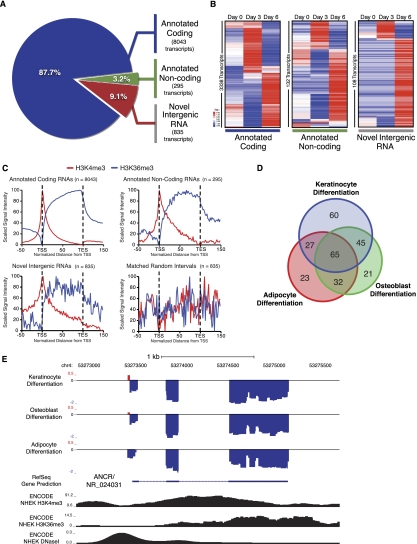
To define transcriptional dynamics during terminal differentiation, we performed paired-end, high-throughput RNA-Seq of primary human keratinocytes during days 0, 3, and 6 of calcium-induced differentiation. Reads were aligned to the human genome using the split-read mapper TopHat (Trapnell et al. 2009), generating between 100 × 106 and 110 × 106 mapped reads per condition. Reads were then assembled using reference annotation-based transcriptome assembly (Roberts et al. 2011) from the Cufflinks suite using RefSeq predicted genes as a reference. Significantly expressed transcripts were categorized based on this assembly into three groups: annotated protein-coding, annotated noncoding, and unannotated novel intergenic RNAs. We defined novel intergenic RNAs as having multiple assembled exons and a total length of >200 bp, and not contained within, or overlapping with, any annotated gene. This analysis and depth of sequencing yielded a differentiation-based transcriptome containing 8043 (87.7%) annotated protein-coding RNAs, 295 (3.2%) annotated noncoding RNAs, and 835 (9.1%) unannotated putative novel intergenic RNAs that were confidently detected (Fig. 1A; Supplemental Tables S1, S2).
Figure 1.
Transcriptome characterization identifies ANCR as a lncRNA suppressed during terminal differentiation. (A) High-throughput paired-end RNA-Seq during three time points (days 0, 3, and 6) of human keratinocyte terminal differentiation was performed. Transcriptome assembly and analysis generated three classes of significantly expressed RNA: annotated coding transcripts, annotated noncoding transcripts, and unannotated novel intergenic RNAs not associated with any annotated gene. (B) Mean-centered, hierarchical clustering of differentially expressed transcripts within the three RNA classes during day 0, day 3, and day 6 of terminal differentiation. (C) Aggregate chromatin signatures of H3K4me3 and H3K36me3 over significantly expressed RNA from the three RNA classes and random genomic intervals that are length- and chromosome-matched to novel transcripts. (TSS) Transcription start site; (TSE) transcription end site. (D) Intergenic K4/K36 domain tiling arrays were performed in matched progenitor and differentiated populations of keratinocytes, adipocytes, and osteoblasts using the assembled terminal differentiation transcriptome and all annotated noncoding RNA as a reference. Significantly differentially expressed transcripts are overlapped. (E) Differential expression data from K4/K36 tiling arrays displayed as log2 fold change during differentiation relative to progenitor cells from the three cell types at the ANCR/NR_024031 genomic locus. Keratinocyte ChIP-seq signal data from H3K4me3 and H3K36me3 and DNaseI hypersensitivity sequencing over this locus are depicted below in black. (Blue) Probes down-regulated during differentiation; (red,) probes up-regulated during differentiation.
We compared these classes of transcripts in terms of their differential expression and chromatin signatures. Transcripts within each class with twofold or greater change at either day 3 or day 6 of differentiation relative to the day 0 progenitor population were mean-centered and hierarchically clustered. All three RNA classes displayed dynamic temporal changes, with clusters of transcripts induced and repressed at each stage of differentiation (Fig. 1B). Interestingly, numerous novel intergenic RNAs are transcriptionally silent until the late stages of terminal differentiation at day 6. Actively transcribed coding and noncoding genes are characterized by a chromatin signature of H3K4me3 enrichment at their transcription start sites and H3K36me3 enrichment over their gene bodies (Guttman et al. 2009). To determine the epigenetic state of discovered novel intergenic RNAs relative to expressed annotated genes, we used publicly available ENCODE chromatin immunoprecipitation (ChIP) sequencing (ChIP-seq) data from primary human keratinocytes. We then generated a plot of the average scaled signal intensity for all transcripts of each class relative to their annotated or predicted transcription start sites. Strikingly similar H3K4me3–H3K36me3 (K4–K36) chromatin signatures were observed for coding, noncoding, and novel intergenic RNAs, but not matched random intergenic intervals (Fig. 1C). The shared epigenetic state between annotated genes and novel intergenic RNAs, together with similar patterns of dynamic regulation, support our classification of novel intergenic RNAs as a distinct gene set, rather than transcriptional noise or read-through events.
We next sought to identify previously uncharacterized RNAs with a functional role in suppressing differentiation within progenitor-containing populations. As a complementary technology that can rapidly assay the intergenic space, we used customized high-resolution tiling arrays designed to assay intergenic K4–K36 chromatin domains (Guttman et al. 2009; Khalil et al. 2009). Using all annotated lncRNAs and novel intergenic RNAs as an underlying reference to refine these broader domains into assembled transcripts, we assayed three human cell types undergoing terminal differentiation. RNA was obtained from keratinocytes, adipocytes, and osteoblasts in the progenitor and differentiated state and hybridized to the tiling arrays. Transcripts that were significantly differentially expressed during differentiation were then overlapped between the cell types (Fig. 1D; Supplemental Table S3), establishing a pool of RNAs that may have a functional role during terminal differentiation.
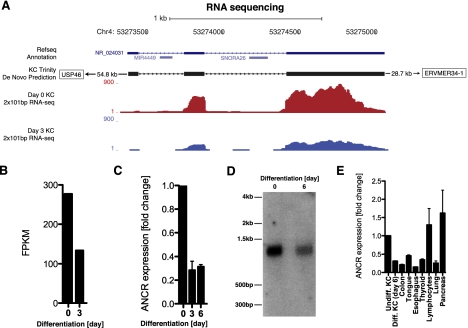
We discovered that an uncharacterized, predicted lncRNA, NR_024031, here termed ANCR, is suppressed upon terminal differentiation of multiple cell types (Fig. 1E). Of the transcripts that are coordinately regulated during differentiation of all three cell types, ANCR is one of the most strongly suppressed RNAs (Supplemental Table S3). The ANCR gene is located on human chromosome 4, with the closest adjacent annotated genes located 54.8 kb upstream of (USP46) and 28.7 kb downstream from (ERVMER34-1) the ANCR locus (Fig. 2A). The ANCR locus consists of three exons and harbors a microRNA (MIR4449) and a snoRNA (SNORA26) in introns 1 and 2, respectively. These small RNAs are not coordinately expressed with ANCR and are not part of the mature ANCR transcript (Supplemental Fig. S1A,B). To analyze ANCR protein-coding potential, we used the PhyloCSF algorithm (Lin et al. 2011). The PhyloCSF score for the ANCR locus, analyzed in three reading frames, was −908.8 decibans, indicating that ANCR is 7.64 × 1090 times more likely to be a noncoding transcript than protein-coding.
Figure 2.
ANCR is a lncRNA repressed during keratinocyte differentiation. (A) Schematic of ANCR genomic locus on chromosome 4, including RefSeq gene prediction for NR_024031. The de novo assembled ANCR transcript region based on keratinocyte RNA-Seq reads using the Trinity algorithm as well as raw RNA-Seq reads plotted for day 0 and day 3 of differentiation. (B) Relative abundance of ANCR transcript in fragments per kilobase of exon model per million mapped fragments (FPKM) during keratinocyte differentiation. (C) ANCR transcript expression is reduced during keratinocyte differentiation as shown by quantitative RT–PCR (qRT–PCR); undifferentiated cells (day 0) and days 3 and 6 of calcium-induced differentiation. (D) Northern blot analysis demonstrating a single 855-kb ANCR RNA species reduced at day 6 of keratinocyte differentiation. (E) ANCR transcript abundance in day 6 differentiated keratinocytes (diff. KC), colon, tongue, esophagus, thyroid, lymphocytes, lung, and pancreas, normalized to ANCR levels in undifferentiated progenitor keratinocytes (undiff. KC).
ANCR produces a single 855-bp RNA transcript that is reduced to 28% and 31% of levels seen in undifferentiated cells by days 3 and 6 of keratinocyte differentiation, respectively (Fig. 2B–D), and is down-regulated in the progenitor compartment, or basal layer, of mature human epidermis (Supplemental Fig. S1C). We then used the Trinity algorithm (Grabherr et al. 2011) for full-length assembly of potential transcripts in the ANCR genomic region based on our keratinocyte RNA-Seq reads without using a reference genome. The de novo assembled 855-bp transcript showed 99.9% identity to the annotated NR_024031 transcript prediction and represented the only significantly expressed RNA in the ANCR locus, containing 95.54% of all RNA-Seq raw reads detected within the ANCR region. ANCR expression is not limited to progenitor keratinocytes, but is widely expressed in a variety of human tissues as well as all ENCODE project cell lines that have available RNA-Seq data (Fig. 2E; Supplemental Fig. S1D). The broad expression pattern of ANCR, in combination with its reduced expression levels in a number of terminally differentiated cell types (keratinocytes, osteoblasts, and adipocytes), raised the possibility of a potential functional role for ANCR in the transition from the progenitor to the differentiated cell state.
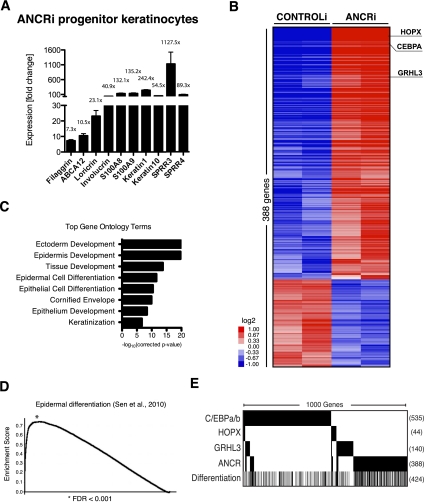
To analyze ANCR function in progenitor keratinocytes, we depleted ANCR expression using RNAi (Supplemental Fig. S2A,B). ANCR depletion by itself triggered robust, spontaneous induction of early and late epidermal differentiation genes, including filaggrin, loricrin, keratin 1, small proline-rich proteins 3 and 4, involucrin, S100 calcium-binding proteins A8 and A9, and ABCA12 (Fig. 3A). To assess the global gene regulatory role of ANCR, we performed oligonucleotide microarray analyses of ANCR loss in progenitor keratinocytes. ANCR depletion disrupted normal expression of 388 annotated genes (Fig. 3B; Supplemental Table S4), including CEBPA, GRHL3, and HOPX, all three of which are transcription factors known to regulate epidermal differentiation (Yu et al. 2006; Lopez et al. 2009; Yang et al. 2010). Gene ontology (GO) analysis of genes differentially expressed upon ANCR depletion produced GO terms strongly enriched for epidermis differentiation, cornification, and keratinization (Fig. 3C). Gene set enrichment analysis (GSEA) (Mootha et al. 2003; Subramanian et al. 2005) of the ANCRi gene set shows highly statistically significant overlap with the calcium-induced keratinocyte differentiation gene signature published previously (Fig. 3D; Sen et al. 2010). These results indicate that ANCR suppresses a genetic program associated with epidermal differentiation.
Figure 3.
ANCR regulates a global gene expression program associated with epidermal differentiation. (A) Increased mRNA expression of early and late differentiation genes in ANCR-depleted progenitor keratinocytes in cultured keratinocytes versus control siRNAs, as shown by qPCR analysis. (B) Heat map depicting transcript profiling of independent biologic replicate progenitor keratinocytes with siRNA-mediated ANCR depletion (ANCRi) versus control siRNA (CONTROLi). Expression changes for each probe are normalized to a mean-centered signal across all samples. (C) Gene ontology terms significantly enriched in the ANCR-depleted gene set. (D) Gene set enrichment analysis (GSEA) shows highly statistically significant overlap of the ANCRi gene set with a calcium-induced keratinocyte differentiation gene signature published previously (Sen et al. 2010). (E) Gene set overlap of ANCRi gene set with published gene signatures from loss of key transcription factors that are dysregulated by ANCR depletion. Black bars indicate significantly differentially expressed genes in the gene set. The 1000 genes total between these four gene sets are compared with each other as well as with the keratinocyte differentiation signature. The total number of genes in each gene signature falling within the 1000-gene set is listed to the right of each row.
We hypothesized that the mechanism by which ANCR affects global gene regulation is through suppression of key transcriptional regulators of epidermal development. To test this hypothesis, we overlapped the 388 genes differentially regulated by ANCR depletion with the published gene signatures from loss of HOPX, CEBPA, and GRHL3—key transcription factors that are dysregulated by ANCR loss. These four gene sets, comprised of 1000 genes in total, were compared with each other as well as with the keratinocyte differentiation signature. We observed that only a small portion (15%) of the ANCRi gene set overlaps with those of known transcription factors regulated by ANCR (Fig. 3E), suggesting that ANCR's impacts on differentiation cannot be explained solely through its impact on these specific transcription factor targets.
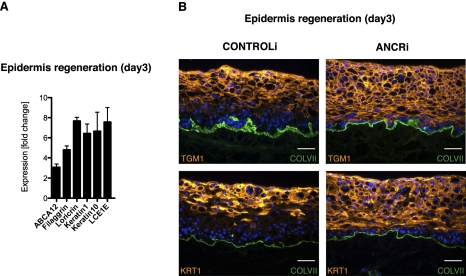
To analyze ANCR function in a human tissue environment, we depleted ANCR expression in regenerated, organotypic epidermal tissue, a setting that recapitulates the histology, tissue structure, and gene expression pattern of human epidermis (Truong et al. 2006; Sen et al. 2008, 2010). ANCR loss in human regenerated epidermis also induced expression of late and early differentiation genes such as filaggrin, loricrin, ABCA12, late cornified envelope protein 1E, and keratins 1 and 10 above normal levels (Fig. 4A). Importantly, depletion of ANCR resulted in expression of differentiation proteins in the epidermal basal layer, a compartment in which differentiation proteins are never normally present (Fig. 4B). ANCR is therefore required to exclude differentiation gene expression from the progenitor cell-containing compartment in epidermal tissue.
Figure 4.
ANCR depletion induces ectopic differentiation in the epidermal progenitor compartment. (A) Increased differentiation gene mRNA expression as a function of ANCR depletion, as shown by qPCR analysis from whole-tissue RNA extracts. (B) Ectopic expression of epidermal differentiation proteins in the epidermal basal and lower spinous layers in ANCR-depleted regenerating organotypic human epidermis (ANCRi) using ANCR-specific versus scrambled control shRNA constructs (CONTROLi). Differentiation proteins detected by immunofluorescence (orange), basement membrane collagen VII (green)], and nuclear Hoechst 33342 (blue). Bar, 50 μm.
In these studies, a combination of high-throughput RNA-Seq and high-resolution tiling arrays was used to identify transcripts altered during the transition from a progenitor to a differentiated cell population. This approach yielded a gene set that included protein-coding, annotated noncoding, and novel intergenic RNA populations. ANCR was found as a lncRNA that was down-regulated during differentiation of progenitor-containing populations. ANCR is required to repress a substantial portion of the epidermal differentiation program, both in vitro and in regenerated organotypic human epidermal tissue. In the latter setting, ANCR depletion led to enhanced differentiation gene expression as well as ectopic differentiation protein induction in less-differentiated cell layers, including the progenitor-containing epidermal basal layer, a compartment where expression of differentiation proteins is never normally found. Taken together, these data demonstrate a functional requirement for ANCR in maintaining the undifferentiated state that is characteristic of progenitor cells.
lncRNAs have recently been shown to function as RNAs with enhancer-like function by regulating genes in cis (eRNAs) (Orom et al. 2010; Wang et al. 2011). We observed no significant expression changes of the proximal genes flanking the ANCR locus during ANCR loss. This finding, together with the observation that ANCR controlled differentiation genes that are distributed throughout the human genome, indicates that ANCR regulates gene targets in a process distinct from classical eRNA action. A recent study (Cesana et al. 2011) has shown that a lncRNA regulates muscle differentiation as a competing endogenous RNA (ceRNA) directly influencing expression of transcription factors known to regulate muscle differentiation. We observed that, although ANCR influences expression of three known transcription factors associated with epidermal development, the majority of gene dysregulation observed as a result of ANCR depletion appears to be independent of these factors. Further studies will be necessary to determine the precise mechanism of action of ANCR, including a possible role for the hosted small RNAs or association with chromatin or other protein complexes.
Taken together, this work defines a human terminal differentiation gene set consisting of protein-coding as well as newly identified and annotated noncoding genes and defines a novel, functionally nonredundant role for lncRNA-mediated control of human tissue homeostasis.
Materials and methods
Tissue culture
Primary human keratinocytes were isolated from freshly discarded skin surgical samples and grown in a 1:1 mixture of KSF-M (GIBCO) and Medium 154 for keratinocytes (GIBCO), supplemented with epidermal growth factor (EGF) and bovine pituitary extract (BPE). Keratinocytes were cultured at 37°C in a humidified chamber with 5% CO2.
Gene transfer
Primary human keratinocytes (1 × 106) were electroporated with 1 nmol of siRNA oligonucleotide using the Amaxa human keratinocyte nucleofector kit (Lonza) as well as Amaxa nucleofection reagents as described previously (Truong et al. 2006). Retroviral gene transfer was performed as described previously (Sen et al. 2008).
The following siRNAs (synthesized by Dharmacon) and shRNA oligonucleotides (cloned into the pSUPER shRNA expression vector; Oligoengine) were used for this work: siControl (sense sequence), GUAGAUUCAUAUUGUAAGGUU; siANCR (sense sequence), GCAGGTATGTTCCTAGCCT; shControl (sense sequence), GCTTCAATTCGCGCACCTA; and shANCR (sense sequence), GCGTACTAACTTGTAGCAA.
Organotypic regenerated human epidermal tissue
For the generation of organotypic regenerated human epidermis, primary human keratinocytes were transduced with shRNA retroviral constructs and selected with puromycin or nucleofected with siRNAs. Keratinocytes (4 × 105) were seeded onto devitalized dermis and raised to the air/liquid interface to initiate stratification and differentiation of the epidermis culture as described earlier (Truong et al. 2006; Sen et al. 2010).
Immunofluorescence and tissue analysis
Seven-micron-thick frozen skin sections from human organotypic skin cultures were fixed in 100% methanol or acetone for 10 min at −20°C, followed by blocking in PBS with 10% BSA for 30 min. Staining conditions are listed in Supplemental Table S5.
Quantitative RT–PCR (qRT–PCR) analysis
Total RNA from cells was isolated with Trizol reagent (Invitrogen), and genomic DNA was removed using the Turbo DNase kit (Ambion) and quantified by NanoDrop. Total RNA from human regenerated epidermis was isolated with the RNeasy Plus minikit (Qiagen) according to manufacturer's instructions. One microgram of total RNA was reverse-transcribed with the iScript cDNA synthesis kit (Bio-Rad). qPCR was performed using the Maxima SYBR Green qPCR master mix (2×; Fermentas) and the Stratagene Mx3000P (Agilent Technologies) thermocycler. Samples were run in triplicate and normalized to 18s rRNA or ribosomal protein L32. Primer sequences are listed in Supplemental Table S6.
Laser capture microdissection (LCM)
For LCM, 7-μm cryosections were stained with cresyl violet according to the manufacturer's protocol (LCM staining kit; Ambion). A Leica AS LMD system was used to isolate progenitor as well as differentiated epidermal compartments from adult abdominal skin specimens. Total RNA was purified with the RNAqueous microkit (Ambion), and RNA abundance was tested by qRT–PCR analysis.
qRT–PCR analysis with microRNAs
Total RNA from keratinocytes was isolated with Trizol reagent (Invitrogen) and miRNeasy minikit (Qiagen). RNA was treated with Turbo DNase (Ambion) and cleaned up with the miRNeasy minikit. RNA (0.5 ug) was reverse-transcribed with the SuperScript III reverse transcriptase kit (Invitrogen) using microRNA-specific primers. qPCR was performed using Brilliant SYBR Green qPCR master mix (Agilent Technology) with microRNA-specific primers, as listed in Supplemental Table S6. miR-16 was used as a normalization control.
Northern blot analysis
Total RNA from keratinocytes was isolated with Trizol reagent (Invitrogen). An ANCR-specific, radioactive DNA probe with a length of 850 bp was generated using dCTP [α-32P] (Perkin Elmer) and the Megaprime DNA labeling system (GE Healthcare). Hybridization was performed using QuickHyb (Agilent) following the manufacturer′s instructions. 18s and 28s rRNA bands on the Northern blot membrane after transfer were used as loading controls.
RNA-Seq
RNA-Seq sequencing libraries were prepared with the mRNA-Seq sample prep kit (Illumina) with total RNA isolated from differentiated (days 3 and 6) and progenitor (d0) human keratinocytes, following the manufacturer's instructions.
Illumina paired-end sequencing was performed on the HiSeq platform with a read length of 101 nucleotides. Reads were aligned to the human reference sequence NCBI Build 36.1/hg18 with the TopHat algorithm (Trapnell et al. 2009). Reference annotation-based transcript assembly (Roberts et al. 2011), transcriptome comparison, and differential expression analysis were performed with the Cufflinks suite using human RefSeq gene predictions as a reference transcriptome. Annotated coding (prefix NM) and noncoding (prefix NR) transcripts were filtered for significant expression by a fragments per kilobase of exon model per million mapped fragment (FFPKM) cutoff of 5 in at least one time point during epidermal differentiation. Novel intergenic RNAs were selected based on a length cutoff of >200 bp and the presence of at least two contiguously assembled exons. Novel RNAs that overlapped with, or were contained within, annotated genes from RefSeq or University of California at Santa Cruz (UCSC) Known Genes were excluded to select for intergenic transcripts.
Differential expression analysis and chromatin state maps
Cuffdiff (Trapnell et al. 2010) was used to assess differentially expressed transcripts that were significantly expressed at any time point during differentiation. To prevent artificially inflated fold changes, FPKM values of <1 were set to 1 for fold change calculations. Transcripts that were twofold or greater changed in at least one time point were hierarchically clustered using the Cluster program (Eisen et al. 1998) and were visualized in Java Treeview (Eisen et al. 1998).
ChIP-seq data for normal human epidermal keratinocytes from the ENCODE project (The ENCODE Project Consortium 2007) for H3K4me3 and H3K36me3 were downloaded from the UCSC Genome Browser. Average ChIP signal was calculated from a distance beginning upstream by 50% of the gene body and continuing downstream from the transcript by the same 50% distance. Signal intensity for each mark was scaled from local minima to local maxima to allow presentation of H3K4 and H3K36 on the same normalized axis. Random intervals were generated that were length-matched and contained within the same chromosome as the novel intergenic transcripts. These intervals were subjected to the same chromatin signature analysis and scaling.
Tiling array hybridization and analysis
Customized high-resolution tiling arrays designed to assay long intergenic noncoding RNA exons and control regions have been described previously (Khalil et al. 2009). Total RNA from progenitor and differentiated cells, extracted from primary keratinocytes, or purchased from Cell Applications (adipocytes and osteoblasts) was labeled and hybridized as described previously (Rinn et al. 2007). Array images were processed, and the resulting data files were RMA-normalized with Nimblescan (Roche/Nimblegen). Probe-level data within each channel were mapped to the annotated noncoding RNAs and novel intergenic RNAs derived from the reference-based assembly and averaged assessed for fold change between the two channels.
Gene expression profiling
Microarray analysis was performed with biological duplicate samples. Labeling of cDNA derived from control and ANCR-deficient human progenitor keratinocytes was done using the MessageAmp III RNA amplification kit (Ambion) following the manufacturer's instructions. Hybridization to Affymetrix Gene Chip HG-U133 Plus 2.0 arrays was carried out at Stanford's P.A.N. facility. Gene expression analysis was performed as described earlier (Sen et al. 2010).
Accession numbers
Array and sequencing data are publicly available in the NCBI Gene Expression Omnibus (GEO) under the accession number GSE34767.
Acknowledgments
We thank Gordon K. Lee for providing us with skin specimens. This work was supported by the U.S. Veterans Affairs Office of Research and Development and by NIH/NIAMS grant AR49737 to P.A.K.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.182121.111.
References
- Bartolomei MS, Zemel S, Tilghman SM 1991. Parental imprinting of the mouse H19 gene. Nature 351: 153–155 [DOI] [PubMed] [Google Scholar]
- Blanpain C, Fuchs E 2006. Epidermal stem cells of the skin. Annu Rev Cell Dev Biol 22: 339–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL 2011. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25: 1915–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I 2011. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147: 358–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D 1998. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD 2006. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev 20: 1470–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. 2011. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477: 295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR 2007. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet 8: 413–423 [DOI] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. 2009. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci 106: 11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT 2009. Lessons from X-chromosome inactivation: Long ncRNA as guides and tethers to the epigenome. Genes Dev 23: 1831–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Jungreis I, Kellis M 2011. PhyloCSF: A comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27: i275–i282 doi: 10.1093/bioinformatics/btr209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez RG, Garcia-Silva S, Moore SJ, Bereshchenko O, Martinez-Cruz AB, Ermakova O, Kurz E, Paramio JM, Nerlov C 2009. C/EBPα and β couple interfollicular keratinocyte proliferation arrest to commitment and terminal differentiation. Nat Cell Biol 11: 1181–1190 [DOI] [PubMed] [Google Scholar]
- Lyle R, Watanabe D, te Vruchte D, Lerchner W, Smrzka OW, Wutz A, Schageman J, Hahner L, Davies C, Barlow DP 2000. The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nat Genet 25: 19–21 [DOI] [PubMed] [Google Scholar]
- Mao YS, Sunwoo H, Zhang B, Spector DL 2011. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol 13: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A 2007. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 445: 666–670 [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. 2003. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34: 267–273 [DOI] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. 2010. Long noncoding RNAs with enhancer-like function in human cells. Cell 143: 46–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Pimentel H, Trapnell C, Pachter L 2011. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27: 2325–2329 [DOI] [PubMed] [Google Scholar]
- Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA 2008. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev 22: 1865–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA 2010. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature 463: 563–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Sun S, Lee JT 2010. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell 143: 390–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL 2009. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA 2006. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev 20: 3185–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. 2011. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472: 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG 2005. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 309: 1570–1573 [DOI] [PubMed] [Google Scholar]
- Yang JM, Sim SM, Kim HY, Park GT 2010. Expression of the homeobox gene, HOPX, is modulated by cell differentiation in human keratinocytes and is involved in the expression of differentiation markers. Eur J Cell Biol 89: 537–546 [DOI] [PubMed] [Google Scholar]
- Yu Z, Lin KK, Bhandari A, Spencer JA, Xu X, Wang N, Lu Z, Gill GN, Roop DR, Wertz P, et al. 2006. The Grainyhead-like epithelial transactivator Get-1/Grhl3 regulates epidermal terminal differentiation and interacts functionally with LMO4. Dev Biol 299: 122–136 [DOI] [PubMed] [Google Scholar]